Summary
Apaf-1 co-assembles with cytochrome c to form the apoptosome, which then binds and activates procaspase-9. We removed pc-9 catalytic domains from the holo-apoptosome by site-directed thrombinolysis. A structure of the resulting apoptosome-pc-9 CARD complex was then determined at ~9.5Å resolution. In our model, the central hub is constructed like other AAA+ protein rings but also contains novel features. At higher radius, the regulatory region of each Apaf-1 is comprised of tandem 7- and 8-blade β-propellers with cytochrome c docked between them. Remarkably, Apaf-1 CARDs are disordered in the ground state. During activation, each Apaf-1 CARD interacts with a pc-9 CARD and these heterodimers form a flexibly-tethered “disk” that sits above the central hub. When taken together, the data reveal conformational changes during Apaf-1 assembly that allow pc-9 activation. The model also provides a plausible explanation for the effects of NOD mutations that have been mapped onto the central hub.
Introduction
Programmed cell death provides a method to sculpt tissues in developing metazoans and maintain tissue homeostasis (Ferraro et al., 2003). Apoptosis is also used by cells to defend against pathogens and hyper-proliferative cells. Thus, apoptotic pathways are down-regulated in certain cancers and autoimmunity diseases (Song & Steller, 1999; Salvesen & Dixit, 1997; Green & Evan, 2002). Conversely, apoptosis is up-regulated in diseases that destroy cells, such as AIDS, neuro-degeneration and ischemic stroke (Thompson, 1995; Danial & Korsmeyer, 2004).
In chordates, programmed cell death requires cytochrome c release from mitochondria in the intrinsic pathway. This occurs in response to developmental cues, cytotoxic damage or genomic stress (Wang, 2001; Newmeyer & Ferguson-Miller 2003). Cytochrome c binds to the inactive Apaf-1 monomer and triggers apoptosome assembly. This process probably requires conformational changes coupled with nucleotide exchange, while the requirement for nucleotide hydrolysis remains controversial (Reubold et al., 2009; Liu et al., 1996; Hu et al., 1999; Jiang & Wang, 2000; Kim et al., 2005; Yu et al., 2005). The apoptosome binds to procaspase-9 (pc-9) and activates the zymogen to create a platform that mediates proteolytic processing of down-stream procaspases (Li et al., 1997; Srinivasula et al., 1998). Executioner caspases, such as pc-3 and pc-7 (Zou et al., 1997), target certain intra-cellular proteins for proteolysis which leads to an orderly cell death (Danial & Korsmeyer, 2004).
The nucleotide oligomerization domain (NOD) family contains ~23 proteins in human cells that participate in apoptosis, innate immunity and other processes. NOD proteins are members of the AAA+ super-family of ATPases (Inohara and Nunez; 2003; Leipe et al., 2004) and contain a conserved nucleotide binding domain (NBD) and a small helical domain (HD1) that bind nucleotide (Figure 1A, DeLaBarre and Brunger, 2003; Danot et al., 2009; Zhang et al., 2000). In addition, NOD proteins contain a novel winged helix domain (WHD) that is involved in oligomerization, which is followed by a second helical domain (HD2; Figures 1A, 1B; Riedl et al., 2005; Yan et al. 2005). Based on electron cryo-microscopy, the NOD of Apaf-1 is used to assemble a central hub. Seven arms radiate from the hub and are formed by HD2. A regulatory region is located at the end of each arm and contains two β-propellers with cytochrome c sandwiched between them (Yu et al., 2005; Acehan et al., 2002). However, the architecture of the human apoptosome is not clear because a previous map at ~12.8Å resolution did not allow automated and unbiased docking of Apaf-1 domains. Indeed, computer modeling suggested that Apaf-1 may follow a classic AAA+ paradigm for ring formation (Diemand and Lupas, 2006; Propell et. al., 2008), but this remains to be shown experimentally.
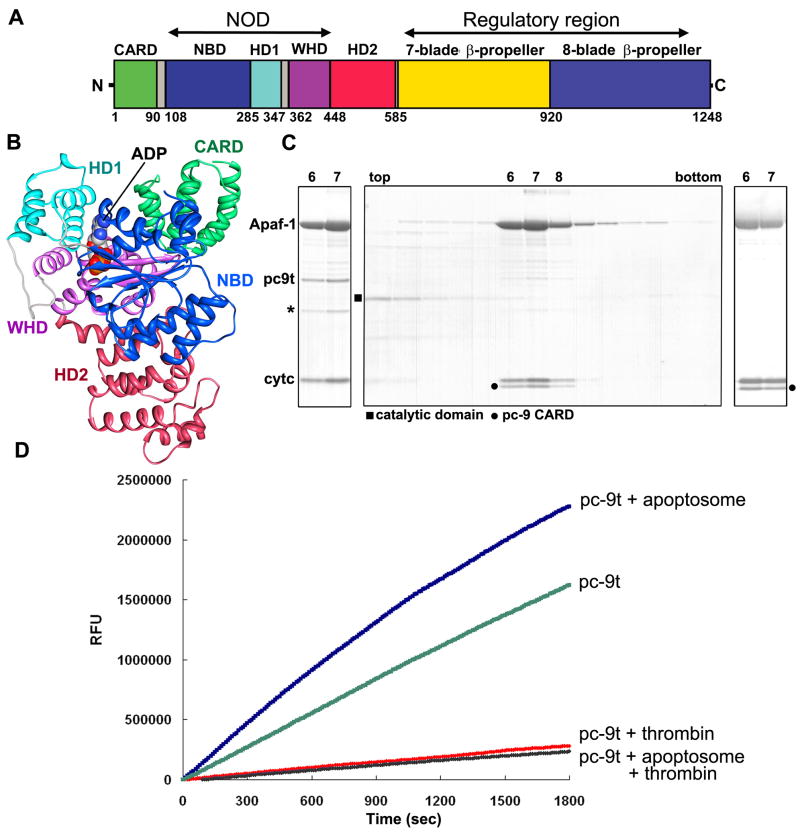
Figure 1. Active apoptosome and apoptosome-pc-9 CARD complexes.
A. A linear diagram of Apaf-1 is shown with color-coded domains and linkers.
B. A ribbon diagram of the Apaf 1-591 crystal structure with bound ADP is shown with color coded domains (Riedl et al., 2005; 1Z6T).
C. (Left panel) Peak fractions for the Apaf-1/pc-9t complex from a glycerol gradient are shown. Some pc-9t clip product is present (*). (middle panel) A complex of Apaf-1 and pc-9t was treated with thrombin. On a glycerol gradient, pc-9 catalytic domains remain at the top (fractions 2–4), while the apoptosome-pc-9 CARD complex migrated in fractions 7–9. (right panel) A mutant of pc-9t with no internal cleavage sites forms a similar apoptosome-pc-9 CARD complex and was used for structural studies.
D. Proteolytic time courses with Ac-LEHD-AFC as a substrate are shown for four conditions described in the text. Apoptosome with bound pc-9t show at least a 10-fold enhancement of proteolytic activity when compared to complexes treated with thrombin.
Procaspase-9 binding to Apaf-1 is mediated by homotypic interactions between CAspase Recognition Domains in each protein (CARDs; Qin et al., 1999). While precise details of pc-9 activation are not known (Rodriguez, J. and Lazebnik, 1999; Zou et al., 1999), it appears that pc-9 zymogens may be disordered when bound to the apoptosome (Acehan et al., 2002). Apical procaspases are monomers in solution (Boatright et al., 2003, Chao et al., 2005; Li and Yuan, 2008), and executioner caspases are constitutive dimers (Riedl et al. 2001). Thus, pc-9 activation may require dimerization mediated by the C-terminal p10 domain (Yin et al., 2006; Pop et al., 2006, Boatright et al., 2003). The resulting pc-9 dimer is functionally asymmetric and contains one active catalytic site and a second, non-functional catalytic site (Renatus et al. 2001). When taken together, these data are consistent with a proximity-induced dimerization model for apical procaspase activation (Boatright et al., 2003; Renatus et al. 2001).
