Abstract
OBJECTIVE
Multiple type 1 diabetes susceptibility genes have now been identified in both humans and mice, yet mechanistic understanding of how they impact disease pathogenesis is still minimal. We have sought to dissect the cellular basis for how the highly protective mouse Idd9 region limits the expansion of autoreactive CD8+ T-cells, a key cell type in destruction of the islets.
RESEARCH DESIGN AND METHODS
We assess the endogenous CD8+ T-cell repertoire for reactivity to the islet antigen glucose-6-phosphatase–related protein (IGRP). Through the use of adoptively transferred T-cells, bone marrow chimeras, and reconstituted severe combined immunodeficient mice, we identify the protective cell types involved.
RESULTS
IGRP-specific CD8+ T-cells are present at low frequency in the insulitic lesions of Idd9 mice and could not be recalled in the periphery by viral expansion. We show that Idd9 genes act extrinsically to the CD8+ T-cell to prevent the massive expansion of pathogenic effectors near the time of disease onset that occurs in NOD mice. The subregions Idd9.2 and Idd9.3 mediated this effect. Interestingly, the Idd9.1 region, which provides significant protection from disease, did not prevent the expansion of autoreactive CD8+ T-cells. Expression of Idd9 genes was required by both CD4+ T-cells and a nonlymphoid cell to induce optimal tolerance.
CONCLUSIONS
Idd9 protective alleles are associated with reduced expansion of IGRP-specific CD8+ T-cells. Intrinsic expression of protective Idd9 alleles in CD4+ T-cells and nonlymphoid cells is required to achieve an optimal level of tolerance. Protective alleles in the Idd9.2 congenic subregion are required for the maximal reduction of islet-specific CD8+ T-cells.
The nonobese diabetic (NOD) mouse model of type 1 diabetes has proven to be extremely valuable in the study of this chronic disease. Tolerance of both the CD8+ and CD4+ T-cell subsets toward islet β-cells is perturbed in the NOD mouse resulting in β-cell destruction. This loss of tolerance is determined by the more than 18 genetic regions, termed Idd regions, identified as contributing to diabetes susceptibility (1). The multiplicity of genes contributing to disease progression in both mice and humans most likely reflects several checkpoints of tolerance that must be bypassed for overt disease to occur.
Expression of the protective B10 alleles of Idd9 in NOD.B10 Idd9 congenic mice (Idd9 mice) reduces the incidence of diabetes from ∼80% in the NOD parental strain to ∼4% in Idd9 mice (2). Interestingly, Idd9 mice still develop extensive insulitis and produce islet-specific autoantibodies, indicating that islet-specific tolerance is not complete and that a late checkpoint of tolerance may be functioning to constrain the autoimmune response (3). The Idd9 region on chromosome 4 is composed of at least three separate intervals, Idd9.1, Idd9.2, and Idd9.3 (2). Fine-mapping, functional, and sequence data from the Idd9.3 region suggest that the gene encoding CD137 (4–1BB) is the most likely candidate gene for Idd9.3 (2,4).
Several studies have examined the contribution of Idd9 genes to disease protection. We have recently shown that CD8+ T-cell tolerance to transgenic hemagglutinin expressed under the rat insulin promoter (InsHA) is restored in Idd9-InsHA mice (5). Unlike mice expressing protective Idd3 and Idd5 alleles, tolerance was not restored upon initial activation of autoreactive CD8+ T-cells in the pancreatic draining lymph nodes (PcLNs) but at an unknown later time. When Idd9 genes were expressed by T-cells from BDC2.5 transgenic mice, the ability of these cells to transfer diabetes into NOD–severe combined immunodeficient (SCID) recipients was reduced (6). Idd9.1 was recently reported to control the suppressive activity of regulatory CD4+ T-cells (7). Idd9 may also contribute to protection of islet β-cells from cytotoxic T-lymphocyte (CTL) killing (8). Idd9/11 (a B6-derived region overlapping Idd9.1 and Idd9.2) was also reported to diminish the diabetogenic potential of CD4+ T-cells (9), as well as normalizing tolerance of pathogenic B-cells (10). In the present study, restored CD8+ T-cell tolerance to the islet-expressed antigen, islet-specific glucose-6-phosphatase–related protein (IGRP), by Idd9 genes was assessed. We have focused on the identification of the cell types that must express Idd9 genes and on the Idd9 subregions responsible for CD8+ T-cell tolerance.
RESEARCH DESIGN AND METHODS
Mice.
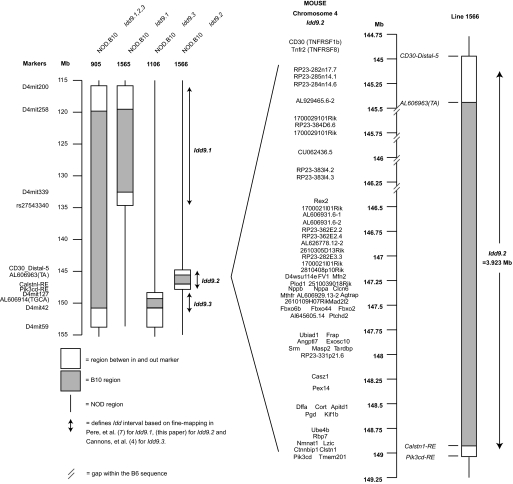
Experimental procedures were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals (protocol number 09–0074). Clone-4 T-cell receptor (TCR) NOD mice and NOD-CD45.2 congenic mice were previously described (5,11). Thy1.1+NOD-8.3-SCID mice were generated by intercrossing NOD-8.3 TCR transgenic mice (The Jackson Laboratory), with NOD-SCID mice (Taconic) and NOD-Thy1.1 mice (5). NOD-MrkTac mice (Taconic) and B10.D2 mice (The Scripps Research Institute breeding colony) were purchased. The NOD.B10 Idd9 congenic strain (Taconic line 905), which contains a continuous B10-derived DNA segment including type 1 diabetes–protective alleles Idd9.1, Idd9.2, and Idd9.3 (Fig. 1), has been previously described (5). The gene content of the congenic intervals present in NOD-Idd9.1 (line 1565) and NOD-Idd9.3 (line 1106) mice has been described previously (4,7), and NOD-Idd9.2 (line 1566) is depicted in Fig. 1. Idd9 (line 905) mice were intercrossed with NOD-SCID−/− mice to generate Idd9-SCID−/− mice, and NOD-Thy1.1+ mice to generate Idd9-Thy1.1+ mice.
