Abstract
Several family-based approaches have been previously proposed to enhance the power for testing genetic association when the traits are measured longitudinally or repeatedly. In this paper, we show that some of these FBAT approaches can be easily extended to accommodate incomplete data and remain unbiased tests. We also show that because of the nature of FBAT approaches, we can impute the missing phenotypes without biasing our tests and achieve higher power. We propose two imputation techniques based on E-M algorithm and the conditional mean model, respectively. Through simulation studies, these two imputation techniques are shown to have correct false positive rate and generally achieve higher power than complete case analysis or simple mean-imputation. Application of these approaches for testing an association between Body Mass Index and a previously reported candidate SNP confirms our results.
Key Words: FBAT, Longitudinal Phenotype, Missing Data
Introduction
For many family-based studies of complex disease, multiple disease-related phenotypes are often measured longitudinally or repeatedly for each subject in the sample. When there are no missing observations, several different family-based approaches have been previously discussed to utilize the multivariate data efficiently to test for genetic association [1, 2].
For many phenotypes, especially those related to complex disease, measurements are often difficult to obtain and record. In practice, we can expect some subjects to have missing data. Many statistical methods for missing data analysis have been reviewed by Little and Rubin [3]. The simplest method to deal with missing data is to use the complete data subset, which means we only use subjects with all phenotypes available and discard all the subjects with any missing observation. Another intuitive method is to modify the existing tests to utilize all the information available. In other words, the original test statistics are appropriately adapted so that they can accommodate a subject's observed phenotypes even when the rest are missing. A third commonly used method is to impute the missing phenotypes in the dataset.
In this paper, we show that some FBAT approaches can be easily extended to accommodate subjects with partially missing phenotypes and remain valid tests. We propose two imputation techniques based on E-M algorithm and the conditional mean model respectively. With simulation studies, we check the false positive rate of these methods and compare their power to the complete data analysis and the mean-imputation technique. Our new imputation techniques are found to be unbiased and generally more powerful than complete case analysis or simple mean imputation. Applications of these methods for handling missingness to the Framingham Heart Study data confirm our results.
Review of Methods for the Complete Data Setting
Suppose there are N families. For simplicity, assume we have parents with one offspring (trios); the results can be easily generalized to other family structures [4]. We denote the vector containing all m phenotypic observations for each offspring by , where Yij is the j-th phenotype for the i-th offspring. The standard biometric model [5] describing a single phenotype as a function of the genotype can be extended as
| (1) |
| (2) |
where is the intercept vector, is the vector of genetic effects, Xi denotes the coding of the marker genotype of the i-th offspring, and VP is the phenotypic residual variance-covariance matrix. The vector containing all traits for each offspring can be expressed as , where Tij is the j-th trait for the i-th offspring. Here Tij is a function of the phenotype Yij, for example, or Yij adjusted for covariates [6].
For the j-th measurement, the univariate family-based association test (FBAT) statistic [4] can be written as
| (3) |
where E(Xi | Pi) and Var(Xi | Pi) (shown in equation 4 below) denote the expectation and variance of the marker score computed under the null hypothesis (no genetic association), conditional on the parental genotypes Pi. With large samples, the vector containing all univariate test statistics asymptotically follows a multivariate normal distribution under H0 [7]. Here is an m-dimensional vector of zeroes and Σ0 is the variance-covariance matrix of those univariate test statistics,
| (4) |
Several approaches have been introduced to utilize the multivariate data efficiently to test for genetic association in family-based studies. Lange et al. developed the FBAT-PC approach [1], which is an expansion of the univariate FBAT for traits that are measured longitudinally or repeatedly over time. Based on generalized principle component analysis, FBAT-PC amplifies the genetic effect of each measurement by constructing an overall phenotype with maximal locus-specific heritability. Ding [2] introduced FBAT-PCM as a modification to FBAT-PC with higher power, along with two other approaches, FBAT-LC and FBAT-LCC, which have more power in some circumstances.
All three of these statistics can be expressed as a weighted combination of those univariate tests Sj, with different approaches used to compute the weights,
| (5) |
| (6) |
| (7) |
where
If no missing observation exists, FBAT-LC has the highest power when the genetic effects are same for all measurement points. When the genetic effect sizes differ, FBAT-LC is more powerful when the phenotypic correlation is low, while FBAT-PCM achieves the highest power when the correlation is high [2].
