Summary
Keratan sulfate was originally identified as the major glycosaminoglycan of cornea but is now known to modify at least a dozen different proteins in a wide variety of tissues. Despite a large body of research documenting keratan sulfate structure, and an increasing interest in the biological functions of keratan sulfate, until recently little was known of the specific enzymes involved in keratan sulfate biosynthesis or of the molecular mechanisms that control keratan sulfate expression. In the last 2 years, however, marked progress has been achieved in identification of genes involved in keratan sulfate biosynthesis and in development of experimental conditions to study keratan sulfate secretion and control in vitro. This review summarizes current understanding of keratan sulfate structure and recent developments in understanding keratan sulfate biosynthesis.
Keywords: Cartilage, cornea, keratan sulfate, proteoglycan
INTRODUCTION
The existence of keratan sulfate has been known since the 1930s and by the 1950s, the basic structure had been elucidated. Development of keratan sulfate-specific endoglycosidases in the 1970s and of antikeratan sulfate monoclonal antibodies in the 1980s provided sensitive detection of keratan sulfate, leading to generation of a body of almost 1,000 research articles in which keratan sulfate is listed as a keyword. Yet despite this venerable history, many aspects of keratan sulfate, particularly the specific enzymes involved in its biosynthesis, remain obscure. The structure and tissue location of keratan sulfate have been reviewed several times in the last decade (1–3). This review focusses primarily on the most recent advances in our understanding of keratan sulfate structure and particularly, on keratan sulfate biosynthesis.
Keratan Sulfate Structure: One Name, Many Molecules
Meyer and coworkers determined keratan sulfate to be a linear polymer of N-acetylactosamine [→3Galβ(1→4)GlcNAcβ(1→] with sulfation occurring on the 6-carbon of both sugar moieties (4). The name “keratan” was coined with reference to the corneal origin of the molecule but a similar polysaccharide was also identified in cartilage. Keratan sulfate from the two tissues differs in the oligosaccharides linking the polymer to protein, leading to the designations of keratan sulfate I and II. Corneal keratan sulfate (KSI) is attached to Asn in core proteins via a complex-type N-linked branched oligosaccharide, whereas in cartilage, KSII is O-linked via GalNAc to Ser or Thr residues via a structure known as the mucin core-2. A third type of linkage to protein via mannose O-linked to Ser was identified more recently in brain. This glycoform can be considered KSIII (5). The structure of these linkage oligosaccharides is presented in Fig. 1.
Figure 1.
Keratan sulfate linkage oligosaccharides. The oligosaccharides linking keratan sulfate to protein define the three classes of keratan sulfate. “KS” refers to the keratan sulfate chain extension as described in the text. KS in parentheses indicates that keratan sulfate may not be present in all cases.
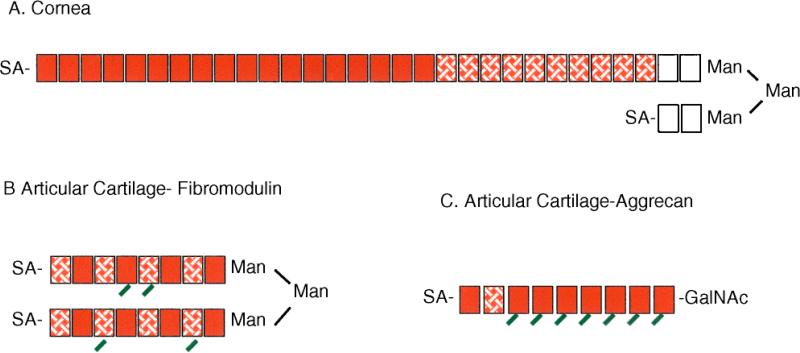
Keratan sulfate exhibits a remarkable variety of length and sulfation patterns. In porcine corneal keratan sulfate, described by Oeben et al. (6) and illustrated in Fig. 2A, keratan sulfate chains extend the C-6 branch of the linkage oligosaccharide but the C-3 branch is terminated in a single lactosamine capped by sialic acid. Sulfation in this porcine corneal keratan sulfate is distributed in a nonrandom pattern with two nonsulfated lactosamine disaccharides nearest the reducing end and 10–12 disaccharides distal to this region sulfated only on the GlcNAc moiety. The nonreducing end of these chains consists of a domain of variable length (8–34) of disulfated disaccharides. Further studies of corneal keratan sulfate from several sources support the idea that a single branch of the linker can be extended (as shown in Fig. 2A), but also indicate the presence of keratan sulfate in which both arms of the biantennary oligosaccharide are extended [reviewed in (2)]. The nonreducing ends of the keratan sulfate chains are capped with neuraminic acid, βGalNAc, or αGal (7, 8).
Figure 2.
The range of keratan sulfate chain structures. Polymer structure is represented graphically for keratan sulfate samples from several sources. Each box represents an N-acetyllactosamine disaccharide repeat [Galβ(1→4)GlcNAcβ(1→3)].  = unsulfated disaccharide,
= unsulfated disaccharide,  = disaccharide sulfated on GlcNAc only,
= disaccharide sulfated on GlcNAc only,  = disaccharide sulfated on both Gal and GlcNAc. Angled tails on the boxes indicate fucosylation. SA represents capping sugars, frequently sialic acid. The fully sulfated portion of the chain in A is variable in length and can be as much as twice as long as shown. The exact distribution of mono- and disulfated monomers in the internal portion of the chain in B has not been determined experimentally.
= disaccharide sulfated on both Gal and GlcNAc. Angled tails on the boxes indicate fucosylation. SA represents capping sugars, frequently sialic acid. The fully sulfated portion of the chain in A is variable in length and can be as much as twice as long as shown. The exact distribution of mono- and disulfated monomers in the internal portion of the chain in B has not been determined experimentally.
KSI is not limited to cornea. N-linked keratan sulfate chains are found in cartilage, modifying the proteins fibromodulin, PRELP, and osteoadherin (9–11). Aggrecan also contains 2–3 N-linked keratan sulfate chains in addition to 20 or more O-linked keratan sulfate chains (12). Proteins of the zona pellucida also carry keratan sulfate-like carbohydrates (13), and N-linked keratan sulfate has been isolated from the dermis of the pacific mackerel (14). The KSI structure in noncorneal tissues differs from that in cornea. As shown in Fig. 2B, KSI in fibromodulin is relatively short (8–9 disaccharides), more highly sulfated than keratan sulfate in cornea (15), and lacks the domain structure of corneal keratan sulfate. Size analysis indicates primarily a biantennary structure (16). Saccharides capping the nonreducing terminus of cartilage fibromodulin KSI resemble those modifying cartilage KSII more than those modifying corneal KSI (15), suggesting that capping may be tissue-specific rather than KS “type” specific.
