Abstract
OBJECTIVE
Thioredoxin interacting protein (Txnip), a regulator of cellular oxidative stress, is induced by hyperglycemia and inhibits glucose uptake into fat and muscle, suggesting a role for Txnip in type 2 diabetes pathogenesis. Here, we tested the hypothesis that Txnip-null (knockout) mice are protected from insulin resistance induced by a high-fat diet.
RESEARCH DESIGN AND METHODS
Txnip gene-deleted (knockout) mice and age-matched wild-type littermate control mice were maintained on a standard chow diet or subjected to 4 weeks of high-fat feeding. Mice were assessed for body composition, fat development, energy balance, and insulin responsiveness. Adipogenesis was measured from ex vivo fat preparations, and in mouse embryonic fibroblasts (MEFs) and 3T3-L1 preadipocytes after forced manipulation of Txnip expression.
RESULTS
Txnip knockout mice gained significantly more adipose mass than controls due to a primary increase in both calorie consumption and adipogenesis. Despite increased fat mass, Txnip knockout mice were markedly more insulin sensitive than controls, and augmented glucose transport was identified in both adipose and skeletal muscle. RNA interference gene-silenced preadipocytes and Txnip−/− MEFs were markedly adipogenic, whereas Txnip overexpression impaired adipocyte differentiation. As increased adipogenesis and insulin sensitivity suggested aspects of augmented peroxisome proliferator–activated receptor-γ (PPARγ) response, we investigated Txnip's regulation of PPARγ function; manipulation of Txnip expression directly regulated PPARγ expression and activity.
CONCLUSIONS
Txnip deletion promotes adiposity in the face of high-fat caloric excess; however, loss of this α-arrestin protein simultaneously enhances insulin responsiveness in fat and skeletal muscle, revealing Txnip as a novel mediator of insulin resistance and a regulator of adipogenesis.
Adipose expansion and plasticity link obesity to insulin resistance (1). Adipocytes either store fatty acids as triglycerides or release them in response to fluctuating states of nutritional intake and energy expenditure; adipocytes also secrete adipokine peptide hormones that regulate the metabolic activity of other tissues. Dysregulation in either process can markedly affect whole-body energy balance and insulin sensitivity (2). Insulin resistance may also result from increased release of inflammatory cytokines or adipokines derived from adipocytes or fat-infiltrating macrophages. However, increased fat is not synonymous with dysregulated metabolism. For example, lipodystrophies are often associated with severe insulin resistance (3–5), whereas subsets of obese humans and mouse models exist with normal metabolic indexes despite markedly increased adipose mass (6).
Thioredoxin interacting protein (Txnip) is one of six members of the mammalian α-arrestin family. The α-arrestins are structurally related to the β-arrestins, which are well-characterized mediators of G-protein–coupled receptor signaling and endocytosis. Furthermore, β-arrestin-2 was recently shown to be a key contributor to insulin signaling (7). Little is known about α-arrestin function, though several family members regulate endocytosis in yeast (8). Txnip was first identified as a binding partner and inhibitor of the antioxidant protein thioredoxin (9). Recent studies show Txnip to be a novel metabolic regulator, with effects on glucose and lipids. Txnip also regulates cellular growth, differentiation, and programmed death (10–14). Txnip expression is markedly glucose responsive (15), and its expression is elevated in the skeletal muscle of diabetic and glucose-intolerant patients (16). Interestingly, Txnip is the only α-arrestin family member to bind thioredoxin (17). Another α-arrestin, Arrdc3, has been linked to development of obesity in humans (18), and both Arrdc4 and a Txnip mutant that does not bind thioredoxin have been shown to alter glucose metabolism (19), supporting α-arrestins as a new class of metabolic regulators that operate independently of thioredoxin.
We previously observed that fasted Txnip-null mice are hypoglycemic and described a functional role for Txnip in enhancing insulin responsiveness and glucose uptake in muscle and fat (11,16). Given those findings, we hypothesized that Txnip deletion would protect against the development of insulin resistance induced by a high-fat diet. Here we assessed insulin action by hyperinsulinemic clamp studies and the regulation of adipogenesis in high-fat diet–fed Txnip-null mice. We report that loss of Txnip leads to a pattern of increased insulin sensitivity despite expansion of adiposity that is suggestive of clinical responses seen with peroxisome proliferator–activated receptor-γ (PPARγ)–activating thiazolidinediones (TZDs), and we furthermore found that Txnip regulates PPARγ activity. These data support the concept that Txnip promotes adipose tissue expansion, which contributes to its role as a metabolic regulator.
RESEARCH DESIGN AND METHODS
Animals.
Txnip−/− mice were generated as described (20). Mice were housed under controlled temperature (22 ± 2°C) and lighting (12 h of light, 0700–1900 h; 12 h of dark, 1900–0700 h) with free access to water and food. Male Txnip−/− and wild-type mice between 12 and 18 weeks old were fed a regular diet (TD2018; Harlan Teklad, Madison, WI) or high-fat diet (HFD; 55% fat by calories; TD93075; Harlan Teklad) ad libitum for 4 weeks, and metabolic parameters and insulin action were measured. Mice were maintained in accordance with the Institutional Animal Care and Use Committees of the Harvard School of Medicine and the Yale University School of Medicine.
Basal study.
Fat and lean body masses were assessed by 1H magnetic resonance spectroscopy before and after 4 weeks of high-fat diet. Comprehensive animal metabolic monitoring system (CLAMS; Columbus Instruments, Columbus, OH) was used to evaluate activity, food consumption, and energy expenditure during a 72-h period, with hourly values averaged to a composite 24-h period, as detailed in the supplementary Methods, available in an online appendix at http://diabetes.diabetesjournals.org/content/early/2010/03/10/db09-1212/suppl/DC1.
Hyperinsulinemic-euglycemic clamp studies.
Hyperinsulinemic-euglycemic clamp studies were conducted as described previously for 140 min with a primed/continuous infusion of human insulin (21 mU/kg prime for 3 min, 3 mU/kg per min infusion) using mice that were fasted overnight. Protocols and calculations are detailed in supplementary Methods.
Biochemical analysis.
Plasma glucose was analyzed by glucose oxidase method. Plasma insulin, glucagon, and adiponectin were measured by radioimmunoassay using kits from Linco. Plasma leptin was measured using the LINCOplex Assay (Linco). Serum tumor necrosis factor and interleukin-6 levels were measured by ELISA (BD Biosciences). Colorimetric assays were used for plasma fatty acid (Wako) and β-hydroxybutyrate concentrations (StanBio Labs). Plasma [3H]glucose was measured by scintillation counting of ZnSO4/Ba(OH)2 deproteinized serum, dried to remove 3H2O.
Adipose tissue biopsy and cell size analysis.
Adipocyte sizing using a Beckman Coulter Multisizer III with a 400-μm aperture, set to count 6,000 particles, was performed on osmium tetroxide–fixed epididymal fat tissue as previously described (21).
MEF isolation and adipogenesis.
Mouse embryonic fibroblasts (MEFs) were isolated from day-14.5 embryos as previously described (22). Adipogenesis was induced in 2-day postconfluent cells (passage 4) by standard dexamethasone, methylisobutylxanthine, and insulin (DMI) induction (16) with rosiglitazone or DMSO vehicle when indicated.
mRNA quantification by real-time PCR.
