Abstract
OBJECTIVE
Common variants in the melatonin receptor type 1B (MTNR1B) locus have been shown to increase fasting plasma glucose (FPG) and the risk of type 2 diabetes. The aims of this study were to evaluate whether nonsynonymous variants in MTNR1B associate with monogenic forms of hyperglycemia, type 2 diabetes, or related metabolic traits.
RESEARCH DESIGN AND METHODS
MTNR1B was sequenced in 47 probands with clinical maturity-onset diabetes of the young (MODY), in 51 probands with early-onset familial type 2 diabetes, and in 94 control individuals. Six nonsynonymous variants (G24E, L60R, V124I, R138C, R231H, and K243R) were genotyped in up to 22,142 Europeans. Constitutive and melatonin-induced signaling was characterized for the wild-type melatonin receptor type 1B (MT2) and the 24E, 60R, and 124I MT2 mutants in transfected COS-7 cells.
RESULTS
No mutations in MTNR1B were MODY specific, and none of the investigated MTNR1B variants associated with type 2 diabetes. The common 24E variant associated with increased prevalence of obesity (odds ratio 1.20 [1.08–1.34]; P = 8.3 × 10−4) and increased BMI (β = 0.5 kg/m2; P = 1.2 × 10−5) and waist circumference (β = 1.2 cm; P = 9 × 10−6) in combined Danish and French study samples. 24E also associated with decreased FPG (β = −0.08 mmol/l; P = 9.2 × 10−4) in the Danish Inter99 population. Slightly decreased constitutive activity was observed for the MT2 24E mutant, while the 124I and 60R mutants displayed considerably decreased or completely disrupted signaling, respectively.
CONCLUSIONS
Nonsynonymous mutations in MTNR1B are not a common cause of MODY or type 2 diabetes among Danes. MTNR1B 24E associates with increased body mass and decreased FPG. Decreased MT2 signaling does apparently not directly associate with FPG or type 2 diabetes.
Genome-wide association (GWA) studies have shown that common variation in the melatonin receptor type 1B (MTNR1B) locus increases the level of fasting plasma glucose (FPG) and the risk of type 2 diabetes (1,2). The intronic rs10830963 variant showed the strongest and most independent signal of association (1,3), and subsequent studies have reported that carriers of the rs10830963 risk allele have reduced insulin secretion after both oral and intravenous glucose challenges (3–5). Thus, it has been proposed that this common MTNR1B variant increases the risk of type 2 diabetes and impaired fasting glycemia as a result of a defect of the pancreatic β-cells.
The neurohormone melatonin is secreted from the pineal gland during the night and functions as a hormonal message of the photoperiod. Nocturnal pineal melatonin secretion is generated by the central clock in the suprachiasmatic nucleus, which is entrained by the 24-h light-darkness cycle (6). Melatonin is particularly involved in regulation of seasonal and circadian rhythms, such as the sleep-wake cycle, locomotor activity, feeding, core body temperature, immune function, and hormone levels (6,7). Circadian rhythms are highly involved in regulation of a wide range of physiological processes including metabolism (8), and circadian disturbances including shift work and sleep disorders increase the risk of both cardiovascular and metabolic disorders (9,10). Moreover, chronic sleep deficit is an independent risk factor for obesity (11), and both circadian rhythms and melatonin secretion are impaired in type 2 diabetic patients (12). Extrapineal sources of melatonin secretion have also been described, e.g., the retina, the pancreas, and the enteroendocrine cells of the gastrointestinal tract (GI-tract). In these peripheral tissues, melatonin may have autocrine and paracrine properties (13,14). Interestingly, the concentration of melatonin within the gastrointestinal tract is considerably higher than in plasma, presumably due to release from the enteroendocrine cells (15,16). This secretion of melatonin into the gastrointestinal tract is suggested to be related to food intake and digestion, independently of the photoperiodic secretion of melatonin from the pineal gland (16,17).
Melatonin mediates its effect through two high-affinity receptors, the MT1 and MT2 receptors, encoded by MTNR1A and MTNR1B, respectively (18,19). Both receptors belong to the family of seven transmembrane (7TM) G-protein–coupled receptors. In humans, MT2 is expressed in the suprachiasmatic nucleus and various brain regions yet also in peripheral tissues, including adipose tissue and the gastrointestinal tract (20,21). Recently, MTNR1B expression was also found in human pancreatic islets and in β-cells (2,4,22).
It has been shown that common variants within a gene can lead to increased risk of type 2 diabetes while rare coding mutations within the same gene lead to monogenic forms of diabetes, e.g., maturity-onset diabetes of the young (MODY) (23). In addition, coding MTNR1B variants have not yet been reported in relation to metabolism. Thus, the aims of this study were to evaluate whether mutations in MTNR1B are responsible for genetically unexplained MODY subtypes (MODY-X) and to investigate the influence of nonsynonymous MTNR1B variants in relation to type 2 diabetes and related metabolic traits. We also aimed to determine the functional effect of selected MTNR1B variants on MT2 receptor signaling.
RESEARCH DESIGN AND METHODS
Detailed characteristics of the screened probands are presented in online appendix Table 1 (available at http://diabetes.diabetesjournals.org/cgi/content/full/db09-1757/DC1). Control individuals (n = 94) for the mutation screening were drawn by random from the population-based Inter99 study.
MODY patients.
MODY patients (n = 47) without mutations in HNF4A, GCK, or HNF1A referred to molecular genetic testing were recruited via the outpatient clinic at Steno Diabetes Center. Inclusion criteria for MODY families were at least one family member with diagnosis of diabetes before the age of 25 years and diabetes in two or more consecutive generations. In addition, the proband should not have been treated with insulin within the first year of diagnosis or should have displayed fasting serum C-peptide >100 pmol/l 1 year after diagnosis. Patients were excluded if they were GAD65 auto-antibody positive.
Early-onset type 2 diabetic patients.
Early-onset type 2 diabetic patients (n = 51) were recruited at Steno Diabetes Center. All patients were diagnosed before the age of 40 years and had a positive family history of type 2 diabetes in two or more consecutive generations. Patients were excluded if they were GAD65 auto-antibody positive or if fasting serum C-peptide was <100 pmol/l at investigation.
Epidemiological studies.
Genetic epidemiological studies were performed in the Danish population-based Inter99 study (n = 6,002) (24,25), the Danish ADDITION screening cohort of individuals at high risk of developing type 2 diabetes (n = 8,576) (26), individuals recruited at Steno Diabetes Center (n = 2,293), the French population–based D.E.S.I.R. study (n = 4,626) (27,28), and in a sample of adults from the French Super Obese (FSO) study (29) (n = 645). Detailed descriptions and clinical characteristics of the study populations are found in appendix Tables 2 and 3.
Case-control studies and quantitative trait analyses.
Case-control studies of type 2 diabetes (n = 3,617 case and n = 4,975 control subjects) included all unrelated type 2 diabetic patients (n = 328) and glucose-tolerant control individuals (n = 4,460) from the Inter99 study, type 2 diabetic patients from the Danish ADDITION intervention study (n = 1,617), and individuals recruited from the outpatient clinic at Steno Diabetes Center (n = 1,672 case and n = 515 control subjects). Normal glucose tolerance and type 2 diabetes were defined according to the World Health Organization 1999 criteria (30), and all control individuals had normal fasting glycaemia and were normal glucose tolerant following a 75-g oral glucose tolerance test (OGTT).
