Abstract
Chromosomal imbalances are the major cause of mental retardation (MR). Many of these imbalances are caused by submicroscopic deletions or duplications not detected by conventional cytogenetic methods. Microarray-based comparative genomic hybridization (array-CGH) is considered to be superior for the investigation of chromosomal aberrations in children with MR, and has been demonstrated to improve the diagnostic detection rate of these small chromosomal abnormalities. In this study we used 1 Mb genome-wide array-CGH to screen 48 children with MR and congenital malformations for submicroscopic chromosomal imbalances, where the underlying cause was unknown. All children were clinically investigated and subtelomere FISH analysis had been performed in all cases. Suspected microdeletion syndromes such as deletion 22q11.2, Williams-Beuren and Angelman syndromes were excluded before array-CGH analysis was performed. We identified de novo interstitial chromosomal imbalances in two patients (4%), and an interstitial deletion inherited from an affected mother in one patient (2%). In another two of the children (4%), suspected imbalances were detected but were also found in one of the non-affected parents. The yield of identified de novo alterations detected in this study is somewhat less than previously described, and might reflect the importance of which selection criterion of patients to be used before array-CGH analysis is performed. However, array-CGH proved to be a high-quality and reliable tool for genome-wide screening of MR patients of unknown etiology.
Introduction
Mental retardation (MR) is a variable and heterogeneous manifestation of central nervous system dysfunction, affecting approximately 3% of the population. In a major part of these patients the etiology is unknown. In half of the cases, MR with or without congenital malformations and dysmorphic features is thought to be caused by chromosomal imbalances. Some of theses imbalances can be explained by gross chromosomal abnormalities, detected by conventional cytogenetic techniques such as GTG-banding. However, microdeletions and duplications are not detectable using those techniques, since the level of resolution is inadequate or unreliable for the detection of subtle copy number changes involving chromosome segments of 5 Mb or less.
Subtelomeric rearrangements contribute significantly to unexplained dysmorphisms, malformations and/or MR and have been detected in approximately 5% of MR patients by fluorescent in situ hybridization (FISH) (Flint et al., 1995; Knight et al., 1999) analysis and multiplex ligation-dependent probe amplification (MLPA) (Schouten et al., 2002; Koolen et al., 2005). As these techniques utilize designed locus-specific probes they only return results specific for the assay, and therefore are not suitable for screening submicroscopic rearrangements on a genome-wide scale. On the other hand, microarray-based comparative genomic hybridization (array-CGH) (Solinas-Toldo et al., 1997; Pinkel et al., 1998) enables genome-wide detection of DNA copy number alterations at high resolution, and has proven to have a major impact on research and diagnostics. In several studies array-CGH has shown to have great potential for the detection of submicroscopic chromosomal abnormalities in children with MR and dysmorphisms, which to date have escaped identification. Recent studies using such genome-wide arrays to investigate patients with MR with and without dysmorphic features have suggested a diagnostic yield of 10–25%, of which de novo findings count for approximately 10% (Vissers et al., 2003; Shaw-Smith et al., 2004; de Vries et al., 2005; Schoumans et al., 2005; Tyson et al., 2005; Menten et al., 2006; Rosenberg et al., 2006).
In this study we have used genome-wide array-CGH, with an average resolution of 1 Mb, to screen 48 children with idiopathic MR and dysmorphic features for chromosomal imbalances not detected by conventional cytogenetic and FISH techniques.
Materials and methods
Patient material
48 patients with idiopathic MR (22 girls and 26 boys; age range 2 to 15 years) were selected for array-CGH analysis. All patients were examined by a clinical geneticist. Apart from MR, patients also presented with additional features like congenital malformations and/or dysmorphism. All patients had at least one major and one minor or at least three minor malformations as previously described by Smith (1982). All patients had a normal karyotype from GTG-banding analysis at the +400-band level and subtelomeric rearrangements were excluded by FISH (Vysis). Genomic DNA was extracted from peripheral blood lymphocytes according to standard procedures. Patient samples were studied with the approval of the local Research Ethics Committee, Faculty of Medicine, Uppsala University.
