Abstract
The cellular process of macromolecular degradation known as macroautophagy has long been known to play a role in the elimination of mitochondria. Over the past decade, much progress has been made in the development of systems by which the nature and mechanism of mitochondria degradation may be studied. Recent findings imply that the degradation of mitochondria via autophagy may be more specific and more tightly regulated than originally thought, and have led to designation of this specific type of autophagy as “mitophagy”. In this review we provide a brief history of the development of mitophagy models and their associated discoveries.
Keywords: Mitophagy, Autophagy, Mitochondria, Adipocyte, Reticulocyte
1. Introduction
Macroautophagy, hereafter referred to as autophagy, is a process by which cells degrade macromolecular intracellular material via sequestration in a double-membrane structure, known as an autophagosome, which then delivers the enclosed material to a lysosome for degradation (Klionsky, 2007). Initially believed to be a system dedicated to the “recycling” of macromolecular material within the cell, autophagy is now known to be involved in a multitude of cellular processes including immunity (Nakagawa et al., 2004; Paludan et al., 2005), tumorigenesis (Klionsky, 2007; Qu et al., 2003; Yue et al., 2003), programmed cell death (Tsujimoto and Shimizu, 2005), the selective degradation of organelles (Kissova et al., 2007), aging (Cuervo et al., 2005) and a host of neurodegenerative conditions (Rubinsztein et al., 2005). There are currently 33 identified Autophagy-related genes (denoted by the Atg prefix) shown to play a role in autophagy in yeast, many with homologs now identified in the mammalian genome (Reggiori and Klionsky, 2002; Tsukada and Ohsumi, 1993).
In the earliest days of autophagy research, the presence of mitochondria in lysosomes and autophagosomes bespoke a significant role for autophagy in the removal and degradation of these important organelles (Ashford and Porter, 1962; Beaulaton and Lockshin, 1977). Ultimately, this process of mitochondria autophagy was dubbed “mitophagy” in deference to its perceived specificity of action (Lemasters, 2005). Since the discovery of mitochondria within autophagosomes in both mammalian cells (Ashford and Porter, 1962; Clark, 1957) and yeast (Takeshige et al., 1992), the issue of whether or not mitochondria are specifically targeted by autophagy has been a source of both persistent debate and intense scientific scrutiny.
Originally it was believed by many that autophagy was a strictly non-selective process, randomly engulfing cytosolic components and subject only to universal up- or down-regulation (Deter and De Duve, 1967; Ericsson, 1969a,b; Kim et al., 2001; Shelburne et al., 1973). Later, the concept of “targeted” or specific autophagy began to evolve in which autophagosomes were observed to preferentially degrade particular macromolecular constituents within the cytosol (Hopgood et al., 1986; Scott et al., 1997). In addition to mitophagy, examples of this process include the targeted degradation of peroxisomes (pexophagy), foreign bodies (xenophagy), protein aggregates (aggrephagy), endoplasmic reticulum (reticulophagy), endosomes (heterophagy), and golgi apparatus (crinophagy) (Klionsky, 2007). Despite the broadening repertoire of functions displayed by autophagy, little progress has yet been made in understanding the targeting mechanisms that may bestow specificity upon this process.
In order to appreciate the advances in our collective understanding of mitophagy and its means of operation, it behooves us to become familiar with the model systems in use. Both yeast and mammal systems have contributed to the exploration of mitophagy, often with a discovery in one system being recapitulated in another. It is our intent to provide the reader with a brief history and background on the development and use of these models, emphasizing key moments of progress in the evolution of the debate over the specificity and functional purpose of mitochondria degradation. Additionally, we propose a new mammalian model system for the investigation of mitochondria degradation based on recent work in our laboratory.
2. Mitophagy in yeast
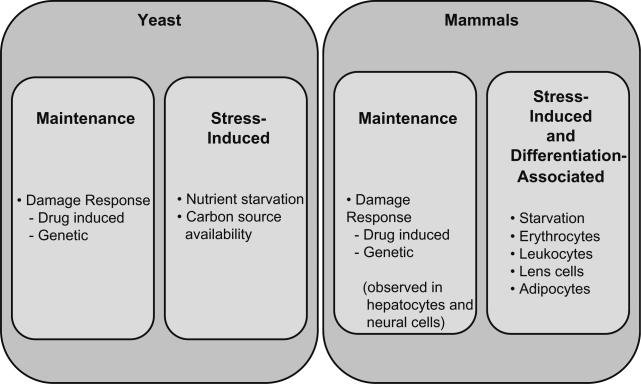
While the initial discovery of autophagic degradation of mitochondria occurred via the observance of mammalian cells (Ashford and Porter, 1962), the bulk of information available on the specificity of mitophagy comes from studies conducted in the yeast Sacchromyces cerevisiae. The fact that this organism utilizes a complex autophagy system, one that has been shown to be largely homologous with that in humans, in addition to its hardiness and ease of manipulation, makes S. cerevisiae an ideal system in which this process may be investigated. The study of mitophagy in yeast thus far can be roughly divided into three areas: (1) the determination of autophagy as the primary clearance mechanism for mitochondria, (2) the types of mitophagy, subdivided here into “maintenance” and “stress-induced” mitophagy that are induced under different conditions (Fig. 1), and (3) the molecular mechanisms that result in the specificity of mitophagy. Remarkably, almost all of the advances in these three areas of study have occurred over the past decade, most as the direct result of labor-intensive high-throughput genetic screens.