FIG. 1.
Map of the Idd9 congenic intervals contained in lines 905, 1565, 1566, and 1106 and the gene content of Idd9.2. Gaps in the assembly of the B6 sequence are marked.
Virus.
Recombinant vaccinia virus expressing the H-2Kd–restricted epitope VYLKTNVFL, amino acid residues 206–214 of murine IGRP (Vac-IGRP), was generated as described (12). Vac-KdHA has been previously described (5). Mice were infected intraperitoneally with 1 × 107 plaque-forming units (pfu) of virus and CD8+ T-cell responses measured in the spleen 7 days later.
Preparation and adoptive transfer of T cells.
TCR transgenic 8.3 or Clone-4 CD8+ T-cells were isolated from the spleen and lymph nodes of TCR transgenic mice (6–8 weeks of age) using a CD8+ T-cell enrichment kit (BD Biosciences). Recipient female mice (6–10 weeks) were injected intravenously with 1 × 104 purified 8.3 or Clone-4 cells mixed with 2 × 106 total NOD or Idd9 spleen cells (syngeneic with the recipient mouse) as carrier cells. SCID mice were reconstituted with spleen and lymph node cell populations derived from 3-to 4-week-old donor mice. Total CD8+ T-cells and CD4+ T-cells were purified with a CD8+ T-enrichment kit and CD4+ T-enrichment kit (BD Biosciences) using double the recommended amount of reagents. In some experiments, the CD4+ T-cells were further purified by sorting for CD4+ Thy1.2+ cells with a FACSAria (BD Biosciences). CD4+ T-cells and CD8+ T-cells were depleted with CD4+ microbeads and CD8+ microbeads (Miltenyi) using double the recommended amount of beads.
In vivo CTLs.
Mice were injected intravenously with 1 × 107 of a 50:50 mixture of carboxyfluorescein succinimidyl esterHigh (CFSEHigh)–IGRP206–214–loaded NOD splenocytes and CFSElow control NOD splenocytes as described (13).
Flow cytometry.
Isolation of islet lymphocytes was performed as previously described (5). CD8+ T-cells were stained with H-2Kd–IGRP206–214–phycoerythrin (PE) tetramers (National Institute of Allergy and Infectious Diseases MHC Tetramer core facility) for 15 min at room temperature, followed by staining with anti–CD8–fluorescein isothiocyanate (FITC) at 4°C for 15 min. Cells were acquired with either a FACSCalibur or LSRII (both from BD Biosciences) and analyzed with FlowJo software.
Bone marrow chimeras.
Bone marrow cells were prepared as previously described (11). Briefly, mice were irradiated (1,200 rad) and reconstituted with 7–8 × 106 bone marrow cells that had been depleted of Thy1+ cells by complement-mediated lysis. Mice were further depleted of residual T-cells with anti-CD4 and anti-CD8 depleting antibodies.
Statistical analysis.
Differences between groups were tested via the Mann-Whitney test and median values are reported in the text unless otherwise noted, with first to third interquartile ranges (IQRs). Mean values are reported as mean ± SD.
RESULTS
Idd9 congenic mice have reduced numbers of IGRP-specific CD8+ T-cells.
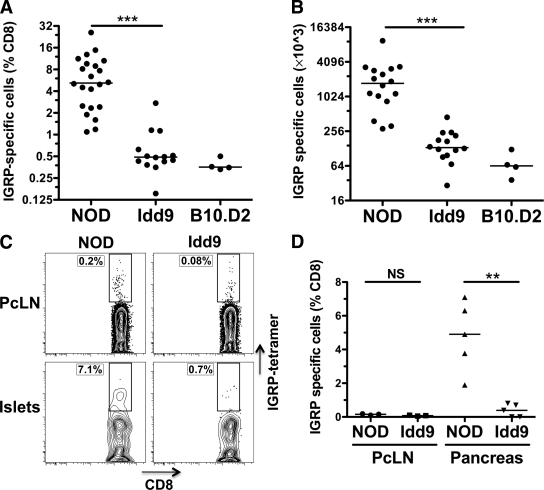
To assess whether tolerance to the endogenous islet antigen IGRP was restored in Idd9 mice (line 905, Fig. 1), we infected NOD and Idd9 female mice with a vaccinia virus encoding the H-2Kd-IGRP206–214 epitope (Vac-IGRP) (Fig. 2A and B). The frequency of IGRP-specific CD8+ T-cells in Idd9 mice (0.5%) was significantly reduced compared with NOD (5.2%) and was similar to that found in B10.D2 control mice (0.4%), indicating that IGRP-specific CD8+ T-cells had been eliminated or were unresponsive in Idd9 and B10.D2 mice (Fig. 2A). The total number of IGRP-specific CD8+ T-cells was also significantly reduced in Idd9 and B10.D2 mice compared with NOD (Fig. 2B). We demonstrated previously that both NOD and Idd9 mice develop robust responses to a non–self-antigen (HA) after infection with Vac-KdHA (5). Furthermore, transferred naive HA-specific Clone-4 cells expand with equal efficiency in NOD and Idd9 mice after infection with Vac-KdHA (supplementary Fig. 1, available in an online appendix at http://diabetes.diabetesjournals.org/content/early/2010/03/10/db09-1801/suppl/DC1). Thus, the reduction in IGRP-specific CD8+ T-cell responses does not signify a comprehensive difference in the viral response between the strains, but rather an antigen-specific response.
FIG. 2.
The frequency of IGRP-specific CD8+ T-cells is significantly reduced in Idd9 congenic mice. NOD, NOD-Idd9, and B10.D2 10- to 14-week-old female mice were infected with 1 × 107 pfu Vac-IGRP i.p. On day 7 after infection, spleens were analyzed by fluorescence-activated cell sorter (FACS) for CD8+ IGRP-tetramer+ cells. Pooled results from three experiments are shown; horizontal lines show median value. A: Frequency P < 0.0001 (***) of NOD (5.2%, IQR: 2.4–9.4%) vs. Idd9 (0.5%, IQR 0.4–0.6%) using the Mann-Whitney test. B: Total cell number P < 0.0001 (***) of NOD (1,731 × 103, IQR 996–2,939) vs. Idd9 (133 × 103, IQR 103–205) using the Mann-Whitney test. C and D: Islet-infiltrating leukocytes were isolated from the pancreas of NOD and Idd9 10- to 14-week-old female mice (three pooled mice). Pancreas leukocytes and PcLN cells were stained with IGRP-H2Kd tetramer anti-CD8+ and propidium iodide. C: Individual FACS plots. D: Collective data from three experiments. Line depicts mean value. PcLN, NOD (0.15 ± 0.04%) vs. Idd9 (0.08 ± 0.03%), P = 0.06; pancreas, NOD (4.8 ± 2.1%) vs. Idd9 (0.4 ± 0.4%), P = 0.0015 (**, mean ± SD, P value calculated using unpaired t test). NS, not significant.
