Abstract
Estrogens are key mediators of neuronal processes in vertebrates. As such, xenoestrogens present in the environment have the potential to alter normal central nervous system (CNS) function. The objectives of the present study were 1) to identify proteins with altered expression in the male fathead minnow telencephalon as a result of low level exposure to 17α-ethinylestradiol (EE2), and 2) to better understand the underlying mechanisms of 17β-estradiol (E2) feedback in this important neuroendocrine tissue. Male fathead minnows exposed to a measured concentration of 5.4 ng EE2/L for 48 hours showed decreased plasma E2 levels of approximately 2-fold. Of 77 proteins that were quantified statistically, 14 proteins were down-regulated after EE2 exposure, including four histone proteins, ATP synthase, H+ transporting subunits, and metabolic proteins (lactate dehydrogenase B4, malate dehydrogenase 1b). Twelve proteins were significantly induced by EE2 including microtubule-associated protein tau (MAPT), astrocytic phosphoprotein, ependymin precursor, and calmodulin. MAPT showed an increase in protein abundance but a decrease in mRNA expression after EE2 exposure, suggesting there may be a negative feedback response in the telencephalon to decrease mRNA transcription with increasing MAPT protein abundance. These results demonstrate that a low, environmentally relevant exposure to EE2 can rapidly alter the abundance of proteins involved in cell differentiation and proliferation, neuron network morphology, and long term synaptic potentiation. Together, these findings provide a better understanding of the molecular responses underlying E2 feedback in the brain and demonstrate that quantitative proteomics can be successfully used in ecotoxicology to characterize affected cellular pathways and endocrine physiology.
Introduction
It is well documented that 17β-estradiol (E2) regulates a number of physiological processes, including reproduction, metabolism, development, and growth. In the vertebrate central nervous system (CNS), E2 can both stimulate and inhibit neuronal cell proliferation (Brännvall et al., 2002; Galea et al., 2006; Mouriec et al., 2008), improve spatial learning and memory (Frye et al., 2007), modulate synaptic plasticity (Sá and Madeira, 2005), mediate neuroendocrine function (Kelly et al., 2002), and provide protection against neurotoxicity (Morissette et al., 2007). Therefore, many of the aforementioned processes that are regulated by E2 are also potentially disrupted by xenoestrogens that are found in the aquatic environment.
The telencephalon is a neuroendocrine tissue in teleost fish that contains the preoptic area and is dense in neurotransmitter producing cells that contain axonal projections to innervate the pituitary (Anglade et al., 1993). Experiments conducted decades ago with tritiated E2 revealed that the teleost neuroendocrine brain is a potential target for E2 feedback. Kim et al. (1978) identified E2 concentrating cells located in the supracommissural area of the telencephalon and preoptic area (POA) of goldfish (Carassius auratus), as well as in regions of the hypothalamus. Similarly, in the oyster toadfish (Opsanus tau), E2 positive cells are located in the posterior nuclei of the ventral telencephalon and nuclei of the POA (Fine et al., 1990). The localization of estrogen receptors (ERs) to telencephalic regions in several species of fish such as rainbow trout (Oncorhynchus mykiss) (Salbert et al., 1991), Atlantic croaker (Micropogonias undulatus) (Hawkins et al., 2005), and European seabass (Dicentrarchus labrax) (Muriach et al., 2008), provides additional evidence that the telencephalon is a target for E2 feedback. There is also a high capacity of the teleost brain to locally produce neuroestrogen, given its uniquely high aromatase (CYP19b) activity (Pasmanik and Callard, 1985) when compared to the mammalian brain. Aromatase converts testosterone (T) into E2, and there is close association between ERs and aromatase mRNA and protein levels in the forebrain of fish (Pellegrini et al., 2007). Therefore, there is good evidence that the telencephalon in fish is sensitive to E2 mediated signaling.
Recent studies have reported on the transcriptomic responses to estrogens in neuroendocrine regions of the teleost brain, such as the telencephalon and hypothalamus (Martyniuk et al., 2006, 2007; Marlatt et al., 2008). Some examples of major biological processes affected by estrogens in the teleostean telencephalon include cell differentiation, development, morphogenesis, signal transduction, and transcription (Martyniuk et al., 2007). In the hypothalamus, E2 affects the biological processes of cell protein metabolism, response to stimuli, transport, nucleobase or nucleotide metabolism, cell organization, and regulation of cellular processes (Marlatt et al., 2008). Although studies have demonstrated that the teleostean neuroendocrine transcriptome can be affected by estrogens, little is known about how changes in the transcriptome correlate to changes at the translational level in the brain.
This study investigated the response of the proteome in the telencephalon of male fathead minnows (FHMs; Pimephales promelas) exposed to 17α-ethinylestradiol (EE2) to better characterize the effects of estrogens in the teleost brain at the molecular level. The widespread inclusion of EE2 in oral contraceptives and subsequent elimination of EE2 from the body through waste products has led to a significant deposition of EE2 into water systems worldwide at concentrations typically ranging between 1 to 40 ng EE2/L (Yin et al., 2002). Most alarming is that continuous exposure to low concentrations (<10 ng EE2/L) can feminize male fish in the natural environment to a point where the population can no longer be sustained, as in the case of the FHM (Kidd et al., 2007). Currently, it is not well understood how environmentally relevant exposures to estrogenic pharmaceuticals and other xenoestrogens affect neuroendocrine signaling in aquatic species. This study provides complementary and unique perspectives on E2-mediated responses in the teleost brain from a mechanistic point of view (i.e. E2 feedback), and identifies putative proteins and cellular pathways potentially altered by environmentally relevant concentrations of a pharmaceutical estrogen.
Material and Methods
Male FHMs and EE2 exposure
Reproductively-mature, pond-reared male FHMs were purchased from Andersen Minnow Farm (Arkansas, USA). Upon arrival, the fish were treated for parasites and bacteria by a prophylactic salt-water dip (3%, 1 min), and were treated for trematodes using Praziquantel (2 mg/ml; once a week for three weeks) with a regime of formalin (Rid-ick Plus). Fish were allowed to recover for approximately three months prior to experimentation.
Males were separated from the general population and acclimated in the treatment aquaria for 48 h prior to exposure. The water used for this study was carbon-filtered, dechlorinated tap water. The exposure system consisted of 40 L glass aquaria and exposures were run in quadruplicate. Each aquarium contained six male FHMs in 25 L of treatment water. Because of the size of the fish, plasma and tissues were limited and different subsets of animals were used to measure plasma steroids and for proteomics. Whenever possible we used tissues from the same animals. In all cases, we used fish from replicate exposure tanks for each measurement. Each dose was prepared in separate 250 L fiberglass tanks the day of exposure. Tanks had fiberglass covers and were fitted with a pump and delivery system. Aquaria were equilibrated with test chemicals for 24 h prior to the introduction of fish to saturate the system. Test waters were renewed (90%) at 24 h by partial draining of the tank. Water temperature was maintained at 25°C, and the photoperiod consisted of a 16 h light:8 h dark cycle. The positions of the tanks were randomized. Fish were not fed the day before or during the experiment. Exposure initiation times were staggered in order to maintain an exposure/sampling interval of 48 h. The 48 h exposures was chosen based on vitellogenin (Vtg) induction from previous experiments using a single E2 injection in male fish in which both Vtg mRNA and plasma Vtg were at a maximum after 48 h (Bowman et al., 2002). In addition, based on other studies in small fish (Hoffmann et al. 2006), there would be a defined physiological response, specifically reduced sex steroids, after 48 hours that could be correlated to potential protein changes in the brain. Lastly, in the study by Martyniuk et al., (2009), 48 h exposure of FHMs was sufficient to alter the liver proteome with an anti-androgen and androgen. Therefore, it was expected that E2-sensitive proteins would also change in abundance within this time point in the brain.
EE2 was purchased from Sigma Chemical Company (St. Louis, MO). Working solutions for EE2 consisted of 1 mg/mL test compound solubilized in ethanol (3 mL) and mixed with a triethylene glycol (TEG) carrier (7 mL). The working solution was further diluted to make stock solutions for the treatment dose (nominal concentration of an environmentally relevant concentration of 10 ng EE2/L), and to maintain a concentration of 50 μl of TEG/L (0.005%) of test water. Control dose without EE2 contained ethanol and TEG at the same concentrations as the treatment dose.