Array-CGH
The BAC arrays used in this study had a resolution of 1 Mb and were produced as previously described (Fiegler et al., 2003). One microgram of genomic test and reference DNA were labeled and hybridized for 16–18 h as previously described by Mantripragada et al. (2006). Dye swap experiments where performed on each case where a copy number change was suspected. A pool of sex-matched DNA from either eight normal male or female blood donors was used for the reference samples.
Image and data analysis
Image acquisition was performed using a GenePix 4000B scanner (Axon Instruments Inc, Union City, CA). Images were quantified using SPOT software (Jain et al., 2002). Further data analysis was performed as previously described (Fiegler et al., 2003). In addition, clones listed as polymorphic in the Database of Genomic Variants (http://projects.tcag.ca/variation/) were excluded from further analysis. To reduce false positive results, the SD was calculated for all autosomal controls. Clones displaying a test/reference ratio value exceeding ±4DELETESD were considered as clinically significant and were subjected to further investigation (Vermeesch et al., 2005). Hybridizations where the SD exceeded 0.1 were considered to have failed and the patient sample was rehybridized. Clones which displayed copy number alterations in the dye swap experiments were confirmed by FISH or MLPA. Parents of the children displaying copy number changes were investigated for the origin of the altered chromosomal region. Cases where chromosomal anomalies were detected were submitted to the DECIPHER database (http://www.sanger.ac.uk/Postgenomics/decipher) after gaining consent from the parents of these children.
FISH
FISH validation experiments were performed in cases 1, 3 and 5 on metaphase spreads prepared from lymphocyte cultures of peripheral blood to confirm deletions identified by array-CGH analysis, as previously described (Wester et al., 2006). BAC clones were obtained from The Wellcome Trust Sanger Institute (Cambridge, UK).
MLPA
Confirmation of case 2 was carried out using the SALSA MLPA kit P064 MR1 (MRC-Holland, Amsterdam, Netherlands). In case 4, six specifically designed synthetic MLPA probes were used to cover the region of interest (Stern et al., 2004). MLPA reactions were performed as previously described (Schouten et al., 2002; Mantripragada et al., 2006). One microliter of the amplification product was analyzed using an ABI 3130XL genetic analyzer (Applied Biosystems) using ROX-500 (ABI) as an internal size standard. Analysis was performed with GeneMapper software 3.7. Data analysis was performed in Microsoft Excel, where each peak area was divided by the sum of all control peak areas of that sample, followed by normalization to the ratio obtained by a control sample.
Results
We analyzed 48 idiopathic MR patients with dysmorphic features by array-CGH, with an average resolution of 1 Mb. All patients had a normal karyotype when investigated by GTG-banding. Telomeric rearrangements had been excluded by subtelomere FISH. In total, five copy number changes – three deletions and two duplications – were detected in five patients (10.4%). Three cases were considered as pathological and of clinical significance (6%) and two cases were inherited from phenotypically normal parents (4%). The estimated maximum sizes of the genomic losses and gains ranged from 0.2 Mb (1 clone) to 6.9 Mb (8 clones). A description of the phenotypes of these patients and a summary of the results is presented in Table 1. In addition, single clone gain or loss was detected in numerous cases in this study. However, a majority of these were suspected to be polymorphic loci as they were either reported as copy number variations (CNV) in the Database of Genomic Variants (http://projects.tcag.ca/variation/) or detected in several cases displaying different phenotypes (Table 2) and therefore disregarded. In four cases a deletion was detected in four clones at Yq11.223 (data not shown). This deleted region was deemed unlikely to contribute to the phenotype; hence these patients were excluded from further analysis.
Table 1.