KSII is exclusively linked to the protein aggrecan, the most abundant proteoglycan of cartilage. The structure of KSII from bovine articular cartilage, illustrated in Fig. 2C, consists of chains of 5–11 highly sulfated disaccharides, consisting almost completely of disulfated monomers interrupted occasionally by single monosulfated monomers (17). Gal in the linkage region can be sialylated. The nonreducing ends of the chains are capped by neuraminic acid at the C-3 or C-6 of the terminal GlcNAc. Alpha-fucose is also present on the C-3 of sulfated GlcNAc throughout the chain but not within four residues of the nonreducing terminus (18). KSII from tracheal cartilage is not fucosylated, and carries only (2→3) linked neuraminic acids at the chain terminus (19, 20). These detailed structural analyses show that tissue-specific factors appear to be a major determinant of keratan sulfate chain structure and capping, rather than the keratan sulfate type as determined by the linkage oligosaccharide.
Monoclonal antibodies recognizing sulfated regions of keratan sulfate react with extracts from most mammalian tissues and currently at least 16 proteins are known to be modified with keratan sulfate (2). In addition to proteoglycans of the extracellular matrix, keratan sulfate-linked proteins can be found associated with cell surfaces and intracellularly [reviewed in (2)]. Whereas all other glycosaminoglycans carry at least one negative charge per disaccharide, the lack of uronic acid in keratan sulfate and variability of sulfation of lactosamine monomers can result in a broad range of size and charge of keratan sulfate-containing molecules compared to other proteoglycans (21). Indeed, a number of proteins are modified with linear poly-N-acetyllactosamine that would be considered keratan sulfate except for absence of sulfation (22). The variable nature of keratan sulfate structure and the wide distribution of keratan sulfate epitopes observed in immunodetection studies suggests a likelihood that the number of molecules modified with keratan sulfate glycoforms in published literature will continue to increase.
KERATAN SULFATE BIOSYNTHESIS
Protein Sequence as a Determinant of Keratan Sulfate Biosynthesis
Keratan sulfate can be identified in a wide range of tissues but only a limited number of proteins in any single tissue is modified with keratan sulfate. The amino acid sequence of proteins modified by keratan sulfate must, consequently, be a determinant of keratan sulfate biosynthesis. The corneal and cartilage proteoglycans have been subjected to the greatest degree of analysis, correlating protein sequence with keratan sulfate presence. The keratan sulfate domain of aggrecan consists of a series of tandemly repeated hexapeptides with a consensus sequence of E(E/L)PFPS. Comparison of aggrecan DNA sequences shows that the number of these repeats varies among different species, 13 in human, 4 in rat and mouse, and none in chicken (23). The number of these repeats corresponds closely to the amount of keratan sulfate present in the aggrecan of the different organisms, demonstrating the dependence of KSII biosynthesis on this particular protein sequence.
In cornea three proteins, lumican, keratocan, and mimecan, are modified by keratan sulfate. The sites in these proteins were studied initially by labeling the linkage oligosaccharides after enzymatic removal of keratan sulfate (24). Analysis of labeled tryptic peptides showed only one of three possible N-linked glycosylation consensus sites was occupied by keratan sulfate in bovine lumican and mimecan. In keratocan, however, three of the potential four sites were modified with keratan sulfate. A subsequent study by Dunlevy et al. (25) used a different approach to isolate and sequence keratan sulfate-linked tryptic peptides from chicken cornea. This study identified keratan sulfate attachment at three of four lumican potential sites and 4 of 5 keratocan sites. The implication of both studies is that protein secondary structure and/or sequence, in addition to the consensus amino acid cluster NX(T/S), is involved in determining extension of N-linked oligosaccharides with keratan sulfate.
Corneal keratan sulfate proteoglycans are members of the Small Leucine Rich Proteoglycan (SLRP) family of proteins (26) in which much of the protein is built up of repetitive β-sheet/α-helix motifs, generating a horseshoe-shaped tertiary structure, with β-sheets on the inner surface and α-helices on the outside. The concave surface of the horseshoe is hypothesized to interact with collagen (27). Correlation of the predicted protein shape with sites of keratan sulfate attachment showed keratan sulfate attachment to be restricted to sites on the hydrophilic convex surface of the folded protein (25). This location is consistent with the role of SLRP proteins in collagen fibrillogenesis; however, such a model cannot account for all of the specificity of keratan sulfate chain initiation. Cornea also contains abundant decorin, a SLRP protein with about 40% sequence identity to lumican and keratocan. Decorin binds collagen, is thought to assume a horseshoe shape, and contains N-linked glycosylation sites (28). However, decorin is not modified with keratan sulfate except in embryonic chicken corneas (29). The ability of the biosynthetic apparatus to distinguish among such similar proteins in initiating keratan sulfate elongation, suggests the presence of specific amino acid signaling sequence(s) in the proteins that contain keratan sulfate. Dunlevy et al. suggest that proline residues within three amino acids of the keratan sulfate attachment sites in chicken lumican and keratocan could play a role (25). Additionally, the N-terminal ends of SLRP proteins modified with keratan sulfate (lumican, mimecan, keratocan, fibromodulin, PRELP, and osteoadherin) all contain one or more consensus sequences for tyrosine sulfation (Asp-Tyr) (2). The presence of sulfate at these sites has been confirmed in fibromodulin (9). Decorin and biglycan, SLRP proteins that lack keratan sulfate chains, on the other hand, lack Asp-Tyr sequences near the N-terminus. Currently there is no evidence for the role of tyrosine sulfation in keratan sulfate initiation, but the correlation of the two features in several of proteins is suggestive of such a role. It is clear from the current literature that protein sequence is important in keratan sulfate biosynthesis, but studies involving expression of proteins with modified sequence will be required to fully define this role.
Linkage Oligosaccharides
Keratan sulfate chain elongation is initiated at the nonreducing terminus of oligosaccharides that link the polymer to protein. The three known KS-linkage oligosaccharides (Fig. 1) can each be terminated with a variety of saccharide moieties. Keratan sulfate, in fact, represents an atypical modification of these oligosaccharides. Biosynthesis of these oligosaccharides is clearly an essential component of keratan sulfate biosynthesis, but there is no evidence at this time that synthesis of oligosaccharides extended with keratan sulfate differs from those that terminate in non–keratan sulfate-containing oligosaccharides.