Water samples were collected at the start of the exposures (0 h) and after the exposure (48 h). A sample of the test solution (1 L) was collected in a brown glass bottle with a teflon cap and stored at 4°C. Water sample measurements of EE2 were performed in triplicate using an enzyme-linked immunosorbent assay (ELISA) kit (Abraxis, Los Angeles, CA) following the manufacturer’s instructions. One liter of the water was concentrated on C18 columns (AccuBond II, Agilent Technologies, Palo Alto, CA, USA), eluted with dichloromethane, and evaporated to dryness with nitrogen. The sample was reconstituted with methanol and then water, resulting in 1.0 mL of a 10% methanol solution (1000 fold concentration factor). The average concentration (± SEM) of EE2 in the tanks was 5.4 ± 1.8 ng EE2/L and was measured in triplicate. There was no detectable EE2 in the control water samples.
All procedures involving live fish were reviewed and approved by the University of Florida Institutional Animal Care and Use Committee (IACUC). At the conclusion of the exposures, fish were euthanized using 100 mg/ml buffered tricaine methane sulfonate with sodium bicarbonate, and blood samples were collected from the caudal sinus into heparinized hematocrit tubes for analysis of E2 and testosterone concentrations. Blood samples were immediately centrifuged at 2000 x g for 10 min. and plasma was stored at −80°C for E2 and T quantification. Telencephali were collected from four fish per tank replicate and were immediately frozen in liquid nitrogen. Tissues were stored at −80°C until protein and RNA extraction.
Plasma steroid levels
For plasma T, individuals (n=9, control; n=8, treatment) were chosen at random from the four tanks. This subset was restricted to only those individuals that had sufficient plasma. Individuals originating from each of the 8 tanks (4 control and 4 treatment tanks) were used in the plasma T analysis. For plasma E2, a smaller random subset of animals was used (n=6, control; n=4, treatment) because not all individuals had sufficient plasma after T analysis. Individuals originating from 5 of the 8 tanks (3 control and 2 treatment tanks) were used in the plasma E2 analysis. For extractions of both E2 and T, plasma samples (10 μL) were added to 50 μL of buffer (50 mM sodium phosphate, 0.1% gelatin, pH 7.6) in 10 × 75 mm borosilicate glass test tubes. To determine extraction and resuspension efficiency for each sample, 65 μL of 50 nCi/mL 3H-estradiol or testosterone in buffer (Amersham, specific activity of 44 and 73 Ci/mmol, respectively) was added and samples were briefly mixed by vortexing. Each sample received 1 mL of 1-chlorobutane (n-butylchloride, Acros Organics, 99+% pure, Cat. # AC15463) and samples were mixed by vortexing vigorously for 30 s. Samples were centrifuged at 200 x g for 3 min. to separate phases. The upper organic phase was removed with a borosilicate glass pipet and placed into a 10 × 75 mm tube. The extraction process was repeated with an additional 1 mL of 1-chlorobutane. Extracts were pooled and evaporated under a gentle stream of nitrogen at 30°C. Dried extracts were reconstituted by addition of 75 μL buffer (50 mM sodium phosphate, 0.1% gelatin, 0.1% RIA grade bovine serum albumin, pH 7.6) followed by vortexing for 30 s. Tubes were sealed with parafilm and shaken overnight at 4°C. Standards (in serum) were extracted using the same methodology.
Extraction and reconstitution efficiency was determined by placing 10 μL of reconstituted sample into scintillation vials and adding 5 mL of scintillation cocktail (Scinti-Safe 30%, Fisher Scientific Inc., Rockford, IL., USA). Samples were counted using the tritium window for 3 min. and recovery was determined by comparison to total counts added. Hormone concentrations in reconstituted samples were analyzed using E2 or T specific ELISA assays purchased from Immuno-Biological Laboratories, Inc (IBL America, Minneapolis, MN) and performed according to manufacturer instructions. The limit of quantitation (LOQ) for E2 and T was established at 20 pg/mL and 25 pg/mL in analyzed samples, respectively. Intraday coefficient of variation (CV) was less than 5% across all standards, ranging from 0 to 4.5% as measured for each assay. Measured values were corrected for dilution and recovery estimations. Both absolute T levels and T log10 transformed were determined not to be normally distributed according to Shapiro-Wilk W test. A Brown & Forsythe’s test of homogeneity of variances was used for steroid data because control and treatment groups were unequal in size. Based on this test, E2 plasma levels in control and EE2 treated groups were determined not to be equal in variance. Therefore, a non-parametric Mann-Whitney U test was determined most appropriate for testing differences between control and treatment groups for both steroids. All analyses were performed in SPSS Statistics v17.0 (SPSS Inc. Chicago, Illinois, USA).
Protein Processing, Mass Spectrometry (LC MS/MS), and Database Searches
Approximately 20 mg of telencephalon from control (n=6) and EE2-treated fish (n=6) were mechanically disrupted in RadioImmuno Precipitation Assay (RIPA) Lysis and Extraction Buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% nonyl phenoxylpolyethoxylethanol-40, 1% sodium deoxycholate and 0.1 % SDS) (Pierce, Thermo Fisher Scientific Inc.) with 10 μl protease inhibitor cocktail (1 mg/mL) (Catalogue # P-8340; Sigma-Aldrich, St. Louis, MO, USA) to prevent degradation of proteins before being reduced and digested. To ensure sufficient amounts of protein, samples were pooled into groups of two individuals to yield three control samples and three treatment samples. Specifically, two telencephali were randomly pooled from the four replicate control tanks to yield three separate samples for iTRAQ labelling. This was repeated for the treatment group, and two individuals were pooled at random to yield three samples from the four treatment tanks. Proteins were precipitated with cold acetone by adding 6 times the volume of each sample (i.e., 200 μl sample and 1.2 ml acetone). Briefly, acetone-precipitated proteins were reconstituted in 20 μl dissolution buffer (iTRAQ® reagent; 0.5M triethylammonium bicarbonate, pH=8.5) and total protein was determined using Coomassie Plus Better Bradford Assay Reagent (Pierce). Each sample was adjusted to 25 μg total protein/sample for peptide labelling. Three iTRAQ labelling reactions were processed separately according to the manufacturer’s protocol (Applied Biosystems Inc, Foster City, CA), using label 114 (control=3) and label 115 (treatment=3).
As per the protocol outlined by Applied Biosystems, proteins were first reduced with 50 mM Tris-(2-carboxyethyl)phosphine and thiol groups subsequently blocked with methyl methanethiosulfonate. Proteins were trypsin digested in a volume of 100 μl with 1 mg trypsin/mL (Sigma) for 16 hours at 37°C. After digestion, peptides were labeled with iTRAQ reagents and the three control (label 114) and the three treatment (label 115) samples were randomly combined into three separate pools, each pool containing one labeled control sample and one labeled treated sample.
The following procedure was performed on each pooled iTRAQ sample separately. The sample was desalted using a macrospin column Vydac Silica C18 (The Nest Group Inc, Southboro, MA) and dried. Peptides were resuspended in 100 μl buffer A (75% 0.01M ammonium formate and 25% acetonitrile; ACN) for off-line strong cation exchange (SCX) fractionation on a polysulfoethyl A column (100 × 2.1 mm, 5 μm, 300 Å). Peptides were eluted using a linear gradient of 0–20% buffer B (75% 0.5M ammonium formate, 25% ACN) over 40 min., followed by a gradient of 20–100% buffer B for 5 min. Peptide peaks were monitored by UV at 280 nm. Approximately twenty peptide fractions were collected for each iTRAQ sample. Fractions were then pooled into 8 fractions of equal peptide content for mass spectrometry analysis. Therefore, a total of 24 total fractions were injected individually into a capillary trap LC Packings PepMap (DIONEX, Sunnyvale, CA). Fractions were desalted for 5 min. with a flow rate of 20 μL/min of 3% ACN, 0.1% acetic acid, 0.01% trifluoroacetic acid (TFA), and then continued onto an in-line LC Packing C18 Pep Map HPLC column (300 μM × 5 mm) connected to a hybrid quadrupole-TOF mass spectrometer QSTAR XL (Applied Biosystems) running at 200 nl/min. The elution gradient of the nano-HPLC column started at 3% solvent B (0.1% acetic acid and 96.9% ACN in water) and finished at 60% solvent B, and proceeded for 2 hours. The focusing potential and ion spray voltage was set to 275 V and 2600 V, respectively. The information-dependent acquisition (IDA) mode of operation was employed, in which a survey scan from m/z 400–1200 was acquired followed by collision induced dissociation of the three most intense ions. Survey and MS/MS spectra for each IDA cycle were accumulated for 1 and 3 s, respectively.