Fig. 1.
Experimental conditions known to trigger mitophagy in yeast and mammals.
2.1. Autophagy as the primary clearance mechanism for mitochondria
The convergence of the study of mitochondria degradation and autophagy can be traced back to the discovery of starvation-induced autophagy in yeast (Takeshige et al., 1992). At that time, it was widely believed that mitochondria entered the vacuole for degradation via some direct method of engulfment (i.e. microautophagy, which is defined as the direct engulfment of macromolecular components by the vacuole itself (Campbell and Thorsness, 1998; Takeshige et al., 1992). The work presented by Takeshige, however, not only revealed that placing yeast under the stressful conditions of nutrient starvation could apparently induce increased autophagic activity, but also proposed a role for autophagy in the clearance of macromolecular cytosolic structures, including mitochondria, in yeast.
The next major finding on the path to defining mitophagy as a discrete process was provided by Campbell and Thorsness 6 years later, when they successfully modified the Pho8delta60 assay (originated by Noda (Noda et al., 1995) for the investigation of vacuolar degradation of cytosolic components) into the mito-Pho8delta60 assay (Campbell and Thorsness, 1998). In this assay, a Pho8 enzyme that is specifically targeted to the mitochondria and is only enzymatically active at low pH was used to measure transport of mitochondria to the vacuole. By inactivating the ATP-dependent metalloprotease of the mitochondria via genetic mutation of a critical subunit, Campbell was able to decrease mitochondrial membrane potential (Δψm) and cause the release of mitochondrial DNA. The result of this mutation was a dramatic increase in the rate of mitochondria degradation, as measured by Pho8 enzymatic activity (Campbell and Thorsness, 1998). While this paper did not draw an explicit connection between autophagy and the transportation of mitochondria to the vacuole, it did establish the link between damage to mitochondria and the subsequent up-regulation of degradation.
Genetic evidence for autophagy as a primary transporter of mitochondria to the vacuole was provided courtesy of Kissova et al. in 2007. In this paper, the authors explain that both the loss of a mitochondrial outer-membrane protein, Uth1p, and the absence of the key macroautophagy protein, Atg5p, resulted in the absence of mitophagy as measured by mito-GFP (explained further later in this paper). The Δuth1 deletion also resulted in an absence of increased turnover of mitochondria in response to rapamycin, which is known to induce overall autophagy levels in cells. However, the authors were able to show that the general macroautophagy levels in the Δuth1 yeast responded with a normal increase in activity, despite a total lack of mitochondria degradation (Kissova et al., 2007). The clear implications of this study are that the process of mitophagy relies upon understood macroautophagic machinery to function and that autophagy is a primary means of transporting mitochondria to the vacuole for destruction.
Subsequent work conducted by our laboratory revealed that the deletion of several genes critical to autophagic function (ATG1, ATG6, ATG8, and ATG12) resulted in yeast strains that were incapable of degrading mitochondria in stationary culture (Zhang et al., 2007). Utilizing electron microscopy to visualize and count mitochondria per cell, as well as Southern blot analysis of mitochondrial DNA to obtain direct evidence of the degradation of mitochondria, Zhang et al. presented the case for autophagy serving as the exclusive removal mechanism for mitochondria. Further circumstantial evidence support this hypothesis, as the autophagy-deficient yeast strains used in the study exhibited defects in mitochondria-dependent growth, increased cell-cycle arrest at the G1 phase, lower oxygen consumption rates, lower mitochondrial membrane potential (Δψm) and higher levels of reactive oxygen species (ROS), all in accordance with a defect in mitophagy. These data, in addition to the works previously cited, underscored the importance of autophagy in the degradation of mitochondria, and thence the importance of autophagy in overall cellular metabolism, and also set the stage for determining markers of specificity for mitophagy.
2.1.1. Conditions that induce mitophagy
Mitophagy in yeast can be empirically categorized into two types: “maintenance” mitophagy and “stress-induced” mitophagy. The justification for this classification scheme is that there are known to be certain circumstances under which large numbers of mitochondria are simultaneously enveloped by autophagosomes for disposal (e.g. starvation, changing substrate from a non-fermentative to fermentative carbon source), and alternate circumstances under which very few or even individual mitochondria are targeted after expressing evidence of damage.
2.1.2. “Maintenance” mitophagy
For the purposes of this paper, “maintenance” mitophagy, is defined as a process in which only damaged mitochondria are specifically targeted for disposal. The earliest evidence linking mitochondrial damage to selective degradation was provided by the previously referenced work by Campbell and Thorsness (1998), in which damaged mitochondria resulted in an increase in vacuolar mitochondria content. Later work by Priault et al. showed that the mutation of fmc1 leading to dysfunctional mitochondrial ATP synthase resulted in the preferential removal of mitochondria under anaerobic conditions, when reverse operation of the ATPase becomes essential to the maintenance of Δψm (Priault et al., 2005). Shortly thereafter Nowikovsky et al. showed that mutations in the MDM38 gene that lead to dysfunctional mitochondrial K+/H+ exchange resulted in osmotic swelling and subsequent morphological damage to mitochondria, followed by a marked increase in mitophagic activity (Nowikovsky et al., 2007). Other mechanism for inducing mitochondrial damage, either by pharmacological agent or by additional mutations, have had much the same effect (Kissova et al., 2007; Priault et al., 2005). The common thread among these findings is that mitochondrial damage is the impetus for the up-regulation of mitophagic activity, thus facilitating the removal of the damaged mitochondria from the cytoplasm and reducing both the waste of cellular resources and the exposure of the organism to potentially damaging reactive oxygen species.