In this paper, we report the structure of a human apoptosome with bound pc-9 CARDs at ~9.5Å resolution. Molecular modeling revealed novel domain interactions within the central hub and provided insights into apoptosome assembly. We also find that the architecture of the heptameric platform in “active” and ground state apoptosomes is similar. In particular, the arrangement of NBDs within the central hub is reminiscent of their packing in AAA+ rings. However, important differences in the NODs may reflect apoptosome function. We also modeled tandem β-propellers in the regulatory region of Apaf-1 and found that they contain 7- and 8-blades, respectively. Remarkably, Apaf-1 CARDs are flexibly tethered to NBDs in the ground state. In the presence of initiator procaspase, Apaf-1 CARDs interact with pc-9 CARDs to form a flexibly-tethered disk that sits above the central hub. This disk could facilitate the proximity-induced dimerization of pc-9. Finally, our model provides a scaffold upon which known NOD mutations can be mapped and understood.
Results
The apoptosome with bound pc-9 CARDs
Previously, we found that apoptosome-pc-9 complexes tended to aggregate during sample preparation for electron cryo-microscopy (Acehan et al., 2002). This may have been due, in part, to high concentrations required to force particles into holes in the carbon support film. In addition, pc-9 molecules were largely disordered on the apoptosome, with the exception of a dome of density located above the central hub. This suggested that few if any interactions would exist between pc-9 and the apoptosome in low salt buffer (LSB), except for the Apaf-1 CARD-pc-9 CARD complex (Qin et al., 1999). To prepare a sample for imaging, we inserted a thrombin site in the linker between the N-terminal CARD and p20 domain of pc-9(D315A) to create pc-9t. We titrated the ratio of pc-9t to Apaf-1 with glycerol gradients to find the saturation point (Figure 1C, left panel). When thrombin was added to the complexes the resulting products included pc-9t catalytic domains, which ran at the top of gradient, and the apoptosome-pc-9 CARD complex (Figure 1C, middle panel). However, pc-9t was susceptible to self proteolysis during over-expression. Hence, we engineered a pc-9t mutant with two additional cleavage sites mutated to alanine (E306A, D330A). The resulting apoptosome pc-9t complex migrated normally on glycerol gradients (Figure 1C, right). Complexes with the pc-9t mutant were assembled and used for structure determination, after releasing the catalytic domains with thrombin. We will refer to the resulting particle as the apoptosome-pc-9 CARD complex.
After modifying the pc-9 linker, we tested the pc-9t zymogen for proteolyic activity using Ac-LEHD-AFC at 100 uM. Cleavage of the susceptible bond following aspartate removes AFC fluorescence quenching. In a time course, pc-9t by itself has appreciable proteolytic activity that may be due to self-aggregation mediated by the pc-9 CARD or the longer linker, as thrombin cleavage brought the activity down to background levels (Figure 1D, compare green and red curves). We then added pc-9t to the apoptosome such that binding was just saturated. Under these conditions, proteolytic activity increased and the true activation level was obtained by comparing apoptosome-pc-9t and apoptosome-pc-9 CARD complexes (Figure 1D, blue and black curves). Complex formation with pc-9t at 100 nM Apaf-1 led to at least a 10-fold activation. We conclude that a single clip in the pc-9 CARD linker is sufficient to release catalytic domains from the apoptosome. This argues against extensive interactions between pc-9 catalytic domains and Apaf-1 in LSB (Acehan et al., 2002). Since pc-9t apoptosomes are active in LSB, the data favor a proximity-induced dimerization model for pc-9 activation (Boatright et al., 2003).
Structure of the apoptosome-pc-9 CARD complex
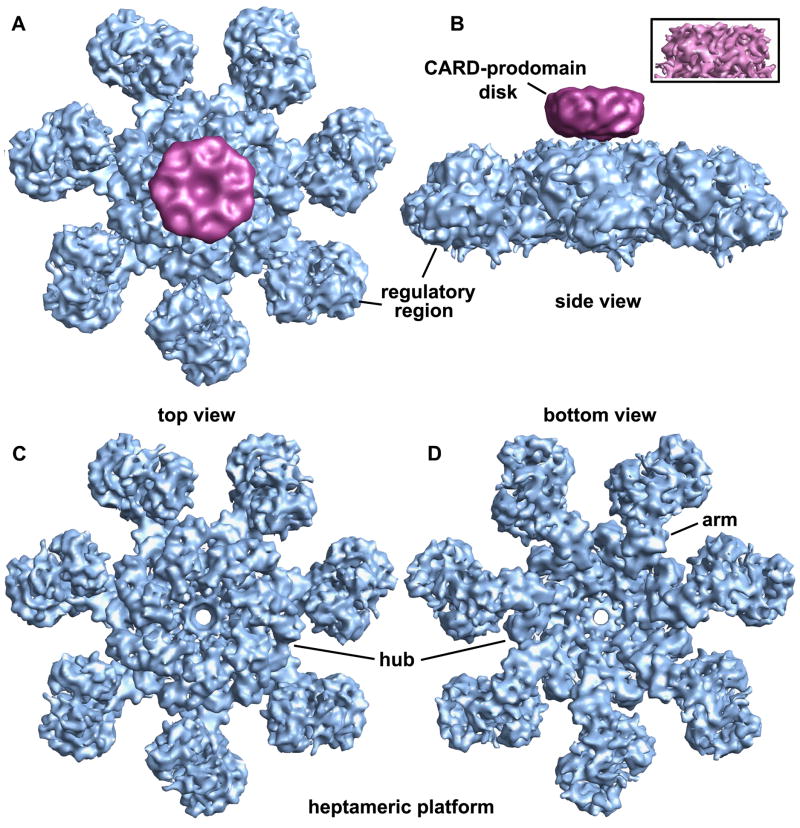
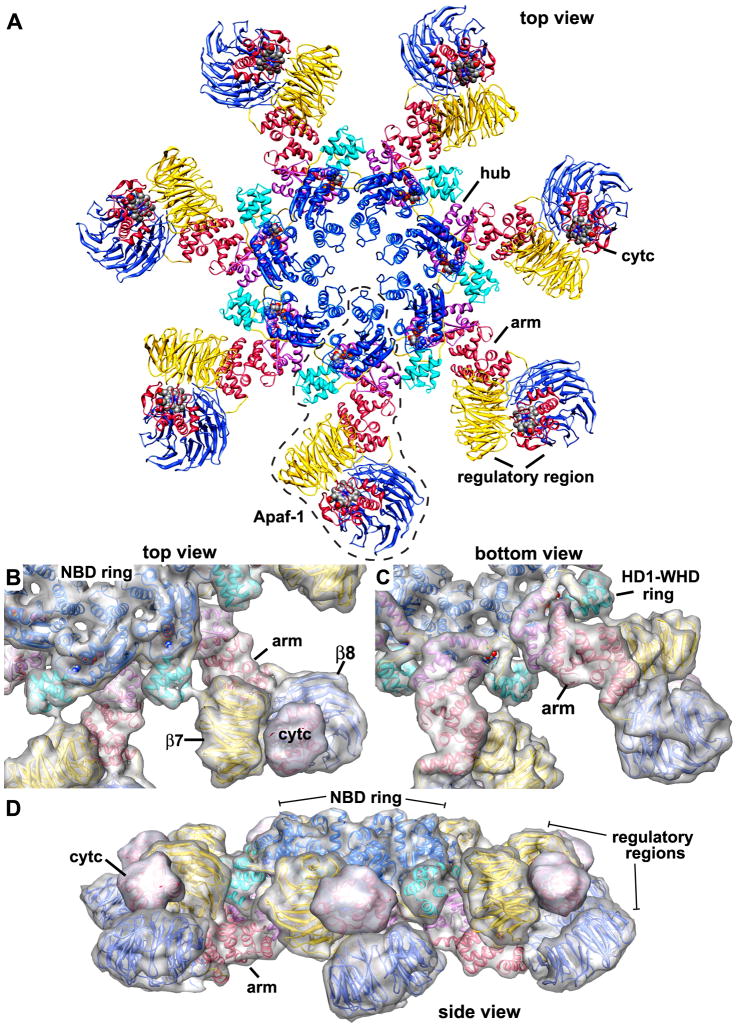
Images of ~42,000 apoptosome-pc-9 CARD complexes in frozen LSB were collected from ~400 micrographs. After refinement with C7 symmetry (EMAN 1.8; Ludtke et al., 1999), we obtained a final map from ~34,000 particles. FSC and Rmeasure curves (Sousa and Grigorieff, 2007) gave 0.5 estimates of 8.3 and 8.9Å resolution, respectively (Figure S1). However, inspection of the map and crystal structure docking with Chimera (see next section) suggests that the actual resolution is ~9–10Å, since individual α-helices are only partly resolved. Thus, we truncated the resolution of the map at 9.5Å resolution. We observed a disk-like density sitting above the central hub of the apoptosome in class averages (not shown). In the final map, this disk was at a lower resolution than the platform and there were no strong connections between these two features. For clarity, we truncated the disk to ~18Å resolution. Top and side views of the map are shown with the platform and disk in blue and magenta, respectively (Figures 2A, 2B). The disk in the original map is shown in the inset (Figure 2B). Disk formation is reminiscent of dome-like density observed in a lower resolution map of the apoptosome-pc-9 complex (Acehan et al., 2002). After removing the disk, top and bottom views reveal a wealth of detail in the central hub, arm and regulatory region (Figures 2C, 2D).