Patients treated for type 2 diabetes were not included in obesity case-control studies or in quantitative trait analyses. Danish case-control studies of obesity included up to 6,828 individuals, comprising 2,608 normal-weight (BMI <25 kg/m2) and 971 obese (BMI ≥30 kg/m2) individuals from the Inter99 study, up to 2,881 obese individuals from the Danish ADDITION screening cohort, and 272 normal-weight and 96 obese individuals from Steno Diabetes Center. French case-control studies of obesity (n = 3,773) included individuals from the population-based D.E.S.I.R. study (normal weight, n = 2,738; obese, n = 390) and obese individuals from the FSO study (n = 645). Quantitative analyses of BMI and waist circumference were performed in individuals from the Danish population-based Inter99 study sample (n = 5,885). Potential associations with BMI and waist circumference for G24E, L60R, and V124I were evaluated in the French population–based D.E.S.I.R. study sample (n = 4,626) and in nonpopulation-based study samples: the Danish ADDITION screening cohort (n = 8,576), samples collected at Steno Diabetes Center (n = 649), and adults from the FSO study (n = 645). Analyses of biochemical variables obtained during an OGTT were performed in individuals from the Danish population-based Inter99 study. All study participants were unrelated and Danish or French by self-report. Informed written consent was obtained from all individuals before participation. The studies were approved by the regional ethical committees and were in accordance with the principles of the Declaration of Helsinki II.
Sequencing.
MTNR1B was divided into four segments covering the two exons, exon-intron boundaries, and untranslated regions (UTRs). The segments were amplified by standard PCR and sequenced directly by the Sanger sequencing method. All primers were designed with Primer3 (http://frodo.wi.mit.edu/). The sequences were analyzed on a 3130XL genetic analyzer (Applied Biosystems), and mutations were detected with SeqScape v2.5 (Applied Biosystems). Details of primers and PCR conditions are provided in online appendix Table 4. DNA was purified from leukocytes obtained from blood samples.
Genotyping.
Genotyping was performed by KASPa® (KBioscience, Hoddesdon).Genotype success rates were >96%, and concordance between 513 duplicate samples was >99%. Genotypes obeyed Hardy-Weinberg equilibrium in all study populations (P > 0.01). For rare variants with minor allele frequencies (MAFs) <0.5% (L60R, V124I, and R138C), mutation carriers and noncalled samples, selected by visual inspection of the genotype plots, were evaluated by direct sequencing according to the mutation detection protocol. No additional carriers were found by resequencing of 384 randomly selected samples. DNA was purified from leukocytes obtained from blood samples.
Statistical analyses.
The statistical analyses were performed using RGui, version 2.8.1 (http://www.r-project.org). A P value of <0.05 was considered significant. Results are reported uncorrected for multiple testing. Statistical analyses were calculated assuming an additive (rs8192552 [G24E]) or dominant (L60R, V124I, rs8192553 [R231H], rs61746674 [R138C], and rs61747139 [K243R]) genetic model. Logistic regression was applied to test for differences in genotype distribution in case-control studies, and linear regression was applied to test quantitative variables for differences between genotype groups. Effect sizes are given as actual values or percentage if the trait was logarithmically transformed. The combined Danish and French case-control studies of obesity and quantitative analyses of BMI and waist circumference were performed assuming a fixed effect while adjusting for nationality (Danish or French) as a factor. All analyses were adjusted for sex and age. Conditional analyses were performed by comparing a model including one variant with an alternative model including two variants adjusted for age, sex, and BMI. In the type 2 diabetes case-control studies, we had >80% statistical power to detect an odds ratio (OR) of 1.65, 1.45, and 1.20, assuming MAFs of 0.5, 1, and 5%, respectively.
MT2 receptor signaling.
Mutations were constructed for G24E, L60R, and V124I by PCR using the overlap extension method as previously described (31). The PCR products were digested, purified, and cloned into the pCMV-Tag(2B) vector. All mutations were verified by restriction endonuclease mapping and subsequent DNA sequence analysis using an ABI 310 automated sequencer. COS-7 cells were grown in Dulbecco's modified Eagle's medium 1885 supplemented with 10% FCS, 2 mmol/l glutamine, and 0.01 mg/ml gentamicin. Cells were transfected with 10 μg cDNA of wild-type or mutant MT2 receptors and 10 μg cDNA of a GαΔ6qi4myr (32) using the calcium phosphate precipitation method with chloroquine addition. The chimeric G-protein allows the Gαi-coupled receptors to signal through the signal transduction pathways known for the Gαq-coupled receptors (32).
One day after transfection, COS-7 cells were incubated for 24 h with 5 μCi of [3H]-myo-inositol (PT6-271; Amersham) in 1 ml medium, washed twice in buffer (20 mmol/l HEPES [pH 7.4]), and were subsequently incubated in 0.5 ml buffer supplemented with 10 mmol/l LiCl at 37°C for 30 min. After stimulation with various concentrations of melatonin for 45 min at 37°C, cells were extracted with 10% ice-cold formic acid followed by incubation on ice for 30 min. The generated [3H]-inositol phosphate was purified on Bio-Rad AG 1-X8 anion-exchange resins. All experiments were done in duplicates and repeated three times. Comparisons at each concentration point of melatonin between wild-type and mutant receptors were made by unpaired t tests.
RESULTS
Mutation detection.
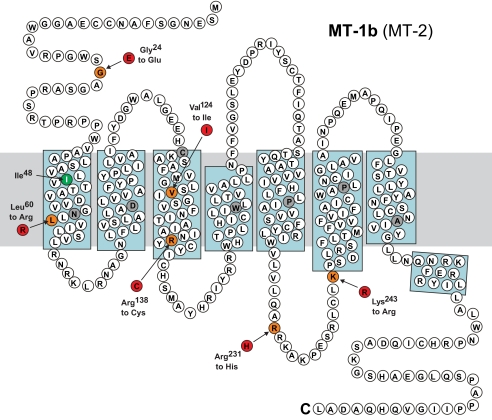
Table 1 and Fig. 1 show a summary of all variants identified in the mutation screening. No novel MTNR1B mutations were found in the 47 MODY-X probands. One novel nonsynonymous mutation, V124I, was found in an early-onset type 2 diabetic patient. One novel synonymous variant, I48I, and one novel 3′UTR variant, exon 2 + 38T>C, as well as seven known variants (dbSNP build 129), were also observed in the screened probands. Four known variants—rs8192552 (G24E), rs61746674 (R138C), rs8192553 (R231H), and rs61747139 (K243R)—changed an amino acid in the protein. In addition, three novel mutations, A13V, A13E, and exon 2 + 107G>T, were identified in three unrelated glucose-tolerant individuals but were not observed in any of the diabetic probands. The previously described L60R mutation was not observed in this screening but was included in the present study because of a reported prevalence of 0.4% in the French population (2). Since no mutations were familyspecific, we did not test for cosegregation in the families.
TABLE 1.
MAFs of MTNR1B variants identified from sequencing of Danish probands from MODY families and families with early-onset type 2 diabetes
| rs | Variant | Position | MAFs |
|||
|---|---|---|---|---|---|---|
| MODY (n = 47) | Early-onset type 2 diabetes (n = 51) | Controls (n = 94) | Genotyping (n = 9,912) | |||
| Novel | c.38C>T, p.13A>V | 92,342,570 (exon 1) | 0 | 0 | 0.005 | – |
| Novel | c.38C>A, p.13A>E | 92,342,570 (exon 1) | 0 | 0 | 0.005 | – |
| rs8192552 | c.71G>A, p.24G>E | 92,342,610 (exon 1) | 0.21 | 0.14 | 0.14 | 0.092 |
| Novel | c.144C>T, p.48I>I | 92,342,683 (exon 1) | 0 | 0.008 | 0 | – |
| rs6483210 | Exon 2 − 18C>T | 92,354,243 (intron 1) | 0.03 | 0.02 | 0.04 | – |
| Novel | c.370G>A, p.124V>I | 92,354,407 (exon 2) | 0 | 0.008 | 0 | 0.004 |
| rs61746674 | c.412C>T, p.138R>C | 92,354,449 (exon 2) | 0 | 0.02 | 0 | 0.001 |
| rs8192553 | c.692G>A, p.231R>H | 92,354,729 (exon 2) | 0 | 0.008 | 0.005 | 0.006 |
| rs61747139 | c.728A>G, p.243K>R | 92,354,765 (exon 2) | 0.01 | 0.02 | 0.04 | 0.033 |
| Novel | Exon 2 + 38T>C | 92,355,164 (3′UTR) | 0 | 0.008 | 0.005 | – |
| Novel | Exon 2 + 107G>T | 92,355,233 (3′UTR | 0 | 0 | 0.005 | – |
| rs1562444 | Exon 2 + 370G>A | 92,355,497 (3′UTR) | 0.59 | 0.55 | 0.49 | – |
| rs12792653 | Exon 2 + 437G>A | 92,355,564 (3′UTR) | 0.59 | 0.55 | 0.48 | – |
| Novel* | c.179T>G, p.60L>R | 92,342,718 (exon 1) | 0 | 0 | 0 | 0.00055 |
Variant locations are displayed as base numbers counting from the p-arm telomere of chromosome 11 (according to the UCSC Genome Browser on Human Mar. 2006 Assembly: http://genome.ucsc.edu/cgi-bin/hgGateway).