Epididymal white adipose tissue total RNA was isolated using Trizol per manufacturer's instructions (Invitrogen). cDNA was reverse transcribed from 1 μg total RNA for real-time PCR as previously described (11) and detailed in supplementary Methods.
Endogenous PPAR and exogenous PPARγ–ligand binding domain reporter assays.
Generation and transduction of Txnip lentivirus is described previously (11). Txnip short hairpin RNA (shRNA) and control shRNA plasmids were purchased from Sigma. 3T3-L1 preadipocyte lines were made with stable overexpression of Txnip, empty vector control, control shRNA, or Txnip shRNA species by puromycin selection after viral transduction. Txnip expression was confirmed by Western blot analysis with anti-Txnip antibodies as described (20). PPAR response element and ligand binding domain assays were performed as described (23), as detailed in supplementary Methods.
Statistical analysis.
A two-tailed Student t test was used to test differences between Txnip−/− and wild-type mice; 2-way ANOVA was performed to test multiple effects during the hyperinsulinemic-euglycemic clamp. One-way ANCOVA analysis was performed using a Web-based calculator provided by Vassar College (http://faculty.vassar.edu/lowry/VassarStats.htm). Values are presented as mean ± SE; P < 0.05 was considered statistically significant.
RESULTS
High-fat feeding increased adiposity in Txnip-null mice.
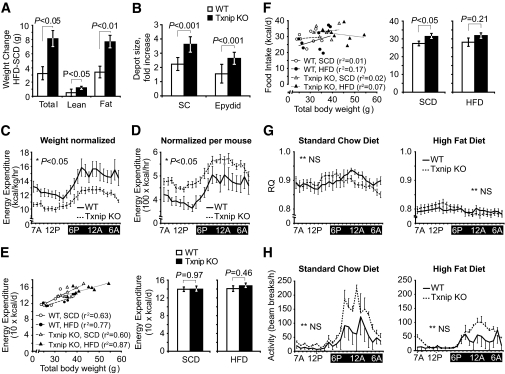
Txnip knockout and wild-type control mice were fed a 4-week high-fat, high-caloric diet (55% fat, 24% carbohydrate), and whole-body metabolism and insulin responsiveness were measured. Baseline body composition was determined prior to high-fat feeding while on standard chow diet (SCD): age-matched Txnip knockout mice were heavier than wild-type littermates (36.7 ± 1.0g vs. 30.5 ± 1.2 g, P < 0.05, supplementary Fig. 1A). Magnetic resonance (MRS) analysis of body composition revealed Txnip knockout mice had greater total fat and lean mass, but the same ratio of fat to total body weight as wild-type controls (supplementary Fig. 1A). After 4 weeks of HFD, Txnip knockout total body weight increased twice as much as control mice (Fig. 1A). MRS indicated this was almost entirely due to fat mass expansion: Txnip knockout mice gained 8.1 ± 1.1 g of fat mass relative to 3.3 ± 1.1 g for wild type (Fig. 1A, P < 0.01), and greater fat mass relative to total body mass (wild type: 23 ± 2%, Txnip knockout: 32 ± 2%, P < 0.05). Both subcutaneous and visceral Txnip knockout fat depots expanded relative to wild-type controls (Fig. 1B).
FIG. 1.
Adiposity, food consumption, metabolic rate, and physical activity in Txnip-null and wild-type control mice before and after high-fat feeding. A: Total mass, lean body mass, and total adipose weight for Txnip knockout and wild-type control mice assessed by 1H magnetic resonance spectroscopy and expressed as After HFD − Before HFD weights. n = 8 per group. B: Mean difference in weight between HFD and SCD excised fat depots. n = 6 per group. C–H: Metabolic activity measurements assessed by CLAMS. Values represent 72 h of monitoring, graphically represented with hourly values averaged to the same 24-h period. n = 8 per group: (C) Energy expenditure indexed to body mass in kilograms. D: Energy expenditure unadjusted for body mass. E, left: Energy expenditure plotted against total body mass with fitted lines of linear regression; (right) energy expenditure adjusted for body mass by ANCOVA. n = 8 per group. F, left: Average food consumption during a 60-h period plotted against total body mass with fitted lines of linear regression; (right) unadjusted comparison of mean food consumption. n = 8 per group. G: Respiratory quotient (RQ). H: Locomotor activity. *P < 0.05. **Not significant (NS) for the comparison of the mean difference between wild-type and Txnip knockout mice integrated during 72 h of monitoring. Epydid, epididymal; KO, knockout; SC, subcutaneous; WT, wild type.
Increased adiposity in HFD Txnip knockout mice was due to increase calorie intake.
Energy balance was determined before and after high-fat feeding using an Oxymax CLAMS system for a 3-day period. Total energy expenditure averaged to a 24 h cycle was markedly influenced by the method of indexing; indexing energy expenditure to body mass indicated Txnip knockout mice expended less energy than control mice, whereas energy expenditure indexed per mouse indicated Txnip knockout mice expended greater energy (Fig. 1C and D, representative tracings on HFD). As this discordance was likely due to the significant differences in body composition, we adjusted the energy expenditure for body mass by ANCOVA, as described by Packard and Boardman (24). Energy expenditure was strongly correlated to body mass with r2 values from 0.6 to 0.87 (Fig. 1E). After adjustment of energy expenditure for differences in body mass by ANCOVA, there were no significant differences between wild-type and Txnip knockout mouse energy expenditure for mice fed either SCD or HFD (Fig. 1E). Adjustment of energy expenditure for lean body mass (using values derived from MRS) gave similar results (data not shown).
Food intake was also compared with total body mass. No significant correlation was seen for either genotype on either diet, with r2 values from 0.01 to 0.17 (Fig. 1F), hence ANCOVA was not performed. Txnip knockout mice consumed 14% more calories per day than wild-type mice on SCD (P < 0.05); a similar trend, although not significant, was seen during HFD (Fig. 1F). From these data, we conclude that the development of obesity in Txnip knockout mice was most attributable to primary excess in food consumption.
Txnip-null mice use the same fuel source as wild-type mice.
Previous ex vivo tissue studies of substrate oxidation in different Txnip-deficiency models indicated both carbohydrate and lipid oxidation are diminished (25,26). We examined respiratory quotient profiles for mice on both diets to determine whether Txnip deletion altered carbohydrate versus lipid substrate oxidation in vivo. The 24-h averaged respiratory quotient for Txnip knockout mice was the same as for wild-type controls on each diet (Fig. 1G, SCD: 0.90 ± 0.01 for both; HFD: 0.80 ± 0.01 for both). Respiratory quotient was equally reduced for both mice after 4 weeks of HFD feeding, consistent with increased fat oxidation. A subtle variance in light/dark respiratory quotient values was discernable between each group, with Txnip-null mice exhibiting a greater trend toward carbohydrate oxidation during relatively inactive light periods, and a more pronounced shift to lipid oxidation during more active nighttime periods. Txnip knockout mice trended toward greater locomotor activity on both diets relative to control mice, most evident during dark cycles, although the differences were not statistically significant when averaged over a 24-h period (Fig. 1H).
Txnip knockout mice are protected from insulin resistance after prolonged high-fat feeding.