Biosynthesis of N-linked KSI is inhibited by tunicamycin (30, 31), indicating that as with other N-linked oligosaccharides, the protein is initially glycosylated from a dolichol-linked high-mannose oligosaccharide donor substrate (32). Biosynthesis of N-linked keratan sulfate of fibromodulin is blocked by castanospermine and 1-(+)deoxymannojirimycin, inhibitors of processing of the high-mannose oligosaccharide to the biantennary form on which the keratan sulfate is extended (33). Swain-sonine, an inhibitor of processing of the C6 arm of the precursor, did not block addition of keratan sulfate to fibromodulin. In both control and swainsonine-treated cells, one or two N-linked oligosaccharides on each protein molecule were modified with keratan sulfate containing 4–6 repeating disaccharide units. Single short chains were added either to the C-3 or the C-6 branch in control cultures, whereas only the C-3 branch was substituted in the presence of swainsonine. Corneal KSI biosynthesis is similarly not blocked by swainsonine (31). These results indicate that the well-characterized pathways of processing of the high-mannose precursor oligosaccharide are involved in the biosynthesis of KSI. These studies also support the idea that polylactosamine addition to the precursor can extend either branch of the biantennary core depicted in Fig. 2A. It is not known, however, if extension of the C6 and/or C3 branches is site-specific with respect to the several N-linked glycosylation sites occurring in each keratan sulfate-linked protein.
KSII linkage region is identical to the mucin core-2 structure. A β-1,6-N-acetylglucosaminyltransferase designated as C2GnT is essential for formation of the core-2 oligosaccharide in a number of cell types. Its overexpression in CHO cells induces biosynthesis of polylactosamine (essentially nonsulfated keratan sulfate) on leukosialin, a protein not normally so modified (34). These results suggest that availability of the linkage oligosaccharide may represent a limiting step in the initiation of keratan sulfate addition to aggrecan.
Keratan sulfate in brain (KSIII) is linked to protein via 2-O-mannose (Fig. 1C). Although rare in most tissues, 2-O-mannose glycopeptides are present in as many as 30% of glycopeptides released by alkali-borohydride treatment of brain glycoproteins (35). Typically brain 2-O-mannose structures contain one or more nonsulfated lactosamine disaccharides, thus keratan sulfate in brain represents extension and sulfation of a glycoform that is common in brain but rare in other tissues. Enzymes directing O-mannosylation of mammalian proteins have not been well characterized, but appear homologous to a family of enzymes in yeast with specificities that depend on individual protein acceptors (35). Addition of GlcN to the mannosylated proteins is catalysed by protein-O-mannose β-1,2-N-acetylglucosaminyltransferase, known as POMGnT1. Mutations in the gene for this protein appear to be the cause of muscle-eye-brain disease, an autosomal recessive disorder characterized by congenital muscular dystrophy, ocular abnormalities, and lissencephaly (36).
Elongation of Keratan Sulfate
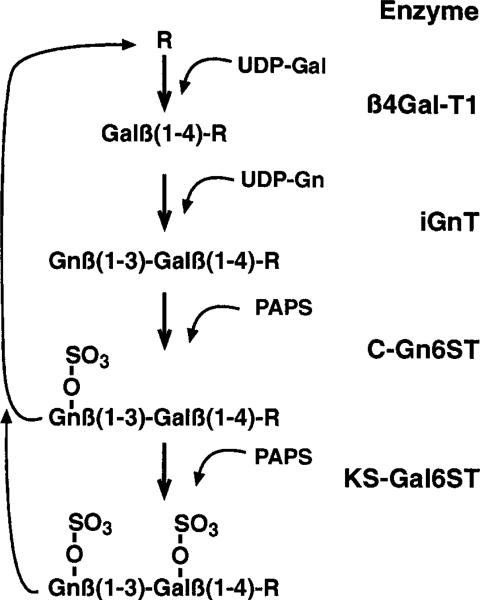
Extension of the keratan sulfate chain occurs via the action of glycosyltransferases that alternately add Gal and GlcNAc to the growing polymer (Fig. 3). Galactosyltransferase extracted from cornea resembles the β-1,4-galactosyltransferase enzyme (β4Gal-T1) abundant in serum and milk (37, 38). The gene for this common enzyme has been cloned and its expression found to be widespread. In many tissues βGal-T1 is considered a “housekeeping” gene and is expressed at levels independent of cell activity. During chick development a corneal β4Gal-T transcript was found to increase simultaneously with developmental upregulation of corneal keratan sulfate synthesis. Expression of this enzyme was maintained at unusually high levels in adult corneal cells (39). Interestingly, β4Gal-T activity continues at elevated levels in cultured corneal cells that have lost keratan sulfate synthesis (37).
Figure 3.
Enzymes involved in keratan sulfate biosynthesis. This illustrates the sequence of addition of sugars and sulfate to the growing chain (R). Sulfation of GlcNAc must occur before further elongation and may be required for chain elongation. Sulfation of Gal can occur after polymerization and appears optional. βGal-T1, β-1,4-galactosyltransferase; iGnT, beta-1,3-N-acetylglucosaminyltransferase; C-GlcNAc6ST, corneal N-acetylglucosaminyl-6-sulfotransferase; KS-Gal6ST, keratan sulfate galactosyl-6-sulfotransferase. Further description of these enzymes is provided in the text.
The β4Gal-T1 gene exhibits different modes of expression based on alternative transcription start sites. In most tissues, constitutive transcription results in moderate mRNA pools that do not fluctuate greatly. In mammary epithelial cells with high levels of lactose biosynthesis, a second transcriptional start site, regulated by a stronger, mammary gland-restricted promoter is involved. The transcript produced is distinguished from its housekeeping counterpart by the absence of approximately 180 nucleotides of 5′-untranslated sequence. This truncated transcript is translated more efficiently relative to the major transcript expressed in other somatic tissues (40). Promoter usage and transcript size of βGal-T1 in the keratan sulfate-producing cells are yet to be determined.
Screening of EST databases has resulted in discovery of least six additional genes showing close sequence similarity to the common β4Gal-T1 (41). Comparison of substrate specificity of recombinant forms of these enzymes has shown the β4Gal-T1 enzyme to be most efficient at synthesis of linear polylactosamine chains in N-linked oligosaccharides; however, the transferase β4Gal-T4 appears to be essential in generating the initial branches found in core-2 O-linked polylactosamine (42). The structural identity between N-linked polylactosamine and KSI chains suggests that the same enzymes are likely to be involved in their elongation, adding to the body of evidence pointing to β4Gal-T1 as the galactosyltransferase involved in keratan sulfate elongation. Null mutations in β4Gal-T1 gene in mice produce severe physiological problems and a high level of prenatal mortality (43). Glycoproteins in these mice are not completely devoid of 4-linked βGal residues, however, indicating that Galβ-T1 activity is replaced or supplemented by other family members in specific functions. Analysis of keratan sulfate in these mice would help confirm a central role for this gene in biosynthesis of keratan sulfate.
