Abstract
Molecular cytogenetic studies have been instrumental in defining the nature of numerical and structural chromosome changes in human cancers, but their significance remains to be fully understood. The emergence of high quality genome assemblies for several model organisms provides exciting opportunities to develop novel genome-integrated molecular cytogenetic resources that now permit a comparative approach to evaluating the relevance of tumor-associated chromosome aberrations, both within and between species. We have used the dog genome sequence assembly to identify a framework panel of 2,097 bacterial artificial chromosome (BAC) clones, selected at intervals of approximately one megabase. Each clone has been evaluated by multicolor fluorescence in situ hybridization (FISH) to confirm its unique cytogenetic location in concordance with its reported position in the genome assembly, providing new information on the organization of the dog genome. This panel of BAC clones also represents a powerful cytogenetic resource with numerous potential applications. We have used the clone set to develop a genome-wide microarray for comparative genomic hybridization (aCGH) analysis, and demonstrate its application in detection of tumor-associated DNA copy number aberrations (CNAs) including single copy deletions and amplifications, regional aneuploidy and whole chromosome aneuploidy. We also show how individual clones selected from the BAC panel can be used as FISH probes in direct evaluation of tumor karyotypes, to verify and explore CNAs detected using aCGH analysis. This cytogenetically validated, genome integrated BAC clone panel has enormous potential for aiding gene discovery through a comparative approach to molecular oncology.
Introduction
The correlation of non-random karyotypic aberrations with the clinical behavior of human neoplasms is widely recognized as an approach for developing novel means for diagnosis, prognosis and therapeutic design. The expanding field of cancer cytogenetics has stimulated the development of a spectrum of techniques and resources with which such genomic changes can be detected and characterized (for example Cowell et al., 2004; Ishkanian et al., 2004). Despite this, the ability to translate the multitude of reported tumor-associated structural and numerical chromosome abnormalities into meaningful advances in clinical management has yet to reach its full potential. In part this stems from the challenge of establishing which aberrations are biologically significant versus those that are consequences of the disregulation of cell proliferation inherent to tumorigenesis. Since many human cancers have direct counterparts in other species, a comparative approach to molecular oncology presents a means by which to address this challenge. Detection of evolutionarily-conserved tumor-associated chromosome aberrations in different species adds weight to their association with the disease process, whilst the comparative study of the specific sequences involved in recurrent genomic aberrations may clarify how chromosome structure and organization influences the transition from a normal to a malignant karyotype.
The immense value of the domestic dog as a model system for human clinical research is particularly well recognized, a consequence of the similarity in their physiology, their shared environment and the multitude of common and spontaneously-occurring conditions with an overtly genetic basis. Most importantly, the genomic homogeneity of many purebred domestic dog populations provides tremendous opportunities to unearth the underlying etiology of shared genetic traits that have thus far been obscured in human studies by the vast heterogeneity of human populations. We have previously identified evolutionarily-conserved chromosome aberrations in a range of dog cancers that may suggest an ancestrally retained pathogenetic mechanism of cancer (Dunn et al., 2000; Thomas et al., 2003c, 2005; Breen and Modiano, 2008). More importantly, however, these studies have also revealed genomic changes in the canine counterpart that are thus far unreported in the human literature, perhaps as a result of the confounding effects of ‘noise’ in the human genome. In order to understand the biological and comparative relevance of these genomic changes it is essential to be able to interpret cytogenetic observations in direct context with their relevance to gene involvement, both within and between species. The ability to perform fully informative translational studies of canine cancer cytogenetics has, however, been hindered by the use of dog genomics resources that have, by necessity, been reliant primarily on anonymous genetic markers (Mellersh et al., 2000; Guyon et al., 2003; Thomas et al., 2003b, 2005; Breen et al., 2004).
The release of a high-quality, annotated 7.6× genome sequence assembly for the domestic dog (Lindblad-Toh et al., 2005) provides tremendous new opportunities to develop and apply more sophisticated resources and techniques for comparative studies. We are able, for the first time, to compare the DNA sequence of any given chromosome region with the annotated dog genome assembly to establish precisely its gene content, and in turn to correlate it directly with the orthologous sequence in the genome assemblies of human, mouse and an increasing number of other species. By localizing that DNA sequence to a defined cytogenetic location, we can establish with ease and accuracy how structural and numerical chromosome abnormalities affect the status of key genes. The utilization of the genome sequence assembly in context with the cytogenetic map of the dog therefore represents a powerful combination for investigating the range, frequency and implications of both structural and numerical chromosome abnormalities. Previously we developed a low-resolution panel of BAC clone markers distributed at 10 Mb intervals along each dog chromosome, as a means to aid chromosome identification and to permit rapid, first-pass analysis of genomic aberrations in dog tumors, prior to more detailed evaluation (Thomas et al., 2007). As a means to fully integrate the dog cytogenetic map with the genome sequence assembly, we have now developed a framework panel of 2097 cytogenetically validated bacterial artificial chromosome (BAC) clones selected at intervals of approximately one megabase (1 Mb) from the ∼2.4 Gb of euchromatic sequence in the dog genome assembly. We demonstrate the excellent correlation between the cytogenetic map and the genome assembly, and discuss the wide range of potential applications of this BAC panel in canine and comparative genomics analysis. Finally, we present the development of this BAC clone set into a 1 Mb resolution, genome-anchored microarray for comparative genomic hybridization (aCGH) analysis. We demonstrate how this tool enables direct translation of dog tumor-associated chromosome copy number changes into DNA sequence, revealing the consequences of genomic aneuploidy on gene dosage, and providing exciting new opportunities for detailed comparative cancer studies.
Materials and methods
Identification of a 1 Mb resolution BAC panel from the dog genome assembly
Unless otherwise stated, all clones described in the present study were derived from the CHORI-82 dog BAC library (http://bacpac.chori.org, BACPAC Resources, Children's Hospital Oakland Research Institute, Oakland, CA), derived from a female Boxer, and from which the 7.6× dog genome assembly was constructed (Lindblad-Toh et al., 2005). BAC paired-end read placements were used to identify clones distributed evenly along each of the 39 different chromosomes in the female dog, by reference to the genome assembly (version 2.0, May 2005, Lindblad-Toh et al., 2005). For clones with ambiguous assembly placements the paired-end reads were aligned to the genome using BLAST. Only clones with high quality positioning of both ends in the genome, and with accurate relative positioning (50–400 kb apart), were candidates for inclusion in the map.