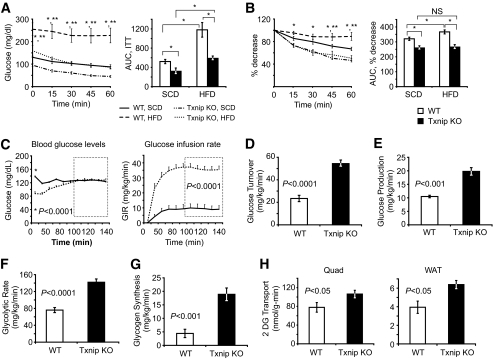
Insulin responsiveness was first assessed in SCD-fed and HFD-fed mice by intraperitoneal insulin tolerance testing. SCD-fed Txnip knockout mice had lower baseline blood glucose levels and a more pronounced insulin-stimulated blood glucose drop than SCD-fed wild-type controls, both by absolute and relative measures of insulin response (Fig. 2A and B). After HFD feeding, Txnip knockout blood glucose levels were markedly lower than wild-type mice (Fig. 2A). HFD-fed wild-type insulin responsiveness was blunted relative to SCD-fed wild-type mice, but HFD-fed Txnip knockout mouse insulin response was unchanged relative to SCD-fed Txnip knockout mice (Fig. 2B).
FIG. 2.
Effects of insulin stimulation on glucose utilization during hyperinsulinemic-euglycemic clamp studies after 4 weeks of HFD feeding. A and B: Intraperitoneal insulin tolerance test (ITT), 0.25 mU insulin/g body wt. A: Absolute blood glucose levels. B: Relative blood glucose levels indexed to t = 0 levels in each group. n = 6–7 per group. *P < 0.05 between HFD-fed wild-type and Txnip knockout mice, **P < 0.05 between HFD-fed wild-type and SCD-fed wild-type mice. C: Basal glucose and glucose infusion rates (GIRs) for wild-type and Txnip knockout mice during euglycemic-hyperinsulinemic clamp studies. GIR was averaged over the final 60 min of the clamp (inset box), P < 0.0001. n = 7 per group. *P < 0.0001 for basal glucose levels prior to insulin infusion. D–H: Labeled glucose precursor whole-body glucose utilization studies. D: Whole-body glucose disposal. E: Basal hepatic glucose output. F: Whole-body glycolytic rate. G: Whole-body glycogen synthesis rates. Parameters were determined by 3H-glucose infusion throughout the euglycemic-hyperinsulinemic clamp protocol. n = 7 per group for each. H: 2DG uptake into skeletal muscle (quad) and WAT. 14C-2DG was infused during the final 55 min of the 140-min euglycemic-hyperinsulinemic clamp protocol, and individual tissues were excised for scintillation counting. n = 7 per group. AUC, area under the curve.
Fasting plasma glucose levels were 37% lower and fasting insulin levels were ∼70% lower for HFD-fed Txnip-null mice than wild-type controls (Fig. 2C and Table 1; P < 0.01 and P = 0.05, respectively). Fasting glucagon levels were essentially unchanged, despite Txnip knockout relative hypoglycemia (Table 1). Interestingly, despite increased adiposity, Txnip knockout leptin serum levels were lower by 40% (P < 0.05) and adiponectin levels were lower by 20% (P < 0.05, Table 1). HFD-fed Txnip knockout mice had increased triglycerides (176%, P < 0.05), free fatty acids (137%, P < 0.05), and serum ketones (3.7-fold, P < 0.0001) relative to wild-type controls after an overnight fast, reflecting a similar fasting physiology to SCD-fed Txnip knockout fasted mice (11).
TABLE 1.
High-fat–fed serum hormone and serum metabolite levels
| Wild type | Txnip KO | P value | |
|---|---|---|---|
| Serum hormone | |||
| Insulin (pg/ml) | 70.0 ± 19.4 | 28.3 ± 4.0 | 0.05 |
| Glucagon (pg/ml) | 30.4 ± 3.0 | 29.4 ± 5.1 | 0.92 |
| Leptin (pg/ml) | 611 ± 110 | 360 ± 52 | 0.03* |
| Adiponectin, HFD (ng/ml) | 9.1 ± 4.2 | 4.2 ± 0.6 | <0.01* |
| Adiponectin, SCD (ng/ml) | 5.7 ± 0.9 | 6.8 ± 0.5 | 0.33 |
| Serum metabolites | |||
| Glucose (mg/dl) | 139 ± 9 | 87 ± 6 | <0.001* |
| Triglycerides (mg/dl) | 146 ± 32 | 258 ± 31 | 0.02* |
| FFA (before clamp, mEq/ml) | 0.97 ± 0.07 | 1.33 ± 0.16 | 0.04* |
| Δ FFA (after clamp, % change) | −3.3 ± 8.9 | −31.8 ± 6.2 | 0.02* |
| β-hydroxybutyrate (mmol/l) | 1.57 ± 0.26 | 5.88 ± 0.47 | <0.0001* |
Unless indicated, values reflect serum levels after an overnight fast.
*Statistical significance by Student t test.
Txnip knockout mice required nearly fourfold higher glucose infusion rates to maintain euglycemia compared with wild-type HFD-fed littermates during hyperinsulinemic-euglycemic clamp studies (Fig. 2C, P < 0.0001). This corresponded to a 2.1-fold increase in insulin-stimulated whole-body glucose turnover (Fig. 2D, P < 0.0001). Basal hepatic glucose output (HGO) was 189% higher in the Txnip-null mice (Fig. 2E, P < 0.01). Hepatic glucose production was not suppressed by insulin in either wild-type or Txnip knockout animals (supplementary Fig. 2). Increases in Txnip knockout whole-body glucose uptake were associated with significant increases in whole-body glycolysis (182%) and glycogen synthesis (429%) (Fig. 2F and G).
Peripheral glucose disposal is augmented in Txnip-deficient mice.
Insulin-stimulated glucose transport into skeletal muscle was increased by 36% in Txnip knockout mice (Fig. 2H, quad). Glucose uptake into white adipose tissue (WAT) was similarly increase by 40% (Fig. 2H, WAT). No differences were seen in brown adipose glucose transport levels (315 ± 56 vs. 302 ± 72 nmol/g/min). Txnip knockout mice exhibited nearly a 10-fold greater postclamp reduction in free fatty acid (FFA) levels than wild-type controls (31.8% vs. 3.3%, respectively, P < 0.05, Table 1). Reduced FFA levels after insulin stimulation are an index of adipose insulin sensitivity, reflecting decreased adipocyte lipolysis and increased adipose triglyceride synthesis from FFA reesterification. Together, these data point to globally enhanced peripheral insulin sensitivity in Txnip-null mice, protecting against high-fat diet–induced insulin resistance.
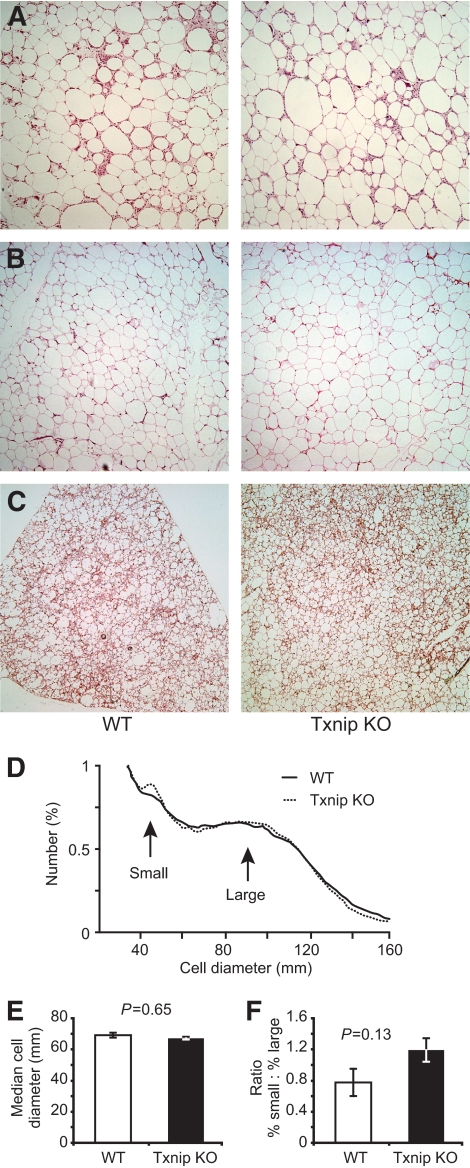
High-fat diet induces adipogenesis in the Txnip-null mice.