Tandem mass spectra were extracted by Analyst (v1.1, Applied Biosystems). In order to better assign peptides to proteins, a previously constructed ray-finned fish database (Martyniuk et al., 2009) containing trypsin digested peptides was searched using MS/MS data interpretation algorithms within Protein Pilot™ (Paragon™ algorithm, v 2.0, Applied Biosystems). All iTRAQ data were analyzed as a group to increase the power of the analysis. The Paragon algorithm from Protein Pilot™ was set up to search iTRAQ 4-plex samples as variable modifications, with methyl methanethiosulfonate as a fixed modification. The Protein Pilot™ algorithm was selected to search automatically for biological modifications such as homocysteines (Shilov et al., 2007). The confidence level for protein identification was set at 1.3 (95%), which is the default for the detected protein threshold in Paragon™. To calculate a false discovery rate (FDR) for peptide-protein assignments, Proteomics System Performance Evaluation Pipeline (ProteomicS PEP, Applied Biosystems) in Protein Pilot™ was used to create a reversed ray-finned fish database. The differential expression ratios for protein quantitation were obtained from Protein Pilot™, which calculates protein ratios using only ratios from the spectra that are distinct to each protein and excluding the shared peptides of protein isoforms. Peptides with low spectral counts are also excluded from the calculation of averages by setting the intensity threshold for the sum of the signal-to-noise ratio for all the peak pairs at >9. All the quantitative ratios were then automatically corrected for bias by Protein Pilot™ when processing the data to create the Pro Group™ algorithm result. Each protein that was quantified was done so with a minimum of two spectra. To calculate differential expression ratios, all high quality spectra from a protein were used to obtain an average protein ratio relative to the control label (i.e. fold change) using an error factor which represents 95% of the uncertainty range for a reported ratio. The p-value is then calculated based on the 95% confidence interval.
Pathway Analysis of Regulated Proteins
For proteins with altered abundance, pathway analysis was performed to identify cellular processes affected by EE2 (Pathway Studio® (v5.0), Ariadne Genomics, Rockville, MD, USA) (Nikitin et al., 2003). Human homologs (NCBI) for FHM proteins were obtained (RefSeq in NCBI), and Entrez Gene identifiers were retrieved using the ID mapping service of MD Anderson GeneLink (University of Texas, Houston, TX, USA). Pathways were built by finding the shortest paths between selected entities using the Resnet5 database in Pathway Studio®. All protein entities were used to identify common cellular processes that were modulated by EE2.
SYBR Green Real-Time PCR
Steady state mRNA levels for proteins representing diverse biological functions were measured by real-time PCR to evaluate the relationship between mRNA and protein levels. All primers for real-time PCR were designed using Primer3 (Rozen and Skaletsky, 2000) and were synthesized by MWG Biotech (Eurofin MWG Operon, Huntsville, AL) (Table 1). Optimal annealing temperature was between 58–60 °C and primers were designed to amplify sequences between 181–238 bp. Gene cloning strategies and cDNA synthesis protocol (Superscript-II, Invitrogen, Carlsbad, CA, USA) are described in Martyniuk et al. (2009). Accession numbers for partial FHM gene sequences cloned in this study are as follows: lactate dehydrogenase B4 (Ldhb4) (FJ755477), microtubule associated protein Tau (Mapt) (FJ755478), myelin protein zero (Mpz) (FJ755481), and Cu/Zn superoxide dismutase (Sod1) (FJ030938). We also investigated Cyp19b (AJ277866) steady state mRNA levels because of evidence that Cyp19b mRNA is regulated by estrogens in the teleost brain. The FHM amplicon aligned with other teleostean brain aromatase nucleotide sequences (ClustalW, NCBI).
Table 1.
Real-time PCR primers used to quantify mRNA levels in the telencephalon. Note that the forward primer is 5′-3′ in relation to the 5′-3′ coding strand and the reverse primer is 5′ – 3′ in relation to the 5′ – 3′ complimentary strand direction (anti-sense).
| Gene | Forward 5′-3′ Direction | Reverse 5′ to 3′ Direction | Amplicon Size (base pairs) |
|---|---|---|---|
| Cytochrome P450 aromatase (Cyp19b) | GGC ATC ATC TTC AAC AGC AA | CGC TCA GTG GGA TTT TTA GG | 238 |
| Lactate dehydrogenase B4 (Ldhb) | TGA ACT GGC TCT GGT TGA TG | TCT GAG GGA TGA TGT GCT TG | 233 |
| Microtubule associated protein Tau (Mapt) | GGC TCC AAA GAC AAT ATC AAA CA | AAT CCT CCT GTT GCC TCC TC | 234 |
| Myelin protein zero (Mpz) | TGG TGT AGT GTT GGG ACT GCT | AAT GAA GGG TGG GGA TGG | 181 |
| Superoxide dismutase (Sod1) | GCA CTT CAA CCC TCA CAC AC | TCT TCA TTG CCT CCC TTA CC | 213 |
| 18S ribosomal | CGG TTC TAT TTT GTG GGT TTC T | CCT CCG ACT TTC GTT CTT G | 212 |
Gene expression analysis (n=5 control; n=5 EE2) was performed on a subset of fathead minnows for which plasma T was also collected. Real-time PCR reactions were conducted using 1X iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA), 1 μl 10 mM gene specific primers, and 100–150 ng first-strand cDNA derived from DNase treated RNA samples. Transcripts were assayed on an iCycler Thermal Cycler (Bio-Rad). 18S rRNA was used to normalize gene expression (IQ Supermix, Bio-Rad). Serial dilutions (n=8) from a calculated starting copy number of 108 (to 101) were used to construct standard curves for each gene of interest as per the following equation; number of copies = (X * 6.022 × 1023)/(Y * 1×109 * 650), where X is the template amount (ng of vector + insert), Y is the template length (bp vector + insert), and 650 Da is the average weight of a base pair. The two step thermal cycling parameters were as follows: initial 1 cycle Taq activation at 95°C for 3 min, followed by 40 cycles of 95°C for 15 seconds, and 60°C for 1 min. After 40 cycles, a dissociation curve was produced starting at 55°C (+1°C/30 seconds) to 95°C. Reaction efficiencies for real-time PCR reactions ranged between 99.8–102.9% and R2 > 0.990. As negative controls, two independent samples of cDNA reactions without reverse transcriptase (no RT control) and two independent samples without cDNA template were included in all real-time PCR reaction plates. All samples and controls were run in duplicate. All gene expression data were normally distributed after log10 transformation except expression data for Mpz according to Shapiro-Wilk W test. A Levene’s test of homogeneity of variance revealed that gene expression data in control and EE2 treated groups were not equal in variance for Ldhb4 and Sod1. Therefore, it was determined that a non-parametric Mann-Whitney U test was most appropriate for evaluating differences in Ldhb4, Mpz and Sod1 mRNA steady state levels. A one-way ANOVA was performed on log transformed data for Cyp19b and Mapt. All analyses were performed in SPSS Statistics v17.0.
Results
EE2 decreased plasma E2 in FHM males
EE2 significantly decreased plasma E2 levels in male FHMs (p<0.01) (Figure 1A). Plasma E2 levels ranged from 482.8 to 878.5 pg/mL in control males (n=6) with a mean (±SEM) of 672.0 ± 58.1 pg/mL and ranged from 333.0 to 468.6 pg/mL in EE2 exposed males (n=4), with a mean (±SEM) of 392.31 ± 28.1 pg/mL.
Figure 1.
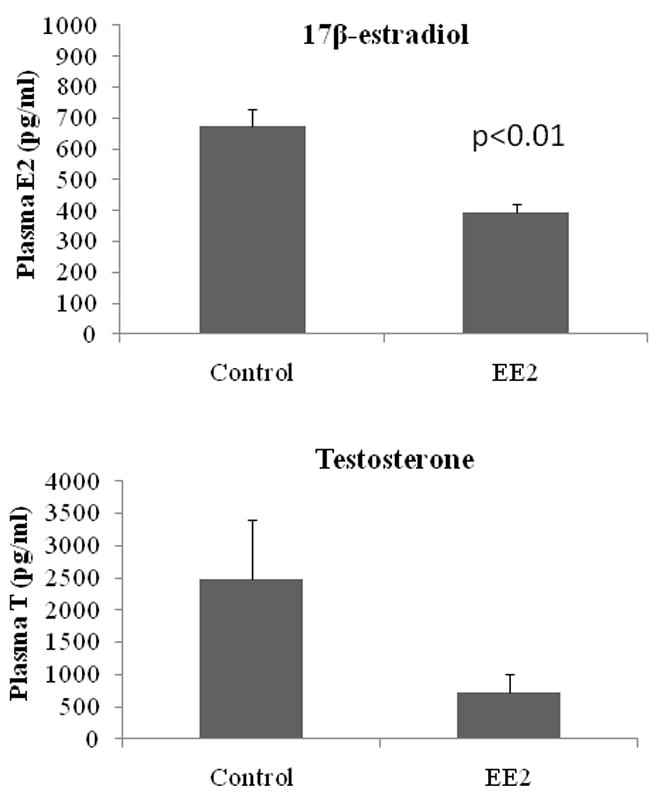
Plasma levels of A) 17β-estradiol (E2); (control, n=6; EE2, n=4) and B) testosterone (T) (control, n=9; EE2, n=8) in male fathead minnow after 48 h exposure to 5.4 ng 17α-ethinylestradiol (EE2)/L. Steroid levels were analyzed using a non-parametric Mann-Whitney U test but data are presented as mean plasma steroid concentration (±SEM). A significant difference was considered p<0.05.