This difference in frequency of IGRP-specific CD8+ T-cells was also detectable in the islets prior to expansion of the IGRP-specific population with virus (Fig. 2C and D). IGRP-specific CD8+ T-cells were found infiltrating the islets of NOD but not Idd9 mice (4.8% vs. 0.4% of CD8+ cells, respectively, P = 0.0015). A small reduction in the frequency of tetramer+ cells was also observed in the PcLN of Idd9 (0.08%) compared with NOD mice (0.15%, Fig. 2B); however, the frequencies were extremely low in the PcLN and this difference did not reach significance (P = 0.06). One previous study reported a reduction in the frequency of CD8+ T-cells in the islet infiltrates of Idd9 mice (8); however, we did not observe this difference (supplementary Fig. 2). We conclude that NOD and Idd9 mice have a profoundly altered CD8+ T-cell functional repertoire to IGRP, observable both with and without prior expansion by Vac-IGRP infection.
Idd9.2 and Idd9.3 region genes contribute to restored CD8+ T-cell tolerance.
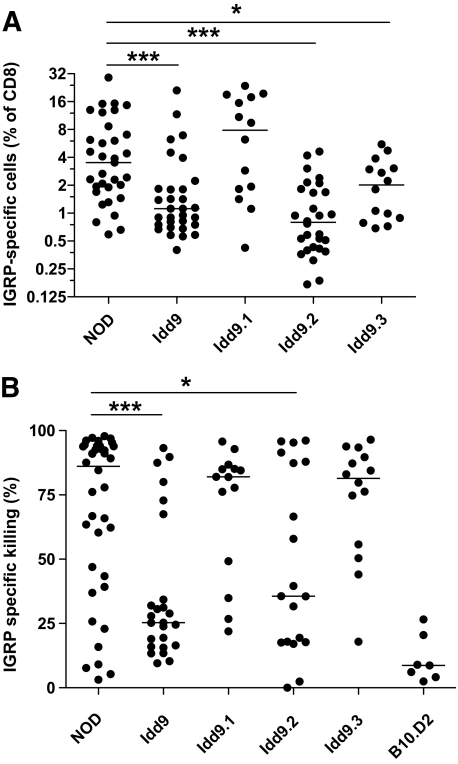
The Idd9 region has been reported to contain at least three separate subregions (Idd9.1, Idd9.2, and Idd9.3, Fig. 1). Congenic strains having only one of the protective regions are each partially protected from diabetes (2,4,7). To determine which of these regions contributes to restored tolerance to islet IGRP, subcongenic mice were infected with Vac-IGRP and the frequency of IGRP-specific tetramer+ cells was assessed (Fig. 3A). As expected from the results in Fig. 2A, Idd9 mice were highly tolerant compared with NOD mice. Idd9.1 mice contained very high percentages of IGRP-specific cells (7.9%). Idd9.2 mice exhibited a greatly reduced frequency of IGRP-specific cells (0.8%) that was similar to Idd9 mice (1.1%). Idd9.3 mice showed an intermediate frequency (2.0%). Therefore, at least two genes within the Idd9 region contribute to tolerance of IGRP-specific CD8+ T-cells and these are within the Idd9.2 and Idd9.3 regions.
FIG. 3.
Idd9.2 and Idd9.3 genes contribute to restored tolerance of IGRP-specific CD8+ T-cells. NOD, Idd9, Idd9.1, Idd9.2, and Idd9.3 mice (10–14 weeks of age) were infected with Vac-IGRP and 7 days later (A) IGRP-tetramer+ CD8+ T-cells were measured in the spleen. Pooled data from three experiments are shown. The median line is shown for each group. NOD (3.5%, IQR 1.9–7.0%) vs. Idd9.2 (0.8%, IQR 0.4–1.7%), P < 0.0001 (***); and NOD vs. Idd9.3 (2.0%, IQR 0.9–3.0%), P = 0.047 (*), using the Mann-Whitney test. B: Infected and naive control mice were injected with CFSEHigh IGRP206–214–loaded NOD (or B10.D2) splenocytes and CFSElow control NOD (or B10.D2) splenocytes. Killing was assessed 16 h later in the spleen by FACS. Specific killing was calculated by the formula: 100 − [(percentage of CFSEhigh/percentage of CFSElow)/Ratio naive) × 100]. Pooled data from three experiments are shown. Line depicts median value. NOD (85%, IQR 43–94%) vs. Idd9 (27%, IQR 16–43%), P = 0.0004 (***); NOD vs. Idd9.2 (36%, IQR 18–88%), P = 0.027 (*).
We also assessed the effector function of the IGRP-specific CD8+ T-cells that expanded after infection with Vac-IGRP by measuring in vivo CTL activity (Fig. 3B). NOD mice had a very high efficiency of IGRP-specific killing (85%), whereas Idd9 mice had low levels of killing (27%). Idd9.2 mice were intermediate between NOD and Idd9 (36%), but neither Idd9.1 (82%) nor Idd9.3 (83%) showed any reduction. We interpreted this to mean that mice with an intermediate frequency of IGRP-specific tetramer+ CD8+ T-cells (such as Idd9.3) had sufficient numbers of CTLs to efficiently kill the available in vivo targets. These data further verify that among the three Idd9 regions, Idd9.2 has the greatest capacity to protect from the expansion of pathogenic islet-specific CTLs.
Idd9 genes expressed in the host prevent the late expansion of autoreactive CD8+ T-cells in the periphery.