Due to the presence of highly repetitive sequences at the centromere that are intractable to genome sequence assembly, the most proximal clone available on each of the dog autosomes was typically located approximately 3 Mb distal to the centromere (mean 3.17 Mb, range 3.01–3.33 Mb). From this point, BAC clones were selected from the assembly at approximately 1 Mb intervals along the length of each dog chromosome to the distal end of the available genome sequence assembly, resulting in a total of 2122 clones. For practical reasons clone selection was restricted to the smallest number of library master plates that could be accessed in order to result in a clone set of approximately 1 Mb. The BAC panel also included clones representing canine orthologues of 53 human genes associated with a range of cancers. All BAC clone addresses and their assembly positions are provided in the supplementary online material associated with this report (located at http://www.cvm.ncsu.edu/mbs/breen_matthew.htm - tables A and B). Locations of BAC clones are denoted herein according to their chromosome of origin and then their Mb position on that chromosome, according to the dog genome assembly (for example, CFA1; 3 Mb).
Cytogenetic validation of BAC clones and construction of the dog 1 Mb array
BAC DNA was isolated from 2.5 ml bacterial cultures of each clone using a Qiagen REAL Prep 96 kit (Qiagen, Valencia, CA). Cytogenetic mapping was performed according to our routine multicolor FISH protocols (Breen et al., 2004) in groups of five consecutive, differentially-labeled fluorescent probes, beginning with the most proximal clone on each chromosome (Thomas et al., 2007). Cytogenetic assignments are reported according to the DAPI-banded dog karyotype nomenclature of Breen et al. (1999a). In order to be included in microarray construction, clones were required to generate a unique FISH hybridization signal consistent with their position in the genome assembly. Clones that did not meet these criteria were replaced by alternate selections from the assembly. If the alternate clone also failed to meet the required standards, that locus was excluded from all downstream procedures.
Microarray construction was performed as described previously using the optimized panel of BAC clones (Thomas et al.,2005). Array performance was first investigated by performing self-self and sex-mismatch hybridization of peripheral blood DNA from clinically healthy donors. We then performed aCGH analysis of DNA isolated from a histiocytic tumor biopsy from a female Flat-Coated Retriever. Tumor DNA was labelled with Cyanine3-dCTP (Perkin Elmer Life Sciences, Boston, MA) using a BioPrime Array CGH Labelling System (Invitrogen, Carlsbad, CA). A reference sample representing equimolar pools of DNA isolated from the peripheral blood of ten clinically-healthy female Flat-Coated Retrievers was similarly labelled with Cyanine5-dCTP (Perkin Elmer), and aCGH analysis was performed as described previously (Thomas et al., 2007). A dye-swap analysis was also performed, in which the test and reference samples were labelled with Cyanine5-dCTP and Cyanine3-dCTP respectively. Regions of genomic imbalance were identified using the aCGH-smooth algorithm (Jong et al.,2004), with threshold limits for gain and loss set at log2 values equivalent to 1.15:1 (gain) and 0.85:1 (loss) of tumor vs. reference ratios respectively.
Evaluation of genomic imbalances by single-locus probe FISH analysis of tumor cells
Chromosome preparations were generated from a fresh biopsy specimen from the histiocytic malignancy by direct harvest of tumor cells using conventional techniques of colcemid arrest, hypotonic treatment and methanol:glacial acetic acid fixation. For consistency, these cells were isolated from the same biopsy specimen from which the tumor DNA was isolated for aCGH analysis. FISH analyses of chromosome and interphase preparations from the tumor sample were carried out using single locus BAC probes (SLPs) representing selected clones from the 1 Mb panel, chosen from regions showing a range of normal and aberrant copy number ratios in aCGH. The same probes were also applied to normal dog chromosome preparations and images were acquired from a minimum of 30 representative cells in each instance. The copy number status of each probe was scored by two independent investigators with no prior knowledge of the corresponding aCGH data.
Results
Cytogenetic analysis of the 1 Mb dog BAC panel
Initial (phase I) FISH analysis of our original selected panel of 2122 BAC clones resulted in successful placement of 1941 clones (91.5%) to the expected, unique chromosome location in full concordance with their position in the dog genome assembly, satisfying our criteria for use as downstream cytogenetic markers. Results of FISH analysis are summarized in Table 1, and Fig. 1 demonstrates our FISH analysis approach, using combinations of BAC clones positioned at intervals of 10 Mb and 1 Mb on dog chromosome 10 (Canis familiaris, CFA). Of the 181 clones that were rejected since they did not meet the necessary standard, 108 (60%) mapped to the expected chromosome location but showed hybridization signals at additional genomic sites that would confound downstream analysis. These included 106 clones that showed one or more secondary signals that had no obvious association with the observed primary signal. Two further clones mapped to the centromeres of all chromosomes.
Table 1.
FISH analysis results
| Phase I |
Phase II |
|||
|---|---|---|---|---|
| # BACs | % | # BACs | % | |
| Unique location consistent with assembly | 1941 | 91.5% | 2097 | 98.8% |
| Unique location on expected chromosome but position not consistent with assembly | 3 | 0.1% | 1 | 0.0% |
| Location consistent with assembly but with additional weak secondary signal | 43 | 2.0% | 3 | 0.1% |
| Location consistent with assembly but with additional strong secondary signal | 49 | 2.3% | 3 | 0.1% |
| Location consistent with assembly but with multiple secondary signals | 14 | 0.7% | 3 | 0.1% |
| Mapped to multiple centromeres | 2 | 0.1% | 0 | 0.0% |
| Location not consistent with assembly | 27 | 1.3% | 8 | 0.4% |
| No visible FISH signal after multiple attempts on unrelated dogs | 40 | 1.9% | 7 | 0.3% |
| Selected clone repeatedly failed in culture | 3 | 0.1% | 0 | 0.0% |
| Total | 2122 | 100.0% | 2122 | 100.0% |
Fig. 1.