To avoid biasing the significance level of any subsequent tests, Lange et al. [8, 9] proposed the Conditional Mean Model (CMM) to estimate the unknown variables in these FBAT statistics. In equation (1), we replace the observed marker score xi by the expected marker score E(Xi | Pi), and estimate αj separately by ordinary least square estimation,
Methods
There can be various reasons for missing phenotypic information. For example, a participant may drop out of the study or fail to appear on a follow-up visit, or part of the data may be lost during the data transfer process. For simplicity, we assume offspring are not missing the genotypic data, i.e., all Xi are observed.
When missing observations occur, the phenotypes for the i-th subject can be rewritten as
| (9) |
where is the vector of observed phenotypes, is the vector of missing phenotypes. Here Ii is obtained from an identity matrix Im × m by removing the rows corresponding to the missing observations, and Ji is made up of those removed rows. It is useful to classify the missing-data mechanism in order to understand the performance of different approaches, under different condition [3, 10]. In our setting, when the probability of the missing phenotype Yij is independent of either or the genotype Xi, the outcomes are called to be missing completely at random (MCAR). We say our phenotypes are missing at random (MAR), if the missingness is independent of conditional on and Xi. Furthermore, if the missing probability depends upon given and Xi, the missing-data mechanism is non-ignorable.
We consider several simple, easily implemented and commonly used strategies to deal with the missing data problem. The simplest strategy, known as complete case analysis, is to remove all the subjects with any missing value and only analyze the complete data subset. In other words, all will be discarded if Ii ≠ Im × m. Assuming N* out of the N subjects do have all the observations, the analysis will be applied to the data subset with sample size equal to N*. Alternatively, we can apply the analysis to all the observed data , i = 1, …, N, called all available case analysis. A third strategy is to replace missing phenotypes with some appropriate values, which is also known as imputation analysis [3].
Note that all the FBAT statistics in equations (5)–(7) are calculated conditional on all the phenotypic information and only Xi are considered as random variables. Therefore, with each of these three strategies to deal with missing phenotypes, the validity of these FBAT approaches always holds for both MCAR and MAR, provided that the imputation is independent of the offspring's genotypes, Xi, and the missingness is also independent of Xi. In general, this will be a reasonable assumption. Even when the missingness does depend upon the offspring's genotype, our simulations show that the FBAT approaches can still be valid if the traits are mean-centered, which is generally true in practice. Furthermore, the power of FBAT approaches might be affected by both the underlying missing mechanism and the strategy chosen to handle missingness.
Extending FBAT-LC and LCC to Use All Available Data
Theoretically, FBAT-PCM (as well as FBAT-PC) can be extended to analyze incomplete data [1]. Since the overall phenotypes have to be constructed separately for subjects with different missing patterns, the computation is complex and the interpretation is no longer straightforward. Therefore we do not discuss the extension of FBAT-PCM here. On the contrary, test statistics of FBAT-LC and FBAT-LCC in equation (5) and (6) can easily be extended to use all available phenotypic information.
For the j-th measurement, assume only nj out of the N phenotypes (Y1j, …, YNj) are actually observed, the rest of them are missing. Letting the set Oj = (i1, i2, …, inj) denote the indexes of the nj subjects whose j-th phenotype is available, the univariate FBAT based on all observed data can be written as
| (10) |
where Tij, i ∈ Oj are nj traits corresponding to those observed phenotypes Yij, i ∈ Oj at the j-th measurement time.
Similar to the case when there is no missing [Lange et al., 2003b], under the null hypothesis (no association between Yij and Xi), we have E(S*j) = 0 and
Note that this is true under H0 regardless of the missing-data mechanism, provided the missingness of phenotype is independent of the offspring's genotype.
For i ∈ (1, …, N), j ∈ (1, …, m), we define
| (11) |
Via simple algebra, it is easy to show that equation (10) can be rewritten as
| (12) |
and the variance-covariance matrix for vector can be written as
In addition, the conditional mean model in equation (8) can easily be extended to incomplete data as
| (14) |
Letting
where is obtained via equation (14), FBAT-LC and FBATLCC statistics based on the observed data can be rewritten as
| (15) |
| (16) |
Imputing the Missing Values
With imputation techniques, we estimate the unobserved phenotypes by , and then apply FBAT approaches to the imputed complete data
i = 1, …, N. Since the univariate FBAT statistic in equation (3) is conditional on not only the parental genotypes, but also the offspring's phenotypes, all the FBAT tests shown in equation (5)–(7) are conditional on , i = 1, …, N. Therefore, all the FBAT approaches based on the carefully imputed data will not be biased under the null hypothesis of no genetic association, provided the imputation of does not depend on Xi and the traits are chosen to be mean-centered.