Figure 2. A 3D map of the apoptosome-pc-9 CARD complex.
A. A top view of the apoptosome-pc-9 CARD complex is shown as a surface rendering. The platform is shown in blue and the low pass filtered disk is colored magenta.
B. A side view of the apoptosome-pc-9 CARD complex shows the disk, which sits above the platform without visible connections. The unfiltered disk from the original map is shown in the inset.
C. A top view is shown of the platform with the disk removed.
D. The platform is shown in a bottom view along the 7-fold axis.
Modeling the apoptosome
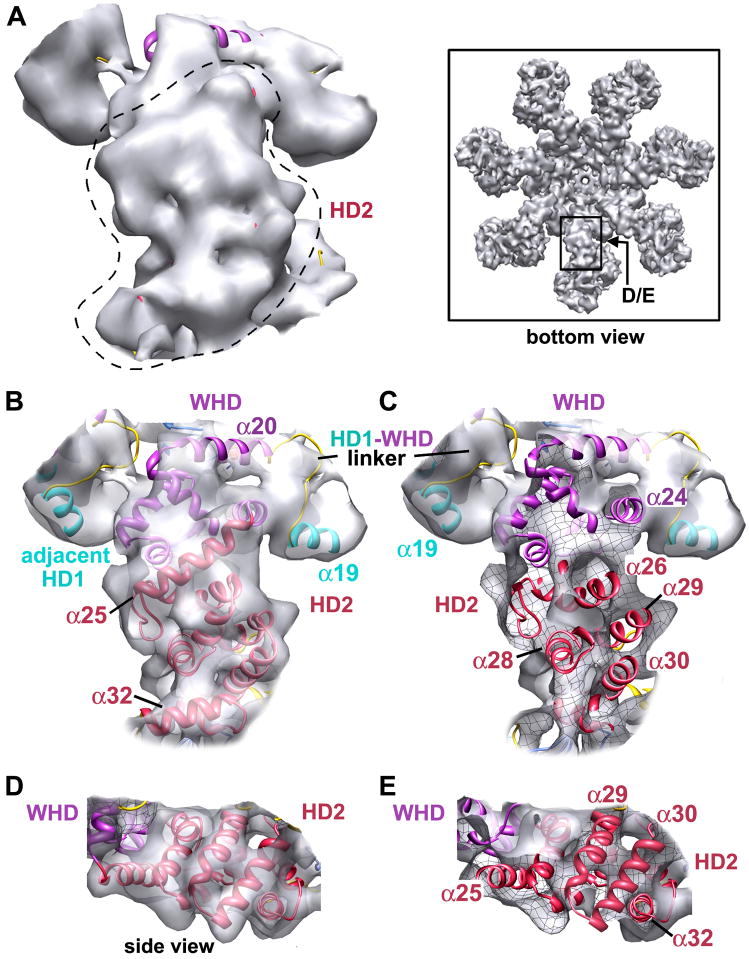
We used a crystal structure of Apaf 1-591 with bound ADP (Figure 1B, PDB code: 1Z6T) to dock NBD, HD1, WHD and HD2 domains into the map, as well as homology models of the C-terminal β-propellers and a crystal structure of bovine cytochrome c (2B4Z). In this work, Apaf-1 domains are color-coded (Figures 1A, 1B), and the nomenclature is based on a structure-based sequence alignment (Figure S2, Riedl et al., 2005). All domains were initially docked as rigid bodies using Chimera (Goddard et al., 2005). They were then re-connected within the map and their fit was refined using Flex-EM (Topf et al, 2008; Supplementary Methods). At this resolution, we did not model all movements of helices within domains. We started with HD2, since its position in the arm was known. Based on the quality of the docking, we were able to ascertain the correct handedness of the 3D map. This was borne out by the fit of domains in the density which gave cross-correlation scores between 0.85–0.95 (Table S1). A close-up of an arm with docked HD2 is shown in Figure 3A, as viewed from below the apoptosome. An icon view of this region is also shown. The fit of HD2 in the arm is shown within a semi-transparent map in Figure 3B and the front surface is cut-away, to reveal the docking of helices and loops within high density features (Figure 3C). A mesh-work has been drawn on the inside of the iso-surface, which allows one to differentiate between inner and outer surfaces. Finally, a side view of the HD2 arm highlights the excellent fit of helices in the density (Figures 3D, 3E).
Figure 3. Helical domain 2 within the arm of the apoptosome.
A. A bottom view is shown of the arm and HD2 is indicated by a dashed outline. The map is rendered in silver and an icon view is shown in the boxed area.
B. The view in panel A is shown with HD2 and adjacent features docked within a semi-transparent map. HD2 is red, HD1 is cyan and the WHD is purple.
C. A view like that in panel B is shown with the front cut by a clipping plane to show helices and loops docked within high density regions. The internal face of the iso-surface can be identified by a visible black meshwork.
D. A side view of the HD2 arm highlights the fit of several parallel helices.
E. A view like that in panel D is shown with a front clipping plane that reveals helices of HD2.
In the next step, we docked WHD-HD2 as a rigid body since there is an extensive interface and a short linker between these domains in the crystal structure (Riedl et al., 2005). We also docked the NBD-HD1 pair as a rigid body, since they form the nucleotide binding pocket and the linker between them is also short (~5 residues). We then relaxed these restraints to dock domains separately within the map. A small rotation was apparent between domains in each of the pairs. The WHD in the context of its neighboring HD1 is shown in a bottom view (Figures S3A, S3B). The β-hairpin “wing” in the winged helix domain is easily recognizable in side and top views (Figures S3B and S3D–S3F). In addition, helix α18 from HD1 packs against the β-hairpin and the HD1-WHD linker may interact with α23 of WHD. There is good density for the central β-sheet and dATP/ATP in the NBD, but local conformational changes were not modeled. Finally, the 4 helix bundle of HD1 fits snugly between adjacent WHDs.
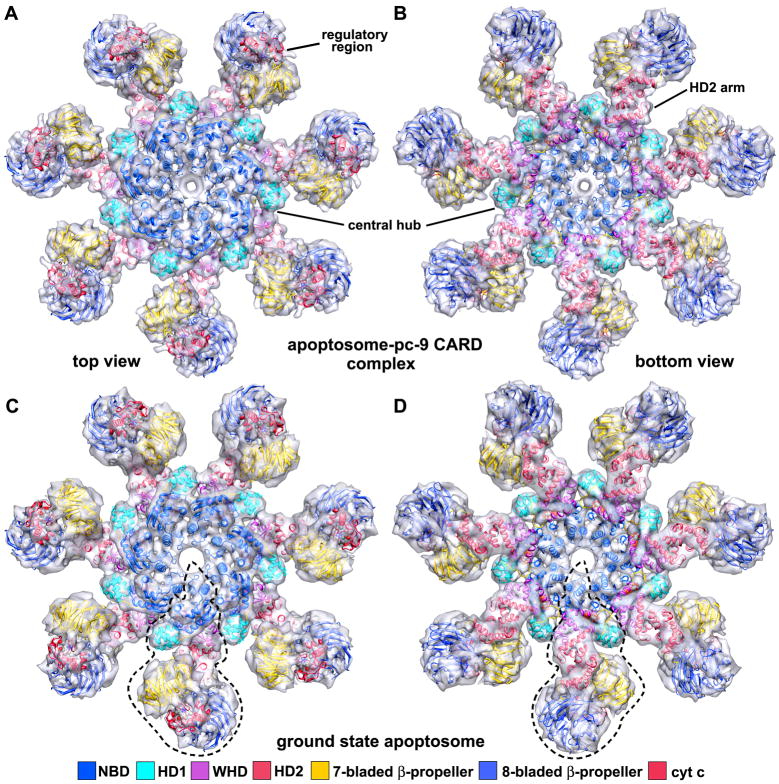
The regulatory region was constructed by docking homology models of 7- and 8-blade β-propellers (Supplmentary Methods) and cytochrome c (Mirkin et al., 2008, 2B4Z) into the density. Ramifications of our modeling of the regulatory region will be discussed in the next Section. After refinement of the entire Apaf-1/cytochrome c model, a pdb with a complete subunit was docked automatically into 7 equivalent positions in the map without major clashes between chains. The quality of the final fit is indicated by the high cross-correlation (0.91; Table S1). An over-view of the final platform model is shown in top and bottom views (Figures 4A, 4B), with the model viewed within a semi-transparent map.