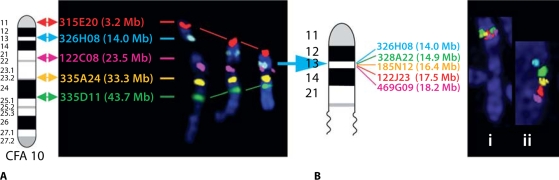
FISH analysis of clones from CFA10. (A) The chromosome location of five differentially labeled clones from the previously reported 10 Mb resolution dog BAC set (Thomas et al., 2007), starting with the most centromeric clone. The text color indicates the fluorochrome with which each clone was labeled for FISH analysis, and the Mb position on CFA10 is shown after the corresponding BAC address. To the left is the CFA10 ideogram, and to the right, three examples of these five probes hybridized to CFA10 at increasingly later stages of metaphase. Accurate assignment of clones at 10 Mb resolution is clearly possible in early metaphase, and probe order is easily ascertained. (B) Five probes at intervals of approximately 1 Mb, starting from the second clone in the previous 10 Mb set (326H08). In metaphase (Bi) these five probes at 1 Mb intervals can be accurately assigned to a chromosome band, but their relative order cannot be readily resolved due to extensive chromosome contraction. Accurate ordering is thus reliant on interphase mapping (Bii).
The remaining 73 (40%) clones failed to hybridize to the expected location. These included three clones that failed to revive in culture, most likely due to a fault during library replication, or loss of the BAC vector resulting in loss of antibiotic resistance. Forty clones did not demonstrate any detectable hybridization signal after multiple attempts on metaphase chromosome preparations from a minimum of two unrelated dogs. A total of 27 clones mapped to a location inconsistent with their position in the genome assembly. Nineteen of these anomalies were resolved by selecting alternate clones from the assembly that share contiguous sequence with the original BAC, and thus were unlikely to reflect major inconsistencies between the cytogenetic map and the genome assembly. Finally, three clones (182G07, 307O16 and 376D10) mapped to the expected chromosome but at a location inconsistent with the genome assembly, and were further investigated. For clones 182G07 and 307O16 (from CFA1 and 6 respectively), the anomaly was resolved by selecting alternate clones from the genome assembly, which mapped to the expected location (see Fig. 2 for details). The observed correlation between genome sequence and FISH data for these alternate clones, positioned 0.3 Mb and 0.1 Mb respectively from the original selections, implies that the phase I findings represent minor discrepancies in the placement of individual clone sequences within the genome assembly for CFA1 and CFA6 rather than extensive discordance between the assembly and the cytogenetic map. A third clone, 376D10, originally placed at CFA2; 4.3 Mb according to BLAST of BAC end sequences, mapped significantly more distally than expected, and through co-hybridization with clones from this region, was resolved to approximately CFA2; 87 Mb. This finding was subsequently attributed to misplacement in the assembly of one end of the BAC clone sequence at CFA2; 4.3 Mb, most likely due to the presence of repetitive sequence, whilst the other end was correctly positioned at CFA2; 86.7 Mb. This region of CFA2, extending from 4 Mb to 11 Mb from the centromere, showed a unique profile of FISH results that reflect the existence of an extensive region of tandemly repeated DNA sequences, an example of which is given in Fig. 3. Details of clones with anomalous FISH data are provided in the supplementary online material associated with this report (http://www.cvm.ncsu.edu/mbs/breen_matthew.htm - table C).
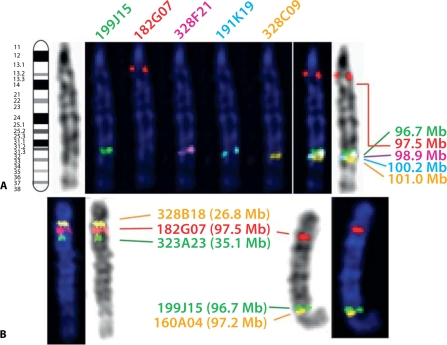
Fig. 2.
Use of FISH analysis to resolve mapping anomalies. Clone 182G07 (CFA1; 97.5 Mb) mapped by FISH analysis to a site significantly more proximal to that suggested by the genome assembly (A). Co-hybridization of this BAC with two clones from this more proximal region of CFA1 resolved the location of 182G07 to approximately CFA1; 30 Mb (B, left image), whilst the gap left by this clone was successfully filled by clone 160A04 (CFA1; 97.2 Mb), selected from the assembly (B, right image). Using the same strategy, the location of clone 307O16 (CFA6; 61.9 Mb) was revised to a more proximal location (CFA6; 45 Mb, data not shown) and the consequent gap was filled successfully by clone 502O13 (CFA6; 62.0 Mb).
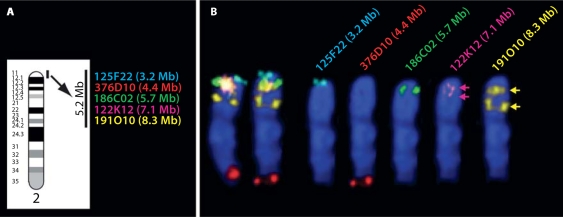
Fig. 3.
FISH analysis of five BAC clones from the CFA2qcen region of the genome assembly. The five most proximal clones selected for analysis in phase I are shown in A against the ideogram of CFA2, spanning a 5.2 Mb region of the genome assembly. (B) Left, the composite results of FISH analysis of these clones on both CFA2 homologues from one cell and right, the five separate planes of fluorescent signal corresponding to each differentially labeled clone are shown individually for one of these homologues. Two clones (125F22 and 186C02) showed unique signal in the expected chromosome location. Clones 122K12 and 191O10 also mapped where expected, but showed additional hybridization sites further down the chromosome, which were of equal strength to the primary signal. These pairs of probe signals (arrowed) suggested the presence of tandem repeat sequences in this region of CFA2, which was supported by the identification of a similar pattern of results for the next distal marker from this region (199D17, CFA2; 8.9 Mb, data not shown). Clones distal to this marker mapped to the expected unique location, thus the region of repetitive sequence appears to extend to approximately 9 Mb from the start of the CFA2 assembly. Note that clone 376D10 (CFA2; 4.3 Mb according to the assembly) mapped significantly more distally than expected, close to the CFA2 telomere. Through co-hybridization of 376D10 with clones from this region (following the strategy outlined in Fig. 2), the cytogenetic location of this BAC was resolved to approximately CFA2; 87 Mb. This finding was subsequently attributed to misplacement of one end of the BAC clone sequence at CFA2; 4.3 Mb, most likely due to the presence of repetitive sequence, whilst the other end was correctly positioned at CFA2; 86.7 Mb.