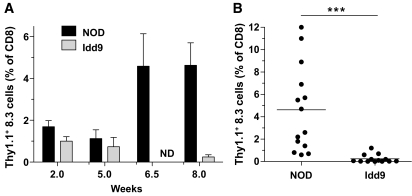
Successful peripheral tolerance of autoreactive CD8+ T-cells in Idd9 mice could be due to expression of tolerance-inducing Idd9 genes either by the CD8+ T-cell or by other cells, or both. We used TCR transgenic NOD-8.3 CD8+ T-cells, specific for IGRP206–214 to assess this issue (14). NOD-Thy1.1+ 8.3 cells (1 × 104) were transferred into either NOD or Idd9 recipients, and 2, 5, 6.5, or 8 weeks later the mice were infected with Vac-IGRP and the frequency of Thy1.1+ 8.3 cells was determined (Fig. 4). At 2 and 5 weeks, no significant difference in the frequency of 8.3 cells was observed between NOD and Idd9 mice (Fig. 4A). However, 6.5 and 8 weeks later the 8.3 T-cells expanded in the NOD mice (mean 4.6 ± 3.9% at 8 weeks) but were not detected in Idd9 (mean 0.2 ± 0.4% at 8 weeks, Fig. 4A and B).
FIG. 4.
Deletion of NOD-8.3 CD8+ T-cells is restored in Idd9 mice. NOD-Thy1.1+ 8.3 T-cells (1 × 104) were transferred into 7- to 10-week-old NOD or Idd9 mice. A: After 2, 5, 6.5 or 8 weeks, mice were infected with Vac-IGRP, and the frequency of Thy1.1+ CD8+ T-cells was determined in the spleen 7 days later. Data from between 3 and 14 mice are pooled per time point. Mean ± SEM is shown. ND, not done. B: Individual mice from the 8-week time point are shown. NOD vs. Idd9 at 8 weeks, P < 0.0001 (***) using the Mann-Whitney test. The mean line is shown for each group.
To verify that NOD CD8+ T-cells transferred into Idd9 mice were not deleted because of an allodifference emanating from the Idd9 congenic interval, we transferred NOD HA-specific Thy1.1+ Clone-4 CD8+ T-cells (1 × 104) into NOD and Idd9 mice. After 2, 5, 6.5, or 8 weeks, the remaining Clone-4 cells were expanded with Vac-KdHA. A similar proportion of Clone-4 cells expanded in both NOD and Idd9 mice at all time points (supplementary Fig. 1), indicating that NOD CD8+ T-cells are not rejected in Idd9 mice.
Intrinsic expression of protective Idd9 alleles in CD8+ T-cells does not establish CD8+ T-cell tolerance.
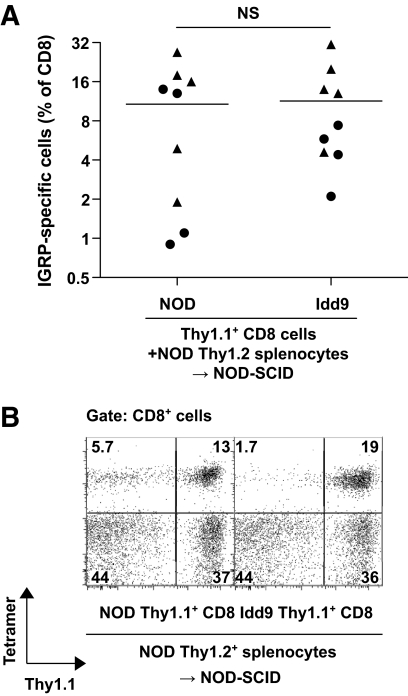
The fact that NOD-8.3 CD8+ T-cells were tolerized in Idd9 hosts suggested that tolerance was caused by expression of Idd9 genes in the host. It was also possible that intrinsic expression of Idd9 genes by CD8+ T-cells contributed to tolerance. To test this possibility, CD8+ T-cells from 3-week-old Idd9-Thy1.1+ mice (4 × 106) were purified and adoptively transferred into NOD-SCID hosts together with CD8+-depleted (90% reduction) NOD-Thy1.2+ splenocytes (30 × 106), from 3-week-old mice. Splenocytes from 3-week-old NOD and Idd9 mice contained only background frequencies of IGRP-specific CD8+ T-cells as assessed by tetramer staining. A second group of mice was prepared similarly but with CD8+ T-cells from NOD-Thy1.1+ donors. After 10 weeks, the mice were infected with Vac-IGRP and the presence of IGRP-tetramer binding CD8+ T-cells determined (Fig. 5A and B). In both groups of mice, approximately half of the CD8+ T-cells were Thy1.1+ at the conclusion of the experiment (supplementary Fig. 3A). We also confirmed that the majority of the CD4+ T-cell population was of NOD-Thy1.2+ origin (supplementary Fig. 3B). Thus, Idd9-derived cells contributed significantly only to the CD8+ T-cell population and not to other cell types. When Idd9-Thy1.1+ CD8+ T-cells were mixed with NOD-Thy1.2+ spleen and lymph node cells, a high frequency of IGRP-specific CD8+ T-cells expanded (Fig. 5A). In both groups, the IGRP-specific cells were derived from both the Thy1.1+ and Thy1.2+ populations, as shown in Fig. 5B. Although individual mice differed in whether the IGRP+ cells were predominantly derived from the Thy1.1+ or Thy1.2+ population, the overall frequency of IGRP-specific cells was comparable between the groups (Fig. 5A). Thus, the peripheral repertoire of 3-week-old Idd9 mice contains autoreactive IGRP-specific CD8+ T-cells that are able to expand and survive when transferred into mice that express susceptible NOD alleles on all other cell types. These results directly demonstrate that intrinsic expression of Idd9 genes within the CD8+ T-cells does not prevent their expansion in a NOD host.
FIG. 5.
Intrinsic expression of Idd9 genes within the CD8+ T-cell does not mediate CD8+ tolerance by Idd9 genes. Purified NOD or Idd9 Thy1.1+ CD8+ T-cells (4 × 106) from 3-week-old donor mice were cotransferred with partially (90%) CD8+ T-cell–depleted Thy1.2+ NOD spleen and LN cells (30 × 106) into NOD-SCID mice. After 10 weeks, mice were infected with Vac-IGRP and IGRP-tetramer+ CD8+ T-cells measured in the spleen 7 days later. A: Frequency of IGRP-specific cells among all CD8+ T-cells; pooled data from two experiments are shown and indicated by circles and triangles; horizontal lines depict mean values. B: Thy1.1 and IGRP-specific tetramer staining of CD8+ T-cells; one representative animal shown.
Idd9 bone marrow–derived cells are sufficient to maintain CD8+ T-cell tolerance in a NOD host.