*L60R was not identified in this screening but identified in a French population recently (ref. 2).
FIG. 1.
Capital letters are abbreviations of amino acids. Orange-colored circles represent the wildtype amino acid in the mutated position, and red-colored circles represent the mutant amino acid. Green-colored circles represent silent mutations. Gray-colored circles are the highly conserved residues in the 7TM rhodopsine family. The gray bar represents the cell membrane, and blue squares indicate α-helices.
Epidemiological studies.
Six nonsynonymous variants (L60R, V124I, rs8192552 [G24E], rs61746674 [R138C], rs8192553 [R231H], and rs61747139 [K243R]) were initially genotyped in the population-based Inter99 population (n = 6,002), screen-detected type 2 diabetic individuals from the Danish ADDITION study (n = 1,617), and additional nondiabetic individuals (n = 621) and type 2 diabetic patients (n = 1,672) recruited from the Steno outpatient clinic. MAFs in the Danish population (n = 9,912) are shown in Table 1.
Case-control studies of type 2 diabetes and obesity.
Potential associations with susceptibility to type 2 diabetes were evaluated in Danish case-control studies (n = 8,592) (Table 2). The investigated MTNR1B nonsynonymous variants did not associate with type 2 diabetes (Table 2) and also did not after the analyses were adjusted for BMI (data not shown).
TABLE 2.
Danish case-control studies of type 2 diabetes for six MTNR1B variants in 3,617 case and 4,975 control individuals
| Normal glucose tolerance | Type 2 diabetes | OR (95%CI) | P | |
|---|---|---|---|---|
| G24E (rs8192552) | 4,019/831/33 | 2,935/598/32 | 1.00 (0.87–1.15) | 0.97 |
| L60R | 4,891/3/0 | 3,548/2/0 | 2.58 (0.32–21.07) | 0.38 |
| V124I | 4,883/35/0 | 3,484/30/0 | 1.27 (0.69–2.35) | 0.44 |
| R138C (rs61746674) | 4,898/9/0 | 3,525/5/0 | 0.91 (0.23–3.63) | 0.89 |
| R231H (rs8192553) | 4,853/53/1 | 3,498/41/0 | 0.98 (0.58–1.63) | 0.92 |
| K243R (rs61747139) | 4,572/331/4 | 3,310/214/5 | 0.95 (0.76–1.19) | 0.67 |
Data are presented as wildtype/heterozygous/homozygous unless otherwise indicated. ORs and P values were calculated assuming a dominant (L60R, V124I, R138C, R231H, and K243R) or additive (G24E) genetic model adjusted for age and sex.
Potential associations with susceptibility to obesity were evaluated in Danish (up to 6,828 subjects) and French (n = 3,773) case-control studies. Codons 24E and 124I were initially associated with increased risk of obesity (G24E per allele OR 1.27 [95% CI 1.08–1.48], P = 0.003; V124I 2.17 [1.01–4.67], P = 0.05) in a Danish case-control study (n = 4,837). No obese individuals were carriers of L60R, and no associations with obesity were observed for R138C, R231H, or K243R (Table 3). Following genotyping of additional Danish obese case subjects (n = 1,991), the association between G24E and obesity remained significant in Danish analyses (1.21 [1.05–1.39), P = 0.008) (Table 3). In a French case-control study, 24E similarly associated with increased risk of obesity (1.21 [1.00–1.46], P = 0.05) (Table 3), and in combined analyses of Danish and French individuals the association was substantiated (1.20 [1.08–1.34], P = 8 × 10−4) (Table 3). When these analyses were restricted to the population-based samples Inter99 and D.E.S.I.R., results were similar (Inter99 1.37 [1.15–1.64], P = 5 × 10−4; D.E.S.I.R. 1.48 [1.13–1.93], P = 0.004; and combined 1.40 [1.21–1.43], P = 9 × 10−6). Following additional genotyping, L60R and V124I did not associate with obesity in Danish, French, or combined study samples (Table 3).
TABLE 3.
Case-control studies of obesity for six MTNR1B variants in Danish (up to n = 6,828) and French (n = 3,773) individuals
| Danish |
French |
Combined (n = 10,601) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Normal weight | Obesity | OR (95%CI) | P | Normal weight | Obesity | OR (95%CI) | P | OR (95%CI) | P | |
| G24E* | 2,365/442/17 | 3,168/678/33 | 1.21 (1.05–1.39) | 0.0081 | 2,276/353/14 | 859/159/8 | 1.21 (1.00–1.46) | 0.046 | 1.20 (1.08–1.34) | 8.3 × 10−4 |
| L60R* | 2,822/3/0 | 3,868/1/0 | 0.12 (0.01–1.4) | 0.091 | 2,708/8/0 | 1,019/4/0 | 1.29 (0.39–4.33) | 0.68 | 0.66 (0.21–2.06) | 0.48 |
| V124I* | 2,824/12/0 | 3,833/31/0 | 1.74 (0.83–3.67) | 0.14 | 2,691/3/0 | 1,016/2/0 | 1.78 (0.29–10.81) | 0.53 | 1.74 (0.91–3.35) | 0.095 |
| R138C | 2,822/6/0 | 1,906/3/0 | 0.89 (0.21–3.82) | 0.87 | ||||||
| R231H | 2,809/26/1 | 1,888/20/0 | 1.0 (0.53–1.9) | 0.99 | ||||||
| K243R | 2,647/185/2 | 1,786/124/3 | 0.99 (0.77–1.27) | 0.93 | ||||||
Data are presented as wild-type/heterozygous/homozygous unless otherwise indicated. ORs and P values were calculated assuming a dominant (L60R, V124I, R138C, R231H, and K243R) or additive (G24E) genetic model adjusted for age and sex. Nationality has been applied as an adjusting factor in the combined analyses.
*Additional obese cases (n = 1,991) were genotyped for G24E, L60R, and V124I. Normal weight was defined as BMI < 25 kg/m2. Obesity was defined as BMI ≥ 30 kg/m2.
Quantitative analyses of BMI and waist circumference.
Quantitative analyses of BMI and waist circumference were performed in the Danish population–based Inter99 study sample (n = 5,885) (Table 4). Associations for G24E, L60R, and V124I were evaluated in the French D.E.S.I.R. population-based study sample (n = 4,626) (Table 4), as well as in nonpopulation-based samples, i.e., Danish ADDITION screening cohort (n = 8,576), samples collected at Steno Diabetes Center (n = 649), and FSO adults (n = 645) (online appendix Table 7).
TABLE 4.