2.1.3. “Stress-induced” mitophagy
Due to the adaptive nature of their metabolism, certain circumstances exist under which yeast are required to engage in the rapid and specific degradation of a large number of mitochondria. The most obvious example of this is generalized nutrient starvation, under which the maintenance of excess organelles is an unjustifiable expense to the cell. Indeed, under starvation conditions, we see both a generalized increase in macroautophagic activity and an increase in autophagic envelopment of mitochondria (Takeshige et al., 1992). The specific nature of mitophagy under nutrient starvation was made apparent when comparisons between fermentable and non-fermentable carbon sources were used during starvation. Under growth on non-fermentable carbon sources, like lactate, glycerol, pyruvate, or ethanol, yeast rely upon mitochondria to obtain energy (Damsky, 1976). Consequently, an unusual pattern of initial microautophagy of mitochondria is seen, followed by an increase in macro-mitophagy when yeast are subjected to prolonged nitrogen starvation under these conditions (Kissova et al., 2007). However, the level of macro-mitophagy in these conditions is highly attenuated when compared to prolonged nitrogen starvation in a fermentable carbon such as glucose. When yeast are provided with a fermentable carbon source, their need for mitochondria is decreased and subsequent nitrogen starvation will result in a considerable up-regulation of mitophagic activity, while macroautophagic activity is similar in both types of carbon sources. (Kanki et al., 2009a). The removal of mitochondria is part of a generalized cellular response to adverse conditions, but even under this “physiologic” condition mitochondria are still removed in an orderly and specific fashion.
2.2. The molecular mechanisms that confer specificity on mitophagy
The works cited thus far provide substantial support for the concept of mitophagy as a discrete and targeted process, but the keystone to this hypothesis is the existence of genetic evidence of specificity. One gene shown by an early study to play a role in the targeting of autophagosomes to mitochondria was UTH1 (Kissova et al., 2007). In this paper, Kissova et al. showed that the deletion of UTH1, a mitochondrial outer-membrane protein, resulted in a virtual cessation of mitochondria degradation in yeast. Shortly thereafter, the mitochondrial inner-membrane protein Aup1 was also shown to be necessary for the targeted degradation of mitochondria via autophagy (Tal et al., 2007). More recently, massive genetic screens conducted by two groups have revealed multiple new candidate genes thought to confer specificity on mitophagy, two of which have now been designated ATG32 and ATG33 (Kanki et al., 2009b,c; Okamoto et al., 2009) in addition to a small group of yet-to-be-characterized candidate genes. Both ATG32 and ATG33 encode yeast mitochondrial proteins, and although mammalian homologs remained to be identified, ATG32 in particular has shown great potential as the leading candidate for the primary receptor in the mediation of mitophagy (Ishihara and Mizushima, 2009; Kanki and Klionsky, 2009; Okamoto et al., 2009; Tolkovsky, 2009).
2.3. Where can yeast take us from here?
The preponderance of evidence leads us to believe that mitophagy is a specific, targeted removal of mitochondria from the cell via autophagic transport to the vacuole. However, a host of questions remain to be answered. First and foremost, while we now have a considerable body of data supporting the specificity of mitophagy and a small but rapidly growing roster of proteins that may confer this specificity on the otherwise non-specific autophagy machinery, we still lack insight into the actual mechanisms that drive this process. One hypothesis that has been proposed is that the mitophagy-specific proteins discovered thus far must likely interact with the yeast autophagy protein Atg11 (He et al., 2006; Kanki and Klionsky, 2008). Furthermore, there are conflicting data for some of these proteins (namely Aup1 and Uth1 noted above) that remain to be resolved (Ishihara and Mizushima, 2009). Also, we do not yet know whether maintenance and stress-induced mitophagy utilize the same targeting mechanism, or even the same autophagic machinery. Finally, the methods used to induce mitophagy have varied widely from study to study, and while we now understand some of the general conditions under which mitophagy is activated (namely mitochondrial damage, carbon source fermentability, and glutathione pool depletion), little effort has been made to evaluate potential differences between the removal of damaged mitochondria and the removal of excess, yet otherwise normal, mitochondria in yeast. Some information on the nature of this difference is addressed by the mammalian models described later in this manuscript, but these issues must be addressed in the yeast models as well if we are to significantly further our understanding of mitophagy.