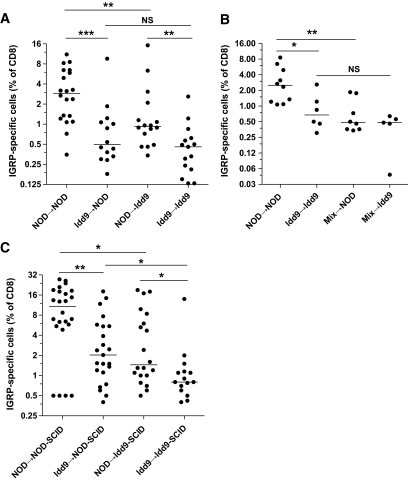
Radiation chimeras were produced to determine which host cell types must express Idd9 genes for maintenance of tolerance. Thy1.2+ Idd9 or NOD recipients were irradiated and reconstituted with Thy1.1+ Idd9 or NOD bone marrow cells and rested for 12 weeks. Reconstitution with Thy1.1+ donor T-cells varied between 71 and 88%, with no difference observed in reconstitution by NOD versus Idd9 bone marrow (supplementary Fig. 4). After infection with Vac-IGRP, the frequency of tetramer+ cells in the NOD→NOD chimeras was significantly higher than that found in the Idd9→Idd9 chimeras (2.9 vs. 0.5%, Fig. 6A). When Idd9 bone marrow was used to reconstitute NOD hosts, IGRP-specific tolerance was well maintained compared with NOD→NOD (0.5%, Fig. 6A). Thus, an Idd9 hematopoietic cell is able to restore CD8+ T-cell tolerance in an NOD host. NOD→Idd9 chimeric mice (0.9%) had significantly fewer IGRP-specific cells than NOD→NOD, yet significantly more than Idd9→Idd9 (Fig. 6A). This may be due to tolerance by expression of Idd9 genes within a host parenchymal cell, or the 20% residual Idd9 hematopoietic cells may have a sufficiently strong dominant effect to afford some tolerance, or both.
FIG. 6.
Expression of Idd9 genes by both hematopoietic cells and nonlymphocytes contributes to CD8+ T-cell tolerance. Recipient female Thy1.2+ mice were irradiated with 1200 rad and reconstituted with 7 × 106 T-cell–depleted bone marrow cells from Thy1.1+ donors (A) or a 50:50 mix of NOD and Idd9 bone marrow (B). After 12 weeks, the mice were infected with 1 × 107 pfu Vac-IGRP. Splenocytes were stained 7 days later with anti-CD8-FITC and IGRP-tetramer-PE. Pooled data from three experiments are shown. Horizontal line depicts median value. A: NOD→NOD vs. Idd9→NOD P = 0.0006 (***), NOD→NOD vs. NOD→Idd9 P = 0.006 (**), Idd9→NOD vs. Idd9→Idd9 P = 0.24, NOD→Idd9 vs. Idd9→Idd9 P = 0.004 (**), NOD→NOD vs. Idd9→Idd9 P < 0.0001 (Mann-Whitney test). B: NOD→NOD vs. mix→NOD P = 0.005 (**), NOD→NOD vs. Idd9→Idd9 P = 0.016 (Mann-Whitney test). C: NOD-SCID and Idd9-SCID mice were reconstituted with 2 × 107 spleen and LN cells from 3- to 4-week-old NOD or Idd9 mice. After 10 weeks, the mice were infected with Vac-IGRP, and IGRP-tetramer+ CD8+ T-cells were measured in the spleen 7 days later. Pooled data from three experiments are shown.
To determine whether Idd9 hematopoietic cells were dominant for tolerance, chimeras were made as described above, but a 50:50 mixture of NOD:Idd9 (or NOD-Thy1.1+:Idd9) bone marrow was used to reconstitute NOD or Idd9 irradiated hosts (Fig. 6B). The mixed chimeras in both hosts maintained IGRP-specific CD8+ T-cell tolerance well and were not significantly different from each other or Idd9→Idd9 control chimeras. We concluded that the mechanism of tolerance established by Idd9 hematopoietic cells is dominant as it overrides the presence of cells expressing NOD genes in the same animal.
Expression of protective Idd9 alleles by a nonlymphocyte population contributes to restoration of CD8+ T-cell tolerance.
To learn more about the cell types responsible for tolerance induction, Idd9-SCID or NOD-SCID mice were reconstituted with total spleen and lymph node cells isolated from 3- to 4-week-old NOD or Idd9 donor mice. After 10 weeks of reconstitution, we infected the mice with Vac-IGRP and assessed the spleen for IGRP-tetramer+ cells (Fig. 6C). NOD→NOD-SCID reconstituted mice had many IGRP-specific CD8+ T-cells (12.9%), significantly more than NOD→Idd9-SCID reconstituted mice (1.5%). Idd9→Idd9-SCID reconstituted mice had very few IGRP-tetramer+ cells (0.8%, Fig. 6C). This frequency was significantly increased in Idd9→NOD-SCID reconstituted mice (1.5%). A comparison of NOD-SCID mice reconstituted with NOD or Idd9 spleen and lymph node cells revealed that this population contains at least one cell type contributing to tolerance (P = 0.01). Likewise, Idd9-SCID mice reconstituted with NOD cells were less tolerant than when reconstituted with Idd9 cells (P = 0.04). Thus, expression of susceptible NOD alleles in both the donor cells and a nonlymphocyte host cell (either a parenchymal cell or a nonlymphocyte hematopoietic cell, e.g., an antigen-presenting cell) inhibits tolerance induction by protective Idd9 alleles.
Expression of protective Idd9 alleles in CD4+ T-cells contributes to restoration of CD8+ T-cell tolerance.