Figure 4. The global fit of Apaf-1 and cytochrome c within “active” and ground state apoptosomes.
A. A molecular model for the platform is shown docked within a semi-transparent top view of the apoptosome-pc9 CARD complex. Individual domains are shown as ribbons and are colored coded as indicated.
B. A bottom view is shown of the 3D map with the molecular model docked within the density.
C. The molecular model for the platform is shown docked within a density map of the ground state apoptosome. An individual Apaf-1 subunit is marked with a dashed outline in this top view.
D. A bottom view is shown of the ground state apoptosome with the docked molecular model.
At this stage, there was no room to fit seven Apaf-1 CARDs within the platform. Hence, the disk must contain Apaf-1 CARD-pc-9 CARD complexes (Qin et al., 1999). However, the size and low resolution prevented us from modeling 7 CARD-CARD heterodimers within the disk. We surmise that Apaf-1 CARDs are flexibly-linked to their respective NBDs within the platform, which leads to a blurring of the disk. This idea is consistent with the location of linker helix α8 (Figure S3E), as the N-terminus of this helix points up towards the disk. Intriguingly, one Apaf-1 isoform with a shorter CARD-NBD linker is still functional (aa99-109 missing in Apaf-1L; Saleh et al., 1999). This suggests that a longer link between the disk and platform may not be required for full function.
We then took the final platform model with 49 domains and docked it as a rigid body into a density map of the ground state apoptosome (Yu et al., 2005). The overall fit was excellent with no significant unfilled regions (Figures 4C, 4D). Hence, the platform probably does not change significantly when pc-9 is bound, with the possible exception of CARD-NBD linker movements.
Interactions within the apoptosome
A ribbon diagram of structural elements in the apoptosome is shown in a top view (Figure 5A), and a single Apaf-1 monomer is surrounded by a dashed boundary line. Additional views and subunit contacts are shown in Figures S4 and S5, respectively. The close-packing of domains is shown in Figures 5B–5D, with a transparent surface overlayed onto the apoptosome model to give a perception of depth. The hub is comprised of two rings: an inner ring containing NBDs encircled by an outer ring that contains alternating HD1 and WHDs (Figures 5B, 5C). The critical unit for assembling the hub is comprised of an NBD-HD1 pair and a WHD.
Figure 5. The heptameric platform of the apoptosome.
A. A top view is shown of the platform in the human apoptosome. Domains are shown as ribbons and are color coded as indicated in Figure 4. Hub, arm and regulatory regions are labeled and a single Apaf-1 monomer is outlined. ATP and heme groups are shown as spheres with cpk colors.
B. A top view is shown of two adjacent subunits within the platform. A calculated surface has been over-layed on the model to provide depth perception.
C. A similar bottom view is shown of the platform.
D. The heptameric platform is shown in a side view after a small tilt about the horizontal axis.
The hub model differs significantly from a previously published domain model that was based on a lower resolution map (Yu et al., 2005). In the earlier work, it was thought that Apaf-1 CARDs may play an integral role in apoptosome assembly, since these domains tended to self-aggregate and CARD-CARD interactions seemed likely (Yu et al., 2006; Shiozaki et al., 2002). In addition, the resolution of the previous map was not sufficient to allow unambiguous, computational docking of small domains, so manual methods were used. When these factors were combined, it resulted in a local minimum being chosen in which NBD and HD1 positions were flipped, because a ring of Apaf-1 CARDs was modeled erroneously at the center of the hub. It is now clear that Apaf-1 CARDs are flexibly-linked to the platform. In addition, higher resolution coupled with more robust docking programs has allowed an unambiguous solution.
In general, the hub has an architecture that is reminiscent of rings formed by other AAA+ ATPases (Diemand and Lupas, 2006; Riedl et al., 2005). However, significant differences arise from the presence of an initiator specific motif in the NBD and from the packing of WHDs in the hub. These differences may reflect the fact that Apaf-1 forms a stable platform instead of performing mechanical work like many AAA+ ATPase rings. This difference is also reflected in the low intrinsic ATPase activity of the apoptosome (Jiang and Wang, 2000; Kim et al., 2005; Yu et al., 2005).
ISM and NBD
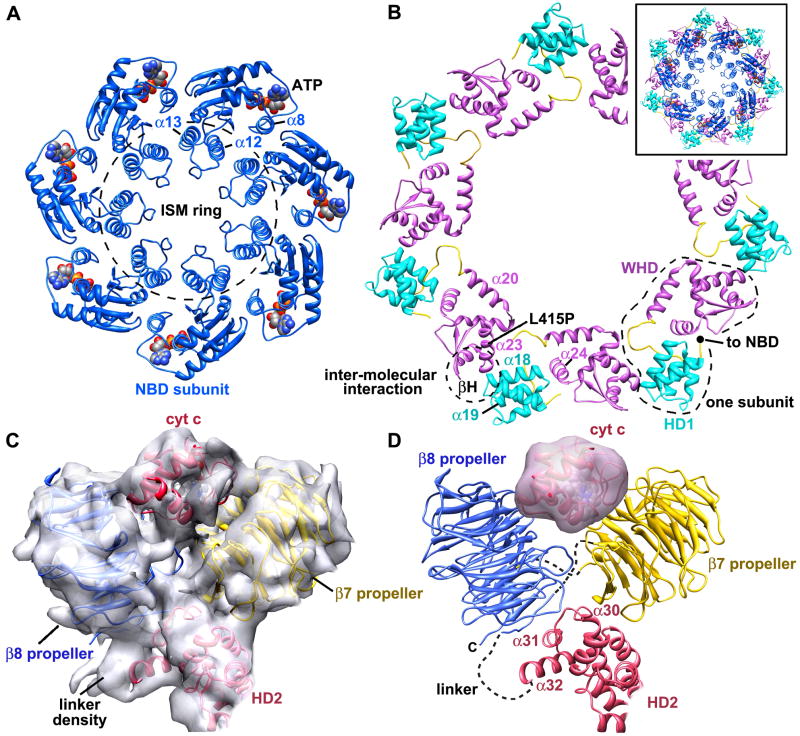
The NBD plays a critical structural role because it is centrally located in the apoptosome and may nucleate ring assembly. Two major changes within the NBD are apparent at this resolution. First, the initiator-specific motif (ISM, helix α12) is present in members of the initiator clade, which are most similar to NOD proteins in the AAA+ superfamily. The ISM of Apaf-1 is inserted between β2 and α13 in the NBD. Together, α12 and α13 form a helix-loop-helix motif. Seven of these paired α-helical motifs align in a nearly vertical orientation to form a ring that encircles the central pore of the apoptosome (Figure 6A; ISM ring). Second, the α8 linker helix in one NBD packs against an adjacent NBD. Nucleotide triphosphate is bound near the subunit interface in a deep pocket between the NBD-HD1 pair and there is no obvious entrance or exit path (Figures S3E, S3F). Density for the “arginine finger” in the NBD (actually K277 at the end of β5) of the adjacent subunit is weak, so it is hard to judge whether NTP hydrolysis could occur in the apoptosome. Neither the vertical packing of the α12-α13 pair to form the ISM ring (Figure S6), nor the re-arrrangement of linker helix α8 could have been predicted using previous AAA+ ring structures (Diemand and Lupas, 2006).
Figure 6. Critical interactions in the heptameric platform.
A. A top view is shown of α12-α13 and NBD rings within the central hub. Molecules of ATP are shown as spheres in cpk colors and the α8 linker helix is also labeled.
B. A top view is shown of the HD1-WHD ring within the central hub. Domains from a single Apaf-1 are outlined and relevant α-helices are labeled. The inset shows a superposition of the NBD and HD1-WHD rings to form the central hub.