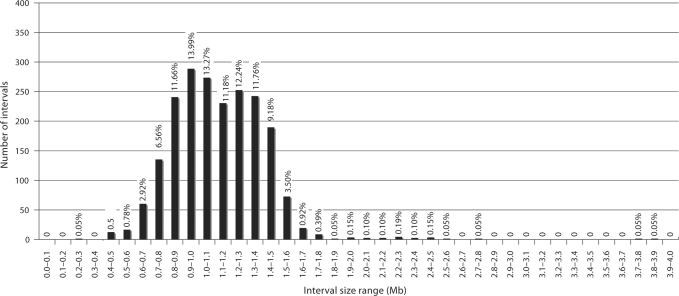
After completion of phase II, following investigation of anomalous data and selection of alternates to replace clones rejected in the first phase of the study, the total number of clones that mapped to a unique chromosomal location in concordance with their position in the dog genome assembly was increased to 2097 (98.8%). This optimized clone set provides a mean resolution of 1.12 Mb throughout the female dog genome (median 1.10 Mb, range 0.28–3.28 Mb), with 94.4% of intervals being ≤1.5 Mb in size. Figure 4 shows the mean, maximum and minimum intervals between clones for each chromosome, and Fig. 5 summarizes the distribution of interval sizes throughout the genome. Detailed results of FISH analysis are provided in the supplementary online material associated with this report.
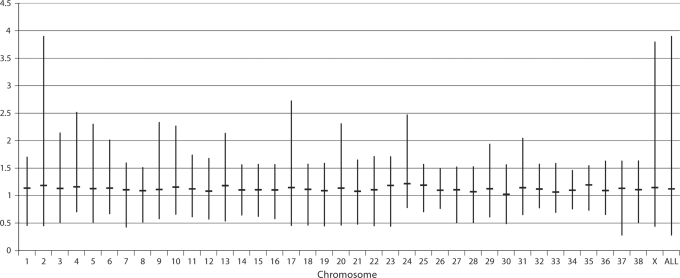
Fig. 4.
Distance between consecutive clones from the optimized 1 Mb clone set on each dog chromosome. The top and bottom of each vertical bar indicate the maximum and minimum interval respectively (in Mb) in the optimized clone panel for each chromosome. The horizontal line bisecting each bar shows the mean interval size on that chromosome. The final bar summarizes interval data for the whole genome, demonstrating the mean genome-wide interval of 1.12 Mb. Note the large maximum interval encountered on CFA2, which is a consequence of the inability to identify BAC clones with unique FISH signals due to the presence of highly repetitive DNA sequence near the centromere. The large maximum interval for CFAX is due to under-representation of the CFAX centromere in the dog genome sequence, which is also attributed to extensive tracts of repetitive sequence in this region.
Fig. 5.
Distribution of intervals between consecutive clones from the optimized 1 Mb clone set. Vertical bars show the number of intervals within the Mb size ranges indicated on the x-axis. The percentage of intervals in each size range is shown above the corresponding bar, showing that 94.4% of clones map at intervals ≤1.5 Mb.
Application of the BAC panel in tumor chromosome analysis
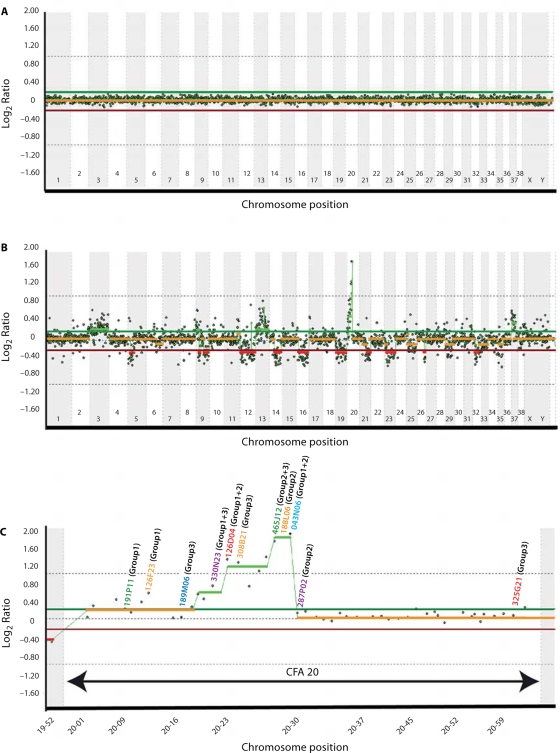
The optimized panel of 2097 cytogenetically-validated BAC clones was used to generate a genome-wide microarray resource. Results of self-self aCGH analysis of genomic DNA isolated from the peripheral lymphocytes of a clinically normal donor (Fig. 6A) were consistent with the expected 1:1 ratio of Cyanine3 versus Cyanine5 hybridization throughout the genome. Subsequent aCGH analysis of a histiocytic tumor isolated from a female Flat-Coated Retriever dog revealed a wide range of whole and partial chromosome copy number changes throughout the genome (Fig. 6B, C). Genomic gains were observed on CFA3, 9, 13, 20 and 37, and copy number losses included regions of CFA5, 9, 12, 14, 16, 19, 21, 23, 26, and 32. aCGH analysis revealed copy number changes involving several known cancer genes, including gain of FES (CFA3), TSC2 (CFA6), and MYC and KIT (CFA13), and loss of BRCA1, NF1 and ABL1 (CFA9) and PTEN (CFA26). A region of high-level amplification was observed on CFA20, approximately 28–30 Mb from the centromere. This region was investigated further by CGH-directed single locus probe (SLP) analysis of the tumor using 11 BAC clones from CFA20, revealing four derivative chromosome structures containing regions corresponding to this chromosome. Enumeration of probe signals for each BAC clone supported their copy number ratios as determined by aCGH analysis. These data are summarized with examples in Fig. 7.
Fig. 6.