Summary of phenotypes and copy number changes detected by array-CGH
| Case | Age | Karyotype | Phenotypea | Array-CGH resultsb | Confirmation method | Origin | Estimated size (Mb) min. / max. |
|---|---|---|---|---|---|---|---|
| 1 | 6 | 46,XY | severe MR, failure to thrive, microcephaly, micropenis, retentio testis, dysmorphic features | del of 7 clones at 20q13.12→q13.13 | FISH | de novo | 3.2 / 5.1 |
| 2 | 6 | 46,XX | severe growth and MR, failure to thrive, dysmorphic features | dup of 8 clones at 17p12→p11.2 | MLPA | de novo | 3.6 / 6.9 |
| 3 | 8 | 46,XX | moderate MR, severe growth retardation, failure to thrive, hyperopia, dysmorphic features | del of 4 clones at 4q28.3→q31.21 | FISH | maternally inherited, mother with MR and similar dysmorphism | 3.8 / 6.9 |
| 4 | 6 | 46,XY | severe MR, failure to thrive, dysmorphic features | dup of 1 clone at 2p25.3 | MLPA | maternally inherited | 0.2 / 0.9 |
| 5 | 3 | 46,XY | moderate MR, severe growth retardation, ventricular septal defect, cleft lip and palate, inguinal hernia, a benign facial histiocytoma, hyperopia, dysmorphic features | del of 1 clone at 7p22.2 | FISH | paternally inherited | 0.09 / 0.2 |
MR = mental retardation.
del = deletion; dup = duplication.
Table 2.
Clone list with suspected polymorphic loci
| Chromosome | Clone ID | Gain | Loss |
|---|---|---|---|
| 19 | CTC-251H24 | 8 | 3 |
| 19 | CTC-260F20 | 9 | 3 |
| 19 | CTC-325L16 | 1 | 1 |
Case reports
Case 1: 46,XY, arr cgh 20q13.12q13.13 (RP5-1049G16→RP5-1071L10)×1 dn. This 6-year-old boy is the third child of healthy parents with two healthy siblings. He was born after a normal pregnancy at 37 weeks and six days gestation. His weight was 3.6 kg, length 51 cm and head circumference 34 cm at birth. At the outset from birth he was muscularly hypertonic. He had severe failure to thrive and got a gastric tube at 5 months of age. At the age of 1 year severe MR was obvious with microcephaly. He started to walk at 2 years of age. He could not speak but gave good eye contact. At the age of 2 years and 3 months the following features were found: He was microcephalic with a head circumference −3 SD. He had epicanthic folds, low set ears with thick down folded helixes, a hypoplastic nasal bridge, a short upturned nose with hypoplastic alas, a long philtrum, clinodactyly of fifth fingers bilaterally, bilateral syndactylies between the 2nd and 3rd toes, a very small penis and scrotum, and retention testis. Chromosomal investigation, subtelomere FISH and Fragile-X analysis were all normal.
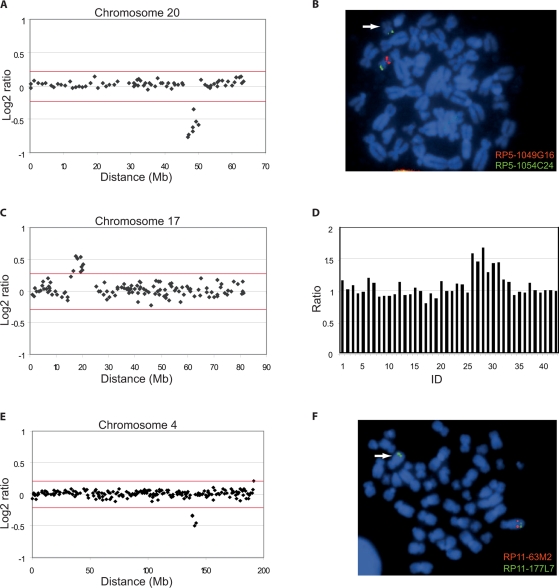
As shown in Fig. 1A, an interstitial deletion spanning seven clones, RP5-1049G16 through RP5-1071L10, was detected at 20q13.12→q13.13, which could be verified by FISH using the two different BAC clones, RP5-1049G16 and RP5-1071L10 located at the detected breakpoints (Fig. 1B, and data not shown). This deletion could not be detected in the parents and was considered de novo and consequently of clinical significance. A fraction of this region, covering RP5-1049G16, has been reported in the Database of Genomic Variants (Tuzun et al., 2005; McCarroll et al., 2006; Redon et al., 2006).