3. Mitophagy in mammalian systems
In mammalian systems, mitophagy has been observed under a variety of circumstances. Here, we divide these circumstances into two classes: (a) “maintenance” mitophagy and (b) mitophagy associated with stress or cellular differentiation. The first class consists of mitophagic activity that takes place in response to damaged mitochondria, resulting in the targeted removal and degradation of those mitochondria. The second class consists of normal physiological conditions under which mitophagy plays a role in the process of cellular differentiation or function. While relatively little progress has yet been made in identifying the specific factors involved in the regulation and targeting of mitophagy in mammalian cells, the data currently available support the concept of mitophagy operating as a specialized process, independent from the regulation of indiscriminate general macroautophagy. These findings imply that mitophagy may play a critical role in both the normal development and maintenance and in the pathophysiological responses of a variety of mammalian tissues (Fig. 1).
3.1. “Maintenance” mitophagy
In the early 1990s John J. Lemasters began his exploration of the conditions that induce mitophagy in hepatocytes, thus linking disturbances in mitochondrial membrane permeability known as the “mitochondrial permeability transition” (MPT) with the up-regulation of autophagic degradation of mitochondria in these cells (Lemasters et al., 1998a,b). Since that time, his laboratory has made many noteworthy contributions to our understanding of mitophagy through the development of improved experimental assays (Lemasters et al., 1998a; Rodriguez-Enriquez et al., 2006), and the provision of evidence supporting both the specific nature of mitophagy (Kim et al., 2007; Lemasters, 2005) and the importance of mitophagy in normal hepatocellular physiology (Elmore et al., 2001; Lemasters, 2005; Rodriguez-Enriquez et al., 2009). The key conclusion to be drawn from this body of work is that mitophagy appears to operate as a mechanism of homeostasis in this tissue, removing damaged mitochondria from otherwise normal cells in a rapid, targeted fashion. From a cellular survival standpoint this finding supports the concept of mitophagy both as a mechanism for reducing cellular exposure to increased reactive oxygen species (ROS) produced by damaged mitochondria and as a way to economize on cellular energy by eliminating the need to support ineffective organelles.
3.2. Mitophagy and cellular differentiation
Mitophagy has been clearly identified as playing a role in reticulocyte development. In the early 1980s initial investigations which established the removal of mitochondria via autophagy as a key process in the development of mature erythrocytes were being conducted by Heynen and Verwilghen (Heynen et al., 1985). More recently, a bevy of papers have implicated the BH3-only protein Nix as an inducer of Δψm destabilization which is believed to lead to selective mitophagy in the developing red blood cell (Chen et al., 2008; Sandoval et al., 2008; Schweers et al., 2007; Zhang and Ney, 2008).
Consistent with the notion that general macroautophagic machinery is critical for mitophagy, another gene, Ulk1, is also believed to be necessary for physiological mitophagy in erythrocytes. Ulk1 was previously identified as one of the two mammalian homologs of the yeast autophagy protein Atg1, which is a protein kinase believed to be essential for normal autophagosome formation (Chan et al., 2007; Kundu et al., 2008). Interestingly, in their most recent publication Kundu et al. show that while the mammalian Ulk1 is apparently required for the elimination of ribosomes and mitochondria from the nascent red blood cell, it is not required for general macroautophagic activity (Kundu et al., 2008).
It is also worth noting here that a recent paper put forth by a key reticulocyte mitophagy laboratory suggests that Nix-dependent mitochondria degradation may not occur due to Nix-induced mitochondrial membrane depolarization, but instead by an as yet unidentified mechanism (Zhang et al., 2009a). Furthermore, this study also determines that depolarized mitochondria fail to accumulate in Atg7 –/– cells as one would expect if Nix was responsible for depolarizing the mitochondrial membrane prior to mitochondria degradation and that Atg7 –/– reticulocytes are still capable of eliminating excess mitochondria in the putative absence of functional autophagy. These findings imply that Nix may drive mitochondria degradation by a unique, undiscovered pathway unrelated to mitochondrial membrane depolarization and that mitochondria are not being exclusively degraded via an autophagy-dependent pathway.
In an interesting coincidence, a letter to Nature in October of 2009 presented the data that mammalian cells lacking Atg5 and Atg7 can still engage in some form of “alternative” macroautophagy. When the information on Nix and mitophagy presented by Zhang and Ney is considered in the light of this “alternative autophagy” data, it seems possibly that Nix-induced depolarization of mitochondrial membranes may indeed trigger mitophagy, but perhaps a mitophagy that is dependent on this “alternative autophagy” machinery rather than on the traditional Atg7-dependent variety.
Recent efforts have also shown that erythrocytes are not the only blood cells to require mitophagy for normal development. Fresh data from experiments on T-lymphocytes shows that, upon exiting the thymus, these cells rely upon mitophagy to remove excess mitochondria (Pua et al., 2009). The implications of this finding have not been fully explored, but it does suggest that mitophagy plays a role in T-cell development. Additionally, there is evidence of the importance of mitochondria clearance in lens cells as well (Bassnett, 2009) during periods of differentiation.