C. The 7- and 8-blade β-propellers are shown in the density map along with cytochrome c.
D. A side view is shown of a ribbon model for the regulatory region. Linkers are indicated with dashed lines.
HD1-WHD ring
The AAA+ proteins do not contain a winged helix domain; instead NBD-HD1 pairs interact with neighbors to form rings and oligomers (Danot et al., 2009; Hanson and Whiteheart, 2005). Hence, the WHD is a major addition to NOD proteins. In the central hub, the WHD interacts with a neighboring NBD-HD1 pair (Figure 6B). This generates an outer ring with alternating WHD and HD1s within the hub. The HD1-WHD linker was not modeled explicitly, but density for this feature is present between HD1 and WHD (Figures 3B, 3C). Within an Apaf-1 monomer, the end of helix α19 in HD1 and the HD1-WHD linker interact with α24 in the WHD. In the formation of the HD1-WHD ring, the loop between α18 and α19 of HD1 contacts the β-hairpin of WHD (Figures 6B, S3D). The WHD also interacts strongly with the HD2 arm which positions the regulatory region of each subunit at higher radius (Figure 5C).
Regulatory region
A more detailed sequence analysis of the regulatory region revealed 15 WD40 motifs rather than the 13 repeats identified previously (Figure S7, Experimental Procedures). This allowed us to model tandem 7- and 8-blade β-propellers within the regulatory region. The resulting model is reminiscent of the crystal structure of actin interacting protein 1 (1PGU; Voegtli et al., 2003), except that an 8-blade propeller in Apaf-1 replaces the second 7-blade propeller in Aip1. The docking of β-propellers within the map shows that the V-shape of the regulatory region is supported at the base by helices α30, α31 and α32 in HD2 (Figures 6C, S8). The model is strongly constrained by the length of the HD2-propeller linker and by links between the two β-propellers. A novel feature is that the HD2-linker forms a d-strand in the last blade of the 8-blade propeller before continuing on to form the 7-blade propeller and the rest of the 8-blade propeller (Figures 6D, S7). This prediction remains to be verified with higher resolution data.
We then docked a crystal structure of cytochrome c into the map (2B4Z). However, several related orientations were found with similar docking scores. This was due to the paucity of longer helices and the presence of large loops in cytochrome c, which are not well defined at the current resolution. Therefore, we chose the orientation with the highest score, but this has to be verified. Even so, each docking placed the heme group in a protected environment between the two β-propellers, consistent with data which shows that the heme is not solvent accessible (Purring-Koch & McLendon, 2000). Several residues on cytochrome c were identified previously through mutations which interfere with Apaf-1 mediated procaspase activation (Yu et al., 2001; Hao et al., 2005). In general, our fit for cytocrhome c places most of these residues, including lysines, in orientations where they could interact with the β-propellers.
Discussion
In this work, we determined a structure of the apoptosome-pc-9 CARD complex at ~9.5Å resolution. We then used molecular docking with crystal structures to create a model for Apaf-1 subunits within the active apoptosome. Our model shows how the central hub is constructed by the NOD and provides insights into conformational changes that occur during assembly. The model also provides a rationale for the roles of cytochrome c and ATP/dATP in assembly and helps to explain phenotypes that arise in certain NOD mutations. Importantly, we find that flexibly-linked Apaf-1 CARDs in the active apoptosome interact with pc-9 CARDs to form a disk that may play a role in procaspase-9 activation. Finally, the model provides a blueprint for other NOD proteins which form activation platforms.
The human apoptosome
We determined the structure of an apoptosome-pc-9 CARD complex after removing the pc-9 catalytic domains with a single thrombin clip in each subunit. Hence, pc-9 catalytic domains are not tightly bound to Apaf-1 in LSB. We also find that pc-9 is flexibly-tethered to the apoptosome by Apaf-1 CARD-pc-9 CARD heterodimers, which form a “disk” that sits above the platform. Thus, our model is representative of the heptameric platform at ~9.5Å resolution. Based on the model, we provide insights into construction of the central hub, arm and regulatory region. In particular, the central hub is reminiscent of other AAA+ rings in which the NBD-HD1 pair mediates assembly through extensive lateral interactions between adjacent subunits. In addition, nucleotide triphosphate is located at the interface between adjacent NBDs. However, NBD-NBD interactions are modified in the apoptosome by the ISM helix (α12, which partners with helix α13 to form a helical pair motif. This helix pair associates with 6 copies of itself to form a ring at the center of the hub that encircles the central pore. At the same time, linker helix α8 between the CARD and NBD is repositioned between neighboring NBDs, near bound ATP/dATP. In addition, helix domain 1 in the NBD-HD1 pair rotates a bit, while maintaining contact with the nucleotide and interacts with the WHD from an adjacent Apaf-1 during assembly. This novel HD1-WHD interaction forms an outer ring within the hub.
Formation of the HD1-WHD ring positions seven HD2 arms and their attached regulatory regions, which extend from the central hub. While we have achieved an accurate model of the central hub and arm, the regulatory region remains somewhat blurred because we don’t have atomic structures for Apaf-1 β-propellers. Even so, we were able to model tandem 7- and 8-blade β-propellers using extensive sequence information and pdb structures, including Aip1, which has two β-propellers with a similar V-like configuration (Voegtli et al., 2003). Finally, we placed cytochrome c between the two β-propellers in an orientation that agrees with known data.
Our model is consistent with functional and mutation studies of Apaf-1 (summarized in Table S2 and Figure S9). For example: Apaf-1-ALT in a prostate cancer cell line is not functional (Ogawa et al., 2003). This isoform is terminated after HD1 and thus, is missing the WHD, arm and regulatory regions. WHD loss could weaken or prevent the assembly of an Apaf-1-ALT ring. Additional studies revealed two loss of function mutants, Apaf-1 (L415P) and 437ter (Table S2; Harlan et al. 2006). When leucine 415 is replaced by proline, helix α23 in WHD would be disrupted (Figure 6B). This mutation may change the conformation of this important region and prevent assembly. For Apaf-1 437ter, termination occurs before a conserved histidine (His438) in the WHD, which may partly destabilize the inactive monomer (Danot et al., 2009). However, the greatest effect of this mutation would be the elimination of helix α24, which forms an important contact to HD1 in the ring. The loss of α24 would also destabilize α20 and α21 in the WHD. Hence, the 437ter mutation is predicted to prevent assembly due to loss of the HD1-WHD ring. Alternatively, incomplete assembly might lead to aggregation and block pc-9 activation.
Our model can be extended to other NOD proteins (Inohara and Nunez, 2003). However, specific details will vary as NOD proteins contain different N-terminal effector and C-terminal regulatory regions. For example, Nod2 is associated with Crohn’s disease, Blau syndrome and early on-set sarcoidosis (Hugot et al., 2001; Miceli-Richard et al., 2001; Kanazawa et al., 2005; van Duist et al., 2005). Crohn’s disease may be due to loss of function mutants whereas gain of function mutations may be responsible for Blau syndrome and early on-set sarcoidosis (Wilmanski et al., 2008). Several loss of function mutants, including C333Y and S344T, along with Crohn’s disease mutants L348V and H352R, are located in the ISM helix region (Table S2 and Figure S9). These mutations may disrupt interactions that are critical to nucleation and assembly of the central hub. Another loss of function mutant, V492E, is located in sensor-2 helix, which is equivalent to helix α18 in Apaf-1. This region mediates interactions between HD1 and a WHD from an adjacent subunit. Mutations in this feature could disrupt assembly or lead to an unstable complex.
Conformational changes during assembly
Platform assembly in the apoptosome is the first step in pc-9 activation (Hill et al., 2004; Zou et al., 1997, 1999; Rodriguez and Lazebnik, 1999; Acehan et al., 2002; Yu et al., 2005). Apaf-1 exists in solution as an inactive monomer and binds cytochrome c, in a series of steps that leads to an open conformation. The open form then promotes apoptosome assembly (Acehan et al., 2002). We do not have a crystal structure of the complete Apaf-1 monomer; instead we have a model with bound ADP that is missing the C-terminal regulatory region (Apaf 1-591; Riedl et al., 2005). The compact nature of the N-terminal region suggests that it may be a good first approximation for a closed, inactive Apaf-1. However, some uncertainty remains about the nature of bound nucleotide in the inactive monomer. Thus, Apaf-1 purified from sf21 cells contained dATP and hydrolysis of the γ-phosphate occurred upon cytochrome c binding (Jiang and Wang, 2000; Kim et al., 2005). Conversely, Apaf-1 purified from Hi-Five/sf21 insect cells and from bacteria contained bound ADP (Bao et al., 2007; Reubold et al., 2009; Riedl et al., 2005). In this case, ATP hydrolysis was not required for apoptosome assembly (Bao et al., 2007; Reubold et al., 2009). However, cytochrome c binding and nucleotide exchange were necessary to stabilize the monomer in an assembly competent form.