Fig. 1.
Array-CGH profiles of detected chromosomal abnormalities and confirmatory FISH and MLPA analysis. The array-CGH profiles of DNA derived from three cases versus sex matched reference DNA are shown in panels A, C and E. Panels B and F show the corresponding FISH analysis for each patient, and Panel D the corresponding MLPA analysis. The x-axis displays the distance in Mb along the chromosome from the p to the q telomere, and the y-axis the log2 ratios at each locus. The red lines indicate the threshold for a deletion or a duplication (±4DELETESD). (A) Array-CGH profile of a 7-clone deletion at 20q13.12→q13.13 in case 1. (B) Confirmatory FISH analysis of case 1 using BAC clone RP5-1049G16 (red) located in the deletion and control clone RP5-1054C24 (green). (C) Array-CGH profile of an 8-clone duplication at 17p12→p11.2 in case 2. (D) Profile from confirmatory MLPA analysis of case 2, displaying a duplication of six probes (ID 26 through 31) located in the critical region of dup(17)(p11.2p11.2) syndrome. ID 1–25 and 32–42 display chromosomal locations associated with other syndromes. (E) Array-CGH profile of a 4-clone deletion at 4q28.3→q31.21 in case 3. (F) Confirmatory FISH analysis of case 3 using BAC clone RP11-63M2 (red) located in the deletion and control clone RP11-177L7 (green).
Case 2: 46,XX, arr cgh 17p12p11.2 (RP11-219A15→RP11-121A13)×3 dn. This 6-year- and 6-month-old girl has one healthy older sibling and non-consanguineous parents. She was born after a normal pregnancy in a normal delivery. Her birth weight was 2.9 kg. She had severe failure to thrive and was fed from birth by tube. At one year of age she was growth retarded (−3.0 SD) and got a gastric tube which allowed her to grow and her height at 4.5 years of age was 1.0 m (–1.0 SD). From the age of 1 year it was obvious that she was mentally retarded. She started to walk and speak the first words at two years of age. At 5.5 years of age she had the following features: She was very thin, her face was triangular, she had hypertelorism with down-slanted palpebral fissures, a divergent strabismus, and long eye lashes. Brainstem audiometrics, eye examinations as well as a CT scan of the brain were all normal. Chromosome and subtelomere FISH analyses were normal.
Array-CGH analysis revealed a duplication spanning eight clones, RP11-219A15 through RP11-121A13, at 17p11.2 (Fig. 1C). This duplication overlaps with the previously described dup(17)(p11.2p11.2) syndrome (Potocki et al., 2000). The duplication was verified by six different probes in the SALSA MLPA kit P064 MR1 (Fig. 1D; ID 26–31), located in the critical region of the Smith-Magenis and dup(17)(p11.2p11.2) syndrome. By the same method this duplication could not be detected in the parents. Consequently this duplication was considered de novo and presumed to be the cause of the phenotype of this girl.
Case 3: 46,XX, arr cgh 4q28.3q31.21 (RP11-63M2→RP11-69O1)×1 mat. This 8-year-old girl was born after a normal pregnancy at 39 weeks gestation. Her weight was 2.98 kg at birth. From 5 months of age she had failure to thrive and she received a gastrostomy at 3 years of age. She was moderate mentally and growth retarded (–2 SD). She was severely hyperopic (+7 dioptres) and got eye glasses at 3 years of age. At 5 years of age she had the following features: short stature, frontal bossing, short neck and hypertelorism. The following investigations gave normal results: hearing investigation, CT scan of the brain, electromyography and muscle biopsy (because of elevated serum levels of CK), chromosomal analysis, subtelomere FISH investigation, and a methylation test for Prader-Willi syndrome.