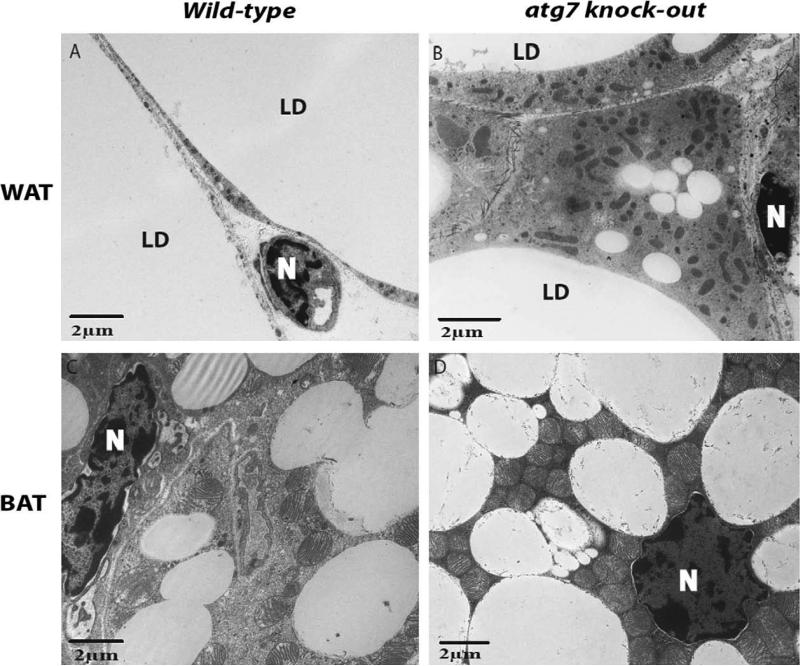
Finally, through recent work in our laboratory we have stumbled upon what we believe to be yet another potential model for investigating the importance of mitophagy in cellular differentiation. In normal mice, energy-storage-associated white adipose tissue consists of cells occupied almost entirely by a single lipid droplet, with minimal presence of observable cytoplasm or organelles other than the nucleus. Alternatively, heat-generating brown adipose tissue is made up of comparatively smaller cells containing many small lipid droplets, observable amounts of cytoplasm, and a considerable number of mitochondria (Fig. 2A and C). By cross-breeding Atg7/flox/flox conditional knock-out mice developed by Komatsu et al. (Komatsu et al., 2005) with mice harboring aP2 (Fabp4)-Cre, we were able to generate mice that selectively lacked autophagic activity in fat tissue (Zhang et al., 2009b). The resulting phenotype exhibited white adipocytes that possessed multiple small lipid droplets (instead of the normal single large lipid droplet observed in normal white adipocytes), contained much more cytoplasm than their wild-type counterparts, and perhaps more importantly, possessed significantly more mitochondria than their wild-type counterparts (Fig. 2A and B). Additionally, a striking phenotype was also observed in brown adipose tissue, where the cytoplasm of the mutant brown adipocytes was saturated with mitochondria (Fig. 2C and D). Our findings are supported by a similar study on Atg7 conditional knock-out mice (Singh et al., 2009) and it appears clear to us that Atg7-dependent autophagy is essential for normal mitochondria homeostasis in adipocytes.
Fig. 2.
Mitophagy is critical for normal adipocytes differentiation. Mice with a targeted deletion of the essential autophagy-related gene Atg7 in adipose tissue show increased mitochondria content. LD: lipid droplet; N: nucleus; WAT: white adipose tissue; BAT: brown adipose tissue.
3.3. Mitophagy-related cellular pathology and diseases
Neurons rely heavily upon mitophagy for normal development and function. These cells depend upon both general autophagy for protein quality control and also upon mitophagy for the removal of damaged mitochondria (Chen and Chan, 2009). In this context, autophagy is believed to serve a neuroprotective role. However, dysregulation of both general and mitochondria-targeted autophagy may result in neurodegeneration (Jellinger, 2009; Lee et al., 2009; Madeo et al., 2009).
Until recently, the connection between mitophagy and neuropathology had been predominantly explored with respect to Parkinson's disease. It is believed that mitochondrial dysfunction may play a key role in the pathogenesis of this disease, and many works have shown an apparent relationship between the disease-associated protein Parkin and the maintenance of normal mitochondrial function (Abou-Sleiman et al., 2006; Park et al., 2009; Yao and Wood, 2009). A recent paper by Narendra et al. has provided direct evidence that Parkin is selectively recruited to the membranes of mitochondria that were rendered dysfunctional via pharmacologic treatment with a mitochondrial uncoupler, and that this recruitment results in the up-regulation of mitophagy (Narendra et al., 2008). The practical implication of these findings is that defective mitophagy (via a lack of mitochondria targeting by Parkin protein) may be to blame for much of the pathology observed in this disease.
Additional work on the relationship between mitophagy and neuron homeostasis has been conducted on Purkinje cell degradation (PCD) mice. These mice suffer a profound neurologic ataxia due to the progressive loss of over 99% of their Purkinje cell neurons in the first three weeks of age (Mullen et al., 1976). Autophagy is up-regulated in the Purkinje cells of these mice, and it appears to be targeted selectively towards the degradation of mitochondria (Chakrabarti et al., 2009). The authors of this study posit the hypothesis that affected Purkinje cells experience a generalized increase in autophagic activity, accompanied by a concurrent increase in mitophagy, and that this increase in mitophagy may lead to the energetic collapse of Purkinje cells, which require a great deal of energy to function normally. However, the authors also note that mitochondria morphology is irregular in the affected cells, which may imply that some sort of mitochondrial dysfunction precedes the up-regulation of mitophagy. In fact, we believe that a more plausible hypothesis in this case is the latter, and that mitophagy is up-regulated in a neuroprotective effort to protect the Purkinje cells from excessive damage due to an accumulation of dysfunctional mitochondria. Regardless, the existing research in both Purkinje cell degeneration and Parkinson's disease models underscores the importance of a selective mechanism for the degradation of mitochondria in the milieu of the multicellular organism.