Extension of keratan sulfate involves transfer of both β-4-galactose and β-3-N-acetylglucosamine to the growing chain (Fig. 3). The β-1,3-N-acetylglucosaminyltransferase (β3GlcNAcT) enzyme responsible for keratan sulfate synthesis has not been unambiguously identified. A number of β3Glc-NAcT enzymes have been described, of which one works efficiently with β4Gal-T1 in synthesis of linear polylactosamine (known as the “i” antigen) (42). This enzyme, designated iGnT, exhibits a broad tissue distribution (44). RNA transcripts for iGnT are enriched in brain, a tissue in which keratan sulfate is actively synthesized. Polylactosamine synthesis in variant clones of PC12 nerve cells correlated with iGnT expression levels, implying a central role for this enzyme in polylactosamine elongation (45). Recently a family of three additional β3GlcNAcT enzymes has been identified and cloned, one of which can also participate in polylactosamine synthesis (46). In terms of keratan sulfate, however, it remains to be demonstrated which (if any) of this group of enzymes acts in concert with β4GalT1 in the elongation of the keratan sulfate chain, and whether identical combinations of enzyme(s) are involved in generation of the variety of keratan sulfates present in different tissues.
Sulfation of Keratan Sulfate
Sulfation of keratan sulfate occurs at the 6-position of both sugar moieties in the polymer. In cornea, galactose sulfation is often only partial, whereas sulfation of glucosamine is more nearly complete. An initial study using corneal cells identified two different proteins that catalyzed transfer of sulfate to keratan sulfate (47). Subsequent studies characterized and cloned cDNA for two different sulfotransferase enzymes active on keratan sulfate (48, 49). One of these enzymes (chondroitin-6-sulfotransferase, C6ST) transfers sulfate to GalNAc moieties of chondroitin sulfate, and also to Gal in keratan sulfate in vitro. This is a widely distributed enzyme that, in vitro, exhibits a 10- to 30-fold greater activity for chondroitin sulfate over keratan sulfate. Mice with knockout mutations for C6ST show a 90% loss of 6-sulfation of chondroitin sulfate but no loss of keratan sulfate in the spleen, as determined by immunohistology (50). These results suggest that C6ST may not be involved in keratan sulfate in vivo. The second enzyme shown to catalyze galactose sulfation of keratan sulfate (KS-Gal6ST) exhibits little activity toward other carbohydrates (48). This gene shows enhanced expression in brain and cornea, tissues with active keratan sulfate biosynthesis. It would therefore appear likely that this sulfotransferase is one of the enzymes involved in keratan sulfate biosynthesis. As indicated in Fig. 3, KS-Gal6ST transfers sulfate to keratan sulfate after polymerization.
Keratan sulfate is also sulfated on the GlcNAc moieties. Nakazawa and coworkers (51) have demonstrated that N-acetylglucosaminyl-6-sulfotransferase (GlcNAc6ST) activity in keratocyte extracts, transfers sulfate only to terminal N-acetylglucosamine, whereas Gal moieties at internal locations were sulfated by keratocytes. As shown in Fig. 3, restriction of GlcNAc sulfation to the terminal sugar requires this sulfation to occur simultaneously with elongation. Five genes for enzymes that transfer sulfate to nonreducing terminal GlcNAc have been identified. These enzymes differ in substrate specificity and in tissue location (52). Expression of one of these enzymes (C-GlcNAc6ST) is highly restricted to cornea, whereas a very closely related gene (I-GlcNAc6ST) is primarily expressed in intestine. These two genes are immediately adjacent to one another on human chromosome 16q22 (53, 54).
The essential role of C-GlcNAc6ST in corneal keratan sulfate synthesis was discovered by genetic linkage analysis studies of the autosomal recessive genetic disease macular corneal dystrophy (MCD). In the second decade of life MCD patients develop corneal opacities, a pathology that is correlated with undersulfation or complete absence of sulfation of corneal keratan sulfate. Two forms of the disease are observed, type I shows absence of keratan sulfate in the serum as well as in cornea (55, 56). In type II MCD, keratan sulfate is absent in the cornea but detected in the serum. In type I MCD, a variety of mutations have been identified all of which occur in the reading frame of the C-GlcNAc6ST protein (57–59). Analysis of a type II patient revealed an apparent homologous recombination between I-GlcNAc6ST and C-GlcNAc6ST genes, replacing the corneal promoter with sequence from the intestinal form of the gene. In this case, keratan sulfate was present in the serum but not in the cornea (59).
The essential role of C-GlcNAc6ST in keratan sulfate biosynthesis was demonstrated when this enzyme together with KS-Gal6ST was expressed in HeLa cells (60). Expression of both enzymes simultaneously resulted in secretion of high molecular weight glycoproteins detected with the anti-keratan sulfate antibody 5-D-4. Introduction of GlcNAc6ST cDNA bearing missense mutations identical to those found in MCD patients abolished keratan sulfate biosynthesis by these cells. Human I-GlcNAc6ST expression could not replace C-GlcNAc6ST in driving keratan sulfate biosynthesis, but cDNA for the highly homologous protein mouse I-GlcNAc6ST did participate in keratan sulfate synthesis. Mouse I-GlcNAc6ST and human C-GlcNAc6ST protein sequences contain alanine at position 217 but the inactive human I-GlcNAc6ST has isoleucine at this location. One of the characterized MCD missense mutations, inactive during in vitro keratan sulfate sulfation, contains a single amino substitution of threonine at position 217. These findings suggest that amino acid 217 may be important in determining sulfotransferase activity of the C/I-GlcNAc6ST enzymes toward keratan sulfate. Interestingly, the mouse I-GlcNAc6ST ortholog of human C-GlcNAc6ST was expressed in mouse cornea, suggesting that in mouse, this gene may be the GlcNAc6ST responsible for keratan sulfate biosynthesis (60).
The specificity of the GlcNAc6ST enzyme predicts that keratan sulfate GlcNAc sulfation may occur simultaneously with elongation and only on the terminus of the growing chain (61, 62). The concept of coordinated elongation and sulfation of keratan sulfate is supported by biosynthetic studies with cell-free corneal extracts showing a coordinate change in the Vmax for both elongation and sulfation activities with respect to keratan sulfate chain length (63). A recent analysis of keratan sulfate structure from MCD corneas using fluorophore-assisted, carbohydrate electrophoresis (64, 65), demonstrated the expected loss of GlcNAc sulfation in keratan sulfate from MCD I and II corneas, but also found elimination or reduction in Gal sulfation and a reduction of keratan sulfate chain length. These results help confirm the involvement of GlcNAc sulfation in chain elongation of corneal keratan sulfate and also implicate GlcNAc6ST activity in Gal sulfation as well. Thus, a growing body of evidence suggests a central role of C-GlcNAc6ST in corneal keratan sulfate biosynthesis. The relationship of these findings to keratan sulfate in other tissues is yet to be determined.