BMI and waist circumference in Danish (n = 5,885) and French (n = 4,626) population–based individuals stratified according to six MTNR1B genotypes
| Danish Inter99 |
French D.E.S.I.R. |
Combined (n = 10,510) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| N | Age (years) | BMI (kg/m2) | Waist (cm) | N | Age (years) | BMI (kg/m2) | Waist (cm) | BMI (kg/m2) | Waist (cm) | |
| G24E (rs8192552) | ||||||||||
| Wild type | 4,736 | 46 ± 8 | 26.1 ± 4.5 | 86 ± 13 | 3,874 | 47 ± 10 | 24.6 ± 3.8 | 82.8 ± 11.5 | ||
| Heterozygous | 995 | 46 ± 8 | 26.6 ± 4.7 | 88 ± 13 | 657 | 46 ± 10 | 25.0 ± 4.2 | 84.3 ± 12.5 | ||
| Homozygous | 36 | 45 ± 10 | 27.1 ± 5.2 | 86 ± 14 | 26 | 46 ± 8 | 25.4 ± 4.3 | 84.4 ± 10.8 | ||
| Effect size | 0.5 kg/m2 | 1.1 cm | 0.5 kg/m2 | 1.3cm | 0.5 kg/m2 | 1.2 cm | ||||
| 95% CI | 0.2 to 0.7 | 0.4–1.8 | 0.2–0.7 | 0.6–2.0 | 0.3–0.7 | 0.7–1.7 | ||||
| Padd | 0.0019 | 0.0029 | 0.0011 | 3.7 × 10−4 | 1.2 × 10−5 | 9.4 × 10−6 | ||||
| L60R | ||||||||||
| Wild type | 5,764 | 46 ± 8 | 26.2 ± 4.5 | 86 ± 13 | 4,641 | 47 ± 10 | 24.6 ± 3.7 | 83.0 ± 11.6 | ||
| Heterozygous | 4 | 48 ± 9 | 21.9 ± 3.9 | 80 ± 13 | 15 | 47 ± 10 | 25.0 ± 4.1 | 86.9 ± 11.0 | ||
| Effect size | −4.4 kg/m2 | −7.1 cm | 0.1 kg/m2 | 0.8 cm | −0.9 kg/m2 | −1.0 cm | ||||
| 95% CI | −8.7 to 0.0 | −18.2 to 4.1 | −1.8 to 2.0 | −4.2 to 5.9 | −2.8 to 1.0 | −6.0 to 4.0 | ||||
| Pdom | 0.051 | 0.21 | 0.92 | 0.75 | 0.36 | 0.69 | ||||
| V124I | ||||||||||
| Wild type | 5,767 | 46 ± 8 | 26.2 ± 4.5 | 87 ± 13 | 4,621 | 47 ± 10 | 24.6 ± 3.7 | 83 ± 12 | ||
| Heterozygous | 41 | 46 ± 8 | 27.0 ± 4.1 | 89 ± 12 | 9 | 47 ± 9 | 25.3 ± 3.4 | 84 ± 11 | ||
| Effect size | 0.8 kg/m2 | 1.5 cm | 0.8 kg/m2 | 1.9 cm | 0.8 kg/m2 | 1.6 cm | ||||
| 95% CI | −0.6 to 2.1 | −20.0 to 50.0 | −1.6 to 3.1 | −4.1 to 8.0 | −0.4 to 1.9 | −1.3 to 4.5 | ||||
| Pdom | 0.28 | 0.41 | 0.52 | 0.53 | 0.19 | 0.29 | ||||
| R138C (rs61746674) | ||||||||||
| Wild type | 5,781 | 46 ± 8 | 26.2 ± 4.5 | 87 ± 13 | ||||||
| Heterozygous | 12 | 45 ± 8 | 25.3 ± 4.0 | 82 ± 10 | ||||||
| Effect size | −0.9 kg/m2 | −4.9 cm | ||||||||
| 95% CI | −3.5 to 1.6 | −11.3 to 1.6 | ||||||||
| Pdom | 0.47 | 0.14 | ||||||||
| R231H (rs8192553) | ||||||||||
| Wild type | 5,733 | 46 ± 8 | 26.2 ± 4.5 | 86.5 ± 13.2 | ||||||
| Heterozygous | 61 | 47 ± 7 | 26.4 ± 4.0 | 86.2 ± 13.9 | ||||||
| Homozygous | 1 | 55 | 23.8 | 74 | ||||||
| Effect size | 0.2 kg/m2 | 0.0 cm | ||||||||
| 95% CI | −0.9 to 1.3 | −2.9 to 2.8 | ||||||||
| Pdom | 0.74 | 1.0 | ||||||||
| K243R (rs61747139) | ||||||||||
| Wild type | 5,418 | 46 ± 8 | 26.2 ± 4.5 | 86.5 ± 13.2 | ||||||
| Heterozygous | 379 | 46 ± 7 | 26.2 ± 4.6 | 86.8 ± 13.4 | ||||||
| Homozygous | 3 | 48 ± 12 | 24.6 ± 1.6 | 82.0 ± 2.7 | ||||||
| Effect size | 0.0 kg/m2 | 0.4 cm | ||||||||
| 95% CI | −0.5 to 0.4 | −0.8 to 1.5 | ||||||||
| Pdom | 0.89 | 0.55 | ||||||||
Data are means ± SD stratified according to genotypes of MTNR1B G24E, L60R, and V124I. Effect sizes and P values are calculated assuming an additive (Padd) or dominant (Pdom) genetic model of inheritance adjusted for age and sex. Nationality has been applied as an adjusting factor in the combined analyses.
Codon 24E associated with a dose-dependent increase in BMI (β = 0.5 kg/m2 [0.2–0.7], P = 0.002) and waist circumference (β = 1.1 cm [0.4–1.8], P = 0.003) in the Danish Inter99 population (Table 4). In the French D.E.S.I.R. population-based study sample, similar associations were observed between 24E and increased BMI (β = 0.5 kg/m2 [0.2–0.7], P = 0.001) and waist circumference (β = 1.3 cm [0.6–2.0], P = 4 × 10−4) (Table 4). In combined Danish and French population–based analyses, the findings were substantiated (BMI β = 0.5 kg/m2 [0.3–0.7], P = 1 × 10−5; waist β = 1.2 cm [0.7–1.7], P = 9 × 10−6) (Table 4). However, G24E did not show any significant associations with BMI and waist circumference in non–population-based study samples, i.e., the Danish ADDITION screening cohort, the samples collected at Steno Diabetes Center, and FSO adults (online appendix Table 7).
L60R was found in only four individuals from the Inter99 population. Codon 60R was borderline associated with decreased BMI (β = −4.4 kg/m2 [−8.7 to 0.0], P = 0.05) in the Danish Inter99 population (Table 4). No significant associations with BMI and waist circumference were observed in the French population–based D.E.S.I.R. cohort (Table 4), in the combined Danish and French analyses (Table 4), or in any of the selected cohorts (online appendix Table 7).
V124I did not associate with BMI or waist circumference in Inter99, in the D.E.S.I.R. cohort, or in combined Danish and French analyses (Table 4). Sporadic associations with increased BMI were found in the Danish ADDITION screening cohort of individuals in high risk of undiagnosed type 2 diabetes and in the French FSO study (online appendix Table 7). R138C, R231H, and K243R did not show any significant association with BMI or waist circumference in the Danish Inter99 population (Table 4).
Quantitative analyses of biochemical variables obtained from an OGTT.
Analyses of anthropometric measurements and biochemical variables during an OGTT were performed in the Danish population–based Inter99 (Table 5). Codon 24E associated with a decrease in FPG (per allele β = −0.06 mmol/l [−0.1 to −0.01], P = 0.02) (Table 5). This association was strengthened when analyses were adjusted for the difference in BMI (β = −0.08 mmol/l [−0.13 to −0.03], P = 9 × 10−4), but no significant interactions with BMI were observed (P = 0.2). Conditional analyses of MTNR1B intronic variant rs10830963, previously reported to increase FPG (1–4), and rs8192552 (G24E) were performed. The linkage disequilibrium between the two variants were D′ = 0.99, r2 = 0.04 (online appendix Table 5). After inclusion of rs10830963 in the model, rs8192552 (G24E) still had an effect on FPG (P = 0.02) (online appendix Table 6).