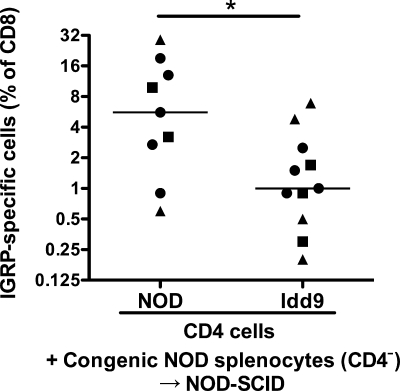
A likely candidate for an Idd9-expressing lymphoid cell that restores tolerance in SCID recipients is the CD4+ T-cell. To test this directly, we reconstituted NOD-SCID mice with highly purified CD4+ T-cells from 4-week-old NOD or Idd9 mice cotransferred with CD4-depleted NOD spleen and lymph node cells expressing a congenic allomarker (either Thy1.1 or CD45.2). At 10 weeks after reconstitution, the CD4+ T-cell population comprised a mean of 70 ± 25% of cells derived from the purified NOD or Idd9 CD4+ noncongenic population (supplementary Fig. 5A). The CD8+ T-cell population, however, was primarily derived from the NOD congenic splenocyte population (supplementary Fig. 5B). Thus, Idd9-derived cells contributed significantly to the CD4+ T-cell population but not to other cell types. The mice were infected with Vac-IGRP and IGRP-specific CD8+ T-cells assessed (Fig. 7). Control mice reconstituted with NOD CD4+ T-cells mixed with CD4+-depleted NOD spleen and lymph node cells developed high frequencies of IGRP-specific CD8+ T-cells (5.6%, IQR: 2.7–13%). In contrast, Idd9 CD4+ T-cells prevented the loss of CD8+ T-cell tolerance to islet IGRP (1%, IQR: 0.7–2.1%). Tolerance occurred even in mice in which only 50–60% of the CD4+ T-cells expressed protective alleles of Idd9 genes, which is consistent with the dominant tolerance observed in the mixed chimeric mice (Fig. 6B).
FIG. 7.
CD4+ T-cells expressing Idd9 genes are sufficient to restore CD8+ tolerance in NOD mice. Purified NOD or Idd9 Thy1.2/CD45.1 CD4+ T-cells (6 × 106) were cotransferred with 1 × 107 NOD-Thy1.1 or NOD-CD45.2 congenic CD4+-depleted spleen and LN cells, all from 4-week-old donor mice, into NOD-SCID mice. After 10 weeks, mice were infected with Vac-IGRP and IGRP-tetramer+ CD8+ T-cells measured in the spleen 7 days later. Pooled data from three experiments are shown; NOD (5.6%, IQR 2.7–13%) vs. Idd9 (1%. IQR 0.7–2.1%) P = 0.027 (*). In the second (circles) and third (squares) experiments the CD4+ T-cells were purified by FACS sorting. Horizontal line depicts median value.
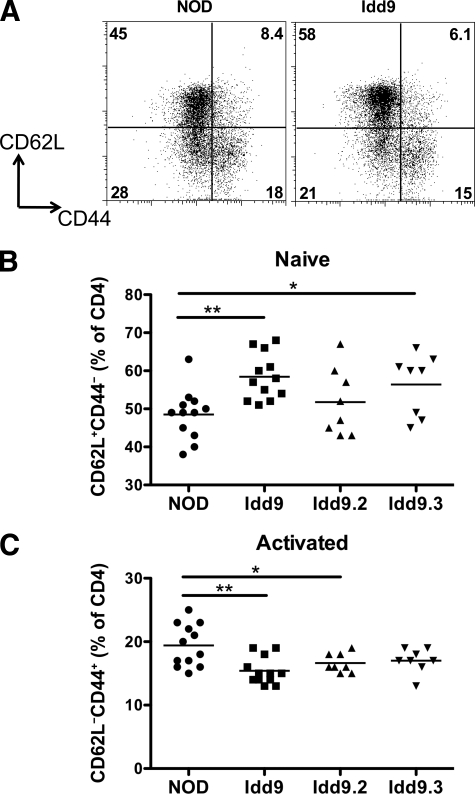
One way in which dominant tolerance may be implemented is via regulatory T-cells that may be increased in number or function. We did not observe any difference in the frequency or number of regulatory T-cells between Idd9 and NOD mice (supplementary Fig. 6). We did, however, find a notable change in the ratio of naive to effector phenotype CD4+ T-cells between the two strains (Fig. 8). NOD mice had fewer naive cells and increased effector cells than Idd9 mice as assessed by staining for CD62L and CD44.
FIG. 8.
The frequency of naturally occurring naive and activated phenotype CD4+ cells differs between NOD and Idd9 mice. Spleens of female 8- to 10-week-old mice were analyzed for expression of CD4, CD62L, and CD44 by flow cytometry. A: Representative FACS plots. B: Frequency of CD4+CD62L+CD44− naive cells. C: Frequency of CD4+CD62L−CD44+ activated phenotype cells. Three pooled experiments are shown. Horizontal line depicts mean value. Groups compared by Student t test. A: NOD (48.5 ± 6.6%) vs. Idd9 (58.4 ± 6.1%), P = 0.0009 (**). NOD vs. Idd9.3 (56.4 ± 8.1%), P = 0.02 (*). B: NOD (19.4 ± 3.3%) vs. Idd9 (15.4 ± 22%), P = 0.002 (**), NOD vs. Idd9.2 (16.6 ± 1.5%), P = 0.04 (*) (mean ± SD).
DISCUSSION
Diabetes susceptibility genes may contribute to disease pathogenesis through alterations of distinct phases of disease such as thymic or peripheral deletion, expansion of self-reactive cells and immune regulation, or effects on apoptosis of the β-cells (15). We hypothesized that many key genes will regulate the expansion of pathogenic CD8+ T-cells, the cell type ultimately responsible for the destruction of the islets. We previously reported that protective alleles of several Idd regions act to restore CD8+ T-cell tolerance to islet antigens in NOD mice (5,16). Here we have further dissected the cell types through which Idd9-protective alleles achieve CD8+ T-cell tolerance.
We chose to examine CD8+ T-cells specific for IGRP, as these T-cells are found at high frequencies in islet infiltrates, and increased frequencies of these cells in the peripheral blood are predictive of diabetes progression in individual NOD mice (17). Although it is unlikely that IGRP is the autoantigen that triggers disease (18), tolerance to this antigen serves as a general model for tolerance to islet proteins. Tolerance to a surrogate islet antigen, transgenic HA, was also restored by Idd9 genes (5); however, we cannot exclude that other islet-specific CD8+ T-cells may exist that do not conform to this principle. We found that the frequency of IGRP-specific CD8+ T-cells was dramatically reduced in Idd9 mice compared with NOD. Markedly few IGRP-specific cells were found in the islets, despite the presence of other CD8+ T-cells and lymphocyte infiltrates, suggesting Idd9 restores islet antigen–specific tolerance. We had previously shown that adoptively transferred naive NOD TCR transgenic CD8+ T-cells became activated to an islet antigen in the PcLNs and the highly proliferated cells infiltrated the islets of Idd9 mice on day 4 after transfer (5,16). The absence of endogenous islet-specific CD8+ T-cells in Idd9 islets suggests that these cells undergo tolerance after they enter the islets. CD8+ T-cells of other specificities were found in the pancreatic infiltrates. These cells may be specific for non–self-antigens, such as environmental antigens, as we have observed in other models (12).