Metabolic Control of Keratan Sulfate Biosynthesis
Keratan sulfate biosynthesis is often markedly altered in response to metabolic, pathologic, or developmental changes in tissues. Typically, keratan sulfate biosynthesis is lost in wound healing and in vitro. Cell types that secrete the most abundant keratan sulfate in vivo (neural cells, chondrocytes, and keratocytes) are quiescent in vivo, but when grown in culture, chondrocytes and keratocytes often revert to fibroblastic morphology and secrete little keratan sulfate. In corneal wounds, keratocytes are activated to cell division, adopt a fibroblastic phenotype similar to cultured corneal fibroblasts, and synthesize little keratan sulfate. Subacute or chronic pathological conditions of the corneal stroma also frequently lead to loss of keratan sulfate in the tissue (66, 67). Typically, loss of corneal keratan sulfate is associated with inflammation, suggesting a role for proinflammatory cytokines in the downregulation of keratan sulfate biosynthesis.
Studies with cultured bovine keratocytes showed expression of all three of the KSI-linked proteins but found them to be modified with short oligolactosamine chains with minimal sulfation (21). These results suggest that downregulation of keratan sulfate biosynthesis in vitro (and by implication in healing wounds) relates to activities of keratan sulfate-specific glycosyl- and/or sulfotransferases. This idea is supported by a study in which freshly isolated chicken keratocytes lost GlcNAc sulfotransferase activity after several days of culture (38). Specific enzymes required for polymerization and sulfation of keratan sulfate may, therefore, be key regulators of keratan sulfate biosynthesis in vitro and possibly in vivo as well. Development of culture methods for chondrocytes and keratocytes allowing biosynthesis of fully sulfated keratan sulfate for extended periods in vitro provide important tools for identification of signals mediating expression of keratan sulfate-specific biosynthetic enzymes (68). Keratan sulfate biosynthesis by keratocytes cultured using this method was downregulated by fetal bovine serum and by transforming growth factor β (TGFβ)(69), but fibroblast growth factor 2 (FGF2) acted to stimulate keratan sulfate synthesis in vitro (70). The recent elucidation of genes coding for the enzymes of keratan sulfate biosynthesis, and the availability of isolated cell cultures in which keratan sulfate biosynthesis can be modulated, provides the opportunity to investigate the molecular mechanisms that control expression of this long-elusive biomolecule.
CONCLUSION
The first six decades of research on keratan sulfate has produced a wealth of information on the structure, location, and biosynthesis of this complex family of glycoconjugates. The next few years are likely to witness positive identification of the remaining genes involved in keratan sulfate biosynthesis and an understanding of how expression of these genes is controlled by signals both outside and inside the cell. The sensitive and subtle patterns of keratan sulfate expression in various tissues during development and wound healing suggests complex biological roles for these molecules. Unraveling these mysteries may not take another 60 years, but it seems highly likely that at least half of the story of keratan sulfate is yet to be told.
ACKNOWLEDGEMENTS
The author thanks Martha Funderburgh for critical reading of the manuscript. This work was supported by National Institutes of Health Grant EY09368, Research to Prevent Blindness, and the Eye and Ear Foundation of Pittsburgh. JLF is a Jules and Doris Stein Research to Prevent Blindness Professor.
Abbreviations
- CHO
Chinese hamster ovary
- C6ST
chrondoitin-6-O-sulfotransferase
- EST
expressed sequence tags
- FGF2
fibroblast growth factor 2
- Gal
D-galactose
- Gal-6ST
galactosyl 6-O-sulfotransferase
- β4Gal-T
β1,4-galactosyltransferase
- GlcNAc
N-acetyl-d-glucosamine
- GlcNAc6ST
N-acetylglucosaminyl 6-O-sulfotransferase
- β3GlcNAcT
β1,3-N-acetylglucosaminyltransferase
- KS
keratan sulfate
- MCD
macular corneal dystrophy
- PRELP
proline arginine-rich end leucine-rich protein
- POMGnT
protein-O-mannose β-1,2-N-acetylglucosaminyl transferase
- SRLP
small leucine-rich protoeglycan
- TGFβ
transforming growth factor β
REFERENCES
- 1.Funderburgh JL, Funderburgh ML, Mann MM, Conrad GW. Physical and biological properties of keratan sulphate proteoglycan. Biochem. Soc. Trans. 1991;19:871–876. doi: 10.1042/bst0190871. [DOI] [PubMed] [Google Scholar]
- 2.Funderburgh JL. Keratan sulfate: structure, biosynthesis, and function. Glycobiology. 2000;10:951–958. doi: 10.1093/glycob/10.10.951. [DOI] [PubMed] [Google Scholar]
- 3.Greiling H. Structure and biological functions of keratan sulfate proteoglycans. EXS. 1994;70:101–122. doi: 10.1007/978-3-0348-7545-5_7. [DOI] [PubMed] [Google Scholar]
- 4.Meyer K, Linker A, Davidson EA, Weissman B. The mucopolysaccharides of the cornea. J. Biol. Chem. 1953;205:611–616. [PubMed] [Google Scholar]
- 5.Krusius T, Finne J, Margolis RK, Margolis RU. Identification of an O-glycosidic mannose-linked sialylated tetrasaccharide and keratan sulfate oligosaccharides in the chondroitin sulfate proteoglycan of brain. J. Biol. Chem. 1986;261:8237–8242. [PubMed] [Google Scholar]
- 6.Oeben M, Keller R, Stuhlsatz HW, Greiling H. Constant and variable domains of different disaccharide structure in corneal keratan sulphate chains. Biochem. J. 1987;248:85–93. doi: 10.1042/bj2480085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tai GH, Nieduszynski IA, Fullwood NJ, Huckerby TN. Human corneal keratan sulfates. J. Biol. Chem. 1997;272:28227–28231. doi: 10.1074/jbc.272.45.28227. [DOI] [PubMed] [Google Scholar]
- 8.Tai GH, Huckerby TN, Nieduszynski IA. Multiple non-reducing chain termini isolated from bovine corneal keratan sulfates. J. Biol. Chem. 1996;271:23535–23546. doi: 10.1074/jbc.271.38.23535. [DOI] [PubMed] [Google Scholar]
- 9.Antonsson P, Heinegard D, Oldberg A. Posttranslational modifications of fibromodulin. J. Biol. Chem. 1991;266:16859–16861. [PubMed] [Google Scholar]
- 10.Sommarin Y, Wendel M, Shen Z, Hellman U, Heinegard D. Osteoadherin, a cell-binding keratan sulfate proteoglycan in bone, belongs to the family of leucine-rich repeat proteins of the extracellular matrix. J. Biol. Chem. 1998;273:16723–16729. doi: 10.1074/jbc.273.27.16723. [DOI] [PubMed] [Google Scholar]
- 11.Bengtsson E, Neame PJ, Heinegard D, Sommarin Y. The primary structure of a basic leucine-rich repeat protein, PRELP, found in connective tissues. J. Biol. Chem. 1995;270:25639–25644. doi: 10.1074/jbc.270.43.25639. [DOI] [PubMed] [Google Scholar]
- 12.Barry FP, Rosenberg LC, Gaw JU, Koob TJ, Neame PJ. N- and O-linked keratan sulfate on the hyaluronan binding region of aggrecan from mature and immature bovine cartilage. J. Biol. Chem. 1995;270:20516–20524. doi: 10.1074/jbc.270.35.20516. [published erratum appears in J. Biol. Chem. (1995) Dec 29;270(52):31414].