Plasma T levels were not significantly altered after a 48 h exposure of FHMs to EE2 (p=0.11) (Figure 1B). Plasma T levels ranged from 144.4 to 8513.9 pg/mL in control males (n=9) with a mean (±SEM) of 2472.0 ± 924.9 pg/mL and ranged from 79.0 to 1830.1 pg/mL in EE2 exposed males (n= 8), with a mean (±SEM) of 733.3 ± 269.7 pg/mL.
Telencephalic proteins and cell processes altered by EE2
A list of all detected proteins (NCBI NR accession), the confidence of the peptide-protein identification, the peptide sequence, modifications to the peptide, and missed peptide cleavages, are provided in Appendix 1. The total number of peptide spectra detected in this study, including post-translational modifications, was 5315. The number of distinct peptides was 1523. Of all the distinct peptides, 39.3% could be assigned to a known protein with confidence. The number of proteins identified in each iTRAQ experiment was 122, 116, and 121. Therefore, there was good overlap in the number of proteins identified across iTRAQ samples, but in some cases, each iTRAQ experiment did detect peptides from different proteins. The total number of unique proteins identified when considering all three iTRAQ experiments was 192 (cut off 1.3; 95%) and 77 proteins were quantifiable with a minimum of two spectra. Identification of false positives in the reverse databases were 10 (iTRAQ 1), 3 (iTRAQ 2), and 5 (iTRAQ 3) respectively, corresponding to a FDR of 8.2%, 2.6%, and 4.1% in each iTRAQ experiment (average FDR = 5.0%).
Of the 77 proteins that were statistically quantified, 14 were significantly decreased in abundance and 12 were significantly increased in abundance after EE2 treatment (Table 2). Four of these proteins were considered to be potentially altered by EE2 (p<0.10), and are also included in Table 2 for comparison. However, it is noted that proteins with the highest probability of being affected by EE2 are those that are identified in multiple iTRAQ experiments, those that have been quantified using multiple spectra, and those that are significant at FDR<5%. Proteins listed in Table 2 are organized by the magnitude of the fold change for both down-regulated and up-regulated proteins. When available, the protein abbreviation is also provided and used in the text throughout the manuscript. Proteins down-regulated by EE2 included four histone proteins, ATP synthase, H+ transporting subunits (ATP5A1 and ATP5B), and metabolic proteins (LDHB4 and MDH1A). Proteins induced by EE2 included MAPT, astrocytic phosphoprotein, EPD, and CAM. Pathway analysis of proteins affected by EE2 in the telencephalon indicated that these proteins are involved in oxidative phophorylation, proliferation, differentiation, apoptosis, neuron network morphology, and long term potentiation (LTP) (Figure 2).
Table 2.
Differentially expressed proteinsa identified by iTRAQ and LC MS/MS (QSTAR) after 17α-ethinylestradiol (EE2) treatment (FDR=5%). All protein fold changes are relative to control. All high quality peptides identified from three independent iTRAQ experiments (each iTRAQ experiment was comprised of a protein pool of two control and two treated males) were used to quantify each protein and generate an error factor which is a 95% uncertainty range for a reported ratio that is used to generate a p-value. Included in the table are the NCBI sequence records (gi), the % coverage of protein using all spectra identified, and the number of spectra used in the quantification of the protein. Number of spectra used in quantitation includes all peptides with selected modifications and all charge states from all three iTRAQ experiments combined.
| NCBI sequence identifier | Protein Nameb | % Coverage of Protein | # Spectra used in Quantitation | Relative Fold Change | p-value |
|---|---|---|---|---|---|
| Down-regulated | |||||
| gi|47221340 | PREDICTED: similar to Histone H1.2 * (HIST1H2) | 20.65 | 7 | −4.49 | 0.005 |
| gi|999097 | Myelin protein zero * (MPZ) | 22.77 | 26 | −3.44 | 0.002 |
| gi|47224014 | Golli-mbp isoform 1 *** (MBP) | 36.87 | 128 | −2.72 | 0.017 |
| gi|125808645 | Histone protein Hist2h3c1 ** (HIST2H3C1) | 59.18 | 28 | −2.36 | <0.001 |
| gi|47225212 | Histone 1, H4c *** (HIST1H4C) | 42.31 | 43 | −2.28 | <0.001 |
| gi|68438153 | PREDICTED: similar to histone cluster 1, H2bb *** (HIST1H2BB) | 50.00 | 18 | −1.93 | 0.004 |
| gi|110351026 | 14 kDa apolipoprotein *** | 67.91 | 24 | −1.92 | <0.001 |
| gi|50344982 | Cytochrome c oxidase subunit Vb **(COX5B) | 29.37 | 17 | −1.39 | 0.071 |
| gi|41053939 | Malate dehydrogenase 1a, NAD (soluble) *(MDH1A) | 23.93 | 8 | −1.37 | 0.030 |
| gi|2208895 | Beta-globin ***(HBB) | 91.22 | 12 | −1.36 | 0.001 |
| gi|47207317 | ATP synthase, H+ transporting, mitochondrial F1 complex alpha subunit 1, cardiac muscle *** (ATP5A1) | 33.94 | 14 | −1.31 | 0.006 |
| gi|47218629 | ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide *** (ATP5B) | 44.68 | 18 | −1.26 | 0.020 |
| gi|56207279 | Lactate dehydrogenase B4 *** (LDHB) | 25.15 | 25 | −1.23 | 0.019 |
| gi|45767777 | Tubulin, alpha 8 like 4 *** (TUBA8L4) | 46.00 | 31 | −1.13 | 0.043 |
| Up-regulated | |||||
| gi|66472750 | Dihydropyrimidinase-like 3 *** (DPYSL3) | 26.28 | 11 | 1.31 | 0.097 |
| gi|3907633 | Beta-1 tubulin ** (TUBB1) | 48.99 | 4 | 1.44 | 0.004 |
| gi|71152827 | Purkinje cell protein 4 *** (PCP4) | 80.95 | 7 | 1.80 | 0.079 |
| gi|37590349 | Enolase 1, (alpha) *** (ENO1) | 51.16 | 3 | 2.14 | 0.037 |
| gi|62955673 | Putative stathmin 1/oncoprotein 18 variant 5 *** (STMN1) | 31.54 | 17 | 2.15 | 0.010 |
| gi|68356454 | Unknown Protein *** | 40.85 | 10 | 2.97 | 0.071 |
| gi|78099194 | Calmodulin (CaM) *** (CAM) | 73.83 | 136 | 3.12 | <0.001 |
| gi|47086029 | Myristoylated alanine-rich C kinase substrate 2 ** (MARCKS) | 27.05 | 8 | 3.26 | <0.001 |
| gi|125818422 | Unknown Protein *** | 10.91 | 5 | 3.69 | 0.035 |
| gi|47215443 | Astrocytic phosphoprotein * | 35.38 | 9 | 3.71 | 0.026 |
| gi|125812189 | Microtubule-associated protein tau * (MAPT) | 8.77 | 5 | 6.07 | 0.002 |
| gi|950228 | Ependymin precursor ** (EPD) | 41.28 | 3 | 7.83 | 0.004 |
All proteins in the table were significantly altered (p<0.05) except four proteins (p<0.10) which we also present for comparison. Seventy-seven proteins were quantifiable.
Asterisks indicates the number of iTRAQ labelling (one, two, or three) experiments in which peptides for the protein was identified. Quantitative data for all proteins are provided in the Appendix.
Figure 2.