A deletion spanning four clones, RP11-63M2 through RP11-69O1, at 4q28.3→q31.21 was identified by array-CGH, which could be verified by FISH analysis (Fig. 1E and F). An identical deletion was detected by array-CGH in the mother, who has a mild MR, severe congenital unilateral hearing deficit, impaired social interaction with severe speech problems and with a phenotype similar to her daughter. Considering this information the detected deletion is considered as pathogenic. One part of this region, spanning RP11-425J20, is reported in the Database of Genomic Variants (Redon et al., 2006).
Case 4: 46,XY, arr cgh 2p25.3 (RP11-168K7)×3 mat. This is a 6-year-old boy of healthy non-consanguineous parents. He was born in a normal delivery after a 40-week normal pregnancy. His weight was 4.24 kg and length was 50 cm at birth. He had severe failure to thrive and received a gastrostomy before the age of six months. He had an abnormal motor development with jerky movements. He had a pathological electroencephalogram but no convulsions. He has severe MR and could not sit without support at 2 years of age and still has no spoken language. At 2 years of age he had the following features: flat occipital region, high vaulted and narrow palate, dysmorphic ears, a flat philtrum, epicanthic folds, speckled iris, and broad thumbs. Brain scan (MRT) and an examination of vision and hearing revealed normal results. Chromosomal and subtelomere FISH analyses were normal. Angelman and Williams-Beuren syndromes were excluded by methylation test and FISH analysis, respectively.
Array-CGH analysis detected a single clone duplication of RP11-168K7 at 2p25.3. The duplication was verified with six specifically designed synthetic MLPA probes, three for each of the two genes PXDN and MYT1L, situated within the duplicated BAC clone (data not shown). However, as an identical duplication was detected by array-CGH and MLPA in the phenotypically normal mother, this aberration was considered to be benign.
Case 5: 46,XY, arr cgh 7p22.2 (RP11-348A21)×1 pat. This is the first child of Turkish parents who are double first cousins. The boy was born at full term after a normal pregnancy. His weight was 3.0 kg, length 48 cm and head circumference 37 cm at birth. He had severe postnatal feeding difficulties and received a gastric tube at the age of 1 year. He was severely growth retarded at the age of 2 years and 4 months with a weight of 9.6 kg (–4 SD) and a height of 80 cm (–4 SD). He had a ventricular septal defect, bilateral retentio testis, and bilateral cleft lip and palate. He had an inguinal hernia which was surgically corrected at eight months of age. He was bilaterally hyperopic and requires eye glasses (+5 dioptres). At 9 months of age he was operated upon because a benign histiocytoma on his right chin. At the age of 2 years and 4 months he had the following features: He is mildly mentally retarded and has just learned to walk and can speak a few words. His ears are big. He has hypertelorism and bilateral ptosis, his eye lashes are long and the lateral parts of the palpebral fissures are inverted. The nasal bridge is high and broad. Kabuki syndrome was suspected, but his finger pads as well as nails were normal. Chromosomal and subtelomere FISH analyses were both normal.
A single clone deletion, RP11-348A21, was detected at 7p22.2 by array-CGH and was further verified by FISH. However, as a deletion of the same size was detected by array-CGH in the phenotypically normal father, this aberration was considered benign. Recently a deletion polymorphism was identified just upstream of this BAC clone, RP11-348A21, involving the same gene, SDK1 (Conrad et al., 2006; McCarroll et al., 2006; Redon et al., 2006). Consequently the present deletion is most likely a deletion polymorphism. Since the parents are related an autosomal recessive syndrome might be suspected.
Discussion
The diagnostic tools for patients with dysmorphic features, malformations and/or MR have been exhausted in many cases. Consequently, the diagnosis and the underlying genetic lesions remain unclear. With the long-term goal to identify the genetic basis behind most constitutional phenotypes associated with chromosomal imbalances, the establishment of array-CGH analysis is necessary. The array-CGH technique has demonstrated the potential to clarify unexplained phenotypes at the genomic level and when extended, to identify candidate gene regions and candidate genes behind any condition with alterations in gene copy number.