- 13.Noguchi S, Nakano M. Structure of the acidic N-linked carbohydrate chains of the 55-kDa glycoprotein family (PZP3) from porcine zona pellucida. Eur. J. Biochem. 1992;209:883–894. doi: 10.1111/j.1432-1033.1992.tb17361.x. [DOI] [PubMed] [Google Scholar]
- 14.Ito M, Kitamikado M, Yamagata T. Isolation and characterization of an asparagine-linked keratan sulfate from the skin of a marine teleost, Scomber japonicus. Biochim. Biophys. Acta. 1984;797:221–230. doi: 10.1016/0304-4165(84)90125-9. [DOI] [PubMed] [Google Scholar]
- 15.Lauder RM, Huckerby TN, Nieduszynski IA. The structure of the keratan sulphate chains attached to fibromodulin from human articular cartilage. Glycoconj. J. 1997;14:651–660. doi: 10.1023/a:1018552913584. [DOI] [PubMed] [Google Scholar]
- 16.Lauder RM, Huckerby TN, Nieduszynski IA, Plaas AH. Age-related changes in the structure of the keratan sulphate chains attached to fibromodulin isolated from articular cartilage. Biochem. J. 1998;330:753–757. doi: 10.1042/bj3300753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nieduszynski IA, Huckerby TN, Dickenson JM, Brown GM, Tai GH, Bayliss MT. Structural aspects of skeletal keratan sulphates. Biochem. Soc. Trans. 1990;18:792–793. doi: 10.1042/bst0180792. [DOI] [PubMed] [Google Scholar]
- 18.Brown GM, Huckerby TN, Abram BL, Nieduszynski IA. Characterization of a non-reducing terminal fragment from bovine articular cartilage keratan sulphates containing alpha(2-3)-linked sialic acid and alpha(1-3)-linked fucose. A sulphated variant of the VIM-2 epitope. Biochem. J. 1996;319:137–141. doi: 10.1042/bj3190137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nieduszynski IA, Huckerby TN, Dickenson JM, Brown GM, Tai GH, Morris HG, Eady S. There are two major types of skeletal keratan sulphates. Biochem. J. 1990;271:243–245. doi: 10.1042/bj2710243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickenson JM, Huckerby TN, Nieduszynski IA. A non-reducing terminal fragment from tracheal cartilage keratan sulphate chains contains alpha (2-3)-linked N-acetylneuraminic acid. Biochem. J. 1991;278:779–785. doi: 10.1042/bj2780779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Funderburgh JL, Funderburgh ML, Mann MM, Prakash S, Conrad GW. Synthesis of corneal keratan sulfate proteoglycans by bovine keratocytes in vitro. J. Biol. Chem. 1996;271:31431–31436. doi: 10.1074/jbc.271.49.31431. [DOI] [PubMed] [Google Scholar]
- 22.Hanisch FG, Uhlenbruck G, Peter-Katalinic J, Egge H, Dabrowski U, Dabrowski J. Unbranched polylactosamino-O-glycans on human skim milk mucins exhibit Gal beta(1-4)GlcNAc beta(1-6) repeating units. Symp. Soc. Exp. Biol. 1989;43:155–162. [PubMed] [Google Scholar]
- 23.Barry FP, Neame PJ, Sasse J, Pearson D. Length variation in the keratan sulfate domain of mammalian aggrecan. Matrix Biol. 1994;14:323–328. doi: 10.1016/0945-053x(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 24.Funderburgh JL, Funderburgh ML, Mann MM, Conrad GW. Unique glycosylation of three keratan sulfate proteoglycan isoforms. J. Biol. Chem. 1991;266:14226–14231. [PubMed] [Google Scholar]
- 25.Dunlevy JR, Neame PJ, Vergnes JP, Hassell JR. Identification of the N-linked oligosaccharide sites in chick corneal lumican and keratocan that receive keratan sulfate. J. Biol. Chem. 1998;273:9615–9621. doi: 10.1074/jbc.273.16.9615. [DOI] [PubMed] [Google Scholar]
- 26.Iozzo RV. The family of the small leucine-rich proteoglycans: key regulators of matrix assembly and cellular growth. Crit. Rev. Biochem. Mol. Biol. 1997;32:141–174. doi: 10.3109/10409239709108551. [DOI] [PubMed] [Google Scholar]
- 27.Scott JE. Proteodermatan and proteokeratan sulfate (decorin, lumican/fibromodulin) proteins are horseshoe shaped. Implications for their interactions with collagen. Biochemistry. 1996;35:8795–8799. doi: 10.1021/bi960773t. [DOI] [PubMed] [Google Scholar]
- 28.Weber IT, Harrison RW, Iozzo RV. Model structure of decorin and implications for collagen fibrillogenesis. J. Biol. Chem. 1996;271:31767–31770. doi: 10.1074/jbc.271.50.31767. [DOI] [PubMed] [Google Scholar]
- 29.Blochberger TC, Cornuet PK, Hassell JR. Isolation and partial characterization of lumican and decorin from adult chicken corneas. A keratan sulfate-containing isoform of decorin is developmentally regulated. J. Biol. Chem. 1992;267:20613–20619. [PubMed] [Google Scholar]
- 30.Hart GW, Lennarz WJ. Effects of tunicamycin on the biosyn-thesis of glycosaminoglycans by embryonic chick cornea. J. Biol. Chem. 1978;253:5795–5801. [PubMed] [Google Scholar]
- 31.Ziegler C, Mersmann G. Influence of effectors of the complex-type-oligosaccharide biosynthesis on the formation of proteokeratan sulfate in bovine cornea. Biochim. Biophys. Acta. 1984;799:203–208. doi: 10.1016/0304-4165(84)90262-9. [DOI] [PubMed] [Google Scholar]
- 32.Helenius A, Aebi M. Intracellular functions of N-linked glycans. Science. 2001;291:2364–2369. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]
- 33.Plaas AH, Wong-Palms S. Biosynthetic mechanisms for the addition of polylactosamine to chondrocyte fibromodulin. J. Biol. Chem. 1993;268:26634–26644. [PubMed] [Google Scholar]
- 34.Fukuda M, Tsuboi S. Mucin-type O-glycans and leukosialin. Biochim. Biophys. Acta. 1999;1455:205–217. doi: 10.1016/s0925-4439(99)00067-8. [DOI] [PubMed] [Google Scholar]