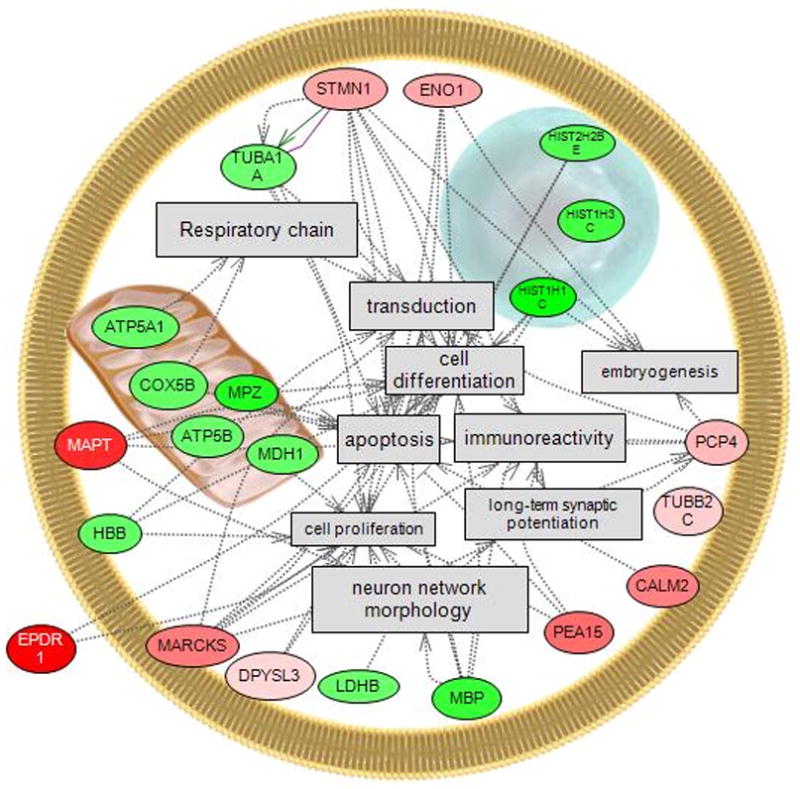
Pathway analysis of significantly altered proteins in the telencephalon after 48 h exposure to 17α-ethinylestradiol (EE2). Proteins altered in abundance by EE2 were involved in cell processes that included proliferation, differentiation, apoptosis, oxidative phosphorylation, neuron network morphology, and long term potentiation. Red indicates an induction in protein abundance and green indicates a reduction in protein abundance. Symbols include  extracellular/intracellular proteins and receptors. Except for oxidative phosphorylation, all cell processes shown are affected by >3 of the regulated proteins. Abbreviations are as follows; ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1, ATP5A1; ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide, ATP5B; calmodulin 2, CALM2; cytochrome c oxidase subunit Vb, COX5B; dihydropyrimidinase-like 3, DPYSL3; UCC1; enolase 1, alpha non-neuron, ENO1; ependymin related protein 2, hemoglobin, beta adult minor chain, HBB; histone 1, H2bl, H2BFQ; histone 1, H1c, HIF2; lactate dehydrogenase B, LDHB; malate dehydrogenase 1, NAD (soluble), MDH1; microtubule-associated protein tau, MAPT; myelin basic protein, MBP; myelin protein zero, MPZ; phosphoprotein enriched in astrocytes 15, PEA15; Purkinje cell protein 4, PCP4; similar to CG31613-PA, H3FC; similar to Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate 80 kDa protein), MARCKS; stathmin 1, STMN1; tubulin, alpha 1, TUBA3; tubulin, beta 2c, TUBB2.
extracellular/intracellular proteins and receptors. Except for oxidative phosphorylation, all cell processes shown are affected by >3 of the regulated proteins. Abbreviations are as follows; ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit, isoform 1, ATP5A1; ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide, ATP5B; calmodulin 2, CALM2; cytochrome c oxidase subunit Vb, COX5B; dihydropyrimidinase-like 3, DPYSL3; UCC1; enolase 1, alpha non-neuron, ENO1; ependymin related protein 2, hemoglobin, beta adult minor chain, HBB; histone 1, H2bl, H2BFQ; histone 1, H1c, HIF2; lactate dehydrogenase B, LDHB; malate dehydrogenase 1, NAD (soluble), MDH1; microtubule-associated protein tau, MAPT; myelin basic protein, MBP; myelin protein zero, MPZ; phosphoprotein enriched in astrocytes 15, PEA15; Purkinje cell protein 4, PCP4; similar to CG31613-PA, H3FC; similar to Myristoylated alanine-rich C-kinase substrate (MARCKS) (Protein kinase C substrate 80 kDa protein), MARCKS; stathmin 1, STMN1; tubulin, alpha 1, TUBA3; tubulin, beta 2c, TUBB2.
Transcript steady state levels in the telencephalon for selected proteins
There were no significant changes in mRNA steady state levels of Cyp19b, Ldhb4, Mpz, and Sod1 (Figure 3A, B, D, E). Transcript levels of Mapt (Figure 3C) were decreased in the telencephalon after EE2 treatment (d.f.=1; F=4.23; p=0.07). CYP19b protein was not detected with our proteomics method. SOD1 protein was detected in our analysis but did not have the minimum number of suitable spectra for confident quantification. MAPT protein was significantly increased and MPZ and LDHB4 were both significantly decreased after EE2 exposure. In the case of MAPT, MPZ and LDHB4, transcript levels did not correspond to changes at the protein level.
Figure 3.
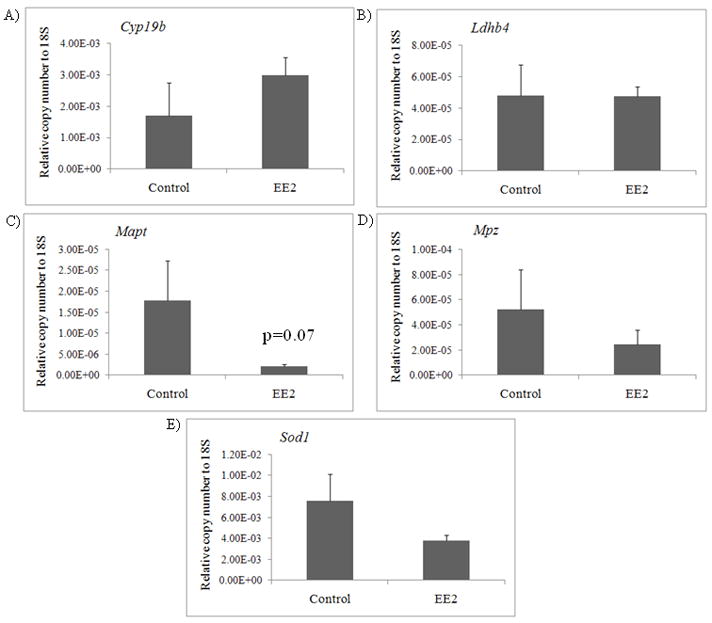
Real-time PCR (control n=5; EE2 n=5) for A) Cyp19b mRNA B) B) lactate dehydrogenase B4 (Ldhb4) mRNA C) MAP Tau (Mapt) mRNA D) myelin protein zero (Mpz) mRNA, and E) Cu/Zn superoxide dismutase (Sod1) mRNA. Data are presented as mean copy number (±SEM). A significant difference was considered p<0.05.
Discussion
EE2 significantly decreases plasma E2 in FHM males
In the present study, exposure of male FHMs to 5.4 ng EE2/L for 48 h resulted in a significant 2-fold reduction in plasma E2 levels. Testosterone levels between control and EE2-treated males were not statistically significant due to the high variation in circulating T levels among control males. The high variation in T levels may reflect complex socio- responses because many species of fish tend to have both dominant and subordinate males, which influence circulating steroid levels (Cardwell et al., 1996).
Environmentally relevant concentrations of EE2 have previously been shown to reduce circulating levels of plasma steroids in fish. In male goldfish, Martyniuk et al. (2006) observed a significant 10-fold reduction in plasma E2 and a 3-fold decrease in plasma T after 2 weeks exposure to 30 ng EE2/L. Male FHMs treated with 10, 20 or 40 ng EE2/L for 21 days showed significant decreases in E2, T, and 11-ketotestosterone of approximately 2-fold, 4-fold, and 10-fold respectively (Salierno and Kane, 2009). The relatively rapid inhibition of sex steroids by EE2 has also been observed in female zebrafish (Danio rerio). Exposure of zebrafish to 15, 40, and 100 ng EE2/L for 48 h significantly reduced circulating E2, while T was significantly reduced by 40 and 100 ng EE2/L (Hoffmann et al. 2006). The reduction in plasma E2 and T were between 2–3 fold. This study and others suggests that environmentally relevant levels of EE2 can rapidly reduce (within 48 h) E2 and T levels in small teleost fish. Reductions in circulating plasma levels of sex steroids have been associated with decreased population size over time in FHMs using a model-based approach (Ankley et al., 2008), suggesting that low exposures to xenoestrogens such as EE2 can potentially have a significant negative impact on local populations by altering plasma sex steroid levels.