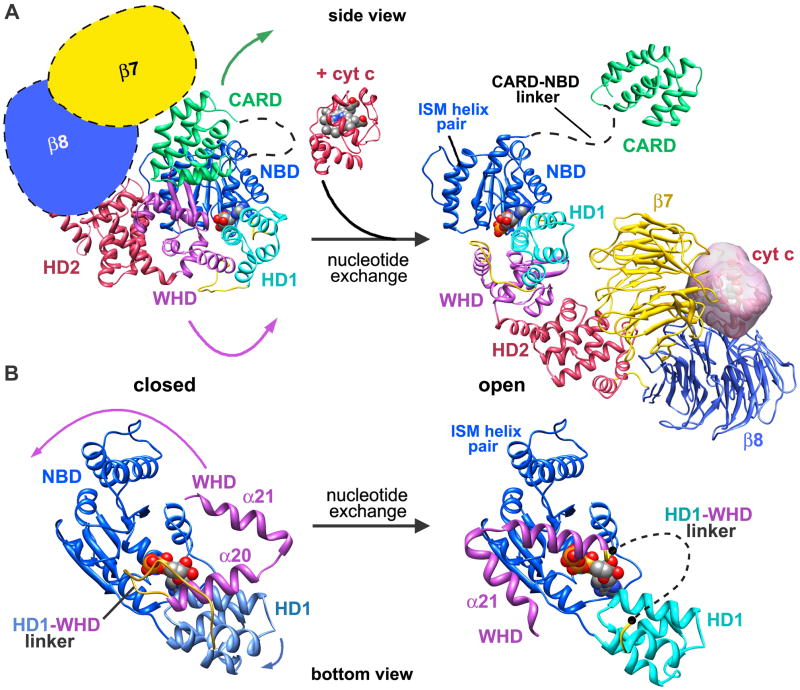
Beta propellers in the regulatory region are thought to lock Apaf-1 in an inactive conformation. To emulate this state, we placed “cartoon” β-propellers at the C-terminus of folded Apaf 1-591. This created a compact monomer while retaining the rough geometry of the regulatory region (Figure 7A, left panel). Note that we kept the NBD in a fixed position throughout this analysis. In the first stage, cytochrome c may bind between two β-propellers with a geometry like that observed in the apoptosome. This would trigger an opening of the compact Apaf-1 monomer with the regulatory region moving away from the NOD region. During this step, the WHD and regulatory region would swing ~120° about a pivot point located between HD1 and WHD. In the crystal structure of Apaf 1-591 (Riedl et al., 2005; Danot et al., 2009), the WHD is bent back towards ADP and a conserved histidine (His438) forms a hydrogen bond with ADP to help stabilize the inactive monomer. This hydrogen bond would be broken when the Apaf-1 monomer opens up.
Figure 7. Assembly of the human apoptosome.
A. Conformational changes are shown that may occur in the transition from a closed to an open Apaf-1 conformation. See text for details. The position of the NBD has been fixed during the modeling and the CARD-NBD linker is shown as a dashed line in both states.
B. A bottom view is shown of the NBD to highlight changes that occur during nucleotide exchange which include large movements of HD1, the HD1-WHD linker and WHD.
In the second stage, nucleotide diphosphate is exchanged for dATP/ATP to presumably lock the Apaf-1 monomer in an open and assembly competent form (Figure 7A, right panel; Acehan et al., 2002). The overall effect of these large domain movements is two-fold. The rearrangements would expose the α12-α13 helix pair along with the rest of the NBD, so that the NBD ring in the central hub could assemble. Changes in the vicinity of bound ATP/dATP must be responsible for the transition from a closed to an open form, as shown in a bottom view of the NBD that compares closed and open conformations (Figure 7B). In our model, there is room in the binding pocket to accommodate ATP. However, nucleotide exchange and divalent cation binding probably alter interactions with the CARD-NBD linker, which may trigger a repositioning of helix α8 during assembly. At the same time, HD1 rotates by ~12° relative to the NBD and the HD1-WHD linker flips through an angle of ~50–60° to help form a contact with an adjacent Apaf-1 subunit. Although we could not accurately model the HD1-WHD linker (see dashed line, Figure 7B, right panel), there is clear density for this feature in the 3D map. Linker movement is coupled with a rotation of the WHD by ~120° (for clarity, only helices α20 and α21 are shown). This would position WHDs and HD1s to form the outermost ring in the hub, as NBDs in each Apaf-1 associate to form the inner ring. The relative geometry of the WHD-HD2 pair remains fixed during this large conformational change and creates the extended arm which supports the regulatory region. Clearly, more precise details of nucleotide exchange and its function as a trigger for assembly will require a higher resolution structure of the apoptosome.
The major goal of Apaf-1 assembly is the formation of a stable platform while releasing CARDs from interactions with their regulatory regions. This frees up seven Apaf-1 CARDs that remain flexibly-linked to the platform through CARD-NBD linkers which are ~17 residues long. Indeed, the CARD-NBD linker is also disordered in the crystal structure of Apaf1-591 (Riedl et al., 2005). There is also a precedent for CARD flexibility in a crystal structure of the CED4-CED9 complex (Yan et al., 2005). In this case, a CED4 CARD was missing because it did not make stabilizing contacts in the crystal.
We tested the idea of CARD flexibility by constructing Apaf-1 with a thrombin site in the CARD-NBD linker. After apoptosome assembly, Apaf-1 CARDs were readily removed by thrombin (data not shown), consistent with these domains being flexibly-tethered to the platform. Thus, flexibly-linked CARDs in the apoptosome are free to interact with pc-9 CARDs. This allows the formation of a disk that is likely comprised of Apaf-1 CARD-pc-9 CARD heterodimers (Qin et al., 1999). Disk assembly may be driven by the high local concentration of CARDs in the region above the central hub. The importance of this disk will be described in a forthcoming paper.
A conserved platform architecture
Apoptosomes are present in chordates and arthropods while a smaller protein, CED4, is present in C. elegans. Conservation of the NOD suggests that apoptosomes will have a similar architecture for the central hub. However, in Drosophila the apoptosome contains 8 subunits per ring, rather than the seven subunits found in the human complex. Even so, the general arrangement of protein domains appears to be similar in human and Drosophila apoptosomes (data not shown; Yu et al., 2006).
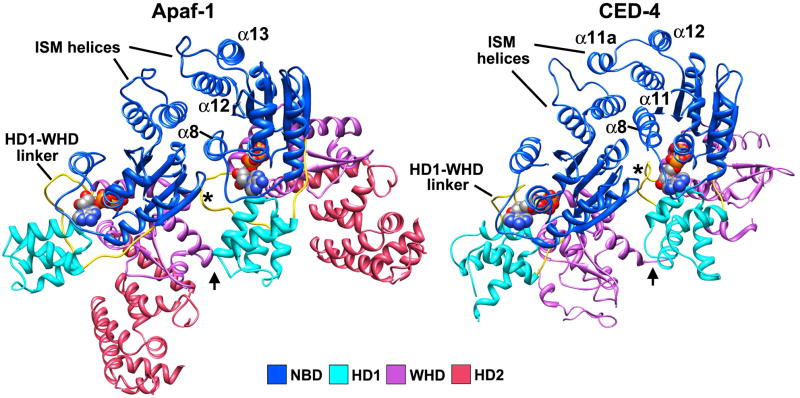
A crystal structure of an inhibited CED4-CED9 complex revealed a non-symmetric CED4 dimer (Yan et al., 2005). We compared the CED4 dimer to a pair of Apaf-1 subunits extracted from our apoptosome model. This revealed an excellent correspondence between structures when they were aligned on the NBD of the leftmost subunit (Figure 8). In the CED4 dimer, there are three major interfaces. In the first site, the ISM helix (α11) and α11a (or a loop) are packed against α12 (equivalent to α13 in Apaf-1) of the adjacent molecule. In addition, linker helix α8 is packed between the NBDs, although its length and position differs from the equivalent helix in the human apoptosome. At the second site, the HD1-WHD linker from one subunit interacts with the adjacent NBD (asterisk in Figure 8, right panel). At the third site, HD1 interacts with a WHD from the adjacent molecule (arrow in Figure 8, right panel). These three contact sites are similar, though not identical, in a lateral Apaf-1 dimer extracted from the apoptosome (Figure 8, left panel). Hence, the CED4 dimer could represent a building block of the C. elegans apoptosome in which CED9 blocks the oligomerization interface of one CED4 molecule. This predicts that a CED4 apoptosome may contain 7 or 8 subunits. However, the CED4 subunits have a closer lateral spacing which arises in part, from differences in the HD1-WHD linker (see asterisk in Figure 8) and linker helix α8. This might favor a CED4 apoptosome with 8 subunits when the dimeric nature of the putative building block is factored in. Intriguingly, CED4 binds ATP and does not have ATPase activity so nucleotide exchange may not drive assembly. Instead, the removal of CED9 by EGL1 may be sufficient to promote assembly of this apoptosome from dimeric CED4 building blocks (Yan et al., 2005).