Another possible mechanism for targeted mitochondria degradation in neuronal tissue that is currently under investigation is the activity of protein kinases. Recent work utilizing models of Parkinson's disease and related parkinsonian disorders has identified several kinases, including c-Jun N-terminal kinases (JNKs) (Rumora et al., 2007; Zablocka et al., 2003), leucine rich repeat kinase 2 (LRRK2) (Biskup et al., 2006), PTEN-induced kinase 1 (PINK1) (Dagda et al., 2009), and the extracellular signal-related kinase complexes (ERK1/2) (Dagda et al., 2008; Kulich et al., 2007; Zhu et al., 2007) that may be involved in this process. Deregulation of these critical kinases might contribute to a host of neurodegenerative diseases through their impact on mitophagy (Dagda et al., 2009; Horbinski and Chu, 2005; Pagliarini and Dixon, 2006; Thomas and Cookson, 2009).
4. Concluding remarks
Since the study of autophagy at the molecular level began recently, our knowledge of the regulation, influence, and mechanisms of this process has expanded exponentially, leading to the remarkably complex system in place today. In the case of mitochondrial clearance, autophagy presents serves as the ideal mechanism for the clearance of damaged or excess mitochondria. Whether serving as a maintenance mechanism to protect the cell from damaged mitochondria, operating as a mechanism of energy homeostasis through the judicious regulation of the number of mitochondria, or playing a role in the complex progression of tissue differentiation, mitophagy has established itself as an increasingly important process. Future work will no doubt provide better insight into the regulation of mitophagy and into the mechanisms which endow it with its specificity. These findings will enhance our understanding of both autophagy and mitochondrial function within the cell, and will no doubt lead to advances in both the basic sciences and, hopefully, in the clinical environment as well.
References
- Abou-Sleiman PM, Muqit MM, McDonald NQ, Yang YX, Gandhi S, Healy DG, Harvey K, Harvey RJ, Deas E, Bhatia K, Quinn N, Lees A, Latchman DS, Wood NW. A heterozygous effect for PINK1 mutations in Parkinson's disease? Ann. Neurol. 2006;60:414–419. doi: 10.1002/ana.20960. [DOI] [PubMed] [Google Scholar]
- Ashford TP, Porter KR. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 1962;12:198–202. doi: 10.1083/jcb.12.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassnett S. On the mechanism of organelle degradation in the vertebrate lens. Exp. Eye Res. 2009;88:133–139. doi: 10.1016/j.exer.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulaton J, Lockshin RA. Ultrastructural study of the normal degeneration of the intersegmental muscles of Antheraea polyphemus and Manduca sexta (Insecta, Lepidoptera) with particular reference of cellular autophagy. J. Morphol. 1977;154:39–57. doi: 10.1002/jmor.1051540104. [DOI] [PubMed] [Google Scholar]
- Biskup S, Moore DJ, Celsi F, Higashi S, West AB, Andrabi SA, Kurkinen K, Yu SW, Savitt JM, Waldvogel HJ, Faull RL, Emson PC, Torp R, Ottersen OP, Dawson TM, Dawson VL. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann. Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Campbell CL, Thorsness PE. Escape of mitochondrial DNA to the nucleus in yme1 yeast is mediated by vacuolar-dependent turnover of abnormal mitochondrial compartments. J. Cell Sci. 1998;111(Pt. 16):2455–2464. doi: 10.1242/jcs.111.16.2455. [DOI] [PubMed] [Google Scholar]
- Chakrabarti L, Eng J, Ivanov N, Garden GA, La Spada AR. Autophagy activation and enhanced mitophagy characterize the Purkinje cells of pcd mice prior to neuronal death. Mol. Brain. 2009;2:24. doi: 10.1186/1756-6606-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EY, Kir S, Tooze SA. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 2007;282:25464–25474. doi: 10.1074/jbc.M703663200. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics – fusion, fission, movement, and mitophagy – in neurodegenerative diseases. Hum. Mol. Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Sandoval H, Wang J. Selective mitochondrial autophagy during erythroid maturation. Autophagy. 2008;4:926–928. doi: 10.4161/auto.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SL., Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 1957;3:349–362. doi: 10.1083/jcb.3.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 2005;1:131–140. doi: 10.4161/auto.1.3.2017. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Kulich SM, Chu CT. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress: implications for Parkinson's disease. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagda RK, Zhu J, Chu CT. Mitochondrial kinases in Parkinson's disease: converging insights from neurotoxin and genetic models. Mitochondrion. 2009;9:289–298. doi: 10.1016/j.mito.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky CH. Environmentally induced changes in mitochondria and endoplasmic reticulum of Saccharomyces carlsbergensis yeast. J. Cell Biol. 1976;71:123–135. doi: 10.1083/jcb.71.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. J. Cell Biol. 1967;33:437–449. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore SP, Qian T, Grissom SF, Lemasters JJ. The mitochondrial permeability transition initiates autophagy in rat hepatocytes. FASEB J. 2001;15:2286–2287. doi: 10.1096/fj.01-0206fje. [DOI] [PubMed] [Google Scholar]