TABLE 5.
Biochemical measurements obtained from an oral glucose tolerance test in 5,885 non-diabetic or treatment-naïve individuals from the Danish Inter99 study stratified according to 6 MTNR1B genotypes
| N | Age (years) | Plasma glucose (mmol/l) |
Serum insulin (pmol/l) |
Homa-IR | Insulinogenic index | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fasting | 30 min | 120 min | Fasting | 30 min | 120 min | |||||
| G24E (rs8192552) | ||||||||||
| Wild type | 4,736 | 46 ± 8 | 5.5 ± 0.8 | 8.7 ± 1.9 | 6.2 ± 2.1 | 34 (24–51) | 243 (174–349) | 155 (96–253) | 8.3 (5.6–12.8) | 24.3 (16.8–35.9) |
| Heterozygous | 995 | 46 ± 8 | 5.5 ± 0.6 | 8.7 ± 1.8 | 6.2 ± 2.0 | 35 (24–53) | 263 (181–368) | 160 (95–277) | 8.3 (5.7–13.1) | 25.1 (17.1–38.4) |
| Homozygous | 36 | 45 ± 10 | 5.3 ± 0.8 | 8.1 ± 1.6 | 6.8 ± 2.3 | 38 (24–53) | 257 (1,799–380) | 168 (117–295) | 9.2 (5.4–12.1) | 28.6 (20.5–39.0) |
| Effect size | −0.06 mmol/l | 0.01 mmol/l | 0.04 mmol/l | 1% | 4% | 3% | 0% | 4% | ||
| 95% CI | −0.1 to −0.01 | −0.1 to 0.1 | −0.09 to 0.2 | −3 to 5 | 1 to 8 | −2 to 8 | −4 to 4 | 0 to 8 | ||
| Padd | 0.017* | 0.91 | 0.53 | 0.62 | 0.018 | 0.25 | 0.96 | 0.040 | ||
| L60R | ||||||||||
| Wild type | 5,764 | 46 ± 8 | 5.5 ± 0.8 | 8.7 ± 1.9 | 6.2 ± 2.1 | 34 (24–51) | 244 (175–354) | 156 (96–256) | 8.3 (5.6–12.9) | 24.4 (16.8–36.3) |
| Heterozygous | 4 | 48 ± 9 | 5.6 ± 0.5 | 9.1 ± 1.8 | 5.5 ± 2.6 | 17 (15–21) | 147 (109–173) | 95 (79–122) | 4.2 (3.6–5.3) | 12.1 (9.3–14.3) |
| Effect size | 0.01 mmol/l | 0.3 mmol/l | −0.8 mmol/l | −66% | −67% | −58% | −66% | −83% | ||
| 95% CI | −0.7 to 0.7 | −1.4 to 2.0 | −2.8 to 1.2 | −124 to −9 | −130 to −4 | −136 to 20 | −127 to −4 | −152 to −13 | ||
| Pdom | 0.98 | 0.76 | 0.45 | 0.023 | 0.036 | 0.15 | 0.037 | 0.020* | ||
| V124I | ||||||||||
| Wild type | 5,767 | 46 ± 8 | 5.5 ± 0.8 | 8.7 ± 1.9 | 6.2 ± 2.1 | 34 (24–51) | 245 (175–354) | 156 (96–256) | 8.3 (5.7–12.9) | 24.5 (16.8–36.5) |
| Heterozygous | 41 | 46 ± 8 | 5.6 ± 0.7 | 9.1 ± 1.7 | 6.4 ± 2.6 | 31 (21–44) | 235 (145–321) | 140 (74–234) | 7.7 (5.2–10.6) | 22.9 (15.8–30.9) |
| Effect size | −0.0 mmol/l | 0.4 mmol/l | 0.2 mmol/l | 7% | 14% | 15% | 7% | 13% | ||
| 95% CI | −0.2 to 0.2 | −0.1 to 0.9 | −0.4 to 0.8 | −25 to 11 | −31 to 3 | −39 to 11 | −27 to 12 | −33 to 6 | ||
| Pdom | 0.95 | 0.16 | 0.54 | 0.42 | 0.12 | 0.26 | 0.47 | 0.18 | ||
| R138C (rs61746674) | ||||||||||
| Wild type | 5,781 | 46 ± 8 | 5.5 ± 0.8 | 8.7 ± 1.9 | 6.2 ± 2.1 | 34 (24–51) | 245 (175–353) | 156 (96–256) | 8.3 (5.7–12.9) | 24.5 (16.8–36.4) |
| Heterozygous | 12 | 45 ± 8 | 5.5 ± 0.6 | 8.2 ± 1.7 | 6.1 ± 1.1 | 33 (23–38) | 240 (197–276) | 167 (121–212) | 8.0 (5.2–9.0) | 25.3 (17.8–32.1) |
| Effect size | −0.09 mmol/l | −0.6 mmol/l | −0.1 mmol/l | 22% | 8% | 3% | 23% | 2% | ||
| 95% CI | −0.5 to 0.3 | −1.6 to 0.4 | −1.3 to 1.1 | −55 to 11 | −42 to 27 | −42 to 49 | −59 to 12 | −37 to 40 | ||
| Pdom | 0.69 | 0.26 | 0.86 | 0.19 | 0.66 | 0.88 | 0.20 | 0.92 | ||
| R231H (rs8192553) | ||||||||||
| Wild type | 5,733 | 46 ± 8 | 5.5 ± 0.8 | 8.7 ± 1.9 | 6.2 ± 2.1 | 34 (24–51) | 245 (175–353) | 156 (97–256) | 8.3 (5.7–12.9) | 24.4 (16.8–36.3) |
| Heterozygous | 61 | 47 ± 7 | 5.6 ± 0.9 | 8.8 ± 2.1 | 6.0 ± 2.6 | 37 (27–48) | 294 (202–376) | 151 (63–230) | 8.9 (6.2–12.0) | 29.1 (18.7–39.5) |
| Homozygous | 1 | 55 | 5.2 | 6.9 | 4.2 | 37 | 212 | 65 | 8.6 | 25.4 |
| Effect size | 0.0 mmol/l | 0.0 mmol/l | −0.4 mmol/l | 2% | 9% | 24% | 2% | 3% | ||
| 95% CI | −0.2 to 0.2 | −0.4 to 0.5 | −0.9 to 0.2 | −13 to 18 | −6 to 23 | −44 to −4 | −14 to 19 | −14 to 20 | ||
| Pdom | 1.0 | 0.91 | 0.17 | 0.78 | 0.24 | 0.021* | 0.78 | 0.75 | ||
| K243R (rs61747139) | ||||||||||
| Wild type | 5,418 | 46 ± 8 | 5.5 ± 0.8 | 8.7 ± 1.9 | 6.2 ± 2.1 | 34 (24–51) | 246 (175–353) | 156 (96–256) | 8.3 (5.6–12.9) | 24.5 (16.8–36.3) |
| Heterozygous | 379 | 46 ± 7 | 5.5 ± 0.7 | 8.6 ± 1.8 | 6.1 ± 2.0 | 35 (24–50) | 232 (177–358) | 154 (95–239) | 8.3 (5.7–13.0) | 24.5 (17.3–36.6) |
| Homozygous | 3 | 48 ± 12 | 5.8 ± 1.3 | 9.2 ± 2.4 | 8.5 ± 4.8 | 27 (26–28) | 313 (190–318) | 146 (146–250) | 6.0 (5.8–6.2) | 37.0 (36.0–37.9) |
| Effect size | −0.08 mmol/l | −0.09 mmol/l | −0.1 mmol/l | 1% | 0% | 3% | 0% | 2% | ||
| 95% CI | −0.2 to −0.01 | −0.3 to 0.1 | −0.3 to 0.1 | −5 to 8 | −6 to 6 | −11 to 6 | −7 to 7 | −5 to 9 | ||
| Pdom | 0.035* | 0.32 | 0.24 | 0.66 | 0.95 | 0.51 | 0.98 | 0.57 | ||
Data are means ± SD or median (interquartile range) unless otherwise indicated, stratified according to genotypes of six MTNR1B variants. Values of serum insulin, HOMA-IR, and insulinogenic index were logarithmically transformed before statistical analyses. Effect sizes and P values are calculated assuming an additive (Padd) or dominant (Pdom) genetic model of inheritance adjusted for age and sex.