Although expanded, Txnip-null visceral, subcutaneous, and brown adipose tissues were structurally similarly to wild type (Fig. 3A–C). CD68+ inflammatory infiltrate was observed in the epididymal WAT of both strains (supplementary Fig. 3A), and no differences were observed in serum levels of tumor necrosis factor or interleukin-6 between the two groups after HFD feeding (supplementary Fig. 3B). HFD-fed Txnip knockout and wild-type control epididymal adipose cell size distribution was determined with a Beckman Coulter Multisizer III. Multisizing revealed wild-type and Txnip knockout adipocyte median size and size distribution were equal (Fig. 3D and E), and the fat depots contained equivalent proportions of small to large adipocytes (Fig. 3F). Given Txnip knockout's greater net fat accumulation, this implied adipogenesis was occurring at pace with lipid storage.
FIG. 3.
Txnip deletion does not alter adipose structure or cell size. A–C: Representative histologic sections from various adipose depots after HFD feeding: (A) visceral fat, (B) subcutaneous fat, and (C) brown fat. Each section is at ×10 magnification. D–F: Adipocyte sizing by multisizer analysis after high-fat feeding: (D) mean relative frequency distribution of epididymal adipocyte cell sizes; n = 6 per genotype. E: Median cell diameter of the large adipocyte population, derived from the Gaussian profile of large adipocyte cell size distribution. F: The fractional ratio of small to large adipocytes. White bars = wild type, black bars = Txnip knockout. (A high-quality digital color representation of this figure is available in the onlime issue.)
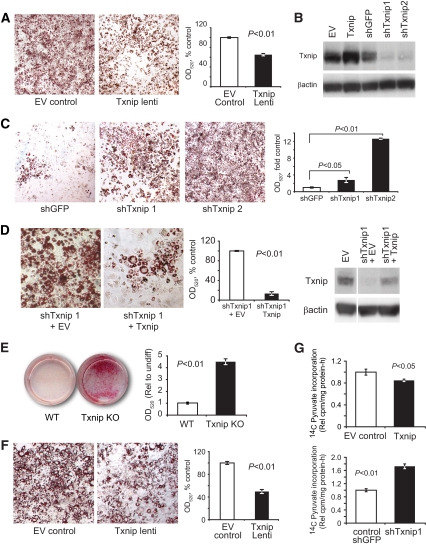
To test directly the hypothesis that Txnip deficiency stimulated adipogenesis, 3T3-L1 preadipocytes were transduced with lentivirus carrying Txnip cDNA or gene-silencing shRNA and were assessed for impaired or enhanced adipogenesis. Txnip overexpression blunted adipogenesis to 65% of control lentivirus–infected 3T3-L1 cells (Fig. 4A and B). Similarly, two different Txnip shRNA species increased adipocyte formation by 3.2-fold and 12.6-fold relative to control shRNA (shTxnip1 and shTxnip2, Fig. 4B and C). Txnip reintroduction into 3T3-L1 cells stably gene-silenced with shTxnip1, which targeted the endogenous Txnip 3′ untranslated region not present in the cDNA transgene, effectively reversed the effects of Txnip gene-silencing, reducing adipogenesis to less than 20% of empty vector control (Fig. 4D). MEFs derived from Txnip knockout embryos exhibited a similar increased potential to differentiate into mature adipocytes—more than a fourfold increase compared with wild-type control MEFs (Fig. 4E, P < 0.0001; similar results were seen in a second independent line). Reintroduction of Txnip into the Txnip-null MEFs by lentiviral transduction reduced adipocyte formation (Fig. 4F, 54% reduction relative to control-infected cells, P < 0.05). Finally, lentiviral overexpression of Txnip in mature differentiated 3T3-L1 adipocytes reduced de novo glycerol synthesis from labeled pyruvate by 15%, whereas Txnip gene-silencing increased glycerol formation nearly twice that of control transduced cells (Fig. 4F, P < 0.05 and P < 0.01, respectively). Rates of glyceroneogenesis correlate with adipogenesis as adipocytes exhibit nominal glycerol kinase activity and must generate de novo glycerol-3-phosphate for fatty acid esterification during triglyceride synthesis (27). These results support the concept that Txnip intrinsically regulates adipocyte differentiation and lipogenesis.
FIG. 4.
Txnip deletion promotes adipocyte differentiation. A, left: Representative Oil red O (ORO) staining of 3T3-L1 adipocytes after lentiviral overexpression of Txnip or control vectors and standard DMI induction; ×4 magnification. Right: Triglyceride content from alcohol-extracted ORO dye quantified at optical density 520 nm (OD520). n = 4 replicates. B: Representative Txnip protein expression and β-actin loading control levels in 3T3-L1 96 h after lentiviral transduction. C: 3T3-L1 adipocyte formation using ½ standard DMI induction after Txnip silencing by shRNA lentiviral transduction. n = 4 replicates for ORO extraction. D: 3T3-L1 stably transduced with shTxnip1 (shRNA targeting 3′ untranslated region) followed by Txnip cDNA or control vector lentivirus superinfection, then induced to differentiate with ½ standard DMI. Representative images; graph represents ORO extraction with n = 6 replicates. Txnip and β-actin loading control protein expression levels relative to unsilenced 3T3-L1 transduced with empty vector. The white line indicates removed lanes. E: Wild-type and Txnip knockout embryonic fibroblast (day 14.5, untransformed passage 4) at day 9 after adipocyte induction with standard DMI + 50 μmol/l rosiglitazone. n = 6 replicates for ORO extraction. F: Txnip knockout MEFs after Txnip or control vector expression by lentiviral transduction; n = 4 replicates for ORO extraction. G: Rate of 14C-pyruvate incorporation into de novo glycerol formation in differentiated 3T3-L1 adipocytes, 96 h after transduction with Txnip or Txnip shRNA lentivirus and indicated controls. n = 6 replicates. Graphs depict single representative experiments with error bars reflecting replicates; all experiments were repeated with similar results. MEF differentiation was confirmed using an independent untransformed wild-type and Txnip knockout line. Representative color images were nonlinearly color-contrast enhanced using equivalent settings and were not used for quantitative analysis. EV, empty vector; lenti, lentivirus; Rel, relative; undiff, undifferentiated. (A high-quality digial color representation of this figure is available in the online issue.)
Txnip deletion augments PPARγ-stimulated adipogenesis.