Structural proteins are altered by EE2
We have demonstrated recently that the iTRAQ method can be applied successfully to study the effects of aquatic contaminant exposures in teleost fish (Martyniuk et al., 2009). The present study detected 77 proteins that were quantifiable using LC MS/MS and of these, 14 proteins were significantly down-regulated and 12 proteins were significantly up-regulated after EE2 treatment. Alterations in the relative fold change of protein abundance ranged from a 4.5-fold decrease in HIST1H2 to a 7.8- fold increase in EPD, a brain glycoprotein found in the extracellular space of the teleost CNS. Transcript levels for many of the proteins altered by EE2 have previously been shown to be responsive to E2 in the teleostean brain (Martyniuk et al., 2006, 2007; Marlatt et al., 2008). A comparison of our results with the findings reported in these studies is provided later in the discussion.
In the current study, EE2 exposure significantly induced MAPT protein by approximately 6-fold. In the CNS, MAPs are found in high abundance in the axons of neuronal cells and are primarily involved in stabilizing microtubule structure, acting as centers for microtubule association and disassociation. MAPT is also implicated in the etiology of neurodegenerative diseases such as Alzheimer’s disease. MAPT is associated with tangles that form throughout the CNS, and is susceptible to multiple post-translational modifications that prevent the proper formation of cytoskeletal structure (reviewed in Chen et al., 2004). Ferreira and Caceres (1991) demonstrated that E2 can increase MAPT protein in cultures of rat hypothalamic neurons approximately 3-fold between days three and seven of exposure. The increase in MAPT was also associated with enhanced stability of microtubules and neurite length, suggestive of a neuroprotective role for E2. Conversely, during the rat estrous cycle, MAPT was reported not to significantly change in protein abundance in the hippocampus or frontal cortex (Reyna-Neyra et al., 2004) suggesting that MAPT sensitivity to E2 may be different in specific regions of the brain. In the same study, however, microtubule associated protein 2 (MAP2) did show estrous related changes in the hippocampus and was significantly up-regulated during the transition from the proestrous to the metestrous stage. Therefore, related structural MAPs can be differentially expressed throughout reproductive stages.
There is also evidence that MAPT is involved in neuroendocrine function as microtubules are considered to play an important role in the secretion of rat pituitary hormones such as prolactin (Martinez de la Escalera and Weiner, 1992). This role of microtubules may be modulated in part by estrogens. Rats injected every 4 weeks with 5 mg E2 for 7 days showed a significant increase in the abundance of MAPT in the pituitary (Matsuno et al., 1997). Here, we show that EE2 can increase MAPT protein in vivo in the teleostean telencephalon and suggest that there is the potential for neuroendocrine function to be affected by exposure to environmental levels of EE2.
A major function of MAPs involves interacting with multiple forms of tubulins, such as TUBB1 and TUBA8L4, to form the cytoskeleton. We observed an increase in protein levels for TUBB1 and a decrease in protein levels for TUBA8L4 after EE2 treatment. In the goldfish retina, MAPT and TUBB1 transcription are significantly increased and correlated in expression after physical neuronal damage (Neumann et al., 1983). Here we also observed a positive correlation with protein levels of MAPT and TUBB1 in the telencephalon in response to EE2. There is evidence that protein interactions with MAPT are modulated by estrogens. It has been proposed that E2 facilitates the interaction between MAPT and glycogen synthase kinase 3/β-catenin and mediates the hyperphosphorylation of MAPT, a possible neuroprotective mechanism against neurodegeneration by stabilizing cytoskeletal synaptic plasticity (Garcia-Segura et al., 2007). EE2 appears to modulate both the transcription and translation of MAPT in the teleost brain and the regulation of MAPT may be involved in E2-mediated synaptic transmission as well as neuroprotection within the CNS.
Cell differentiation, proliferation, neuron network morphology, and long term potentiation (LTP) are processes affected by EE2
Estrogens have long been identified as hormones that promote neurite growth in vitro (Ferreira and Caceres, 1991; Lustig et al., 1994), a result consistent with the pathway analysis that EE2 regulates cell differentiation and proliferation in the FHM telencephalon. Of specific interest, some proteins regulated by EE2 were also involved in the processes of neuron network morphology and LTP. Many EE2 sensitive proteins including MPZ, MBP, and MAPT are structural proteins that play a key role in maintaining neuron network morphology. This structural network is needed for balancing plasticity and stabilizing synaptic connections. In addition, MBP, a number of MAPs, and TUBB are major proteins found in glial cytoplasmic preparations from human multiple system atrophy, an adult onset neurodegenerative disease (Wenning et al., 2008). E2-mediated changes in the abundance of these structural proteins may lead to increased synaptic stability and may play a role in neuroprotection of estrogens.
Another cell process involving EE2-mediated proteins was LTP. LTP involves repetitive activation of excitatory synapses that increases synaptic strength and prolongs signaling. LTP improves neuronal communication across the synapse and involves calmodulin and Ca2+ mediated pathways, in addition to several other biochemical pathways that include NMDAR, AMPAR, PKA and PKC, to allow for prolonged neuronal activity (reviewed in Dineley et al., 2001). Electrophysiological evidence suggests that estrogens modulate LTP within the mammalian CNS. In rat hippocampal CA1 slices perfused with 100 pmol E2, significant enhancement of LTP was observed after high frequency stimulation (100 Hertz, 1 second, 0.1 ms pulse, 10 second intervals) (Kim et al., 2002). In adult ovariectomized rats, stimulation pulses conforming to an LTP paradigm were applied to stratum radiatum in the CA1 region of both control and E2 treated animals (i.p. injection of 100 μg/kg body weight). E2 treated animals demonstrated a significant longer lasting increase in the measurement of synaptic strength (Córdoba Montoya and Carrer, 1997). Increased production of proteins such as CAM, MARCKS, and STMN1 by EE2 in this study may be involved in E2 modulation of LTP within the CNS. Given the importance of neuronal signaling in behavior, altered protein abundance within the telencephalon due to estrogens may be associated with rapid changes in reproductive behaviors. For example, alterations in nesting and courtship behaviours and reproductive fitness have been observed in male FHMs (Martinovi et al., 2007; Salierno and Kane, 2008) and male zebrafish (Colman et al., 2009) after varying exposure regimes to 17β-estradiol or EE2. However, there are complex and dynamic regulatory processes occurring in the CNS and it is currently speculative whether proteomic changes in the FHM brain precede changes in reproductive behaviors.
Transcript responses to EE2 in the male telencephalon
The expression of Cyp19b mRNA was quantified in this study because this transcript is responsive to E2 treatments in FHMs (Halm et al., 2002) and because in some teleost fish, the promoter of this gene is known to contain E2 response elements (Tchoudakova et al., 2001). However, this study detected no significant change in Cyp19b mRNA in the telencephalon. The lack of response of Cyp19b mRNA to EE2 exposure in the FHM telencephalon may be due to the low dose of EE2 used or short time point examined, as Cyp19b mRNA may be induced at later stages of exposure in order to maintain neuroestrogen synthesis needed for normal brain function in response to decreasing plasma E2 levels. A two week exposure to 300 ng EE2/L in male goldfish significantly induced Cyp19b in the telencephalon (Martyniuk et al., 2006). Peptides arising from Cyp19b protein were not detected using LC MS/MS and may be the result of low abundance or lack of peptide ionization.
Of the other genes examined, mRNA transcripts and their corresponding proteins did not typically follow the same directional fold change after exposure to EE2. In the case of MAPT, mRNA levels were decreased and protein abundance was increased. A lack of correlation between Mapt transcript and protein abundance may be due to differential regulatory mechanisms at the level of the transcriptome and proteome or differences in temporal patterns (i.e. time lag) of expression. The reduction in Mapt mRNA may be a negative feedback response of the genome to increasing protein abundance of Mapt. In a previous study in which male FHMs were exposed to a pharmaceutical androgen and anti-androgen for 48 h, liver mRNA levels also, in general, did not correlate with protein abundance (Martyniuk et al., 2009). It is important to keep in mind that protein regulation is occurring at that translational level and a change in mRNA steady state abundance is not a prerequisite for a change in protein abundance. Conversely, in the brain, there may be relatively rapid transcriptional changes that occur which return to basal mRNA levels after 48 h. Detailed time-course studies are needed to dissect the temporal relationship between gene expression and protein abundance after EE2 treatment.