- Ericsson JL. Studies on induced cellular autophagy. I. Electron microscopy of cells with in vivo labelled lysosomes. Exp. Cell Res. 1969a;55:95–106. doi: 10.1016/0014-4827(69)90462-5. [DOI] [PubMed] [Google Scholar]
- Ericsson JL. Studies on induced cellular autophagy. II. Characterization of the membranes bordering autophagosomes in parenchymal liver cells. Exp. Cell Res. 1969b;56:393–405. doi: 10.1016/0014-4827(69)90030-5. [DOI] [PubMed] [Google Scholar]
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, Klionsky DJ. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J. Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heynen MJ, Tricot G, Verwilghen RL. Autophagy of mitochondria in rat bone marrow erythroid cells. Relation to nuclear extrusion. Cell Tissue Res. 1985;239:235–239. doi: 10.1007/BF00214924. [DOI] [PubMed] [Google Scholar]
- Hopgood MF, Knowles SE, Ballard FJ. Degradation of microinjected glycolytic enzymes in L6 myoblasts. Biomed. Biochim. Acta. 1986;45:1603–1610. [PubMed] [Google Scholar]
- Horbinski C, Chu CT. Kinase signaling cascades in the mitochondrion: a matter of life or death. Free Radical Biol. Med. 2005;38:2–11. doi: 10.1016/j.freeradbiomed.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Mizushima N. A receptor for eating mitochondria. Dev. Cell. 2009;17:1–2. doi: 10.1016/j.devcel.2009.07.003. [DOI] [PubMed] [Google Scholar]
- Jellinger KA. Recent advances in our understanding of neurodegeneration. J. Neural Transm. 2009;116:1111–1162. doi: 10.1007/s00702-009-0240-y. [DOI] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 2008;283:32386–32393. doi: 10.1074/jbc.M802403200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Klionsky DJ. Atg32 is a tag for mitochondria degradation in yeast. Autophagy. 2009;5 doi: 10.4161/auto.5.8.9747. [DOI] [PubMed] [Google Scholar]
- Kanki T, Kang D, Klionsky DJ. Monitoring mitophagy in yeast: the Om45-GFP processing assay. Autophagy. 2009a;5 doi: 10.4161/auto.5.8.9854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Baba M, Bartholomew CR, Lynch-Day MA, Du Z, Geng J, Mao K, Yang Z, Yen WL, Klionsky DJ. A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell. 2009b doi: 10.1091/mbc.E09-03-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009c;17:98–109. doi: 10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr., Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 2001;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Arch. Biochem. Biophys. 2007;462:245–253. doi: 10.1016/j.abb.2007.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissova I, Salin B, Schaeffer J, Bhatia S, Manon S, Camougrand N. Selective and non-selective autophagic degradation of mitochondria in yeast. Autophagy. 2007;3:329–336. doi: 10.4161/auto.4034. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat. Rev. Mol. Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich SM, Horbinski C, Patel M, Chu CT. 6-Hydroxydopamine induces mitochondrial ERK activation. Free Radical Biol. Med. 2007;43:372–383. doi: 10.1016/j.freeradbiomed.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Lindsten T, Yang CY, Wu J, Zhao F, Zhang J, Selak MA, Ney PA, Thompson CB. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Liu L, Gao FB. Autophagy defects contribute to neurodegeneration induced by dysfunctional ESCRT-III. Autophagy. 2009;5:1070–1072. doi: 10.4161/auto.5.7.9823. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005;8:3–5. doi: 10.1089/rej.2005.8.3. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ, Nieminen AL, Qian T, Trost LC, Elmore SP, Nishimura Y, Crowe RA, Cascio WE, Bradham CA, Brenner DA, Herman B. The mitochondrial permeability transition in cell death: a common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta. 1998a;1366:177–196. doi: 10.1016/s0005-2728(98)00112-1. [DOI] [PubMed] [Google Scholar]
- Lemasters JJ, Qian T, Elmore SP, Trost LC, Nishimura Y, Herman B, Bradham CA, Brenner DA, Nieminen AL. Confocal microscopy of the mitochondrial permeability transition in necrotic cell killing, apoptosis and autophagy. Biofactors. 1998b;8:283–285. doi: 10.1002/biof.5520080316. [DOI] [PubMed] [Google Scholar]
- Madeo F, Eisenberg T, Kroemer G. Autophagy for the avoidance of neurodegeneration. Genes Dev. 2009;23:2253–2259. doi: 10.1101/gad.1858009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen RJ, Eicher EM, Sidman RL. Purkinje cell degeneration, a new neurological mutation in the mouse. Proc. Natl. Acad. Sci. USA. 1976;73:208–212. doi: 10.1073/pnas.73.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagy defends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- Nowikovsky K, Reipert S, Devenish RJ, Schweyen RJ. Mdm38 protein depletion causes loss of mitochondrial K+/H+ exchange activity, osmotic swelling and mitophagy. Cell Death Differ. 2007;14:1647–1656. doi: 10.1038/sj.cdd.4402167. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Kondo-Okamoto N, Ohsumi Y. A landmark protein essential for mitophagy: Atg32 recruits the autophagic machinery to mitochondria. Autophagy. 2009;5 doi: 10.4161/auto.5.8.9830. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Dixon JE. Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem. Sci. 2006;31:26–34. doi: 10.1016/j.tibs.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Munz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- Park J, Kim Y, Chung J. Mitochondrial dysfunction and Parkinson's disease genes: insights from Drosophila. Dis. Model Mech. 2009;2:336–340. doi: 10.1242/dmm.003178. [DOI] [PubMed] [Google Scholar]