Upon transfer of 8.3 CD8+ T-cells to NOD versus Idd9 recipients, we observed a dramatic difference in the numbers of cells recovered 6.5–8 weeks after transfer. Both groups were similar at the early time points, consistent with our previous observation that early activation of transferred CD8+ T-cells occurs in both strains (5,16). The time of expansion occurred when the mice were 13–15 weeks of age, which correlates with the first age of onset of diabetes in our colony. Thus, expansion of islet antigen–specific CD8+ cells may accompany disease progression.
The observation that transferred NOD-8.3 T-cells were tolerized in Idd9 mice demonstrates that Idd9 genes expressed by cells in the host restore peripheral tolerance. We have not yet been able to directly determine whether 8.3 cells transferred into Idd9 mice undergo tolerance through anergy or deletion. Because of limitation in antigen availability, it is necessary to transfer a very small number of cells to ensure complete activation in the PcLN. When larger cell numbers are used, a population of naive cells remains in the mice for many weeks (19). The fact that a strong stimulus such as Vac-IGRP does not promote activation suggests deletion rather than anergy may occur.
It is also possible that Idd9 genes contribute to correcting the defective thymic tolerance described in NOD mice (20,21). We did not see any evidence that thymic tolerance significantly contributed to IGRP-specific CD8+ tolerance. Mature CD8+ T-cells isolated from the periphery of both Idd9 and NOD mice contained IGRP-specific CD8+ T-cells that expanded and accumulated when transferred into an NOD environment, also indicating that endogenous expression of Idd9 genes did not promote tolerance. Thus, the Idd9 thymus failed to remove self-reactive IGRP-specific clones from the repertoire. This could be due to low expression of IGRP in the thymus (18), and would not necessarily be the case for all islet antigens or all islet reactive T cells.
Experiments in which purified Idd9 CD4+ cells were found to transfer tolerance into a NOD-SCID host strongly support a role for CD4+ T-cells in disease protection. It is likely that Idd9 genes act intrinsically to the CD4+ T-cell, although it is also possible that expression in another cell type during development permanently altered the Idd9 CD4+ T-cell compartment rendering them tolerogenic. As the cells were transferred 10 weeks before CD8+ T-cell tolerance was assessed in the reconstituted hosts, this seems unlikely. Although we have observed a dominant protection by Idd9 bone marrow, we have not observed evidence of altered frequencies of CD4+ regulatory T-cells in Idd9 mice. We did, however, find a notable change in the ratio of naive to effector phenotype CD4+ T-cells between the two strains: NOD mice had fewer naive cells and increased effector cells. Thus, the presence of NOD susceptible alleles of Idd9 genes appears to support higher numbers of activated CD4+ T-cells, which may then provide increased “help” to CD8+ T-cells, e.g., by licensing of dendritic cells (DCs) or within the target tissue (22,23). This is the subject of ongoing research. It is also possible that Idd9 regulatory T-cells have enhanced suppressive ability, as suggested elsewhere (7).
Dissection of the Idd9 region into three subregions has been previously described (2). We have found that Idd9.2 is the most potent region in regard to restraining the expansion of autoreactive CD8+ T-cells, whereas Idd9.1 had no such effect. However, all three Idd9 regions have similar diabetes frequencies (2,4,7). This implies that the amount of expansion of autoreactive CD8+ T-cells does not correlate directly with disease progression. Idd9.1 has been described to increase the suppressive function of regulatory T-cells (7). This may result in reduced diabetes incidence without altering the frequency of IGRP-specific CD8+ T-cells that expand upon viral infection. We conclude that CD8+ tolerance is an active mechanism strongly modulated by Idd9.2 genes.
Fine-mapping of Idd9.2 is ongoing to define which genes in the current interval are positional candidate genes. One gene in the Idd9.2 region, Fbxo2, was recently reported to have a twofold lower expression level in NOD than protected Idd9.2 mice (24). Idd9.3 is a small congenic interval containing only 15 genes including the primary candidate Cd137 (41bb) (4). CD137 is expressed on activated T-cells as well as activated DCs where it has recently been described to support DC survival after maturation (25). Thus, is it plausible that Idd9.3 could function pleiotropically on both CD4+ T-cells and DCs. We are currently working to identify the cell types in which Idd9.2 and Idd9.3 must be expressed for optimal tolerance induction. Through identification of the protective pathways of genes such as Idd9.2 and Idd9.3, it is possible that targets for future type 1 diabetes therapies or preventative treatments may be identified.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI 070351 from the National Institutes of Health. E.E.H.-W. and X.M. are supported by postdoctoral fellowships from the Juvenile Diabetes Research Foundation (JDRF). L.S.W. is supported by grants from the JDRF and the Wellcome Trust and is a Wellcome Trust Principal Research Fellow. The Cambridge Institute for Medical Research is the recipient of a Wellcome Trust Strategic Award (079895). The availability of NOD congenic mice through the Taconic Farms Emerging Models Program has been supported by grants from the Merck Genome Research Institute, NIAID, and the JDRF.
No potential conflicts of interest relevant to this article were reported.