Neuroendocrine Genes and Proteins Regulated by Estrogens
The proteomic response in the FHM telencephalon was compared to previously conducted genomic studies in neuroendocrine brain of teleosts. Interestingly, there were common genes and proteins identified as being regulated by estrogens. In male zebrafish, Martyniuk et al. (2007) examined the genomic response in the telencephalon after a 3-week exposure to a comparable dose of EE2 (10 ng EE2/L). Histone-related genes such as H3 histone family 3b (Hist3h3b) were induced >3-fold; however, this was opposite to the protein changes of histones in the FHM telencephalon after a 48 h exposure. Microarray analysis in male goldfish hypothalamus after treatment with 30 and 300 ng EE2/L for two weeks revealed significant transcriptional changes in Stmn1 (1.9-fold decrease), Hist2h2 (1.6-fold decrease), and Epd-II (1.3-fold decrease). Accompanying these changes in gene expression was a decrease in circulating plasma E2 levels in EE2-treated male goldfish, similar to our observations here in FHMs. Moreover, Zhang et al. (2009) used fadrozole, a reversible competitive inhibitor of aromatase, to investigate the transcriptional response in the telencephalon to a fadrozole-induced E2 decline in female goldfish. Fadrozole significantly reduced plasma levels of E2 and increased mRNA levels (1.3 to 1.5-fold) of Epd I and EpdII, Eno1, Mbp, Ldhb4, and Stmn1 and decreased mRNA levels of Cam. In the current study, EE2 also decreased plasma levels of E2 and altered protein abundance of EPD, ENO1, MBP, LDHB4 and CAM. The molecular changes that occur in the telencephalon at the transcript and protein level may therefore be an indirect response to the reduction of circulating plasma E2. In addition, physiological E2 action in the goldfish neuroendocrine brain included the regulation of calcium signaling pathway (Zhang et al., 2009), a result that is supported by our observation of inductions in FHM telencephalic proteins such as astrocytic phosphoprotein, MBP, and CAM after EE2 treatment as each of these proteins play a significant role in calcium signaling. Noteworthy is that there is evidence that mammalian ER-α has a CAM binding site, supporting a direct role of E2-mediated calcium signaling (García Pedrero et al., 2002). Lastly, the transcriptional (Martyniuk et al., 2006, 2007; Marlatt 2008) and translational (this study) regulation of apolipoproteins indicates that this may be a common gene and protein family affected by estrogens in the telencephalon. Lipid regulation, metabolism, and transport can be seasonally dependent, suggesting, in part that sex steroids mediate these processes in fish tissues (Jensen and Taylor, 2002).
To conclude, an increasing number of studies in the teleostean neuroendocrine brain are identifying genes and proteins regulated by estrogens. The proteomic data presented here complement genomic data obtained from previous studies with estrogens in neuroendocrine regions of the teleostean brain. There is a lack of information in fish reproductive biology addressing how gene expression relates to protein changes on a broad scale (Martyniuk and Denslow, 2009) and additional studies are required to better elucidate the extensive molecular responses initiated by sex steroids along the reproductive axis. In addition to demonstrating that quantitative proteomics in ecotoxicology may be a useful approach for biomarker discovery, this study provides novel data on sensitive proteins affected by EE2 and contributes to an improved understanding of the E2-mediated molecular cascades underlying neuroendocrine function in fish.
Supplementary Material
Acknowledgments
The authors would like to thank S McClung at the ICBR (UF) and S Alvarez for assistance adapting the iTRAQ method for use in teleost fishes. We also thank MC Croteau for helpful comments on the manuscript. This research was funded by a Natural Science and Engineering Research Council (NSERC) PDF fellowship to CJM and NIH grant # RO1 ES015449 awarded to NDD and DSB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anglade I, Zandbergen T, Kah O. Origin of the pituitary innervation in the goldfish. Cell Tissue Res. 1993;273:345–355. doi: 10.1007/BF00312837. [DOI] [PubMed] [Google Scholar]
- Ankley GT, Miller DH, Jensen KM, Villeneuve DL, Martinovi D. Relationship of plasma sex steroid concentrations in female fathead minnows to reproductive success and population status. Aquat Toxicol. 2008;88:69–74. doi: 10.1016/j.aquatox.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Bowman CJ, Kroll KJ, Gross TG, Denslow ND. Estradiol-induced gene expression in largemouth bass (Micropterus salmoides) Mol Cell Endocrinol. 2002;196:67–77. doi: 10.1016/s0303-7207(02)00224-1. [DOI] [PubMed] [Google Scholar]
- Brännvall K, Korhonen L, Lindholm D. Estrogen-receptor-dependent regulation of neural stem cell proliferation and differentiation. Mol Cell Neurosci. 2002;21:512–520. doi: 10.1006/mcne.2002.1194. [DOI] [PubMed] [Google Scholar]
- Cardwell JR, Sorensen PW, Van der Kraak GJ, Liley NR. Effect of dominance status on sex hormone levels in laboratory and wild-spawning male trout. Gen Comp Endocrinol. 1996;101:333–341. doi: 10.1006/gcen.1996.0036. [DOI] [PubMed] [Google Scholar]
- Chen F, David D, Ferrari A, Götz J. Posttranslational modifications of tau--role in human tauopathies and modeling in transgenic animals. Curr Drug Targets. 2004;5:503–515. doi: 10.2174/1389450043345236. [DOI] [PubMed] [Google Scholar]
- Colman JR, Baldwin D, Johnson LL, Scholz NL. Effects of the synthetic estrogen, 17alpha-ethinylestradiol, on aggression and courtship behavior in male zebrafish (Danio rerio) Aquat Toxicol. 2009;91:346–354. doi: 10.1016/j.aquatox.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Córdoba Montoya DA, Carrer HF. Estrogen facilitates induction of long term potentiation in the hippocampus of awake rats. Brain Res. 1997;778:430–438. doi: 10.1016/s0006-8993(97)01206-7. [DOI] [PubMed] [Google Scholar]
- Dineley KT, Weeber EJ, Atkins C, Adams JP, Anderson AE, Sweatt JD. Leitmotifs in the biochemistry of LTP induction: amplification, integration and coordination. J Neurochem. 2001;77:961–971. doi: 10.1046/j.1471-4159.2001.00321.x. [DOI] [PubMed] [Google Scholar]
- Ferreira A, Caceres A. Estrogen-enhanced neurite growth: evidence for a selective induction of Tau and stable microtubules. J Neurosci. 1991;11:392–400. doi: 10.1523/JNEUROSCI.11-02-00392.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine ML, Keefer DA, Russel-Mergenthal H. Autoradiographic localization of estrogen-concentrating cells in the brain and pituitary of the oyster toadfish. Brain Res. 1990;536:207–219. doi: 10.1016/0006-8993(90)90027-9. [DOI] [PubMed] [Google Scholar]
- Frye CA, Duffy CK, Walf AA. Estrogens and progestins enhance spatial learning of intact and ovariectomized rats in the object placement task. Neurobiol Learn Mem. 2007;88:208–216. doi: 10.1016/j.nlm.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galea LA, Spritzer MD, Barker JM, Pawluski JL. Gonadal hormone modulation of hippocampal neurogenesis in the adult. Hippocampus. 2006;16:225–232. doi: 10.1002/hipo.20154. [DOI] [PubMed] [Google Scholar]
- García Pedrero JM, Del Rio B, Martínez-Campa C, Muramatsu M, Lazo PS, Ramos S. Calmodulin is a selective modulator of estrogen receptors. Mol Endocrinol. 2002;16:947–960. doi: 10.1210/mend.16.5.0830. [DOI] [PubMed] [Google Scholar]
- Garcia-Segura LM, Diz-Chaves Y, Perez-Martin M, Darnaudéry M. Estradiol, insulin-like growth factor-I and brain aging. Psychoneuroendocrinology. 2007;32:S57–61. doi: 10.1016/j.psyneuen.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Halm S, Pounds N, Maddix S, Rand-Weaver M, Sumpter JP, Hutchinson TH, Tyler CR. Exposure to exogenous 17beta-oestradiol disrupts p450aromB mRNA expression in the brain and gonad of adult fathead minnows (Pimephales promelas) Aquat Toxicol. 2002;60:285–299. doi: 10.1016/s0166-445x(02)00011-5. [DOI] [PubMed] [Google Scholar]