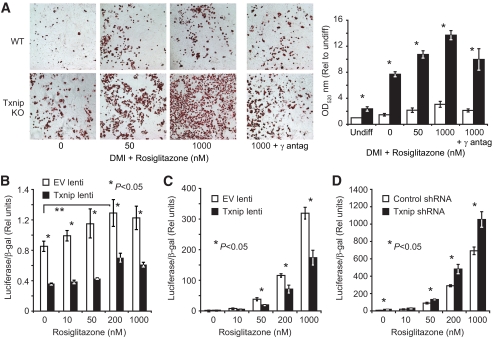
Increased insulin sensitivity, increased adiposity, and augmented adipogenesis are similar to the clinical response of PPARγ agonism, as seen in thiazolidinedione treatment of patients with type 2 diabetes (5,28). To test the hypothesis that Txnip-null cells show increased PPARγ activity, we repeated MEF adipogenesis in the presence of the PPARγ-selective agonist rosiglitazone, and demonstrated PPARγ specificity with the coadministration of the PPARγ-selective irreversible antagonist GW9662 (29). Txnip-null MEFs had approximately fivefold greater adipogenic response than wild-type controls at each dose of rosiglitazone (Fig. 5A, P < 0.0001), which was attenuated with the coapplication of the antagonist GW9662. Net adipogenesis was uniformly greater at each dose of rosiglitazone, suggesting that Txnip did not affect ligand responsiveness.
FIG. 5.
Txnip deletion augments PPARγ-stimulated adipogenesis and PPARγ activity. A: Wild-type and Txnip knockout adipocyte differentiation at day 9 after DMI induction + increasing rosiglitazone concentrations. Total lipid levels are quantified after ORO extraction at OD520 staining. γ-antagonist = 1 μmol/l GW9229. *P < 0.001 for wild-type vs. Txnip knockout at each rosiglitazone concentration; n = 6 replicates per group. The representative color images were nonlinearly color-contrast enhanced using equivalent settings, and were not used for quantitative analysis. White bars = wild type, black bars = Txnip knockout. B: Endogenous PPARγ activation with increasing rosiglitazone dosing in 3T3-L1 cells stably transduced with Txnip or control lentivirus. PPARγ activity was determined by a transfected PPAR response element luciferase reporter stimulated by 18 h of rosiglitazone treatment (48 h after transfection). Transfection efficiency was normalized to β-galactosidase (β-gal) activity from a cotransfected β-gal reporter. n = 4 replicates per group. C and D: PPARγ LBD activation assay. PPARγ-LBD::GAL4 DNA-BD fusion protein was cotransfected with a GAL4 promoter-luciferase reporter into 3T3-L1 preadipocytes expressing Txnip lentivirus (C) or Txnip shRNA lentivirus (D) compared with relevant controls. Luciferase activity was determined after 18-h rosiglitazone stimulation. Transfection efficiency was normalized to β-gal activity from a cotransfected β-gal reporter. *P < 0.05, n = 4 replicates per condition. Graphs depict single representative experiments with error bars reflecting replicates. antag, antagonist. (A high-quality digital color representation of this figure is available in the online issue.)
Txnip expression modulates PPARγ activity level.
Endogenous PPAR activity was assessed in 3T3-L1 cells overexpressing Txnip or control vectors, using a transiently expressed (PPRE)3TKLuc PPAR response element luciferase reporter (PPRE). Txnip overexpression inhibited the PPRE reporter 2.5-fold relative to empty vector control (Fig. 5B, P < 0.05). This was followed by an assessment of Txnip's ability to activate or inhibit the PPARγ-ligand binding domain (LBD) in a standard chimeric Gal4-DBD-hPPARγ-LBD fusion protein/Gal4-luciferase reporter assay. 3T3-L1 cells stably transduced with Txnip or shTxnip1 were compared with relevant controls. Txnip overexpression reduced PPARγ-LBD activation nearly twofold relative to control in response to rosiglitazone (Fig. 5C, P < 0.05). In the reciprocal study, Txnip gene silencing induced PPARγ-LBD activation 1.5-fold more than the shRNA control at each increasing dose of rosiglitazone (Fig. 5D, P < 0.05). Reflecting the MEF adipogenesis results, total PPRE reporter activity was reduced by Txnip overexpression; however, rosiglitazone dose responsiveness was similar to that of the control, suggesting Txnip expression impedes PPARγ activity without inhibiting the response to ligand activation.
PPARγ activation suppresses Txnip expression during adipocyte differentiation.
Txnip expression progressively increased threefold relative to uninduced preadipocytes for 6 days after standard DMI induction (supplementary Fig. 4). The addition of 1 μmol/l rosiglitazone suppressed Txnip mRNA expression during the early phase of adipogenesis (days 0–3) relative to standard DMI differentiation alone (P < 0.01), suggesting PPARγ activation suppresses Txnip expression to impede Txnip's negative feedback inhibition of PPARγ.
Txnip deletion promotes adipogenic PPARγ target gene expression after high-fat feeding.
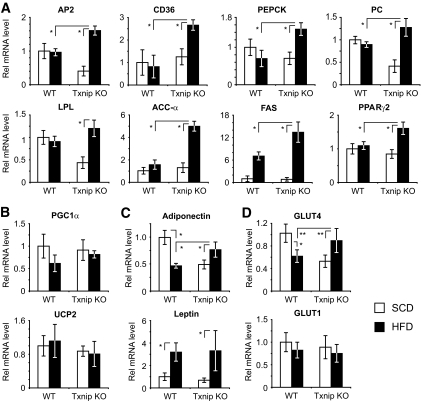
In vivo expression of WAT adipogenic and lipogenic PPARγ target genes was measured in SCD-fed and HFD-fed mice to determine whether HFD potentiated PPARγ activation in Txnip-null mice. AP2, CD36, pyruvate carboxylase, fatty acid synthase, phosphoenolpyruvate carboxykinase, lipoprotein lipase, and acetyl-CoA carboxylase-α are known adipogenic and lipogenic PPARγ target genes (30). After 4 weeks of HFD, these PPARγ targets were uniformly upregulated in Txnip knockout HFD-fed adipose tissue relative to Txnip knockout SCD-fed adipose tissue, and HFD Txnip knockout transcript levels were consistently greater than HFD-fed wild-type expression levels (n = 8–12 per group, P < 0.01, Fig. 6A). PGC1α and UCP2 expressions were measured, as the two genes influence lipid metabolism, adiposity, and energy balance, but are not PPARγ targets (31,32). No significant differences in expression were detected for either gene on SCD or after HFD (Fig. 6B). Although HFD-fed Txnip knockout serum adiponectin levels were lower than wild type, adiponectin transcript levels in Txnip knockout HFD-fed adipose were increased relative to wild-type mice, suggesting regulation of, or a defect in, adiponectin translation, protein stability, and/or secretion (Fig. 6C). Similarly, leptin transcript levels in HFD-fed Txnip knockout and wild-type adipose were equivalent, although Txnip knockout serum leptin levels were lower (Fig. 6C). GLUT mRNA levels for the insulin-regulated (and known PPARγ target) GLUT4 and the constitutively expressed GLUT1 were also determined. GLUT1 expression trended downward for both mice after HFD, but GLUT4 expression was significantly increased in Txnip knockout WAT relative to wild type after high-fat diet. Wild-type GLUT4 levels were reduced by 40% after HFD, whereas Txnip knockout post-HFD levels increased 1.6-fold (P < 0.01, Fig. 6D). Finally, we examined both PPARγ and Txnip expression, as PPARγ expression is influenced by its own activation (33), and Txnip expression is augmented in states of insulin resistance (16). High-fat diet increased PPARγ2 expression in Txnip knockout WAT by 1.7-fold (P < 0.01, Fig. 6A). In wild-type WAT, high-fat feeding increased Txnip expression 1.4-fold (P < 0.01, supplementary Fig. 4B). These data support the concept that Txnip deletion augments adipogenic and lipogenic programs in WAT in vivo after high-fat feeding by preferentially augmenting PPARγ activation. HFD-induced Txnip expression may play a role in suppressing dietary PPARγ activation to impede adipogenesis in the face of positive energy balance, resulting in diet-induced insulin resistance.