Array-CGH has already been shown by several studies to be a reliable tool for genome-wide detection of copy number changes in MR patients (Vissers et al., 2003; Shaw-Smith et al., 2004; de Vries et al., 2005; Schoumans et al., 2005; Tyson et al., 2005; Menten et al., 2006; Rosenberg et al., 2006).
In this study we screened 48 patients with MR and dysmorphic features. All patients had a normal karyotype by standard cytogenetic chromosomal banding and subtelomere FISH analysis. We detected pathogenic copy number changes in three patients (6%), of which two were de novo. All chromosomal imbalances contain genes annotated by Ensembl. Case 1 had a deletion 20q13.12→q13.13. No previous patient has been described with an interstitial deletion of that segment. One patient with severe MR and a terminal deletion del(20)(q13→qter) is reported (Fraisse et al., 1982). That boy had a hypoplastic nasal bridge and a long philtrum similar to the present boy. Case 2 was identified as the newly reported dup(17)(p11.2p11.2) syndrome (Potocki et al., 2000). This girl has a similar phenotype compared to other reported cases with a triangular face, down-slanted palpebral fissures and growth- and MR (Magenis et al., 1986; Kozma et al., 1991; Potocki et al., 2000). Thus, cases 1 and 2 seem to fit phenotypically with previously described patients with similar chromosomal abnormalities. The girl in case 3 has a 6.9-Mb deletion on 4q28.3→q31.21 inherited from the mother with congenital unilateral hearing deficit, which is not present in the daughter. Thirteen genes are located within the deleted region, of which none has been associated with deafness. In addition, the mother has a mild MR, impaired social interaction and verbal communication, and has a phenotype similar to her daughter. Hence, the detected deletion is a good candidate to cause the phenotype of this girl. Unfortunately DNA from the grandparents is not available, and therefore it is not possible to determine the origin of the detected deletion. An overlapping deletion of 11 Mb is reported in the DECIPHER database. This deletion is de novo and considerably larger in size. The phenotypes of these two patients do not overlap (Laurence Colleaux, personal communication).
In two patients (4%) copy number changes inherited from phenotypically normal parents were detected. It is presumed that inherited aberrations do not contribute to a diagnosed phenotype. It is difficult to interpret whether these anomalies are of any clinical significance, or if they contribute to the phenotype by other mechanisms such as non-penetrance, which has been described in 22q11.2 microdeletion and -duplication syndromes (Wilson et al., 1992; Yobb et al., 2005), or by imprinting. Hence, these benign variations need further genotypic and phenotypic evaluation and it is therefore important to report them to databases such as DECIPHER (http://www.sanger.ac.uk/Postgenomics/decipher) to allow comparisons to be drawn with other clinical cases and to generate more insight into the plasticity of the human genome. Hence, as can be concluded from this study and others (Shaw-Smith et al., 2004; de Vries et al., 2005; Menten et al., 2006; Rosenberg et al., 2006), it appears that confirmation of all array-CGH findings with the parents of the respective child is required, as approximately half of the detected findings appear to be inherited from a phenotypically normal parent.
The two inherited aberrations were checked against the DECIPHER database where copy number changes were detected at the same chromosomal region in both cases. A 2.7-Mb de novo deletion has been reported to the DECIPHER database in the same chromosomal region as detected in case 4, 2p25.3. The detected aberration in the present case however is a 0.9-Mb duplication inherited from the mother. The phenotypes of these two patients do not overlap (Rosemary Ekong, personal communication). A 3.6-Mb deletion was reported in the DECIPHER database in the same region as the 0.2-Mb deletion detected in case 5. However, this reported deletion was de novo and the phenotypes of the two cases do not overlap (Farah Zahir and Jan Friedman, personal communication) (Friedman et al., 2006).