*Remains significant after adjusting for the difference in BMI. HOMA-IR and insulinogenic index are calculated as described in online-only appendix.
Four carriers of codon 60R had decreased insulinogenic index (β = −83% [−152 to −13], P = 0.02) (Table 5), which remained nominally significant when analyses were adjusted for the difference in BMI (β = −72% [−140 to −5], P = 0.04).
243R was nominally associated with decreased FPG (β = −0.08 mmol/l [−0.16 to −0.01], P = 0.04) (Table 5). Conditional analyses of rs10830963 and rs61747139 (K243R) in linkage disequilibrium (D′ = 0.99; r2 = 0.01 were performed (online appendix Table 5). After inclusion of rs10830963 in the model, the effect of rs61747139 (K243R) disappeared (P = 0.2) (online appendix Table 6). No associations with anthropometric or biochemical measurements were observed for V124I, R138C, or R231H.
Analyses of MT2 receptor signaling.
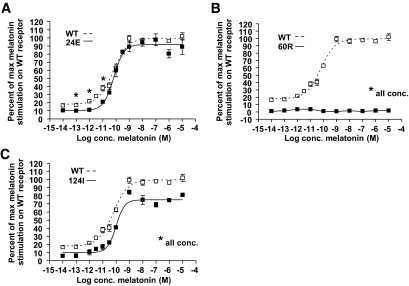
The wild-type MT2 receptor displayed a relatively high degree of constitutive signaling activity and a clear dose-related increase in signaling in response to melatonin (Fig. 2). The dose-response curve for melatonin in the 24E mutant form of the MT2 receptor was similar to that of the wild-type receptor with respect to potency and efficacy; however, the ligand-independent constitutive signaling was slightly decreased (Fig. 2). Functional characterization of the 60R mutant receptor revealed that both constitutive and melatonin-induced signaling were completely disrupted for this mutant (Fig. 2). The 124I mutant receptor had decreased constitutive activity and maximal efficacy compared with the wild-type MT2 receptor.
FIG. 2.
The chimeric G-protein allows the Gαi-coupled receptors to signal through the signal transduction pathways known for the Gαq-coupled receptors. Data are from Inositol turnover after stimulation with various concentrations (conc.) of melatonin (n = 3). □, wild-type (WT) MT2 receptor; ■, 24E, 60R, or 124I mutant MT2 receptors. *P < 0.05 in unpaired t test.
DISCUSSION
In the present study, we found no MODY-specific mutations in MTNR1B. We describe novel associations of G24E with measures of obesity and FPG. None of the investigated MTNR1B variants were associated with type 2 diabetes. Decreased MT2 Gαi-coupled signaling, as observed to various extents for 24E, 60R, and 124I mutant receptors, did not correlate strongly with type 2 diabetes or FPG.
The common 24E variant (MAF 9.2% in the Danish population) increased the risk of obesity and BMI in Danish and French individuals but also decreased FPG by 0.08 mmol/l per allele, as estimated in the Danish population. Conditional analyses showed that this effect on FPG could only be partly explained by linkage to the previously reported intronic rs10830963 variant. An apparent paradox is that FPG is decreased while BMI and waist circumference are increased in carriers of 24E, and we hypothesize that these dual findings could be due to differential regulation in specific target tissues. The effect on FPG may reflect the functionality of pancreatic MT2 receptors as previously suggested for the signal obtained in the GWA studies (4). It has also been suggested based on findings in animal and in vitro experiments that melatonin may act on body weight regulation through activation of central (brain) or peripheral (gastrointestinal tract and adipocyte) receptors (7,33), and the effect we observed on BMI might therefore reflect the function of MT2 receptors in these tissues. When the association between 24E and waist circumference was adjusted for BMI in the Inter99 and D.E.S.I.R. cohorts, no significant associations were observed (data not shown), pointing toward an effect of the genotype on overall fat distribution rather than on specific abdominal obesity. Investigations in populations with detailed measures of body composition would consequently be of interest for future studies.
G24E (rs8192552) has not been reported in large GWA studies of obesity (34–36). The variant has not been genotyped in HapMap, and its coverage by GWA studies is thus unclear. It may therefore be that rs8192552 is not adequately captured by variants analyzed in previous GWA studies. Alternatively, heterogeneity among the investigated GWA populations could be an explanation for the lack of strongly significant association with BMI or obesity in these studies.
G24 is conserved in rhesus and mouse but not in elephant, opossum, or platypus. Functionally, we observed that the MT2 24E mutant had a somewhat decreased constitutive activity but otherwise responded similarly to the wild-type receptor upon melatonin stimulation, as observed in cells cotransfected with a promiscuous G-protein commonly used to study signaling properties of Gαi-coupled receptors (32). The MT2 receptor is believed to signal mainly through Gαi but has been shown to be able to activate a number of intracellular signaling pathways (37,38). The association of the G24E variant with BMI and FPG observed in the present study will justify an in-depth cell biological and molecular pharmacological analysis of this variant in particular; such studies are, however, hampered by lack of appropriate in vitro cell systems that diligently reflect the cell biology of tissues where MT2 is normally expressed. Nevertheless, the normal maximal signaling efficacy observed for the 24E mutant indicates that the amino acid substitution does not lead to misfolding or lack of cell surface expression, which otherwise is a common molecular phenotype for 7TM receptor variants (39). Given that the 24E variant is located in the extracellular part of the receptor, it is plausible that receptor interactions with other ligands or proteins may be modified by the amino acid substitution. Finally, because the variant is common, other yet unidentified regulatory variants in linkage disequilibrium could also be responsible for the observed associations.
L60R was very rare among Danes (MAF 0.056%) where a tendency toward decreased BMI was observed. However, this finding could not be confirmed in a French population, although the variant had a higher frequency (MAF 0.16%) among French individuals. In the Danish Inter99 population, four 60R carriers had on average 83% decreased glucose-stimulated insulin response measured by the insulinogenic index, but still no effect on FPG was observed. It is an important limitation that we have very few 60R carriers. Consequently, we lack statistical power to exclude an effect of L60R, and the results obtained may be spurious. Of particular interest, L60R changes a hydrophobic leucine to a positively charge arginine at position 60, which is highly conserved as an aliphatic hydrophobic residue (82%) in transmembrane segment 1 (TM-I) of 7TM receptors in general (40). It is very likely that the completely eliminated signaling by substitution with the basic arginine residue is caused by disruption of the normal hydrophobic interaction of L60 with the intracellular helix VIII. Although heterozygous knockdown of MT2 signaling revealed no obvious human metabolic phenotype in this study, prospective studies are highly relevant to evaluate whether a phenotype evolves as a function of aging and various environmental pressures. No homozygous mutation carriers have been identified; hence, it is also possible that one functional allele is sufficient to ensure a normal phenotype. Supporting this hypothesis, partial redundancy between the melatonin receptor types has been suggested (37) and both homodimer and MT1/MT2 heterodimer formation is described in vitro, where activation of one binding site is sufficient to induce a conformational change in the dimer (41). In addition, MT2 knockout mice have no obvious metabolic phenotype (42,43).