The easiest way to estimate the missing phenotypes is to replace them by the mean of all observed phenotypes. In other words, if the j-th phenotype for the i-th subject Yij is missing, we can estimate it by the average of all observed phenotypes at the j-th measurement, i.e., .
Furthermore, we can apply the E-M algorithm to the incomplete data [10] to improve our imputation technique by considering the correlation among different measurements for the same subject. Suppose , similar as [3] we get solution of and Σ at the M-step; while at the E-step, we impute the missing part of based on its observed part and the current estimates of , Σ. Iteratively, we can keep updating the imputed values of missing phenotypes iteratively until reaching convergence.
Alternatively, based on conditional mean model, we assume that
| (17) |
where , and Ei = E(Xi ′ Pi). Conditional on the observed phenotypes, the missing part follows multivariate normal distribution
| (18) |
Therefore after we obtain the estimates of , and V, we can impute the missing values by
| (19) |
We can use the N* subjects who have complete m observations to get the ordinary least square estimates (OLS) for
| (20) |
| (21) |
| (22) |
By putting these LSEs into equation (19), we can get an imputed complete dataset, to which we then apply the FBAT approaches for testing.
Note that both the imputation technique based on conditional mean model and the imputation technique based on E-M algorithm impute the missing values without using any genotypic information of the offspring. Therefore when using all the FBAT approaches based on the imputed data we do not need to adjust their p values for using the genotypic data first to impute, then to test.
Simulation
In our simulations, the marker of interest is a bi-allelic locus. Assuming an additive genetic model, the parental genotypes P1 and P2 are independently generated by drawing from a binomial distribution B(2,p) where p is the minor allele frequency (MAF) of the target allele in the population. The genotype X of the offspring is obtained by simulated Mendelian transmission based on the parental genotypes P1 and P2. For each offspring, the same type of phenotype is measured 6 times. The 6-dimensional phenotypic vector is a random sample from a multivariate normal distribution
| (23) |
where VP is the phenotypic variance-covariance matrix, is the phenotypic mean and α1, …, α6 are the genetic effects for measurement 1 to 6, respectively.
The simulation is repeated 5,000 times, in each replicate, 400 trios are generated for analysis. The power of each approach is estimated by the proportion of the number of times when the test statistic is significant at α level = 0.05. We only report results for MAF p = 0.2, as results for other values are very similar. Since the power of a statistical test heavily depends upon the true underlying model, we perform our simulations under several different models for the genetic effects α1, …, α6. In all the models, the variances at each measurement are set to σ2i = 1, i = 1, …, 6, while the correlation matrix CP is chosen to compound symmetry with various correlation values. In other words,
where ρ is the correlation among different measurements for the same subject. Therefore, we have
Model 1: No genetic effect at any measurement point
Under the null hypothesis, there is no genetic association at all (i.e. the genetic effect is zero for any of the six measurement points), so the phenotypes are generated from αi = 0, i = 1, …, 6.
Model 2: Same genetic effects across all measurement points In this model, we assume that αi = αh, i = 1, …, 6, where αh is the genetic effect size that corresponds to the heritability h2 [5], i.e.,
for an additive genetic model. h2 is always set to be 0.01 in model 2 and model 3.
Model 3: Arbitrary effects for different measurement points Here the values of α1, …, 6 are given by
| (24) |
where U is the uniform distribution on the interval. Since the mean of the uniform distribution is αh, the average genetic effect here is also αh, with average univariate heritability equals to 0.01.
Generate Missingness
After the complete dataset is simulated, we consider two different mechanisms to generate the possible missingness. Under MCAR, every phenotype Yij is set to be missing with a fixed probability Pmiss, i.e., each phenotype has a Pmiss chance to be removed from the observed dataset. In addition, we consider both high missing rate (Pmiss = 20%) and low missing rate (Pmiss = 5%).
The other mechanism we considered is missing at random (MAR). For this situation, we assume that the pattern of missing phenotypes depends upon the number of target allele at the marker locus, as well as the previous phenotypic observation. For simplicity, we assume that the first measurement is observed for all subjects, and each following phenotype for the i-th subject Yij, j = 2, …, 6 has a probability Pimiss to be missing. Here Pimiss is modeled by
| (25) |
where a = −0.65626, b = −0.0655 and c = 0.39969 are obtained via logistic regression fitted for missing measurements of body mass index in the Framingham Heart Study.