In a recent literature review of all published 1-Mb screening studies of MR patients the yield was reported to be 8.8% (Menten et al., 2006). The detection rate of de novo findings in our study is somewhat lower (6%). The lower yield detected in our study might be explained by the fact that the patient material was not selected according to the checklist developed by de Vries et al. (2001), but according to the major and minor anomalies described by Smith (1982), commonly used by clinicians. This might suggest that to gain a higher rate of detection, the checklist of de Vries might be more suitable as a selection criterion for array-CGH analysis. However, array-CGH and oligoarrays today are the only diagnostic tools for high-resolution genome scanning, to detect submicroscopic aberrations in these patients. For that reason, all MR patients with or without congenital malformations and/or dysmorphism might benefit from array-CGH.
For clinical settings, an alternative to genome-wide array-CGH could be the targeted genomic microarrays (Shaffer et al., 2006). Compared to other studies the detection rate of these arrays is lower, 5.6% compared to 8.8% (Menten et al., 2006; Shaffer et al., 2006), but they have the advantage of less polymorphic findings. However, they will fail to detect patients with rearrangements not previously described like case 1 and 3 in the present study. In that sense, a 1-Mb genome-wide array with a higher density of large insert clones in known microdeletion and duplication and telomere regions might be more suitable. The detection rate in our study is comparable to the targeted arrays but lower than described in other studies. This might be explained by the fact that our cohort of patients had already been excluded from patients with telomeric rearrangements and known syndromes associated with MR like Prader-Willi, monosomy 1p36 and DiGeorge. Also the number of patients in this study is considerably lower than in some other studies, which affects the statistical significance. Nonetheless, array-CGH still proved to be a high-quality tool for genome-wide screening of MR patients with dysmorphic features where the etiology is unknown.
Clarification of the genetic profile generated by array-CGH analysis may result in detailed follow-ups and, in the long-term, a better overall outcome for these patients. The short-term perspective allows for family counseling and prenatal diagnosis as well as for the identification and functional analysis of candidate gene regions/genes underlying the condition.
Footnotes
This work was supported by grants from the Sävstaholm Society, the Swedish Medical Research Council and by The Wellcome Trust.
References
- Conrad DF, Andrews TD, Carter NP, Hurles ME, Pritchard JK. A high-resolution survey of deletion polymorphism in the human genome. Nat Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- de Vries BB, White SM, Knight SJ, Regan R, Homfray T, et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet. 2001;38:145–150. doi: 10.1136/jmg.38.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries BB, Pfundt R, Leisink M, Koolen DA, Vissers LE, et al. Diagnostic genome profiling in mental retardation. Am J Hum Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- Flint J, Wilkie AO, Buckle VJ, Winter RM, Holland AJ, McDermid HE. The detection of subtelomeric chromosomal rearrangements in idiopathic mental retardation. Nat Genet. 1995;9:132–140. doi: 10.1038/ng0295-132. [DOI] [PubMed] [Google Scholar]
- Fraisse J, Bertheas MF, Frere F, Lauras B, Rolland MO, Brizard CP. [Partial monosomy 20q: a new syndrome. Regional assignment of the ADA locus on 20q132 (author's transl)] Sem Hop. 1982;58:1366–1369. [PubMed] [Google Scholar]
- Friedman JM, Baross A, Delaney AD, Ally A, Arbour L, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am J Hum Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AN, Tokuyasu TA, Snijders AM, Segraves R, Albertson DG, Pinkel D. Fully automatic quantification of microarray image data. Genome Res. 2002;12:325–332. doi: 10.1101/gr.210902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SJ, Regan R, Nicod A, Horsley SW, Kearney L, et al. Subtle chromosomal rearrangements in children with unexplained mental retardation. Lancet. 1999;354:1676–1681. doi: 10.1016/S0140-6736(99)03070-6. [DOI] [PubMed] [Google Scholar]