The initial association between 124I and obesity observed in a Danish case-control study could not be confirmed in combined case-control analyses or in analyses of BMI and waist circumference as quantitative variables in the Danish and French populations. 124I is very rare among French individuals, severely impeding statistical power for replication. We observed, however, an effect of 124I on BMI in the Danish ADDITION screening cohort of individuals in high risk of type 2 diabetes, implying that 124I may increase BMI in selected high-risk groups, although it has no strong effect in the general population. V124 is conserved among mammalian species but not in chicken, lizard, or Xenopus, and it has been proposed that this residue is involved in the binding of melatonin (44) The 124I mutant had impaired MT2 signaling, but because completely disrupted signaling as observed for the 60R mutant does not lead to increased BMI, these results do not support the associations observed for V124I in selected study populations.
We observed various degrees of decreased signaling for the three MT2 mutants, which however, does not seem to associate directly with type 2 diabetes or FPG. This is in line with a recent study by Lyssenko et al. (4) suggesting that increased MTNR1B expression in β-cells may be the pathogenic mechanism by which FPG and the risk of type 2 diabetes are increased, supported by the fact that melatonin inhibits insulin secretion. We did, however, observe impaired insulin release for the four 60R carriers, and it is not known at present whether these individuals will develop hyperglycemia or overt type 2 diabetes over time. The associations observed for G24E may not be ascribed to the slightly lower constitutive activity because more severe disrupted signaling, as observed for 60R and 124I mutants, does not lead to comparable phenotypes.
The putative promoter regions of MTNR1B and potential distant regulatory regions were not targeted in the present study, and these regions may therefore contain yet undiscovered functional variants of importance for the development of type 2 diabetes and related phenotypes.
Throughout this study, effects on BMI and waist circumference observed in selected study samples somewhat differed from those observed in population-based study samples. This heterogeneity may reflect a true differential effect among individual groups, statistical type II errors due to lack of statistical power in the smaller study samples, or statistical type I errors due to the lack of correction for multiple testing. However, we argue that results from large population-based studies are more reliable in the sense that they are largely free of selection bias, whereas results from selected study samples should be analyzed and interpreted separately to evaluate whether the variants have a specific effect in a certain subpopulation of interest. When the results were adjusted for for multiple testing (discovery samples, six SNPs and nine independent tests; replication sample, three SNPs and two independent tests), the associations with BMI and waist circumference for G24E in the French D.E.S.I.R. population and in the combined Danish and French study samples remained significant (P < 0.05). In the present study, the French D.E.S.I.R. population was used to evaluate the obesity-related discovery results obtained in the Danish Inter99 population for G24E, L60R, and V124I. The two populations are comparable with respect to age and the sampling method. The MAF of L60R and V124I differed among Danish and French individuals, implying that population differences across countries exist for rare variants and thus impeding cross-population replication as a result of drift in statistical power.
In conclusion, we find that nonsynonymous variation in MTNR1B is not a common cause of MODY or type 2 diabetes among Danes. MTNR1B 24E associates with increased body mass and decreased FPG. MT2 amino acid variants affect receptor signaling, but the observed effects do not seem to associate strongly with FPG or type 2 diabetes.
Supplementary Material
ACKNOWLEDGMENTS
Research in the laboratory of O.P. and T.H. was supported by grants from the Lundbeck Foundation Centre of Applied Medical Genomics for Personalized Disease Prediction, Prevention and Care, the University of Copenhagen, the Hepatic and Adipose Tissue and Functions in the Metabolic Syndrome (HEPADIP) consortium, the Danish Obesity Research Centre (DANORC) consortium, and the Food Study Group/The Danish Ministry of Food, Agriculture and Fisheries and the Ministry of Family and Consumers Affairs (FOOD). Research in the laboratory of P.F. was supported by Le Conseil Régional Nord Pas de Calais/Fonds Européen de Développement Régional (FEDER), l'Agence Nationale de la Recherche. Research in the laboratory of T.W.S. and B.H. was supported by grants from the Danish Medical Research Council, The Lundbeck Foundation, and University Investment Capital (UNIK): Food, Fitness & Pharma for Health and Disease from the Danish Ministry of Science, Technology and Innovation.
Research in the laboratory of T.W.S. and B.H. was also supported by a grant the Novo Nordisk Foundation. No other potential conflicts of interest relevant to this article were reported.
We thank A. Forman, I.-L. Wantzin, and M. Stendal for technical assistance, A.L. Nielsen for data management, M. Kristensen for scientific administration, and G. Lademann for secretarial support.
Footnotes
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, et al. : Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009; 41: 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, De Graeve F, Chevre JC, Borch-Johnsen K, Hartikainen AL, Ruokonen A, Tichet J, Marre M, Weill J, Heude B, Tauber M, Lemaire K, Schuit F, Elliott P, Jorgensen T, Charpentier G, Hadjadj S, Cauchi S, Vaxillaire M, Sladek R, Visvikis-Siest S, Balkau B, Levy-Marchal C, Pattou F, Meyre D, Blakemore AI, Jarvelin MR, Walley AJ, Hansen T, Dina C, Pedersen O, Froguel P: A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 2009; 41: 89–94 [DOI] [PubMed] [Google Scholar]
- 3.Sparso T, Bonnefond A, Andersson E, Bouatia-Naji N, Holmkvist J, Wegner L, Grarup N, Gjesing AP, Banasik K, Cavalcanti-Proenca C, Marchand M, Vaxillaire M, Charpentier G, Jarvelin MR, Tichet J, Balkau B, Marre M, Levy-Marchal C, Faerch K, Borch-Johnsen K, Jorgensen T, Madsbad S, Poulsen P, Vaag A, Dina C, Hansen T, Pedersen O, Froguel P: G-allele of intronic rs10830963 in MTNR1B confers increased risk of impaired fasting glycemia and type 2 diabetes through an impaired glucose-stimulated insulin release: studies involving 19,605 Europeans. Diabetes 2009; 58: 1450–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spegel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L: Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 2009; 41: 82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staiger H, Machicao F, Schafer SA, Kirchhoff K, Kantartzis K, Guthoff M, Silbernagel G, Stefan N, Haring HU, Fritsche A: Polymorphisms within the novel type 2 diabetes risk locus MTNR1B determine beta-cell function. PLoS One 2008; 3: e3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claustrat B, Brun J, Chazot G: The basic physiology and pathophysiology of melatonin. Sleep Med Rev 2005; 9: 11–24 [DOI] [PubMed] [Google Scholar]
- 7.Barrenetxe J, Delagrange P, Martinez JA: Physiological and metabolic functions of melatonin. J Physiol Biochem 2004; 60: 61–72 [DOI] [PubMed] [Google Scholar]
- 8.Green CB, Takahashi JS, Bass J: The meter of metabolism. Cell 2008; 134: 728–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scheer FA, Hilton MF, Mantzoros CS, Shea SA: Adverse metabolic and cardiovascular consequences of circadian misalignment. Proc Natl Acad Sci U S A 2009; 106: 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knutsson A, Akerstedt T, Jonsson BG, Orth-Gomer K: Increased risk of ischaemic heart disease in shift workers. Lancet 1986; 2: 89–92 [DOI] [PubMed] [Google Scholar]