Whole genome aCGH profiles obtained using the 1 Mb assembly-integrated dog BAC array, showing composite dye-swapped array CGH profiles of (A) a self-self hybridization of differentially labeled male reference DNA and (B) a DNA sample derived from a canine histiocytic tumor co-hybridized with reference DNA. Data are plotted as the median, block-normalized and background-subtracted log2 ratio of the replicate spots for each BAC clone on the 1 Mb array. Log2 ratios representing genomic gain and loss are indicated by horizontal bars above (green line) and below (red line) the midline (orange line) representing normal copy number. The aCGH profile in A shows that the copy number status throughout the genome is reported as normal, as expected for a self-self hybridization. In B this chromosome copy-number status line appears as either red or green in regions where genomic imbalances were apparent (red = loss, green = gain), as determined by the aCGH Smooth algorithm (Jong et al., 2004). The profile indicates whole chromosome gain for CFA3, CFA13 and CFA37, and a high level amplification on CFA20. Genomic losses were detected on CFA5, CFA9, CFA12, CFA14, CFA16, CFA19, CFA21, CFA23, CFA26 and CFA32. The amplification on CFA20 is enlarged to show more detail in C, depicting the copy number ratio for all clones distributed along this chromosome. A subset of these clones (indicated on the CFA20 profile in C) was subsequently used as SLPs for FISH analysis of tumor chromosomes. The color of the text used for each clone address corresponds to the fluorochrome with which it was labeled, and the number shown against each clone address shows which clones were grouped together in FISH analysis (see Fig. 7A–E for details).
Fig. 7.
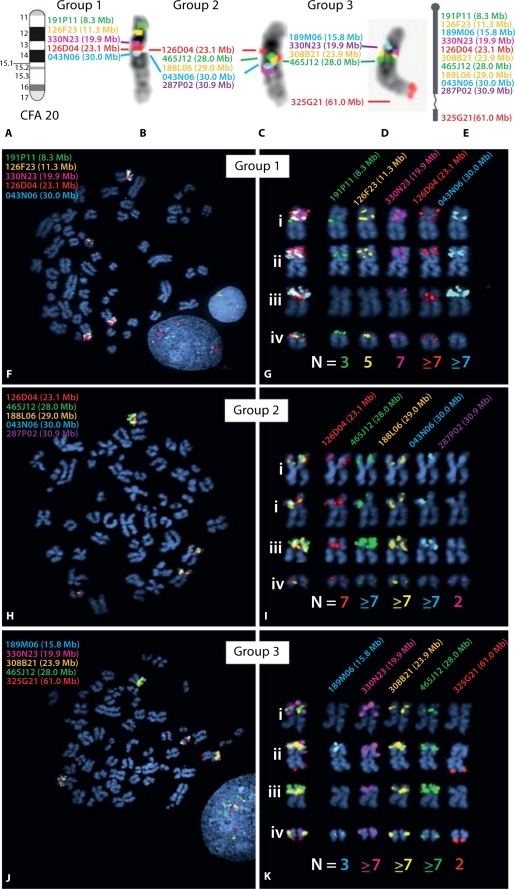
Targeted FISH analysis of CFA20 in the dog histiocytic sarcoma case, using clones from the 1 Mb array. Based on results of aCGH analysis, a panel of 11 BAC clones was selected for detailed characterization of copy number status along CFA20. Probes were combined into three groups of five differentially-labeled probes for FISH analysis (with four probes represented twice). All clones were hybridized onto normal metaphase chromosome preparations from a clinically healthy donor, to confirm the expected probe location relative to the CFA20 ideogram (Fig. 5A). The resulting images of probe signals on normal banded chromosomes are shown in B–D, and these data are summarized schematically in E. The text color corresponds to the fluorochrome with which each BAC probe was labeled, and the Mb position of each clone on CFA20 is also shown. (F–K) Results of FISH analysis using the same three groups of BAC clones on the histiocytic sarcoma case. These data demonstrate four distinct chromosome structures harboring regions corresponding to CFA20, each with a different morphology and probe hybridization profile. These four structures comprised two metacentric chromosomes, one sub-metacentric chromosome and one small acrocentric chromosome whose SLP profile was consistent with that of a grossly normal CFA20. The modal copy number for each probe in all cells analyzed is shown below the corresponding chromosome structure. Note that assessment of copy number status is challenging for probes showing high level amplifications, since apparent tandem duplications often result in large probe signals that cannot be resolved fully even in interphase preparations. Modal copy numbers are therefore based on analysis of both metaphase and interphase chromosomes from >30 cells in each instance.
Discussion
The development of the 7.6× dog genome sequence assembly provides exciting opportunities for increasing the resolution, sophistication and versatility of resources for canine and comparative genomics studies. We have used the assembly to select and validate a genome-wide panel of annotated BAC clones as a tool for genome-integrated molecular cytogenetic analysis, and as a means to investigate the correlation between the assembly-derived position of each clone and its cytogenetic location, through integrated assignment of this framework panel of clones by both techniques. In the first phase of cytogenetic analysis, 91.5% of the 2122 BAC clones selected fulfilled our required criteria, confirming the high quality of the assembled dog genome sequence. In the second phase of cytogenetic analysis we were able to increase the level of concordance to 98.8%, by assessing alternate clones as potential replacements for those rejected during phase I. The cytogenetic distribution of the resulting optimized clone set shows excellent full-length coverage of each dog chromosome, with the known exception of the highly-repetitive centromeric regions. Taking into account the fluctuations in clone positions as the genome assembly has developed since our original clone selection, the distribution of intervals between consecutive clones on each chromosome is highly consistent. Figure 4 shows two major exceptions, namely CFA2 and CFAX, which are deficient in clone density at the centromeres. We have shown previously (Thomas et al., 2007) that clone 283A18 (CFAX; 47.9 Mb) maps by FISH analysis to the centromere of CFAX. This site is proximal to a gap in the genome sequence assembly of approximately 3.04 Mb that is devoid of mapped BAC ends due to the density of repetitive DNA, consistent with its heterochromatic nature. As a consequence, an interval of 3.79 Mb exists in this region of our CFAX BAC map. This is consistent with the mean 3.17 Mb gap in the map of BAC end sequences at the centromere of each autosome in the genome sequence assembly, where, unlike the submetacentric X chromosome, their telocentric structure does not result in interruption of the cytogenetic map. We did, however, identify two clones 326K03 (CFA1; 3.1 Mb) and 330E21 (CFA37; 6.4 Mb) that mapped to the centromeres of all chromosomes, although with variable signal intensity, suggesting that they contain highly repetitive sequence that is more highly represented on certain chromosomes as compared to others.
Tracts of repetitive sequences were not restricted to chromosome ends. Whilst not apparent from the annotation of the genome assembly itself, FISH analysis of clones within the region extending 9 Mb from the centromere of CFA2 showed multiple hybridization signals that were confined to this short length of the chromosome (Fig. 3). These findings revealed the existence of an extensive and previously unreported region of tandemly repeated DNA sequence that precluded the identification of clones with unique FISH profiles, resulting in an interval of 3.89 Mb near CFA2qcen in our genome-integrated BAC panel. This region is now being investigated in more detail in a separate study.