FIG. 6.
High-fat feeding preferentially promotes PPARγ target gene expression in Txnip-null WAT. A: mRNA transcript expression of PPARγ target genes and PPARγ2 in WAT before and after high-fat feeding. n = 8–12 mice per group for each transcript. AP2, fatty acid binding protein 4; PC, pyruvate carboxylase; PEPCK, phosphoenolpyruvate carboxykinase; ACC-α, acetyl-CoA carboxylase-α; FAS, fatty acid synthase; LPL, lipoprotein lipase. B: Transcript expression levels for non-PPARγ target genes PCG1α and UCP2 after HFD. C and D: Adiponectin, leptin, and GLUT transcript expression levels in WAT after SCD vs. HFD. n = 8–12 mice per group.
DISCUSSION
We show here that Txnip plays a central role in regulating insulin sensitivity, energy balance, and adiposity. Previously, we demonstrated Txnip's critical function in glucose homeostasis during nutrient deprivation, in part by regulating liver gluconeogenic capacity and peripheral insulin sensitivity (11). When challenged with high-fat caloric excess, Txnip-deficient mice are protected against insulin resistance, with markedly higher rates of insulin-stimulated glucose disposal. This increased insulin sensitivity occurs despite increasing adiposity. Expanded adipose mass is primarily due to an increased drive for food intake, which may reflect a central defect in satiety or a secondary response to lower circulating factors such as leptin or blood glucose. In concert, cellular storage capacity is expanded through increased adipogenesis.
Our results correspond well with other recent reports using different Txnip-deficient models. Hui et al. showed Txnip-null mice had increased insulin responsiveness primarily from enhanced skeletal muscle insulin sensitivity, which was confirmed using muscle-restricted Txnip-null mice. As a striking observation, they showed high-fat feeding the null mice even further lowered fasting blood glucose (25). Hui et al. also reported increased basal Txnip knockout HGO, which was a surprise given our earlier observations of defective Txnip knockout hepatocyte glucose production (11). We suspect this reflects both increased gluconeogenic substrate delivery to the liver, as Txnip knockout mice have increased serum lactate levels, and diminished HGO suppression from lower endogenous serum insulin levels (10,34). One notable difference is that in the model of Hui et al., fat mass was unchanged and adipose insulin responsiveness was not increased in Txnip-null mice. Strain differences or specific aspects of the gene-targeting approaches may underlie the differences in the two models. In a third model, Chen et al. observed results similar to ours using Hcb-19 mice, which have a naturally occurring Txnip truncation mutation. They reported marked increases in Hcb-19 adiposity relative to controls that developed at an early age, and observed increased glucose tolerance and insulin responsiveness (35). Also in agreement, Chen et al. attributed increased Hcb-19 fat mass to a primary increase in food intake. When the Hcb-19 mice were crossed with the ob/ob leptin-deficient mice, Txnip deletion promoted even greater fat mass expansion, but protected against diabetes development by preventing peripheral insulin resistance and β-cell apoptosis. Collectively, these three different Txnip-deficient models are highly corroborative and underscore Txnip's central importance in metabolic regulation, development of insulin resistance, and global energy balance.
An increased capacity for adipocyte expansion in Txnip-null mice may contribute to the protection against insulin resistance from high-fat feeding. Finite adipose expansion has been proposed to explain the similarities in metabolic dysfunction shared between lipodystrophies and obesity (1,36,37). Once storage capacity is exceeded, toxic metabolite spillover or dysfunctional adipokine and cytokine release may result in insulin resistance (38). Augmented adipogenesis in the null mice may alleviate the damaging metabolic effects of exhausted adipocyte storage capacity, which may similarly underlie the therapeutic benefits of TZD therapy (6,30,36). Other evidence suggests TZDs promote fat redistribution from inflammatory visceral depots to more protective subcutaneous depots. We observe Txnip-null adipose stores are equally expanded without any apparent redistribution from one depot to another. Increased adipose glucose uptake also contributes to the increased insulin-mediated glucose disposal seen in Txnip-null mice. Although Txnip-null brown fat 2-deoxyglucose (2DG) transport rates were not different from wild type, both increased null brown adipose mass and its high glucose transport make it a significant contributor to overall glucose disposal. Based on depot sizes and relative 2DG transport levels, we estimate WAT and brown fat account for 10% of total glucose disposal and ∼1.6-fold greater glucose uptake relative to wild-type fat mass.
Txnip's regulation of PPARγ activity may account for several observations regarding energy balance, adipogenesis, whole-body glucose and lipid homeostasis, and insulin responsiveness. PPARγ is a central regulator of adipogenesis and lipogenesis, and participates in inflammation (30). Many lines of evidence have established PPARγ's role in integrating fat development, metabolic control, and insulin responsiveness (39–41). However, many striking inconsistencies are seen among various mouse PPARγ deletion models, hence a direct comparison with the Txnip-null phenotype is difficult. In addition, several observations here suggest Txnip's alteration of PPARγ function cannot be the sole determinant of the metabolic phenotype. For instance, PPARγ activation is not associated with fasting hypoglycemia, and its activation should suppress triglyceride and free fatty acid levels, however we observed elevated levels. Txnip has recently been shown to regulate PPARα expression and activity (42) and Foxo4 activity in vitro (43), suggesting that Txnip regulates multiple metabolic transcription factors to affect a composite metabolic picture. Txnip likely also more directly affects cellular redox balance and enzymatic functions that mediate triglyceride formation and insulin sensitivity. For example, previous studies show increased ratio of reduced to oxidized nicotinamide adenine dinucleotide (NADH/NAD+) levels in Txnip-null mice, which impair tricarboxylic acid cycle flux and promote fatty acid incorporation into triglycerides (26), whereas Hui et al. demonstrated Txnip deletion altered phosphatase and tensin homolog activity and insulin signaling (25). Further compound mutational studies with Txnip and the different transcriptional regulators of metabolism will be necessary to determine how Txnip regulates metabolism through these pathways, and in which tissues.