- Priault M, Salin B, Schaeffer J, Vallette FM, di Rago JP, Martinou JC. Impairing the bioenergetic status and the biogenesis of mitochondria triggers mitophagy in yeast. Cell Death Differ. 2005;12:1613–1621. doi: 10.1038/sj.cdd.4401697. [DOI] [PubMed] [Google Scholar]
- Pua HH, Guo J, Komatsu M, He YW. Autophagy is essential for mitochondrial clearance in mature T lymphocytes. J. Immunol. 2009;182:4046–4055. doi: 10.4049/jimmunol.0801143. [DOI] [PubMed] [Google Scholar]
- Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot. Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kim I, Currin RT, Lemasters JJ. Tracker dyes to probe mitochondrial autophagy (mitophagy) in rat hepatocytes. Autophagy. 2006;2:39–46. doi: 10.4161/auto.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Enriquez S, Kai Y, Maldonado E, Currin RT, Lemasters JJ. Roles of mitophagy and the mitochondrial permeability transition in remodeling of cultured rat hepatocytes. Autophagy. 2009;5 doi: 10.4161/auto.5.8.9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinsztein DC, DiFiglia M, Heintz N, Nixon RA, Qin ZH, Ravikumar B, Stefanis L, Tolkovsky A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy. 2005;1:11–22. doi: 10.4161/auto.1.1.1513. [DOI] [PubMed] [Google Scholar]
- Rumora L, Lovric J, Sairam MR, Maysinger D. Impairments of heat shock protein expression and MAPK translocation in the central nervous system of follitropin receptor knockout mice. Exp. Gerontol. 2007;42:619–628. doi: 10.1016/j.exger.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Sandoval H, Thiagarajan P, Dasgupta SK, Schumacher A, Prchal JT, Chen M, Wang J. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers RL, Zhang J, Randall MS, Loyd MR, Li W, Dorsey FC, Kundu M, Opferman JT, Cleveland JL, Miller JL, Ney PA. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA. 2007;104:19500–19505. doi: 10.1073/pnas.0708818104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott SV, Baba M, Ohsumi Y, Klionsky DJ. Aminopeptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol. 1997;138:37–44. doi: 10.1083/jcb.138.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne JD, Arstila AU, Trump BF. Studies on cellular autophagocytosis. Cyclic AMP- and dibutyryl cyclic AMP-stimulated autophagy in rat liver. Am. J. Pathol. 1973;72:521–540. [PMC free article] [PubMed] [Google Scholar]
- Singh R, Xiang Y, Wang Y, Baikati K, Cuervo AM, Luu YK, Tang Y, Pessin JE, Schwartz GJ, Czaja MJ. Autophagy regulates adipose mass and differentiation in mice. J. Clin. Invest. 2009 doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal R, Winter G, Ecker N, Klionsky DJ, Abeliovich H. Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 2007;282:5617–5624. doi: 10.1074/jbc.M605940200. [DOI] [PubMed] [Google Scholar]
- Thomas KJ, Cookson MR. The role of PTEN-induced kinase 1 in mitochondrial dysfunction and dynamics. Int. J. Biochem. Cell Biol. 2009;41:2025–2035. doi: 10.1016/j.biocel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolkovsky AM. New gene on the block: Atg32-A specific receptor for selective mitophagy in S. cerevisiae. Autophagy. 2009;5 doi: 10.4161/auto.5.8.10124. [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y, Shimizu S. Another way to die: autophagic programmed cell death. Cell Death Differ. 2005;12(Suppl. 2):1528–1534. doi: 10.1038/sj.cdd.4401777. [DOI] [PubMed] [Google Scholar]
- Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Yao Z, Wood NW. Cell death pathways in Parkinson's disease: role of mitochondria. Antioxid. Redox Signal. 2009 doi: 10.1089/ars.2009.2624. [DOI] [PubMed] [Google Scholar]
- Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablocka B, Dluzniewska J, Zajac H, Domanska-Janik K. Opposite reaction of ERK and JNK in ischemia vulnerable and resistant regions of hippocampus: involvement of mitochondria. Brain Res. Mol. Brain Res. 2003;110:245–252. doi: 10.1016/s0169-328x(02)00653-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy. 2008;4:354–356. doi: 10.4161/auto.5552. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- Zhang J, Randall MS, Loyd MR, Dorsey FC, Kundu M, Cleveland JL, Ney PA. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood. 2009a;114:157–164. doi: 10.1182/blood-2008-04-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. Adiposespecific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proc. Natl. Acad. Sci. USA. 2009b;106:19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]