Results
For various values of the correlation ρ, we examine the type-I error rates of FBAT-PCM, FBAT-LC, FBAT-LCC, as well as ordinary Bonferroni correction [11] under the null hypothesis of no genetic association (model 1). Regardless of the missing mechanism (MCAR or MAR) and the missing rate (Pmiss = 5% or Pmiss = 20%), the type-I error rates are all well maintained for each method discussed in the Methods section. As previously mentioned, this is due to the fact that all the FBAT tests are conditional on the phenotypes and the traits are set to be mean-centered.
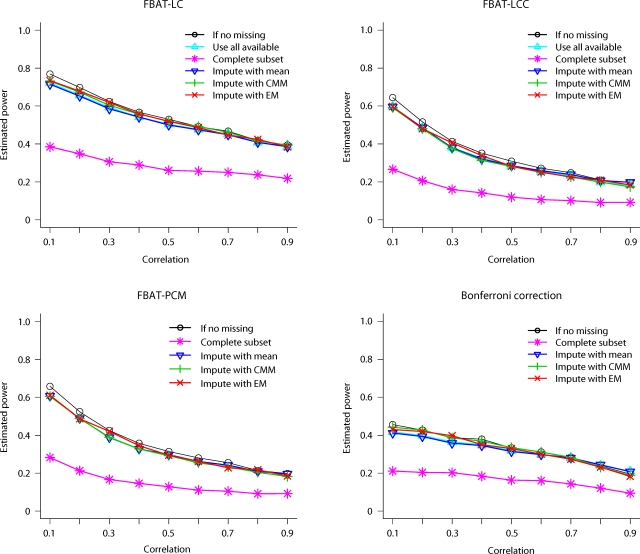
For MCAR and Pmiss = 20%, the estimated power curves of FBAT approaches with different methods to handle missingness are shown in figure 1 and 2, under model 2 and 3, respectively. In figure 1, we see that the complete data analysis suffers a substantial loss of power, compared to any other method. We also find that imputation technique based on the E-M algorithm has a considerable higher power than other ways of handling missingness when FBAT-LC approach is used, which is the most powerful test under model 2.
Fig. 1.
Estimated power of FBAT approaches, when genetic effects are same and the missing rate is high (MCAR).
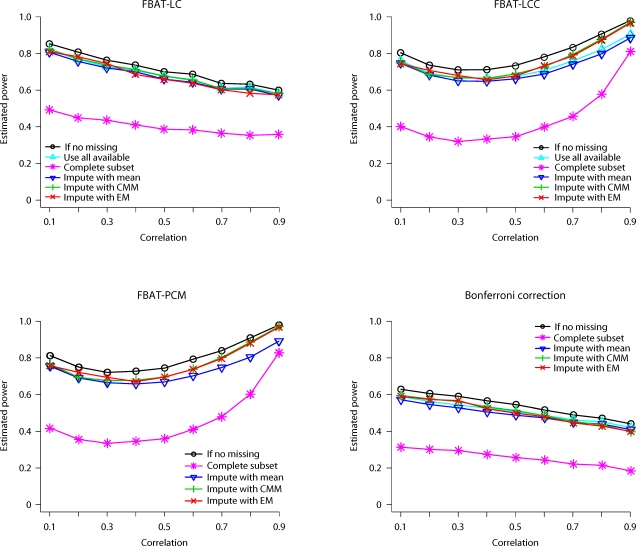
Fig. 2.
Estimated power of FBAT approaches, when genetic effects are uniformly distributed and missing rate is high (MCAR).
Furthermore, as shown in figure 2, the complete data analysis also loses substantial power under model 3. Other methods have almost identical power when the phenotypic correlation is low. On the other hand, when the correlation is high, the imputation technique based on CMM or E-M has substantially higher power than the mean-imputation technique or FBAT-LC/LCC based on all available data.
When the missing rate is relatively low (Pmiss = 5%), the results are quite similar to figure 1 and 2. Discarding all the subjects with any missing observation can still cause a non-negligible loss of power (up to 20%). Other methods to deal with missing data all perform well, especially when the genetic effects are same (all of them almost achieve the power if all phenotypes are actually observed). When the genetic effect sizes differ, imputing the missing values based on E-M algorithm is slightly more powerful than other methods, and the advantage tends to be bigger when the correlation is higher.