Figure 8. A comparison of Apaf-1 and CED4 lateral dimers.
(left panel) A lateral dimer of Apaf-1 from the apoptosome is viewed roughly from the top of the hub. (right panel) The crystal structure of the CED4 dimer is shown without CED9. Models in the two panels were aligned on the leftmost NBDs in each subunit pair. Note that the HD2 region is much smaller in CED4. The α12-α13 helix pair in Apaf-1 and equivalent helices in CED4 are labeled, along with the HD1-WHD linker (asterisk) and the HD1-WHD interaction site (arrow).
In summary, we have presented a comprehensive model of the human apoptosome based on molecular docking into a density map at ~9.5Å resolution. The new model of the platform is consistent with known biochemical properties and can explain the phenotypes of critical mutations in NOD family members. When combined with sequence information, the model provides a blueprint for the structure and assembly of other NOD family members. This work also sets the stage to develop a deeper understanding of the mechanism of pc-9 activation.
Experimental Procedures
Protein preparation and assembly
Recombinant Apaf-1 was expressed and purified from sf21 insect cells (Acehan et al., 2002; Yu et al., 2005). Procasepase-9 mutants were generated with the Quickchange kit (Strategene) and a pc-9t linker mutant was created by inserting a thrombin site (LVPRGS) after Lys100. Procaspase-9 proteins were expressed and purified as described (Chao et al., 2005). Bovine heart cytochrome c (Sigma) was purified by cation-exchange chromatography (HiTrap S, Amersham). To assemble the ground state apoptosome, Apaf-1 in LSB (20 mM HEPES-KOH [pH 7.5], 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM dithiothreitol) was incubated with cytochrome c (in a 1:1 molar ratio) and 0.1 mM dATP at 30 °C for 30 min. For the apoptosome pc-9 complex, Apaf-1 was incubated with cytochrome c, procaspase-9 and 0.1 mM dATP at 30 °C for 30 min. To obtain the apoptosome-pc-9 CARD complex, 3 units of thrombin (Sigma) were added per 50 ug Apaf-1 and the solution was incubated at 30 °C for an additional 30 min. To monitor assembly, apoptosome samples were applied onto a 10–40% glycerol gradient (2.1 ml) volume in LSB and centrifuged at 17°C for 2.5 hrs in an RP55S swinging bucket rotor (55,000 rpm; Sorval). Fractions were collected manually from the top of the tube (14 fractions, 150 ul per fraction), separated on an 8–18% gradient gel by SDS-PAGE and visualized with Coomassie blue.
Procaspase-9 activation assays
A fluorogenic substrate, Ac-LEHD-AFC (Calbiochem), was used for pc-9 proteolytic activity assays. Fluorescence experiments were carried out on a Fluoro-Max2 fluorimeter (Jobin Yvon–SPEX, Instruments SA, Inc.) at room temperature using a 600 μl cuvette with a path length of 1 cm. Samples containing pc-9 or the apoptosome pc-9 complex were diluted with LSB to 600 μl. The ratio of pc-9 to Apaf-1 was obtained from titration experiments with glycerol gradients to insure that no free pc-9 was present. Cleavage reactions were started by adding Ac-LEHD-AFC (100 uM) to the sample. Release of the AFC fluorescent moiety was monitored at 505 nm with an excitation wavelength of 400 nm.
Electron cryo-microscopy, structure determination and modeling
After assembly, the apoptosome pc-9 CARD complex was obtained by thrombin treatment in LSB and concentrated to ~3 mg/ml using YM-30 Microcons (Millipore) in the presence of Nonidet P40 (~0.15% final concentration). Aliquots of the concentrated sample (2 μl) were pipetted onto Quantifoil R1.2/1.3 holey grids, which were then blotted and plunged into liquid ethane, cooled with liquid nitrogen. Plunging was done at 100% humidity and room temperature using a Vitrobot Mark 3 (FEI). Data were collected on film at 62,000X with a Tecnai F20 microscope operated at 120 kV for optimal contrast. Electron micrographs were scanned using a Zeiss scanner at a step size of 7 um and binned to 2.26 Å/pixel. Particles were selected using BOXER (Ludtke et al., 1999) and a hybrid structure factor was generated (Yu et al., 2005). Since the complex was prone to aggregation, only ~100 particles could be selected from each micrograph, which gave a weak signal for CTF fitting. Thus, CTF parameters were determined from the adjacent carbon film with FITCTF, and these values were then used to manually adjust the CTF of the summed particles with CTFIT in EMAN 1.8 (Ludtke et al., 1999). For 3D reconstruction, a ground state apoptosome map was used as a starting model (Yu et al., 2005). About 42,000 particles were selected and after refinement ~34,000 particles were included in the final map with ~238,000 subunits. Map filtration was accomplished by computing a hybrid 1-D structure factor during CTF determination, then applying it to the map during refinement with the setsf option. This was combined with a low-pass filter based on the resolution determined for the final map.
For docking experiments, the four domains (NBD, HD1, WHD and HD2) and helix α8 were taken from Apaf-1 (1-591) (1Z6T; Riedl, et al., 2005). A crystal structure of bovine heart cytochrome c (2B4Z; Mirkin et al., 2008) was also used. Docking was carried out in UCSF Chimera (Goddard et al., 2005) and refinement with Flex-EM (Topf et al, 2008). Further details on the modeling of the two β-propellers and the docking procedures are described in Supplementary Methods. Symmetry related subunits were generated in Moleman2 using 7-fold symmetry by rotating the model of the first subunit about the z-axis by multiples of 51.4° (Kleywegt et al. 2004). Figures were made using Chimera and Adobe Photoshop.