Another well-known region of sequence homology was identified by this study, namely the pseudoautosomal region shared by the sex chromosomes. FISH analysis of clones selected at 1 Mb intervals from the CFAXp chromosome assembly revealed the subset of clones that co-hybridize to the homologous region of CFAYp, and through higher resolution FISH analysis of additional BAC clones selected from the assembly, enabled the precise boundaries of this region to be elucidated. These data are presented elsewhere (Young et al., manuscript in preparation).
The development of a genome-integrated BAC map has thus generated new insights into the structure of domestic dog chromosomes, and provides extensive opportunities for detailed evaluation of genome organization both in normal individuals and also, perhaps more importantly, in aberrant karyotypes. This application has particular relevance in light of the recent explosion in interest in cancer genomics, both in human and veterinary medicine, where the ability to perform translational studies based on comparative analyses has enormous potential for mutual advances in disease diagnosis, prognosis and development of novel therapeutics. Recently we have shown for the first time that human and canine counterparts of the same cancer share evolutionarily conserved chromosome aberrations that result in structural and numerical defects of the same genes (Breen and Modiano, 2008). These observations provide direct evidence for the non-random basis of tumor-associated chromosome arrangement and genomic aneuploidy, although the mechanisms by which they occur are not fully understood. We anticipate that the resource detailed in the present study has enormous potential to contribute towards answering this and similar fundamental questions in molecular oncology. Characterization of dog tumor-associated chromosome aberrations is highly challenging and labor-intensive using classical cytogenetics techniques alone, particularly in view of the similarity in size, morphology and banding pattern of many of the smaller dog chromosomes. The development of a cytogenetically-validated, genome integrated BAC panel for each dog chromosome now overcomes these limitations. We have expanded the ability for comparative tumor cytogenetics studies through the validation by FISH analysis of dog BAC clones representing 53 key cancer-associated genes, which are now being used in our ongoing studies of a wide range of dog tumors and their human counterparts. We limited our selection to CHORI-82 clones that contain the full length of the corresponding gene according to the annotated dog genome browser, which should serve to facilitate future studies directed towards more localized sequence analysis of these key loci. Our own aims, however, are focused heavily on the identification of gene sequences that are associated with the onset and/or behavior of dog tumors, but whose role in human and/or veterinary studies has not previously been described. We demonstrate how our assembly-integrated aCGH and cytogenetic resources might be used towards this goal using the example of a case of histiocytic sarcoma in a female Flat-Coated Retriever, which was recruited into an ongoing study of these tumors by our group. This case has previously been analyzed using our 10 Mb-resolution BAC microarray (Thomas et al., 2007), with which we detected chromosome copy-number changes including gains of CFA3, CFA13 (including the KIT and MYC proto-oncogenes), CFA20 and CFA37, and losses including CFA16, 23 and 26 (including the PTEN tumor-suppressor gene). Most striking was a region of amplification at CFA20q12dist→ q13prox corresponding to BAC clone 126D04 (CFA20; 23.1 Mb). Using aCGH-directed SLP analysis of tumor metaphase preparations, we previously identified four chromosome structures containing regions corresponding to CFA20, each with a different size, morphology and probe hybridization profile resulting from several structural and numerical chromosome changes (Thomas et al.,2007). FISH analysis showed that the amplification corresponding to clone 126D04 was consistent with the presence of six copies of this region distributed across all four of these abnormal chromosome structures. By reference to the dog genome assembly browser (http://genome.ucsc.edu) we identified a gene, FOXP1, less than 1 Mb distal to clone 126D04, for which genomic changes have been associated with prognosis in human cancers (for example, Barranset al., 2004; Fox et al.,2004). We are now investigating this region with particular interest in a wider study of dog tumors.
We elected to use this same case for extended analyses using the 1 Mb resolution microarray in order to demonstrate the degree of additional genomic information that can be extracted as we continue to increase the sophistication of available resources. aCGH analysis of the histiocytic tumor with this higher resolution array confirmed the presence of previously identified copy number changes but also revealed other regions of genomic imbalance, including several additional subchromosomal gains and losses (Fig. 6B), further emphasizing the complex nature of this tumor karyotype. Figure 6C shows a detailed view of the aCGH profile for CFA20. It is evident from this that the peak of amplification corresponding to clone 126D04 (CFA20; 23.1 Mb) identified using the 10 Mb array lies proximal to a region of apparently even greater amplification peaking at the site delineated by clones 465J12, 188L06 and 043N06 (CFA20; 28.0–30.0 Mb), beyond which the distal half of CFA20 appears to be balanced. Clones distributed along CFA20 were assessed in SLP analysis of tumor chromosomes (Fig. 7) and showed close correlation with aCGH data, indicating the presence of more than seven copies of regions corresponding to clones in the region of high-level amplification. We also carried out SLP analysis using clone 308B21, which contains the full length of the FOXP1 gene, and confirmed that over 75% of cells sampled from the histiocytic tumor carry six or more copies of this region.
Clone 465J12 (CFA20; 28 Mb) lies in the region of high-level amplification, with a modal copy number of seven per cell (increasing to ten copies in a small proportion of cells), representing the most extreme copy number increase we have observed in dog cancers to date. By reference to the dog genome assembly browser we established that this clone contains the full coding sequence of gene LRIG1. Tumor-associated chromosomal gain of the evolutionarily conserved region in the short arm of human chromosome 3 (specifically HSA3p14) has previously been correlated with overexpression of the LRIG1 gene (Ljuslinder et al., 2005). In turn the amplified status of the LRIG1 gene has been proposed to confer an improved prognosis in several human cancers (for example, Yang et al., 2006; Lindstrom et al., 2008) by acting as a tumor-suppressor (Hedman et al., 2002; Shattuck et al.,2007). Recently this hypothesis has been revised to suggest that LRIG1 copy number and expression levels may in fact have either positive or negative impacts on prognosis depending on the specific cellular environment (Hedman and Henriksson, 2007). With increasing evidence for the correlation between copy number imbalances and gene expression changes (for example, Hyman et al., 2002; Sweet-Cordero et al., 2006; Lilljebjorn et al., 2007), our interest in this region of CFA20 has increased further as a result of these observations and we are continuing to monitor its frequency and distribution in dog tumors.