Furthermore, the results are still similar when the missing mechanism is MAR instead of MCAR. We find that imputation technique of conditional mean model is still almost identical to the imputation technique of E-M algorithm, and has substantially higher power than other methods. In addition, FBAT-LC-obs and FBAT-LCC-obs also show a noticeable gain of power, compared to mean-imputation or complete data analysis.
Data Analysis
We apply FBAT approaches to test the association between SNP rs7566605 and Body Mass Index (BMI) in the Framingham Heart Study (FHS) offspring cohort.
The Framingham Heart Study is conducted and supported by the National Heart, Lung and Blood Institute (NHLBI) in collaboration with Boston University and the participants are enrolled from the community without ascertainment for a particular trait or disease [12, 13]. SNP rs7566605 is located on chromosome 2q14.2 near the INSIG2 gene and is reported to be associated with obesity in several populations [13]. Six longitudinal measurements of Body Mass Index (BMI) over a follow-up period of 24–25 years, as well as family genotypic information at SNP rs7566605 are provided for study subjects.
Many different family structures exist in the FHS data. For simplicity, we only use the 70 trios (one offspring with the parent-pair) to compare the performance of different methods for handling missingness. For the 70 offspring, there should be 70 × 6 = 420 measurements of BMI, given six per subject. In fact, we have a total of 385 observations, which means the missing rate here is about 8.3%. Furthermore, only 51 offspring have complete six observations. In other words, if we are going to discard subjects with any missing value, our sample size will be only 72.9% of the original size.
For testing approaches FBAT-PCM, FBAT-LC, FBAT-LCC and Bonferroni correction, five different methods to deal with missing values are used here: use the complete data subset, use all available observations, impute the missing by phenotypic mean, impute the missing by conditional mean model, or impute the missing by E-M algorithm. As shown in table 1, due to the small sample size (only 17 out of the 70 trios are informative), after adjusting the p value for multiple comparison, Bonferroni correction does not show any significance, no matter which method is used to handle missingness. In addition, the results for FBAT-LC are basically unaffected by which method is used to handle the missingness.
Table 1.
Testing for association between rs7566605 and BMI in FHS trios data, with missing measurements
| p values |
||||
|---|---|---|---|---|
| FBAT-PCM | FBAT-LCC | FBAT-LC | Bonferroni correction | |
| Use complete subset | 0.083 | 0.045 | 0.035 | 0.18 |
| Use all available | N/A | 0.28 | 0.057 | 0.18 |
| Impute by mean | 0.15 | 0.27 | 0.057 | 0.18 |
| Impute by CMM model | 0.021 | 0.034 | 0.053 | 0.19 |
| Impute by E-M | 0.023 | 0.028 | 0.053 | 0.18 |
p values for FBAT approaches, with different methods to handle missingness.
The p values for imputation technique of CMM are always quite similar to those for imputation technique of E-M. Compared to these two imputation techniques, the mean imputation yields substantially larger p values, since it does not utilize the correlation structure in the data. This is consistent with the result shown in the simulation studies. In addition, When the missing phenotypes are imputed by conditional mean model or E-M algorithm, the most significant results are achieved by FBAT-PCM and FBAT-LCC. This is also consistent with the previous finding that FBAT-PCM and FBAT-LCC tend to have the highest power in the FHS data since the phenotypic correlation is high and the estimated genetic effect sizes show difference over time.
Interestingly, the results of FBAT-LCC and FBAT-LC are also nominally significant when only the complete data subset is used. This is probably due to the fact that the genetic effect for the first BMI measurement is the biggest, and there are no missing observations for the first BMI. In addition, a simple logistic regression model (equation 28) shows that the chance that an offspring's second BMI measurement is missing is significantly associated with the value of his or her first BMI measurement (p = 0.007), as well as genotype at SNP rs7566605 (p = 0.003).
Discussion
Missing phenotypes are a common problem for genetic association studies with longitudinal or repeated measurements. Here we discuss several ways for handling the missingness to improve the power of previously introduced FBAT approaches, because the complete case analysis suffers substantial loss of power even when the missing rate is as low as 5%.
In this paper, we extend FBAT-LC and FBAT-LCC statistics to allow incomplete phenotypes for study subjects. Generally, FBAT-LC-obs and FBAT-LCC-obs based on the observed data outperform the mean-imputation technique, but are not as powerful as other proposed imputation techniques.