Supplementary Material
Acknowledgments
We thank Yigong Shi for providing a pc-9 clone, Fenghe Du for sf21 insect cells and helpful discussions on Apaf-1 purification, and Niko Grigorieff for use of the Zeiss scanner. Computations were done on a Linux cluster at the Scientific Computing and Visualization Group at Boston University. Coordinates for the apoptosome model and the density map have been deposited with the RCSB (xxxx) and EMD (EMD-5186), respectively. The Topf laboratory is supported by an MRC Career Development Award (MT) and the Human Frontier Science Program (RGY0079/2009-C). Wang and Akey laboratories are supported by NIH grants; Dr. Xiaodong Wang was supported by the Welch Foundation and HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abergel C, Bouveret E, Claverie JM, Brown K, Rigal A, Lazdunski C, Benedetti H. Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 Å resolution. Structure. 1999;7:1291–1300. doi: 10.1016/s0969-2126(00)80062-3. [DOI] [PubMed] [Google Scholar]
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW. Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding and activation. Mol Cell. 2002;9:423–432. doi: 10.1016/s1097-2765(02)00442-2. [DOI] [PubMed] [Google Scholar]
- Boatright KM, Renatus M, Scott FL, Sperandio S, Shin H, Pedersen IM, Ricci JE, Edris WA, Sutherlin DP, Green DR, Salvesen GS. A unified model for apical caspase activation. Mol Cell. 2003;11:529–541. doi: 10.1016/s1097-2765(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- Danot O, Marquenet E, Vidal-Ingigliardi D, Richet E. Wheel of life, wheel of death: a mechanistic insights into signaling by STAND proteins. Structure. 2009;17:172–182. doi: 10.1016/j.str.2009.01.001. [DOI] [PubMed] [Google Scholar]
- DeLabarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Ferraro E, Corvaro M, Cecconi F. Physiological and pathological roles of Apaf1 and the apoptosome. J Cell Mol Med. 2003;7:21–34. doi: 10.1111/j.1582-4934.2003.tb00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R, Bohm A, Sigler PB. Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin. Cell. 1996;87:577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- Green DR, Evan GI. A matter of life and death. Cancer Cell. 2002;1:19–30. doi: 10.1016/s1535-6108(02)00024-7. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Ferrin TE. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure. 2005;13:473–482. doi: 10.1016/j.str.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- Harlan J, Chen Y, Gubbins E, Mueller R, Roch J-M, Walter K, Lake M, Olsen T, Metzger P, Dorwin S, Ladror U, Egan DA, Severin J, Johnson RW, Holzman TF, Voelp K, Dacenport C, Beck A, Potter J, Gopalakrishnan M, Hahn A, Spear BB, Halbert DN, Sullivan JP, Abkevich V, Neff CD, Skolnick MH, Shattuck D, Katz DA. Variants in Apaf-1 segregating with major depression promote apoptosome function. Mol Psychiatry. 2006;11:76–85. doi: 10.1038/sj.mp.4001755. [DOI] [PubMed] [Google Scholar]
- Hao Z, Duncan GS, Chang CC, Elia A, Fang M, Wakeham A, Okada H, Calzascia T, Jang Y, You-Ten A, Yeh WC, Ohashi P, Wang X, Mak TW. Specific ablation of the apoptotic functions of cytochrome c reveals a differential requirement for cytochrome c and Apaf-1 in apoptosis. Cell. 2005;121:579–591. doi: 10.1016/j.cell.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Hill MM, Adrain C, Duriez PJ, Creagh EM, Martin SJ. Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes. EMBO J. 2004;23:2134–2145. doi: 10.1038/sj.emboj.7600210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Benedict MA, Ding L, Nunez G. Role of Cytochrome c and dATP/ATP hydrolysis in Apaf-1 mediated caspase-9 activation and apoptosis. EMBO J. 1999;13:3591–3595. doi: 10.1093/emboj/18.13.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Nunez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat Rev Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- Jiang X, Wang X. Cytochrome c promotes caspase-9 activation by inducing nucleotide binding to apaf-1. J Biol Chem. 2000;275:31199–31203. doi: 10.1074/jbc.C000405200. [DOI] [PubMed] [Google Scholar]
- Kim HE, Du F, Fang M, Wang X. Formation of an apoptosome is initiated by cytochrome c induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc Natl Acad Sci U SA. 2005;102:17545–17550. doi: 10.1073/pnas.0507900102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purring-Koch C, McLendon G. Cytochrome c binding to Apaf-1: the effects of dATP and ionic strength. Proc Natl Acad Sci U SA. 2000;97:11928–11931. doi: 10.1073/pnas.220416197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J Mol Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Liu X, Kim CN, Yang J, Jemmerson R, Wang X. Induction of apoptotic program in cell free extracts: requirement for dATP and cytochrome c. Cell. 1996;86:147–157. doi: 10.1016/s0092-8674(00)80085-9. [DOI] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mirkin N, Jaconcic J, Stojanoff V, Moreno A. High resolution X-ray crystallographic structure of bovine heart cytochrome c and its application to the design of an electron transfer biosensor. Proteins. 2008;70:83–92. doi: 10.1002/prot.21452. [DOI] [PubMed] [Google Scholar]
- Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Shiga K, Hashimoto S, Kobayashi T, Horii A, Furukawa T. APAF-1-ALT, a novel alternative splicing form of APAF-1, potentially causes impeded ability of undergoing DNA damage-induced apoptosis in the LNCaP human prostate cancer cell line. Biochem Biophys Res Commun. 2003;306:537–543. doi: 10.1016/s0006-291x(03)00995-1. [DOI] [PubMed] [Google Scholar]
- Pop C, Timmer J, Sperandio S, Salvesen GS. The apoptosome activates caspase-9 by dimerization. Mol Cell. 2006;22:269–275. doi: 10.1016/j.molcel.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Propell M, Riedl SJ, Fritz JH, Rojas AM, Schwarzenbacher R. The Nod-like receptor (NLR) family: a tale of similarities and differences. PLoS One. 2008;3:1–11. doi: 10.1371/journal.pone.0002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Srinivasula SM, Wu G, Fernandes-Alnemri T, Alnemri ES, Shi Y. Structural basis of Procaspase-9 recruitment by the apoptotic protease-activating factor 1. Nature. 1999;399:549–557. doi: 10.1038/21124. [DOI] [PubMed] [Google Scholar]
- Renatus M, Stennicke HR, Scott FL, Liddington RC, Salvesen GS. Dimer formation drives the activation of the cell death protease caspase-9. Proc Natl Acad Sci (USA) 2001;98:14250–14255. doi: 10.1073/pnas.231465798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Wohlgemuth S, Eschenburg S. A new model for the transition of APAF-1 from inactive monomer to caspase-activating apoptosome. J Biol Chem. 2009;284:32717–32724. doi: 10.1074/jbc.M109.014027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Li W, Chao Y, Schwarzenbacher R, Shi Y. Structure of the apoptotic protease-activating factor 1 bound to ADP. Nature. 2005;434:926–933. doi: 10.1038/nature03465. [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lazebnik Y. Caspase-9 and Apaf-1 form an active holoenzyme. Genes Dev. 1999;13:3179–3184. doi: 10.1101/gad.13.24.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh A, Srinivasula SM, Acharya S, Fishel R, Alnemri ES. Cytochrome c and dATP-mediated oligomerization of Apaf-1 is a prerequisite for procaspase-9 activation. J Biol Chem. 1999;274:17941–17945. doi: 10.1074/jbc.274.25.17941. [DOI] [PubMed] [Google Scholar]
- Salvesen GS, Dixit VM. Caspases: intracellular signaling by proteolysis. Cell. 1997;91:443–446. doi: 10.1016/s0092-8674(00)80430-4. [DOI] [PubMed] [Google Scholar]
- Shiozaki EN, Chai J, Shi Y. Oligomerization and activation of caspase-9, induced by Apaf-1 CARD. Proc Natl Acad Sci U S A. 2002;99:4197–4202. doi: 10.1073/pnas.072544399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Steller H. Death by design: mechanism and control of apoptosis. Trend Cell Biol. 1999;12:49–52. [PubMed] [Google Scholar]
- Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J Struct Biol. 2007;157:201–210. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Srinivasula SM, Ahmad M, Fernandes-Alnemri T, Alnemri ES. Autoactivation of procaspase-9 by Apaf-1 mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- Voegtli WC, Madrona AY, Wilson DK. The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization. J Biol Chem. 2003;278:34373–34379. doi: 10.1074/jbc.M302773200. [DOI] [PubMed] [Google Scholar]
- Wang X. The expanding role of mitochondria in apoptosis. Genes Dev. 2001;15:2922–2933. [PubMed] [Google Scholar]
- Yan N, Chai J, Lee ES, Gu L, Liu Q, He J, Wu JW, Kokel D, Li H, Hao Q, Xue D, Shi Y. Structure of the CED-4-CED-9 complex provides insights into programmed cell death in Caenorhabditis elegans. Nature. 2005;437:831–837. doi: 10.1038/nature04002. [DOI] [PubMed] [Google Scholar]
- Yin Q, Park HH, Chung JY, Lin SC, Lo YC, da Graca LS, Jiang X, Wu H. Caspase-9 holoenzyme is a specific and optimal procaspase-3 processing machine. Mol Cell. 2006;22:259–268. doi: 10.1016/j.molcel.2006.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu T, Wang X, Purring-Koch C, Wei Y, McLendon GL. A mutational epitope for cytochrome c binding to the apoptosis protease activation factor-1. J Biol Chem. 2001;276:13034–13038. doi: 10.1074/jbc.M009773200. [DOI] [PubMed] [Google Scholar]
- Yu X, Acehan D, Menetret JF, Booth CR, Ludtke SJ, Riedl SJ, Shi Y, Wang X, Akey CW. A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure. 2005;13:1725–1735. doi: 10.1016/j.str.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Yu X, Wang L, Acehan D, Wang X, Akey CW. Three-dimensional structure of a double apoptosome formed by the Drosophila Apaf-1 related killer. J Mol Biol. 2006;355:577–589. doi: 10.1016/j.jmb.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS. Structure of the AAA+ ATPase p97. Mol Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- Zou H, Li Y, Liu X, Wang X. An Apaf-1 cytochrome c multimeric complex is a functional apoptosome that activates procaspase-9. J Biol Chem. 1999;274:11549–11556. doi: 10.1074/jbc.274.17.11549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.