- Hawkins MB, Godwin J, Crews D, Thomas P. The distributions of the duplicate oestrogen receptors ER-beta a and ER-beta b in the forebrain of the Atlantic croaker (Micropogonias undulatus): evidence for subfunctionalization after gene duplication. Proc Biol Sci. 2005;272:633–641. doi: 10.1098/rspb.2004.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann JL, Torontali SP, Thomason RG, Lee DM, Brill JL, Price BB, Carr GJ, Versteeg DJ. Hepatic gene expression profiling using Genechips in zebrafish exposed to 17alpha-ethynylestradiol. Aquat Toxicol. 2006;79:233–246. doi: 10.1016/j.aquatox.2006.06.009. [DOI] [PubMed] [Google Scholar]
- Jensen B, Taylor M. Lipid transport in female Fundulus heteroclitus during the reproductive season. Fish Physiol Biochem. 2002;25:141–151. [Google Scholar]
- Kelly MJ, Qiu J, Wagner EJ, Rønnekleiv OK. Rapid effects of estrogen on G protein-coupled receptor activation of potassium channels in the central nervous system (CNS) J Steroid Biochem Mol Biol. 2002;83:187–193. doi: 10.1016/s0960-0760(02)00249-2. [DOI] [PubMed] [Google Scholar]
- Kidd KA, Blanchfield PJ, Mills KH, Palace VP, Evans RE, Lazorchak JM, Flick RW. Collapse of a fish population after exposure to a synthetic estrogen. Proc Natl Acad Sci USA. 2007;104:8897–8901. doi: 10.1073/pnas.0609568104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Kim HY, Kim JH, Shin HK, Lee SH, Lee YS, Son H. Enhancement of rat hippocampal long-term potentiation by 17 beta-estradiol involves mitogen-activated protein kinase-dependent and -independent components. Neurosci Lett. 2002;332:65–69. doi: 10.1016/s0304-3940(02)00902-3. [DOI] [PubMed] [Google Scholar]
- Kim YS, Stumpf WE, Sar M. Topography of estrogen target cells in the forebrain of goldfish, Carassius auratus. J Comp Neurol. 1978;182:611–620. doi: 10.1002/cne.901820404. [DOI] [PubMed] [Google Scholar]
- Lustig RH, Hua P, Yu W, Ahmad FJ, Baas PW. An in vitro model for the effects of estrogen on neurons employing estrogen receptor-transfected PC12 cells. J Neurosci. 1994;14:3945–3957. doi: 10.1523/JNEUROSCI.14-06-03945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlatt VL, Martyniuk CJ, Zhang D, Xiong H, Watt J, Xia X, Moon T, Trudeau VL. Auto-regulation of estrogen receptor subtypes and gene expression profiling of 17beta-estradiol action in the neuroendocrine axis of male goldfish. Mol Cell Endocrinol. 2008;283:38–48. doi: 10.1016/j.mce.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Martinez de la Escalera G, Weiner RI. Hypothalamic regulation of microtubule- associated protein phosphorylation in lactotrophs. Neuroendocrinology. 1992;55:327–335. doi: 10.1159/000126133. [DOI] [PubMed] [Google Scholar]
- Martinović D, Hogarth WT, Jones RE, Sorensen PW. Environmental estrogens suppress hormones, behavior, and reproductive fitness in male fathead minnows. Environ Toxicol Chem. 2007;26:271–278. doi: 10.1897/06-065r.1. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Alvarez S, McClung S, Villeneuve DL, Ankley GT, Denslow ND. Quantitative proteomic profiles of androgen receptor signaling in the liver of fathead minnows (Pimephales promelas) 2009;8:2186–2200. doi: 10.1021/pr800627n. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Denslow ND. Towards functional genomics in fish using quantitative proteomics. Gen Comp Endocrinol. 2009;164:135–141. doi: 10.1016/j.ygcen.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Gerrie ER, Popesku JT, Ekker M, Trudeau VL. Microarray analysis in the zebrafish (Danio rerio) liver and telencephalon after exposure to low concentration of 17alpha-ethinylestradiol. Aquat Toxicol. 2007;84:38–49. doi: 10.1016/j.aquatox.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Xiong H, Crump K, Chiu S, Sardana R, Nadler A, Gerrie ER, Xia X, Trudeau VL. Gene expression profiling in the neuroendocrine brain of male goldfish (Carassius auratus) exposed to 17alpha-ethinylestradiol. Physiol Genomics. 2006;27:328–336. doi: 10.1152/physiolgenomics.00090.2006. [DOI] [PubMed] [Google Scholar]
- Matsuno A, Takekoshi S, Sanno N, Utsunomiya H, Ohsugi Y, Saito N, Kanemitsu H, Tamura A, Nagashima T, Osamura RY, Watanabe K. Modulation of protein kinases and microtubule-associated proteins and changes in ultrastructure in female rat pituitary cells: effects of estrogen and bromocriptine. J Histochem Cytochem. 1997;45:805–813. doi: 10.1177/002215549704500605. [DOI] [PubMed] [Google Scholar]
- Morissette M, Jourdain S, Al Sweidi S, Menniti FS, Ramirez AD, Di Paolo T. Role of estrogen receptors in neuroprotection by estradiol against MPTP toxicity. Neuropharmacology. 2007;52:1509–1520. doi: 10.1016/j.neuropharm.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Mouriec K, Pellegrini E, Anglade I, Menuet A, Adrio F, Thieulant ML, Pakdel F, Kah O. Synthesis of estrogens in progenitor cells of adult fish brain: evolutive novelty or exaggeration of a more general mechanism implicating estrogens in neurogenesis? Brain Res Bull. 2008;75:274–280. doi: 10.1016/j.brainresbull.2007.10.030. [DOI] [PubMed] [Google Scholar]
- Muriach B, Carrillo M, Zanuy S, Cerdá-Reverter JM. Distribution of estrogen receptor 2 mRNAs (Esr2a and Esr2b) in the brain and pituitary of the sea bass (Dicentrarchus labrax) Brain Res. 2008;1210:126–141. doi: 10.1016/j.brainres.2008.02.053. [DOI] [PubMed] [Google Scholar]
- Neumann D, Scherson T, Ginzburg I, Littauer UZ, Schwartz M. Regulation of mRNA levels for microtubule proteins during nerve regeneration. FEBS Lett. 1983;162:270–6. doi: 10.1016/0014-5793(83)80770-4. [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio--the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Pasmanik M, Callard GV. Aromatase and 5 alpha-reductase in the teleost brain, spinal cord, and pituitary gland. Gen Comp Endocrinol. 1985;60:244–251. doi: 10.1016/0016-6480(85)90320-x. [DOI] [PubMed] [Google Scholar]
- Pellegrini E, Mouriec K, Anglade I, Menuet A, Le Page Y, Gueguen MM, Marmignon MH, Brion F, Pakdel F, Kah O. Identification of aromatase-positive radial glial cells as progenitor cells in the ventricular layer of the forebrain in zebrafish. J Comp. 2007;501:150–167. doi: 10.1002/cne.21222. [DOI] [PubMed] [Google Scholar]
- Reyna-Neyra A, Arias C, Ferrera P, Morimoto S, Camacho-Arroyo I. Changes in the content and distribution of microtubule associated protein 2 in the hippocampus of the rat during the estrous cycle. J Neurobiol. 2004;60:473–480. doi: 10.1002/neu.20042. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sá SI, Madeira MD. Estrogen modulates the sexually dimorphic synaptic connectivity of the ventromedial nucleus. J Comp Neurol. 2005;484:68–79. doi: 10.1002/cne.20451. [DOI] [PubMed] [Google Scholar]
- Salbert G, Bonnec G, Le Goff P, Boujard D, Valotaire Y, Jego P. Localization of the estradiol receptor mRNA in the forebrain of the rainbow trout. Mol Cell Endocrinol. 1991;76:173–180. doi: 10.1016/0303-7207(91)90271-s. [DOI] [PubMed] [Google Scholar]
- Salierno JD, Kane AS. 17alpha-Ethinylestradiol Alters Reproductive Behaviors, Circulating Hormones, and Sexual Morphology in Male Fathead Minnows (Pimephales promelas) Environ Toxicol Chem. 2009;28:953–961. doi: 10.1897/08-111.1. [DOI] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Tchoudakova A, Kishida M, Wood E, Callard GV. Promoter characteristics of two cyp19 genes differentially expressed in the brain and ovary of teleost fish. J Steroid Biochem Mol Biol. 2001;78:427–439. doi: 10.1016/s0960-0760(01)00120-0. [DOI] [PubMed] [Google Scholar]
- Wenning GK, Stefanova N, Jellinger KA, Poewe W, Schlossmacher MG. Multiple system atrophy: a primary oligodendrogliopathy. Ann Neurol. 2008;64:239–246. doi: 10.1002/ana.21465. [DOI] [PubMed] [Google Scholar]
- Yin GG, Kookana RS, Ru YJ. Occurrence and fate of hormone steroids in the environment. Environ Int. 2002;28:545–551. doi: 10.1016/s0160-4120(02)00075-2. [DOI] [PubMed] [Google Scholar]
- Zhang D, Popesku JT, Martyniuk CJ, Xiong H, Duarte P, Yao L, Xia X, Trudeau VL. Profiling neuroendocrine gene expression changes following fadrozole-induced estrogen decline in the female goldfish. Physiol Genomics. 2009;38:351–361. doi: 10.1152/physiolgenomics.00051.2009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