- 35.Chai W, Yuen CT, Kogelberg H, Carruthers RA, Margolis RU, Feizi T, Lawson AM. High prevalence of 2-mono- and 2,6-di-substituted Manol-terminating sequences among O-glycans released from brain glycopeptides by reductive alkaline hydrolysis. Eur. J. Biochem. 1999;263:879–888. doi: 10.1046/j.1432-1327.1999.00572.x. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida A, Kobayashi K, Manya H, Taniguchi K, Kano H, Mizuno M, Inazu T, Mitsuhashi H, Takahashi S, Takeuchi M, Herrmann R, Straub V, Talim B, Voit T, Topaloglu H, Toda T, Endo T. Muscular dystrophy and neuronal migration disorder caused by mutations in a glycosyltransferase, POMGnT1. Dev. Cell. 2001;1:717–724. doi: 10.1016/s1534-5807(01)00070-3. [DOI] [PubMed] [Google Scholar]
- 37.Christner JE, Distler JJ, Jourdian GW. Biosynthesis of keratan sulfate: purification and properties of a galactosyltransferase from bovine cornea. Arch. Biochem. Biophys. 1979;192:548–558. doi: 10.1016/0003-9861(79)90125-5. [DOI] [PubMed] [Google Scholar]
- 38.Nakazawa K, Takahashi I, Yamamoto Y. Glycosyltransferase and sulfotransferase activities in chick corneal stromal cells before and after in vitro culture. Arch. Biochem. Biophys. 1998;359:269–282. doi: 10.1006/abbi.1998.0897. [DOI] [PubMed] [Google Scholar]
- 39.Cai CX, Gibney E, Gordon MK, Marchant JK, Birk DE, Linsenmayer TF. Characterization and developmental regulation of avian corneal beta-1,4-galactosyltransferase mRNA. Exp. Eye Res. 1996;63:193–200. doi: 10.1006/exer.1996.0108. [DOI] [PubMed] [Google Scholar]
- 40.Shaper NL, Charron M, Lo NW, Shaper JH. Beta1,4-galactosyltransferase and lactose biosynthesis: recruitment of a housekeeping gene from the nonmammalian vertebrate gene pool for a mammary gland specific function. J. Mammary Gland Biol. Neoplasia. 1998;3:315–324. doi: 10.1023/a:1018719612087. [DOI] [PubMed] [Google Scholar]
- 41.Almeida R, Amado M, David L, Levery SB, Holmes EH, Merkx G, Van Kessel AG, Rygaard E, Hassan H, Bennett E, Clausen H. A family of human beta4-galactosyltransferases. Cloning and expression of two novel UDP-galactose:beta-N-acetylglucosamine beta1,4-galactosyltransferases, beta4Gal-T2 and beta4Gal-T3. J. Biol. Chem. 1997;272:31979–31991. doi: 10.1074/jbc.272.51.31979. [published erratum appears in J. Biol. Chem. (1998) Jul 17;273(29):18674].
- 42.Ujita M, Misra AK, Mcauliffe J, Hindsgaul O, Fukuda M. Poly-N-acetyllactosamine extension in N-glycans and core 2- and core 4-branched O-glycans is differentially controlled by i-extension enzyme and different members of the beta 1,4-galactosyltransferase gene family. J. Biol. Chem. 2000;275:15868–15875. doi: 10.1074/jbc.M001034200. [DOI] [PubMed] [Google Scholar]
- 43.Kotani N, Asano M, Iwakura Y, Takasaki S. Knockout of mouse beta 1,4-galactosyltransferase-1 gene results in a dramatic shift of outer chain moieties of N-glycans from type 2 to type 1 chains in hepatic membrane and plasma glycoproteins. Biochem. J. 2001;357:827–834. doi: 10.1042/0264-6021:3570827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasaki K, Kurata-Miura K, Ujita M, Angata K, Nakagawa S, Sekine S, Nishi T, Fukuda M. Expression cloning of cDNA encoding a human beta-1,3-N-acetylglucosaminyltransferase that is essential for poly-N-acetyllactosamine synthesis. Proc. Natl. Acad. Sci. U.S.A. 1997;94:14294–14299. doi: 10.1073/pnas.94.26.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fukuzumi M, Maruyama S, Sano M, Fukui S. Comparison of the expression of cell surface poly-N-acetyllactosamine-type oligosaccharides in PC12 cells with those in its variant PC12D. Glycobiology. 2001;11:481–494. doi: 10.1093/glycob/11.6.481. [DOI] [PubMed] [Google Scholar]
- 46.Shiraishi N, Natsume A, Togayachi A, Endo T, Akashima T, Yamada Y, Imai N, Nakagawa S, Koizumi S, Sekine S, Narimatsu H, Sasaki K. Identification and characterization of three novel beta 1,3-N-acetylglucosaminyltransferases structurally related to the beta 1,3-galactosyltransferase family. J. Biol. Chem. 2001;276:3498–3507. doi: 10.1074/jbc.M004800200. [DOI] [PubMed] [Google Scholar]
- 47.Ruter ER, Kresse H. Partial purification and characterization of 3′- phosphoadenylylsulfate: keratan sulfate sulfotransferases. J. Biol. Chem. 1984;259:11771–11776. [PubMed] [Google Scholar]
- 48.Fukuta M, Inazawa J, Torii T, Tsuzuki K, Shimada E, Habuchi O. Molecular cloning and characterization of human keratan sulfate Gal-6-sulfotransferase. J. Biol. Chem. 1997;272:32321–32328. doi: 10.1074/jbc.272.51.32321. [DOI] [PubMed] [Google Scholar]
- 49.Habuchi O, Hirahara Y, Uchimura K, Fukuta M. Enzymatic sulfation of galactose residue of keratan sulfate by chondroitin 6-sulfotransferase. Glycobiology. 1996;6:51–57. doi: 10.1093/glycob/6.1.51. [DOI] [PubMed] [Google Scholar]
- 50.Uchimura K, Kadomatsu K, Nishimura H, Muramatsu H, Nakamura E, Kurosawa N, Habuchi O, El-Fasakhany FM, Yoshikai Y, Muramatsu T. Functional analysis of the chondroitin 6-sulfotransferase gene in relation to lymphocyte subpopulations, brain development, and oversulfated chondroitin sulfates. J. Biol. Chem. 2002;277:1443–1450. doi: 10.1074/jbc.M104719200. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto Y, Takahashi I, Ogata N, Nakazawa K. Purification and characterization of N-acetylglucosaminyl sulfotransferase from chick corneas. Arch. Biochem. Biophys. 2001;392:87–92. doi: 10.1006/abbi.2001.2422. [DOI] [PubMed] [Google Scholar]