- Koolen DA, Reardon W, Rosser EM, Lacombe D, Hurst JA, et al. Molecular characterisation of patients with subtelomeric 22q abnormalities using chromosome specific array-based comparative genomic hybridisation. Eur J Hum Genet. 2005;13:1019–1024. doi: 10.1038/sj.ejhg.5201456. [DOI] [PubMed] [Google Scholar]
- Kozma C, Meck JM, Loomis KJ, Galindo HC. De novo duplication of 17p [dup(17)(p12–p11.2)]: report of an additional case with confirmation of the cytogenetic, phenotypic, and developmental aspects. Am J Med Genet. 1991;41:446–450. doi: 10.1002/ajmg.1320410413. [DOI] [PubMed] [Google Scholar]
- Magenis RE, Brown MG, Allen L, Reiss J. De novo partial duplication of 17p [dup(17)(p12–p11.2)]: clinical report. Am J Med Genet. 1986;24:415–420. doi: 10.1002/ajmg.1320240304. [DOI] [PubMed] [Google Scholar]
- Mantripragada KK, Thuresson AC, Piotrowski A, Diaz de Stahl T, Menzel U, et al. Identification of novel deletion breakpoints bordered by segmental duplications in the NF1 locus using high resolution array-CGH. J Med Genet. 2006;43:28–38. doi: 10.1136/jmg.2005.033795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, et al. Emerging patterns of cryptic chromosomal imbalances in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of the literature. J Med Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Potocki L, Chen KS, Park SS, Osterholm DE, Withers MA, et al. Molecular mechanism for duplication 17p11.2 – the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat Genet. 2000;24:84–87. doi: 10.1038/71743. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Knijnenburg J, Bakker E, Vianna-Morgante AM, Sloos W, et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: clinical significance of imbalances present both in affected children and normal parents. J Med Genet. 2006;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumans J, Ruivenkamp C, Holmberg E, Kyllerman M, Anderlid BM, Nordenskjold M. Detection of chromosomal imbalances in children with idiopathic mental retardation by array based comparative genomic hybridisation (array-CGH) J Med Genet. 2005;42:699–705. doi: 10.1136/jmg.2004.029637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer LG, Kashork CD, Saleki R, Rorem E, Sundin K, et al. Targeted genomic microarray analysis for identification of chromosome abnormalities in 1500 consecutive clinical cases. J Pediatr. 2006;149:98–102. doi: 10.1016/j.jpeds.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DW. Recognizable patterns of human malformation. Genetic, embryologic and clinical aspects. Third edition. Major Probl Clin Pediatr. 1982;7:1–653. [PubMed] [Google Scholar]
- Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, et al. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Genes Chromosomes Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- Stern RF, Roberts RG, Mann K, Yau SC, Berg J, Ogilvie CM. Multiplex ligation-dependent probe amplification using a completely synthetic probe set. Biotechniques. 2004;37:399–405. doi: 10.2144/04373ST04. [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- Tyson C, Harvard C, Locker R, Friedman JM, Langlois S, et al. Submicroscopic deletions and duplications in individuals with intellectual disability detected by array-CGH. Am J Med Genet A. 2005;139:173–185. doi: 10.1002/ajmg.a.31015. [DOI] [PubMed] [Google Scholar]
- Vermeesch JR, Melotte C, Froyen G, Van Vooren S, Dutta B, et al. Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J Histochem Cytochem. 2005;53:413–422. doi: 10.1369/jhc.4A6436.2005. [DOI] [PubMed] [Google Scholar]
- Vissers LE, de Vries BB, Osoegawa K, Janssen IM, Feuth T, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am J Hum Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester U, Bondeson ML, Edeby C, Anneren G. Clinical and molecular characterization of individuals with 18p deletion: A genotype-phenotype correlation. Am J Med Genet A. 2006;140:1164–1171. doi: 10.1002/ajmg.a.31260. [DOI] [PubMed] [Google Scholar]
- Wilson DI, Goodship JA, Burn J, Cross IE, Scambler PJ. Deletions within chromosome 22q11 in familial congenital heart disease. Lancet. 1992;340:573–575. doi: 10.1016/0140-6736(92)92107-q. [DOI] [PubMed] [Google Scholar]
- Yobb TM, Somerville MJ, Willatt L, Firth HV, Harrison K, et al. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am J Hum Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]