Txnip appears to regulate PPARγ by several mechanisms. First, Txnip is a negative regulator of PPARγ expression. PPARγ activation, in turn, suppresses Txnip expression, reflecting reciprocal feedback inhibition. Several other recent reports describe a similar relationship between Txnip and PPARγ (44,45). Second, Txnip expression blunts PPARγ activity. Recent evidence that Txnip also inhibits PPARα activity suggests its function is more generalized with respect to nuclear hormone receptors (42). The inhibition does not appear to be one of PPARγ ligand responsiveness. Both the PPRE reporter and in vitro differentiation data suggest PPARγ rosiglitazone dose responsiveness is not affected by changes in Txnip expression, hence we speculate that differences in PPARγ activity reflect a tonic change in PPARγ activation. This may reflect a Txnip-mediated shift in PPARγ coactivator recruitment rather than ligand binding. In an analogous example, Foxo4 was recently shown to directly interact with the coactivator p300 through redox-sensitive disulfide bond formation that mediated suppression of Foxo4 activity. The critical cysteine bridge was a thioredoxin target and was regulated by Txnip expression (43). We have been unable to demonstrate any direct interaction between Txnip and PPARγ using coimmunoprecipitation and glutathione S-transferase pull-down techniques (data not shown), hence we hypothesize Txnip regulates PPARγ through more indirect mechanisms. That enhanced PPARγ activation follows high-fat feeding may reflect diet-derived PPARγ-activating factors. In rodents, high-fat diet leads to elevations in PPARγ-activating free fatty acids (and triglyceride lipolysis in certain situations) that have been shown to generate PPARγ ligands and PPARγ activation (46,47). We speculate that HFD simultaneously activates PPARγ while inducing Txnip expression in WAT to represses PPARγ. In the absence of Txnip, high-fat feeding may permit PPARγ-activating factors to operate unopposed, promoting a PPARγ-activated increase in adipogenesis.
In summary, these studies demonstrate that Txnip is an important mediator of energy balance and nutritional sensing. Txnip protects against the insulin-resistant insult of high-fat caloric excess while simultaneously augmenting adipocyte storage capacity. Efforts to regulate Txnip function will shed further insight into metabolic disorders such as diabetes, and may represent a novel strategy for diabetes therapy.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the U.S. Public Health Service (P0–1 HL-048743 to R.T.L., R01 DK-40936 and P30 DK-45735 to G.I.S., K23 RR-17404 to V.T.S., and K08 HL-088977 to W.A.C.) and the Deutsche Forschungsgemeinschaft (German Research Foundation) to A.L.B., and by the Leadership Council for Improving Cardiovascular Care (LCIC) Future Leaders in Cardiovascular Medicine Award (sponsored by Schering-Plough) to W.A.C. G.I.S. is an investigator of the Howard Hughes Medical Institute. V.T.S. also receives support from the Veterans Administration Medical Center (West Haven, CT).
No potential conflicts of interest relevant to this article were reported.
We thank Evan Rosen for his critical insight into the development of this manuscript.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Sethi JK, Vidal-Puig AJ: Thematic review series: adipocyte biology: adipose tissue function and plasticity orchestrate nutritional adaptation. J Lipid Res 2007; 48: 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherer PE: Adipose tissue: from lipid storage compartment to endocrine organ. Diabetes 2006; 55: 1537–1545 [DOI] [PubMed] [Google Scholar]
- 3.Kim JK, Gavrilova O, Chen Y, Reitman ML, Shulman GI: Mechanism of insulin resistance in A-ZIP/F-1 fatless mice. J Biol Chem 2000; 275: 8456–8460 [DOI] [PubMed] [Google Scholar]
- 4.Petersen KF, Oral EA, Dufour S, Befroy D, Ariyan C, Yu C, Cline GW, DePaoli AM, Taylor SI, Gorden P, Shulman GI: Leptin reverses insulin resistance and hepatic steatosis in patients with severe lipodystrophy. J Clin Invest 2002; 109: 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Semple RK, Chatterjee VK, O'Rahilly S: PPAR gamma and human metabolic disease. J Clin Invest 2006; 116: 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, Jelicks LA, Mehler MF, Hui DY, Deshaies Y, Shulman GI, Schwartz GJ, Scherer PE: Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J Clin Invest 2007; 117: 2621–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luan B, Zhao J, Wu H, Duan B, Shu G, Wang X, Li D, Jia W, Kang J, Pei G: Deficiency of a beta-arrestin-2 signal complex contributes to insulin resistance. Nature 2009; 457: 1146–1149 [DOI] [PubMed] [Google Scholar]
- 8.Nikko E, Sullivan JA, Pelham HR: Arrestin-like proteins mediate ubiquitination and endocytosis of the yeast metal transporter Smf1. EMBO Rep 2008; 9: 1216–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nishiyama A, Matsui M, Iwata S, Hirota K, Masutani H, Nakamura H, Takagi Y, Sono H, Gon Y, Yodoi J: Identification of thioredoxin-binding protein-2/vitamin D(3) up-regulated protein 1 as a negative regulator of thioredoxin function and expression. J Biol Chem 1999; 274: 21645–21650 [DOI] [PubMed] [Google Scholar]
- 10.Bodnar JS, Chatterjee A, Castellani LW, Ross DA, Ohmen J, Cavalcoli J, Wu C, Dains KM, Catanese J, Chu M, Sheth SS, Charugundla K, Demant P, West DB, de Jong P, Lusis AJ: Positional cloning of the combined hyperlipidemia gene Hyplip1. Nat Genet 2002; 30: 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chutkow WA, Patwari P, Yoshioka J, Lee RT: Thioredoxin-interacting protein (Txnip) is a critical regulator of hepatic glucose production. J Biol Chem 2008; 283: 2397–2406 [DOI] [PubMed] [Google Scholar]
- 12.Hui TY, Sheth SS, Diffley JM, Potter DW, Lusis AJ, Attie AD, Davis RA: Mice lacking thioredoxin-interacting protein provide evidence linking cellular redox state to appropriate response to nutritional signals. J Biol Chem 2004; 279: 24387–24393 [DOI] [PubMed] [Google Scholar]
- 13.Junn E, Han SH, Im JY, Yang Y, Cho EW, Um HD, Kim DK, Lee KW, Han PL, Rhee SG, Choi I: Vitamin D3 up-regulated protein 1 mediates oxidative stress via suppressing the thioredoxin function. J Immunol 2000; 164: 6287–6295 [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, De Keulenaer GW, Lee RT: Vitamin D(3)-up-regulated protein-1 is a stress-responsive gene that regulates cardiomyocyte viability through interaction with thioredoxin. J Biol Chem 2002; 277: 26496–26500 [DOI] [PubMed] [Google Scholar]
- 15.Schulze PC, Yoshioka J, Takahashi T, He Z, King GL, Lee RT: Hyperglycemia promotes oxidative stress through inhibition of thioredoxin function by thioredoxin-interacting protein. J Biol Chem 2004; 279: 30369–30374 [DOI] [PubMed] [Google Scholar]