Since the test statistics of these FBAT approaches are conditional on the phenotypes, we can impute the missing data without biasing the subsequent tests, provided that the imputation does not involve the offspring's genotypes. We propose an imputation technique that uses the E-M algorithm, whose false positive rate and significance level are always correctly controlled. We also show that this method consistently has higher power than mean-imputation, whose gain of power can be as high as 20%. In addition, if the phenotypic correlation is very high, this method can almost achieve the same power as the no missing situation.
Alternatively, we present another imputation technique which is based on the conditional mean model. This technique is more straightforward to use and involves less computation than the technique using E-M algorithm. Both the simulation studies and the example of FHS data analysis suggest that imputing by conditional mean model is generally as powerful as imputing based on E-M algorithm. We think that this simple imputation technique is practically useful for genetic association studies.
The computation of all these FBAT approaches is straightforward once you have all the univariate FBAT test statistics. In addition, univariate FBAT and FBATLC have been implemented in the software package FBAT and is freely available at http://www.biostat.harvard.edu/∼fbat/default.html; FBAT-PC and FBAT-PCM have been implemented in the software package PBAT and is freely available at http://www.biostat.harvard.edu/∼clange/default.htm.
Acknowledgements
This study was supported by the National Institutes of Health (NIH) grants GM 029745 and MH 05932.
References
- 1.Lange C, Andrew T, MacGregor AJ, Lyon H, Raby B, DeMeo D, Murphy AJ, Silverman AK, Weiss ST, Laird NM. A family-based association test for repeatedly measured quantitative traits adjusting for unknown environmental and/or polygenic effects. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1067. Article 17. [DOI] [PubMed] [Google Scholar]
- 2.Ding X, Lange C, Xu X, Laird NM. ‘New powerful approaches for family-based association tests with longitudinal measurements’. Ann Hum Genet. 2009;73:74–83. doi: 10.1111/j.1469-1809.2008.00481.x. [DOI] [PubMed] [Google Scholar]
- 3.Little RJA, Rubin DB. Statistical Analysis with Missing Data. New York: John Wiley; 1987. [Google Scholar]
- 4.Rabinowitz D, Laird NM. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- 5.Falconer DS, Macky TFC: Introduction to quantitative genetics. Longman, 1997.
- 6.Lunetta KL, Farove SV, Biederman J, Laird NM. Family-based tests of association and linkage using unaffected sibs, covariates and interaction. Am J Hum Genet. 2000;66:605–614. doi: 10.1086/302782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lange C, Silverman EK, Xu X, Weiss ST, Laird NM. A multivariate familybased association test using generalized estimating equations: FBAT-GEE. Biostatistics. 2003;4:195–206. doi: 10.1093/biostatistics/4.2.195. [DOI] [PubMed] [Google Scholar]
- 8.Lange C, Laird NM. Analytical sample size and power calculations for a general class of family-based association tests: Dichotomous traits. Am J Hum Genet. 2003;23:165–180. doi: 10.1086/342406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lange C, Lyon H, DeMeo D, Raby B, Silverman AK, Weiss ST. A new powerful non-parametric two-stage approach for testing multiple phenotypes in family-based association studies. Hum Hered. 2003;56:10–17. doi: 10.1159/000073728. [DOI] [PubMed] [Google Scholar]
- 10.Laird NM: Analysis of longitudinal and cluster-correlated data. NSF-CBMS Regional Conference Series in Probability and Statistics 2004;8.
- 11.Shaffer JP. Multiple hypothesis testing. Anm Rev Psych. 1995;46:561–584. [Google Scholar]
- 12.Kannel WB. The Framingham Study: Its 50-Year Legacy and Future Promise. J Atheroscler Thromb. 2000;6(2):60–66. doi: 10.5551/jat1994.6.60. [DOI] [PubMed] [Google Scholar]
- 13.Herbert A, Gerry NP, McQueen MB, Heid IN, Pfeufer A, Illig T, Wichmann HE, Meitinger T, Hunter D, Hu FB, Colditz G, Hinney A, Hebebrand J, Koberwitz K, Zhu X, Cooper R, Ardlie K, Lyon H, Hirschhorn JN, Laird NM, Lenburg ME, Lange C, Christman MF. A common genetic variant is associated with adult and childhood obesity. Science. 2006;312:279–283. doi: 10.1126/science.1124779. [DOI] [PubMed] [Google Scholar]