- 11.Plante GE: Sleep and vascular disorders. Metabolism 2006; 55: S45–S49 [DOI] [PubMed] [Google Scholar]
- 12.Peschke E, Frese T, Chankiewitz E, Peschke D, Preiss U, Schneyer U, Spessert R, Muhlbauer E: Diabetic Goto Kakizaki rats as well as type 2 diabetic patients show a decreased diurnal serum melatonin level and an increased pancreatic melatonin-receptor status. J Pineal Res 2006; 40: 135–143 [DOI] [PubMed] [Google Scholar]
- 13.Kvetnoy IM: Extrapineal melatonin: location and role within diffuse neuroendocrine system. Histochem J 1999; 31: 1–12 [DOI] [PubMed] [Google Scholar]
- 14.Raikhlin NT, Kvetnoy IM, Tolkachev VN: Melatonin may be synthesised in enterochromaffin cells. Nature 1975; 255: 344–345 [DOI] [PubMed] [Google Scholar]
- 15.Messner M, Huether G, Lorf T, Ramadori G, Schworer H: Presence of melatonin in the human hepatobiliary-gastrointestinal tract. Life Sci 2001; 69: 543–551 [DOI] [PubMed] [Google Scholar]
- 16.Huether G, Poeggeler B, Reimer A, George A: Effect of tryptophan administration on circulating melatonin levels in chicks and rats: evidence for stimulation of melatonin synthesis and release in the gastrointestinal tract. Life Sci 1992; 51: 945–953 [DOI] [PubMed] [Google Scholar]
- 17.Bubenik GA, Pang SF, Cockshut JR, Smith PS, Grovum LW, Friendship RM, Hacker RR: Circadian variation of portal, arterial and venous blood levels of melatonin in pigs and its relationship to food intake and sleep. J Pineal Res 2000; 28: 9–15 [DOI] [PubMed] [Google Scholar]
- 18.Reppert SM, Weaver DR, Ebisawa T: Cloning and characterization of a mammalian melatonin receptor that mediates reproductive and circadian responses. Neuron 1994; 13: 1177–1185 [DOI] [PubMed] [Google Scholar]
- 19.Reppert SM, Godson C, Mahle CD, Weaver DR, Slaugenhaupt SA, Gusella JF: Molecular characterization of a second melatonin receptor expressed in human retina and brain: the Mel1b melatonin receptor. Proc Natl Acad Sci U S A 1995; 92: 8734–8738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ekmekcioglu C: Melatonin receptors in humans: biological role and clinical relevance. Biomed Pharmacother 2006; 60: 97–108 [DOI] [PubMed] [Google Scholar]
- 21.Brydon L, Petit L, Delagrange P, Strosberg AD, Jockers R: Functional expression of MT2 (Mel1b) melatonin receptors in human PAZ6 adipocytes. Endocrinology 2001; 142: 4264–4271 [DOI] [PubMed] [Google Scholar]
- 22.Muhlbauer E, Peschke E: Evidence for the expression of both the MT1- and in addition, the MT2-melatonin receptor, in the rat pancreas, islet and beta-cell. J Pineal Res 2007; 42: 105–106 [DOI] [PubMed] [Google Scholar]
- 23.Hattersley AT, Ashcroft FM: Activating mutations in Kir6.2 and neonatal diabetes: new clinical syndromes, new scientific insights, and new therapy. Diabetes 2005; 54: 2503–2513 [DOI] [PubMed] [Google Scholar]
- 24.Jorgensen T, Borch-Johnsen K, Thomsen TF, Ibsen H, Glumer C, Pisinger C: A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99. Eur J Cardiovasc Prev Rehabil 2003; 10: 377–386 [DOI] [PubMed] [Google Scholar]
- 25.Glumer C, Jorgensen T, Borch-Johnsen K: Prevalences of diabetes and impaired glucose regulation in a Danish population: the Inter99 study. Diabetes Care 2003; 26: 2335–2340 [DOI] [PubMed] [Google Scholar]
- 26.Lauritzen T, Griffin S, Borch-Johnsen K, Wareham NJ, Wolffenbuttel BH, Rutten G: The ADDITION study: proposed trial of the cost-effectiveness of an intensive multifactorial intervention on morbidity and mortality among people with type 2 diabetes detected by screening. Int J Obes Relat Metab Disord 2000; 24( Suppl 3): S6–S11 [DOI] [PubMed] [Google Scholar]
- 27.Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proenca C, Marchand M, Hartikainen AL, Sovio U, De Graeve F, Rung J, Vaxillaire M, Tichet J, Marre M, Balkau B, Weill J, Elliott P, Jarvelin MR, Meyre D, Polychronakos C, Dina C, Sladek R, Froguel P: A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 2008; 320: 1085–1088 [DOI] [PubMed] [Google Scholar]
- 28.Balkau B: [An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome]. Rev Epidemiol Sante Publique 1996; 44: 373–375 [ In French] [PubMed] [Google Scholar]
- 29.Benzinou M, Creemers JW, Choquet H, Lobbens S, Dina C, Durand E, Guerardel A, Boutin P, Jouret B, Heude B, Balkau B, Tichet J, Marre M, Potoczna N, Horber F, Le Stunff C, Czernichow S, Sandbaek A, Lauritzen T, Borch-Johnsen K, Andersen G, Kiess W, Korner A, Kovacs P, Jacobson P, Carlsson LM, Walley AJ, Jorgensen T, Hansen T, Pedersen O, Meyre D, Froguel P: Common nonsynonymous variants in PCSK1 confer risk of obesity. Nat Genet 2008; 40: 943–945 [DOI] [PubMed] [Google Scholar]
- 30.Alberti KG, Zimmet PZ: Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1. Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998; 15: 539–553 [DOI] [PubMed] [Google Scholar]
- 31.Holst B, Hastrup H, Raffetseder U, Martini L, Schwartz TW: Two active molecular phenotypes of the tachykinin NK1 receptor revealed by G-protein fusions and mutagenesis. J Biol Chem 2001; 276: 19793–19799 [DOI] [PubMed] [Google Scholar]
- 32.Kostenis E: Potentiation of GPCR-signaling via membrane targeting of G protein alpha subunits. J Recept Signal Transduct Res 2002; 22: 267–281 [DOI] [PubMed] [Google Scholar]
- 33.Konturek SJ, Konturek PC, Brzozowski T, Bubenik GA: Role of melatonin in upper gastrointestinal tract. J Physiol Pharmacol 2007; 58( Suppl 6): 23–52 [PubMed] [Google Scholar]
- 34.Lindgren CM, Heid IM, Randall JC, Lamina C, Steinthorsdottir V, Qi L, Speliotes EK, Thorleifsson G, Willer CJ, Herrera BM, et al. : Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet 2009; 5: e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, Berndt SI, Elliott AL, Jackson AU, Lamina C, et al. : Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet 2009; 41: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K: Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet 2009; 41: 18–24 [DOI] [PubMed] [Google Scholar]
- 37.von Gall C, Stehle JH, Weaver DR: Mammalian melatonin receptors: molecular biology and signal transduction. Cell Tissue Res 2002; 309: 151–162 [DOI] [PubMed] [Google Scholar]
- 38.Jockers R, Maurice P, Boutin JA, Delagrange P: Melatonin receptors, heterodimerization, signal transduction and binding sites: what's new? Br J Pharmacol 2008; 154: 1182–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson MD, Percy ME, McIntyre Burnham W, Cole DE: G protein-coupled receptors disrupted in human genetic disease. Methods Mol Biol 2008; 448: 109–137 [DOI] [PubMed] [Google Scholar]
- 40.Mirzadegan T, Benko G, Filipek S, Palczewski K: Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry 2003; 42: 2759–2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ayoub MA, Levoye A, Delagrange P, Jockers R: Preferential formation of MT1/MT2 melatonin receptor heterodimers with distinct ligand interaction properties compared with MT2 homodimers. Mol Pharmacol 2004; 66: 312–321 [DOI] [PubMed] [Google Scholar]
- 42.Jin X, von Gall C, Pieschl RL, Gribkoff VK, Stehle JH, Reppert SM, Weaver DR: Targeted disruption of the mouse Mel(1b) melatonin receptor. Mol Cell Biol 2003; 23: 1054–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larson J, Jessen RE, Uz T, Arslan AD, Kurtuncu M, Imbesi M, Manev H: Impaired hippocampal long-term potentiation in melatonin MT2 receptor-deficient mice. Neurosci Lett 2006; 393: 23–26 [DOI] [PubMed] [Google Scholar]
- 44.Mazna P, Berka K, Jelinkova I, Balik A, Svoboda P, Obsilova V, Obsil T, Teisinger J: Ligand binding to the human MT2 melatonin receptor: the role of residues in transmembrane domains 3, 6, and 7. Biochem Biophys Res Commun 2005; 332: 726–734 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.