- 16.Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, Jensen CB, Krook A, Bjornholm M, Tornqvist H, Zierath JR, Ridderstrale M, Altshuler D, Lee RT, Vaag A, Groop LC, Mootha VK: TXNIP regulates peripheral glucose metabolism in humans. PLoS Med 2007; 4: e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patwari P, Higgins LJ, Chutkow WA, Yoshioka J, Lee RT: The interaction of thioredoxin with Txnip: evidence for formation of a mixed disulfide by disulfide exchange. J Biol Chem 2006; 281: 21884–21891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emilsson V, Thorleifsson G, Sainz J, Walters G, Gulcher J, Lamb J, Schadt E: Haplotypes in the human thioredoxin interacting protein homologue (ARRDC3) gene associated with obesity. Geneva: World Intellectual Property Organization, 2005 [Google Scholar]
- 19.Patwari P, Chutkow WA, Cummings K, Verstraeten VL, Lammerding J, Schreiter ER, Lee RT: Thioredoxin-independent regulation of metabolism by the alpha-arrestin proteins. J Biol Chem 2009; 284: 24996–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshioka J, Imahashi K, Gabel SA, Chutkow WA, Burds AA, Gannon J, Schulze PC, MacGillivray C, London RE, Murphy E, Lee RT: Targeted deletion of thioredoxin-interacting protein regulates cardiac dysfunction in response to pressure overload. Circ Res 2007; 101: 1328–1338 [DOI] [PubMed] [Google Scholar]
- 21.McLaughlin T, Sherman A, Tsao P, Gonzalez O, Yee G, Lamendola C, Reaven GM, Cushman SW: Enhanced proportion of small adipose cells in insulin-resistant vs insulin-sensitive obese individuals implicates impaired adipogenesis. Diabetologia 2007; 50: 1707–1715 [DOI] [PubMed] [Google Scholar]
- 22.Samuelson LC, Metzger JM: Isolation and freezing of primary mouse embryonic fibroblasts (MEF) for feeder plates. Cold Spring Harb Protoc 2006; 4482. [DOI] [PubMed] [Google Scholar]
- 23.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM: Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell 1998; 93: 229–240 [DOI] [PubMed] [Google Scholar]
- 24.Packard GC, Boardman TJ: The use of percentages and size-specific indices to normalize physiological data for variation in body size: wasted time, wasted effort? Comp Biochem Physiol A Mol Integr Physiol 1999; 122: 37–44 [Google Scholar]
- 25.Hui ST, Andres AM, Miller AK, Spann NJ, Potter DW, Post NM, Chen AZ, Sachithanantham S, Jung DY, Kim JK, Davis RA: Txnip balances metabolic and growth signaling via PTEN disulfide reduction. Proc Natl Acad Sci U S A 2008; 105: 3921–3926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheth SS, Castellani LW, Chari S, Wagg C, Thipphavong CK, Bodnar JS, Tontonoz P, Attie AD, Lopaschuk GD, Lusis AJ: Thioredoxin-interacting protein deficiency disrupts the fasting-feeding metabolic transition. J Lipid Res 2005; 46: 123–134 [DOI] [PubMed] [Google Scholar]
- 27.Reshef L, Olswang Y, Cassuto H, Blum B, Croniger CM, Kalhan SC, Tilghman SM, Hanson RW: Glyceroneogenesis and the triglyceride/fatty acid cycle. J Biol Chem 2003; 278: 30413–30416 [DOI] [PubMed] [Google Scholar]
- 28.Nolan JJ, Ludvik B, Beerdsen P, Joyce M, Olefsky J: Improvement in glucose tolerance and insulin resistance in obese subjects treated with troglitazone. N Engl J Med 1994; 331: 1188–1193 [DOI] [PubMed] [Google Scholar]
- 29.Bendixen AC, Shevde NK, Dienger KM, Willson TM, Funk CD, Pike JW: IL-4 inhibits osteoclast formation through a direct action on osteoclast precursors via peroxisome proliferator-activated receptor gamma 1. Proc Natl Acad Sci U S A 2001; 98: 2443–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tontonoz P, Spiegelman BM: Fat and beyond: the diverse biology of PPARgamma. Annu Rev Biochem 2008; 77: 289–312 [DOI] [PubMed] [Google Scholar]
- 31.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM: A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998; 92: 829–839 [DOI] [PubMed] [Google Scholar]
- 32.Saleh MC, Wheeler MB, Chan CB: Uncoupling protein-2: evidence for its function as a metabolic regulator. Diabetologia 2002; 45: 174–187 [DOI] [PubMed] [Google Scholar]
- 33.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM: mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev 1994; 8: 1224–1234 [DOI] [PubMed] [Google Scholar]
- 34.Oka S, Liu W, Masutani H, Hirata H, Shinkai Y, Yamada S, Yoshida T, Nakamura H, Yodoi J: Impaired fatty acid utilization in thioredoxin binding protein-2 (TBP-2)-deficient mice: a unique animal model of Reye syndrome. Faseb J 2006; 20: 121–123 [DOI] [PubMed] [Google Scholar]
- 35.Chen J, Hui ST, Couto FM, Mungrue IN, Davis DB, Attie AD, Lusis AJ, Davis RA, Shalev A: Thioredoxin-interacting protein deficiency induces Akt/Bcl-xL signaling and pancreatic beta-cell mass and protects against diabetes. FASEB J 2008; 22: 3581–3594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Savage DB, Petersen KF, Shulman GI: Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol Rev 2007; 87: 507–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shulman GI: Cellular mechanisms of insulin resistance. J Clin Invest 2000; 106: 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan CY, Vidal-Puig A: Adipose tissue expandability: the metabolic problems of obesity may arise from the inability to become more obese. Biochem Soc Trans 2008; 36: 935–940 [DOI] [PubMed] [Google Scholar]
- 39.Agarwal AK, Garg A: A novel heterozygous mutation in peroxisome proliferator-activated receptor-gamma gene in a patient with familial partial lipodystrophy. J Clin Endocrinol Metab 2002; 87: 408–411 [DOI] [PubMed] [Google Scholar]
- 40.Barroso I, Gurnell M, Crowley VE, Agostini M, Schwabe JW, Soos MA, Maslen GL, Williams TD, Lewis H, Schafer AJ, Chatterjee VK, O'Rahilly S: Dominant negative mutations in human PPARgamma associated with severe insulin resistance, diabetes mellitus and hypertension. Nature 1999; 402: 880–883 [DOI] [PubMed] [Google Scholar]
- 41.Hegele RA, Cao H, Frankowski C, Mathews ST, Leff T: PPARG F388L, a transactivation-deficient mutant, in familial partial lipodystrophy. Diabetes 2002; 51: 3586–3590 [DOI] [PubMed] [Google Scholar]
- 42.Oka SI, Yoshihara E, Bizen-Abe A, Liu W, Watanabe M, Yodoi J, Masutani H: Thioredoxin binding protein-2 (TBP-2)/Txnip is a critical regulator of insulin secretion and PPAR function. Endocrinology 2009; 150: 1225–1234 [DOI] [PubMed] [Google Scholar]
- 43.Dansen TB, Smits LM, van Triest MH, de Keizer PL, van Leenen D, Koerkamp MG, Szypowska A, Meppelink A, Brenkman AB, Yodoi J, Holstege FC, Burgering BM: Redox-sensitive cysteines bridge p300/CBP-mediated acetylation and FoxO4 activity. Nat Chem Biol 2009; 5: 664–672 [DOI] [PubMed] [Google Scholar]
- 44.Billiet L, Furman C, Larigauderie G, Copin C, Page S, Fruchart JC, Brand K, Rouis M: Enhanced VDUP-1 gene expression by PPARgamma agonist induces apoptosis in human macrophage. J Cell Physiol 2008; 214: 183–191 [DOI] [PubMed] [Google Scholar]
- 45.Qi W, Chen X, Holian J, Tan CY, Kelly DJ, Pollock CA: Transcription factors Krüppel-like factor 6 and peroxisome proliferator-activated receptor-{gamma} mediate high glucose-induced thioredoxin-interacting protein. Am J Pathol 2009; 175: 1858–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmed W, Ziouzenkova O, Brown J, Devchand P, Francis S, Kadakia M, Kanda T, Orasanu G, Sharlach M, Zandbergen F, Plutzky J: PPARs and their metabolic modulation: new mechanisms for transcriptional regulation? J Intern Med 2007; 262: 184–198 [DOI] [PubMed] [Google Scholar]
- 47.Jump DB, Clarke SD: Regulation of gene expression by dietary fat. Annu Rev Nutr 1999; 19: 63–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.