- 52.Uchimura K, El-Fasakhany FM, Hori M, Hemmerich S, Blink SE, Kansas GS, Kanamori A, Kumamoto K, Kannagi R, Muramatsu T. Specificities of N-acetylglucosamine-6-O-sulfotransferases in relation to l-selectin ligand synthesis and tumor-associated enzyme expression. J. Biol. Chem. 2002;277:3979–3984. doi: 10.1074/jbc.M106587200. [DOI] [PubMed] [Google Scholar]
- 53.Hemmerich S, Lee JK, Bhakta S, Bistrup A, Ruddle NR, Rosen SD. Chromosomal localization and genomic organization for the galactose/N-acetylgalactosamine/N-acetylglucosamine 6-O-sulfotransferase gene family. Glycobiology. 2001;11:75–87. doi: 10.1093/glycob/11.1.75. [DOI] [PubMed] [Google Scholar]
- 54.Bartes A, Bhakta S, Hemmerich S. Sulfation of endothelial mucin by corneal keratan N-acetylglucosamine 6-O-sulfotransferase (GST-4beta). Biochem. Biophys. Res. Commun. 2001;282:928–933. doi: 10.1006/bbrc.2001.4668. [DOI] [PubMed] [Google Scholar]
- 55.Hassell JR, Newsome DA, Krachmer JH, Rodrigues MM. Macular corneal dystrophy: failure to synthesize a mature keratan sulfate proteoglycan. Proc. Natl. Acad. Sci. U.S.A. 1980;77:3705–3709. doi: 10.1073/pnas.77.6.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edward DP, Thonar EJ, Srinivasan M, Yue BJ, Tso MO. Macular dystrophy of the cornea. A systemic disorder of keratan sulfate metabolism. Ophthalmology. 1990;97:1194–1200. doi: 10.1016/s0161-6420(90)32436-3. [DOI] [PubMed] [Google Scholar]
- 57.El-Ashry MF, El-Aziz MM, Wilkins S, Cheetham ME, Wilkie SE, Hardcastle AJ, Halford S, Bayoumi AY, Ficker LA, Tuft S, Bhattacharya SS, Ebenezer ND. Identification of novel mutations in the carbohydrate sulfotransferase gene (CHST6) causing macular corneal dystrophy. Invest. Ophthalmol. Vis. Sci. 2002;43:377–382. [PubMed] [Google Scholar]
- 58.Liu NP, Dew-Knight S, Rayner M, Jonasson F, Akama TO, Fukuda MN, Bao W, Gilbert JR, Vance JM, Klintworth GK. Mutations in corneal carbohydrate sulfotransferase 6 gene (CHST6) cause macular corneal dystrophy in Iceland. Mol. Vis. 2000;6:261–264. [PubMed] [Google Scholar]
- 59.Akama TO, Nishida K, Nakayama J, Watanabe H, Ozaki K, Nakamura T, Dota A, Kawasaki S, Inoue Y, Maeda N, Yamamoto S, Fujiwara T, Thonar EJ, Shimomura Y, Kinoshita S, Tanigami A, Fukuda MN. Macular corneal dystrophy type I and type II are caused by distinct mutations in a new sulphotransferase gene. Nat. Genet. 2000;26:237–241. doi: 10.1038/79987. [DOI] [PubMed] [Google Scholar]
- 60.Akama TO, Nakayama J, Nishida K, Hiraoka N, Suzuki M, McAuliffe J, Hindsgaul O, Fukuda M, Fukuda MN. Human corneal GlcNac 6-O-sulfotransferase and mouse intestinal GlcNac 6-O-sulfotransferase both produce keratan sulfate. J. Biol. Chem. 2001;276:16271–16278. doi: 10.1074/jbc.M009995200. [DOI] [PubMed] [Google Scholar]
- 61.Degroote S, Lo-Guidice JM, Strecker G, Ducourouble MP, Roussel P, Lamblin G. Characterization of an N-acetylglucosamine-6-O-sulfotransferase from human respiratory mucosa active on mucin carbohydrate chains. J. Biol. Chem. 1997;272:29493–29501. doi: 10.1074/jbc.272.47.29493. [DOI] [PubMed] [Google Scholar]
- 62.Uchimura K, Muramatsu H, Kadomatsu K, Fan QW, Kurosawa N, Mitsuoka C, Kannagi R, Habuchi O, Muramatsu T. Molecular cloning and characterization of an N-acetylglucosamine-6-O-sulfotransferase. J. Biol. Chem. 1998;273:22577–22583. doi: 10.1074/jbc.273.35.22577. [DOI] [PubMed] [Google Scholar]
- 63.Keller R, Stuhlsatz HW, Greiling H. Sulphation, chain elongation and chain termination in keratan sulphate biosynthesis. In: Greiling H, Scott JE, editors. Keratan Sulphate, Chemistry, Biology, Chemical Pathology. The Biochemical Society; London: 1989. pp. 39–52. [Google Scholar]
- 64.Plaas AH, West LA, Thonar EJ, Karcioglu ZA, Smith CJ, Klintworth GK, Hascall VC. Altered fine structures of corneal and skeletal keratan sulfate and chondroitin/dermatan sulfate in macular corneal dystrophy. J. Biol. Chem. 2001;276:39788–39796. doi: 10.1074/jbc.M103227200. [DOI] [PubMed] [Google Scholar]
- 65.Plaas AH, West LA, Midura RJ. Keratan sulfate disaccharide composition determined by FACE analysis of keratanase II and endo-beta-galactosidase digestion products. Glycobiology. 2001;11:779–790. doi: 10.1093/glycob/11.10.779. [DOI] [PubMed] [Google Scholar]
- 66.Funderburgh JL, Funderburgh ML, Rodrigues MM, Krachmer JH, Conrad GW. Altered antigenicity of keratan sulfate proteoglycan in selected corneal diseases. Invest. Ophthalmol. Vis. Sci. 1990;31:419–428. [PubMed] [Google Scholar]
- 67.Rodrigues M, Nirankari V, Rajagopalan S, Jones K, Funderburgh J. Clinical and histopathologic changes in the host cornea after epikeratoplasty for keratoconus. Am. J. Ophthalmol. 1992;114:161–170. doi: 10.1016/s0002-9394(14)73980-7. [DOI] [PubMed] [Google Scholar]
- 68.Beales MP, Funderburgh JL, Jester JV, Hassell JR. Proteoglycan synthesis by bovine keratocytes and corneal fibroblasts: maintenance of the keratocyte phenotype in culture. Invest. Ophthalmol. Vis. Sci. 1999;40:1658–1663. [PubMed] [Google Scholar]
- 69.Funderburgh JL, Funderburgh ML, Mann MM, Corpuz L, Roth MR. Proteoglycan expression during transforming growth factor beta-induced keratocyte-myofibroblast transdifferentiation. J. Biol. Chem. 2001;276:44173–44178. doi: 10.1074/jbc.M107596200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Long CJ, Roth MR, Tasheva ES, Funderburgh M, Smit R, Conrad GW, Funderburgh JL. Fibroblast growth factor-2 promotes keratan sulfate proteoglycan expression by keratocytes in vitro. J. Biol. Chem. 2000;275:13918–13923. doi: 10.1074/jbc.275.18.13918. [DOI] [PubMed] [Google Scholar]