The expanding panel of genomics resources for the dog, and now the ease with which we can identify BAC clones representing genes of specific interest from the sequence assembly, provides long-awaited opportunities to evaluate the significance of genomic aneuploidy in canine cancers. Through application of genome-integrated cytogenetic resources in both species it is likely that we will continue to identify evolutionarily related chromosomal defects in human and canine counterparts of the same cancer. Our rationale for developing this canine genomics resource is, however, to permit the detection of novel genetic features in the dog whose role has not yet been evaluated in the human field. The use of the dog genome as a powerful tool for gene discovery has been demonstrated in several prior studies, most notably detection of the HCRTR2 gene disruption responsible for canine narcolepsy (Lin et al., 1999), and the mutation associated with canine hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis (Lingaas et al., 2003), both of which share phenotypic counterparts in human medicine. These studies have set the scene for similar achievements in the detection of novel tumor-associated genes.
We have shown in the present study that the 1 Mb resolution genome-integrated cytogenetic BAC map enables investigation of genome structure, both in normal and malignant cells. The correlation of fragile sites in the human genome with chromosome instability and clinical disorders, including cancer, has long been recognized (see Sutherland and Baker, 2000 for a review). More recently, several sites of non-random chromosome disruption in human tumors have been shown to coincide with known synteny breakpoints in the genomes of a diverse set of species (for example, Darai et al., 2005). This suggests that the regional structural instability of specific chromosome sequences contributes to chromosome evolution in both the long- (between species) and short-term (during tumor development within species). The gross structural relationship between the dog and human genomes has already been established (Breen et al., 1999b, 2004; Yang et al., 1999), revealing the sites of chromosome breakage and fusion that have occurred during divergent evolution from their common ancestral karyotype. Through cytogenetic assignment of domestic dog BAC clones to the chromosomes of other canids, as well as other mammalian species, interruptions in conserved synteny can now be refined at higher resolution. With the ability to identify the precise DNA sequence at these chromosome sites in multiple species we now have an excellent opportunity to elucidate the relationship between evolutionary breakpoints and those associated with tumorigenesis.
The global value of this 1 Mb array extends well beyond its two-fold increase in resolution over our previously described resource (Thomas et al., 2005). Due to the selection of all arrayed targets at uniform intervals directly from within the dog genome assembly, the array offers complete correlation between observed cytogenetic aberrations and the underlying gene sequence without the need to generate DNA sequence information from arrayed targets. This in turn facilitates investigation of the biological consequence of gross genomic alterations in dog cancers, and indeed in any phenotype resulting from chromosome abnormalities, whether structural or numerical, that can be observed using molecular cytogenetic techniques. To our knowledge the development of this CGH microarray now places the dog in a position equivalent to only the traditional rodent model, the laboratory mouse, in terms of the resolution of such reagents (Chung et al., 2004) for non-human species. It now becomes possible to characterize precise gene content at the site of chromosome breakpoints in cancer, and to generate higher-resolution microarrays for subchromosomal regions of specific interest. The technique of array-painting, using microdissected/flow-sorted chromosomes to probe genomic microarrays, will now enable complete characterization of the gene content of complex tumor-associated derivative structures in the dog, which to date has been possible only in the human field (Fiegler et al., 2003; Backx et al., 2007; Gribble et al., 2007). The ultimate goal will be the construction of a tiling-path array for the entire dog genome, comprising overlapping genomic clones with complete coverage of each chromosome, which to date is available only for human studies (Ishkanian et al., 2004).
Supplementary online materials
The following additional data will be provided at the corresponding author's website: http://www.cvm.ncsu.edu/mbs/breen_matthew.htm
SOM Table A: Comprehensive list of mapping data for all clones comprising the 1 Mb resolution genome-integrated and cytogenetically validated BAC panel.
SOM Table B: Mapping data for BAC clones containing specific genes targeted for inclusion in the BAC panel.
SOM Table C: List of BAC clones with anomalous FISH mapping data.
Footnotes
This work was supported by grants from the National Institutes of Health (R21NS051190-01) and the American Kennel Club Canine Health Foundation (CHF-403) awarded to MB. CFL, PE and AE are supported by funds from the Wellcome Trust.
References
- Backx L, Van Esch H, Melotte C, Kosyakova N, Starke H, et al. Array painting using microdissected chromosomes to map chromosomal breakpoints. Cytogenet Genome Res. 2007;116:158–166. doi: 10.1159/000098181. [DOI] [PubMed] [Google Scholar]
- Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood. 2004;104:2933–2935. doi: 10.1182/blood-2004-03-1209. [DOI] [PubMed] [Google Scholar]
- Breen M, Modiano JF. Evolutionarily conserved cytogenetic changes in hematological malignancies of dogs and humans – man and his best friend share more than companionship. Chromosome Res. 2008;16:145–154. doi: 10.1007/s10577-007-1212-4. [DOI] [PubMed] [Google Scholar]
- Breen M, Bullerdiek J, Langford CF. The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res. 1999a;7:401–406. doi: 10.1023/a:1009224232134. [DOI] [PubMed] [Google Scholar]
- Breen M, Thomas R, Binns MM, Carter NP, Langford CF. Reciprocal chromosome painting reveals detailed regions of conserved synteny between the karyotypes of the domestic dog (Canis familiaris) and human. Genomics. 1999b;61:145–155. doi: 10.1006/geno.1999.5947. [DOI] [PubMed] [Google Scholar]
- Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, et al. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:65–75. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung YJ, Jonkers J, Kitson H, Fiegler H, Humphray S, et al. A whole-genome mouse BAC microarray with 1-Mb resolution for analysis of DNA copy number changes by array comparative genomic hybridization. Genome Res. 2004;14:188–196. doi: 10.1101/gr.1878804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell JK, Wang YD, Head K, Conroy J, McQuaid D, Nowak NJ. Identification and characterisation of constitutional chromosome abnormalities using arrays of bacterial artificial chromosomes. Br J Cancer. 2004;90:860–865. doi: 10.1038/sj.bjc.6601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darai E, Kost-Alimova M, Kiss H, Kansoul H, Klein G, Imreh S. Evolutionarily plastic regions at human 3p21.3 coincide with tumor breakpoints identified by the ‘elimination test’. Genomics. 2005;86:1–12. doi: 10.1016/j.ygeno.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Dunn KA, Thomas R, Binns MM, Breen M. Comparative genomic hybridization (CGH) in dogs – application to the study of a canine glial tumour cell line. Vet J. 2000;160:77–82. doi: 10.1053/tvjl.2000.0495. [DOI] [PubMed] [Google Scholar]
- Fiegler H, Gribble SM, Burford DC, Carr P, Prigmore E, et al. Array painting: a method for the rapid analysis of aberrant chromosomes using DNA microarrays. J Med Genet. 2003;40:664–670. doi: 10.1136/jmg.40.9.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox SB, Brown P, Han C, Ashe S, Leek RD, et al. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor alpha and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10:3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- Gribble SM, Kalaitzopoulos D, Burford DC, Prigmore E, Selzer RR, et al. Ultra-high resolution array painting facilitates breakpoint sequencing. J Med Genet. 2007;44:51–58. doi: 10.1136/jmg.2006.044909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyon R, Lorentzen TD, Hitte C, Kim L, Cadieu E, et al. A 1-Mb resolution radiation hybrid map of the canine genome. Proc Natl Acad Sci USA. 2003;100:5296–5301. doi: 10.1073/pnas.0831002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedman H, Henriksson R. LRIG inhibitors of growth factor signalling – double-edged swords in human cancer? Eur J Cancer. 2007;43:676–682. doi: 10.1016/j.ejca.2006.10.021. [DOI] [PubMed] [Google Scholar]
- Hedman H, Nilsson J, Guo D, Henriksson R. Is LRIG1 a tumour suppressor gene at chromosome 3p14.3? Acta Oncol. 2002;41:352–354. doi: 10.1080/028418602760169398. [DOI] [PubMed] [Google Scholar]
- Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–6245. [PubMed] [Google Scholar]
- Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, et al. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- Jong K, Marchiori E, Meijer G, Vaart AV, Ylstra B. Breakpoint identification and smoothing of array comparative genomic hybridization data. Bioinformatics. 2004;20:3636–3637. doi: 10.1093/bioinformatics/bth355. [DOI] [PubMed] [Google Scholar]
- Lilljebjorn H, Heidenblad M, Nilsson B, Lassen C, Horvat A, et al. Combined high-resolution array-based comparative genomic hybridization and expression profiling of ETV6/RUNX1-positive acute lymphoblastic leukemias reveal a high incidence of cryptic Xq duplications and identify several putative target genes within the commonly gained region. Leukemia. 2007;21:2137–2144. doi: 10.1038/sj.leu.2404879. [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Lindstrom AK, Ekman K, Stendahl U, Tot T, Henriksson R, et al. LRIG1 and squamous epithelial uterine cervical cancer: correlation to prognosis, other tumor markers, sex steroid hormones, and smoking. Int J Gynecol Cancer. 2008;18:312–317. doi: 10.1111/j.1525-1438.2007.01021.x. [DOI] [PubMed] [Google Scholar]
- Lingaas F, Comstock KE, Kirkness EF, Sorensen A, Aarskaug T, et al. A mutation in the canine BHD gene is associated with hereditary multifocal renal cystadenocarcinoma and nodular dermatofibrosis in the German Shepherd dog. Hum Mol Genet. 2003;12:3043–3053. doi: 10.1093/hmg/ddg336. [DOI] [PubMed] [Google Scholar]
- Ljuslinder I, Malmer B, Golovleva I, Thomasson M, Grankvist K, et al. Increased copy number at 3p14 in breast cancer. Breast Cancer Res. 2005;7:R719–727. doi: 10.1186/bcr1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellersh CS, Hitte C, Richman M, Vignaux F, Priat C, et al. An integrated linkage-radiation hybrid map of the canine genome. Mamm Genome. 2000;11:120–130. doi: 10.1007/s003350010024. [DOI] [PubMed] [Google Scholar]
- Shattuck DL, Miller JK, Laederich M, Funes M, Petersen H, et al. LRIG1 is a novel negative regulator of the Met receptor and opposes Met and Her2 synergy. Mol Cell Biol. 2007;27:1934–1946. doi: 10.1128/MCB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland GR, Baker E. The clinical significance of fragile sites on human chromosomes. Clin Genet. 2000;58:157–161. doi: 10.1034/j.1399-0004.2000.580301.x. [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Tseng GC, You H, Douglass M, Huey B, et al. Comparison of gene expression and DNA copy number changes in a murine model of lung cancer. Genes Chromosomes Cancer. 2006;45:338–348. doi: 10.1002/gcc.20296. [DOI] [PubMed] [Google Scholar]
- Thomas R, Bridge W, Benke K, Breen M. Isolation and chromosomal assignment of canine genomic BAC clones representing 25 cancer-related genes. Cytogenet Genome Res. 2003a;102:249–253. doi: 10.1159/000075757. [DOI] [PubMed] [Google Scholar]
- Thomas R, Fiegler H, Ostrander EA, Galibert F, Carter NP, Breen M. A canine cancer-gene microarray for CGH analysis of tumors. Cytogenet Genome Res. 2003b;102:254–260. doi: 10.1159/000075758. [DOI] [PubMed] [Google Scholar]
- Thomas R, Smith KC, Ostrander EA, Galibert F, Breen M. Chromosome aberrations in canine multicentric lymphomas detected with comparative genomic hybridisation and a panel of single locus probes. Br J Cancer. 2003c;89:1530–1537. doi: 10.1038/sj.bjc.6601275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Scott A, Langford CF, Fosmire SP, Jubala CM, et al. Construction of a 2-Mb resolution BAC microarray for CGH analysis of canine tumors. Genome Res. 2005;15:1831–1837. doi: 10.1101/gr.3825705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas R, Duke SE, Bloom SK, Breen TE, Young AC, et al. A cytogenetically characterized, genome-anchored 10-Mb BAC set and CGH array for the domestic dog. J Hered. 2007;98:474–484. doi: 10.1093/jhered/esm053. [DOI] [PubMed] [Google Scholar]
- Yang F, O'Brien PC, Milne BS, Graphodatsky AS, Solanky N, et al. A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics. 1999;62:189–202. doi: 10.1006/geno.1999.5989. [DOI] [PubMed] [Google Scholar]
- Yang WM, Yan ZJ, Ye ZQ, Guo DS. LRIG1, a candidate tumour-suppressor gene in human bladder cancer cell line BIU87. BJU Int. 2006;98:898–902. doi: 10.1111/j.1464-410X.2006.06405.x. [DOI] [PubMed] [Google Scholar]