We thank all members of the Sherman laboratory for discussions, Rebecca Trenney, Kristi Marquardt and Judith Biggs for technical assistance and Michelle Gassert for administrative assistance. We also thank the staff of the TSRI Flow cytometry core facility for sorting and assistance.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Maier LM, Wicker LS: Genetic susceptibility to type 1 diabetes. Curr Opin Immunol 2005; 17: 601–608 [DOI] [PubMed] [Google Scholar]
- 2.Lyons PA, Hancock WW, Denny P, Lord CJ, Hill NJ, Armitage N, Siegmund T, Todd JA, Phillips MS, Hess JF, Chen SL, Fischer PA, Peterson LB, Wicker LS: The NOD Idd9 genetic interval influences the pathogenicity of insulitis and contains molecular variants of Cd30, Tnfr2, and Cd137. Immunity 2000; 13: 107–115 [DOI] [PubMed] [Google Scholar]
- 3.Robles DT, Eisenbarth GS, Dailey NJ, Peterson LB, Wicker LS: Insulin autoantibodies are associated with islet inflammation but not always related to diabetes progression in NOD congenic mice. Diabetes 2003; 52: 882–886 [DOI] [PubMed] [Google Scholar]
- 4.Cannons JL, Chamberlain G, Howson J, Smink LJ, Todd JA, Peterson LB, Wicker LS, Watts TH: Genetic and functional association of the immune signaling molecule 4–1BB (CD137/TNFRSF9) with type 1 diabetes. J Autoimmun 2005; 25: 13–20 [DOI] [PubMed] [Google Scholar]
- 5.Martinez X, Kreuwel HT, Redmond WL, Trenney R, Hunter K, Rosen H, Sarvetnick N, Wicker LS, Sherman LA: CD8+ T cell tolerance in nonobese diabetic mice is restored by insulin-dependent diabetes resistance alleles. J Immunol 2005; 175: 1677–1685 [DOI] [PubMed] [Google Scholar]
- 6.Waldner H, Sobel RA, Price N, Kuchroo VK: The autoimmune diabetes locus Idd9 regulates development of type 1 diabetes by affecting the homing of islet-specific T cells. J Immunol 2006; 176: 5455–5462 [DOI] [PubMed] [Google Scholar]
- 7.Yamanouchi J, Puertas MC, Verdaguer J, Lyons PA, Rainbow DB, Chamberlain G, Hunter KM, Peterson LB, Wicker LS, Santamaria P: Idd9.1 locus controls the suppressive activity of FoxP3+CD4+CD25+ regulatory T-cells. Diabetes 2010; 59: 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill NJ, Stotland A, Solomon M, Secrest P, Getzoff E, Sarvetnick N: Resistance of the target islet tissue to autoimmune destruction contributes to genetic susceptibility in Type 1 diabetes. Biol Direct 2007; 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen YG, Scheuplein F, Osborne MA, Tsaih SW, Chapman HD, Serreze DV: Idd9/11 genetic locus regulates diabetogenic activity of CD4 T-cells in nonobese diabetic (NOD) mice. Diabetes 2008; 57: 3273–3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Silveira PA, Chapman HD, Stolp J, Johnson E, Cox SL, Hunter K, Wicker LS, Serreze DV: Genes within the Idd5 and Idd9/11 diabetes susceptibility loci affect the pathogenic activity of B cells in nonobese diabetic mice. J Immunol 2006; 177: 7033–7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton-Williams EE, Martinez X, Clark J, Howlett S, Hunter KM, Rainbow DB, Wen L, Shlomchik MJ, Katz JD, Beilhack GF, Wicker LS, Sherman LA: Expression of diabetes-associated genes by dendritic cells and CD4 T cells drives the loss of tolerance in nonobese diabetic mice. J Immunol 2009; 183: 1533–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong SB, Bos R, Sherman LA: Tumor-specific CD4+ T cells render the tumor environment permissive for infiltration by low-avidity CD8+ T cells. J Immunol 2008; 180: 3122–3131 [DOI] [PubMed] [Google Scholar]
- 13.Hernandez J, Aung S, Redmond WL, Sherman LA: Phenotypic and functional analysis of CD8(+) T cells undergoing peripheral deletion in response to cross-presentation of self-antigen. J Exp Med 2001; 194: 707–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieberman SM, Evans AM, Han B, Takaki T, Vinnitskaya Y, Caldwell JA, Serreze DV, Shabanowitz J, Hunt DF, Nathenson SG, Santamaria P, DiLorenzo TP: Identification of the beta cell antigen targeted by a prevalent population of pathogenic CD8+ T cells in autoimmune diabetes. Proc Natl Acad Sci U S A 2003; 100: 8384–8388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Concannon P, Rich SS, Nepom GT: Genetics of type 1A diabetes. N Engl J Med 2009; 360: 1646–1654 [DOI] [PubMed] [Google Scholar]
- 16.Hamilton-Williams EE, Martinez X, Lyman M, Hunter K, Wicker LS, Sherman LA: The use of idd congenic mice to identify checkpoints of peripheral tolerance to islet antigen. Ann N Y Acad Sci 2007; 1103: 118–127 [DOI] [PubMed] [Google Scholar]
- 17.Trudeau JD, Kelly-Smith C, Verchere CB, Elliott JF, Dutz JP, Finegood DT, Santamaria P, Tan R: Prediction of spontaneous autoimmune diabetes in NOD mice by quantification of autoreactive T cells in peripheral blood. J Clin Invest 2003; 111: 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krishnamurthy B, Dudek NL, McKenzie MD, Purcell AW, Brooks AG, Gellert S, Colman PG, Harrison LC, Lew AM, Thomas HE, Kay TW: Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J Clin Invest 2006; 116: 3258–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan DJ, Kreuwel HT, Sherman LA: Antigen concentration and precursor frequency determine the rate of CD8+ T cell tolerance to peripherally expressed antigens. J Immunol 1999; 163: 723–727 [PubMed] [Google Scholar]
- 20.Lesage S, Hartley SB, Akkaraju S, Wilson J, Townsend M, Goodnow CC: Failure to censor forbidden clones of CD4 T cells in autoimmune diabetes. J Exp Med 2002; 196: 1175–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishimoto H, Sprent J: A defect in central tolerance in NOD mice. Nat Immunol 2001; 2: 1025–1031 [DOI] [PubMed] [Google Scholar]
- 22.Smith CM, Wilson NS, Waithman J, Villadangos JA, Carbone FR, Heath WR, Belz GT: Cognate CD4(+) T cell licensing of dendritic cells in CD8(+) T cell immunity. Nat Immunol 2004; 5: 1143–1148 [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi Y, Lu B, Gerard C, Iwasaki A: CD8(+) T lymphocyte mobilization to virus-infected tissue requires CD4(+) T-cell help. Nature 2009; 462: 510–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kodama K, Butte AJ, Creusot RJ, Su L, Sheng D, Hartnett M, Iwai H, Soares LR, Fathman CG: Tissue- and age-specific changes in gene expression during disease induction and progression in NOD mice. Clin Immunol 2008; 129: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi BK, Kim YH, Kwon PM, Lee SC, Kang SW, Kim MS, Lee MJ, Kwon BS: 4–1BB functions as a survival factor in dendritic cells. J Immunol 2009; 182: 4107–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.