Summary
A technique has been developed for the separation of proteins by two-dimensional polyacrylamide gel electrophoresis. Due to its resolution and sensitivity, this technique is a powerful tool for the analysis and detection of proteins from complex biological sources. Proteins are separated according to isoelectric point by isoelectric focusing in the first dimension, and according to molecular weight by sodium dodecyl sulfate electrophoresis in the second dimension. Since these two parameters are unrelated, it is possible to obtain an almost uniform distribution of protein spots across a two-dimensional gel. This technique has resolved 1100 different components from Escherichia coli and should be capable of resolving a maximum of 5000 proteins. A protein containing as little as one disintegration per min of either 14C or 35S can be detected by autoradiography. A protein which constitutes 10−4 to 10−5% of the total protein can be detected and quantified by autoradiography. The reproducibility of the separation is sufficient to permit each spot on one separation to be matched with a spot on a different separation. This technique provides a method for estimation (at the described sensitivities) of the number of proteins made by any biological system. This system can resolve proteins differing in a single charge and consequently can be used in the analysis of in vivo modifications resulting in a change in charge. Proteins whose charge is changed by missense mutations can be identified. A detailed description of the methods as well as the characteristics of this system are presented.
Polyacrylamide gel electrophoresis has been extremely useful as an analytical tool for the separation and quantification of protein species from complex mixtures. In bacteriophage, where a major proportion of the viral proteins can be resolved, the combination of genetics and analysis by electrophoresis has yielded significant information concerning gene regulation and phage morphogenesis (for example, Refs. 1–5). In systems more complex than bacteriophage the response to pleiotropic effectors, developmental transitions or mutation cannot be adequately analyzed by means of any one-dimensional technique for protein separation unless the analysis of a very restricted subset of the total proteins is acceptable. In order to provide a suitable technique for a more extensive analysis of complex systems, I have developed this technique for the separation of total protein.
In terms of the number of components resolved, previous techniques for two-dimensional electrophoretic protein separation (for example, Refs. 6–11) were not significantly better than one-dimensional separation. Only the procedure of Kaltschmidt and Wittman (12) has been widely used. Although this technique is of limited resolution and applicability, it has been used as the basis for many investigations of ribosomal assembly and structure (for example, Refs. 13–16).
To optimize separation, each dimension must separate proteins according to independent parameters. Otherwise proteins will be distributed across a diagonal rather than across the entire surface of the gel. Isoelectric focusing and a discontinuous SDS1 gel system (1) were chosen because of the high resolution of each system and because they separate proteins according to different properties. Since the procedure is intended for analysis of total proteins, denaturation agents which solubilize most proteins are present during electrophoresis in both dimensions. This system permits simultaneous determination of molecular weights and approximate isoelectric points of proteins. More than 1000 proteins can be resolved and a protein species representing as little as 10−4 to 10−5 of 1% of the total protein can be detected and quantified. Since the position of a spot changes detectably if a single charge is altered, some missense mutations can be detected.
Materials and Methods
Chemicals
Ampholines were obtained from LKB. Several different batch numbers were used during the course of this work. The quality of the gels varied only slightly, but precise reproduction of a separation should not be expected when the Ampholines are changed. Nonidet P-40 (NP-40) was purchased from Imperial Shell. SDS, manufactured by British Drug House Chemical Ltd., was purchased from Gallard-Schlesinger. Acrylamide, N,N′-methylene-bisacrylamide and N,N,N′,N′-tetramethylethylenediamine (TEMED) were purchased from Eastman-Kodak. Stock solutions of acrylamide were prepared and filtered and used without further purification. Glycine and ammonium persulfate were obtained from Fisher Scientific Co. and Tris base from Sigma. Ultrapure grade of urea was purchased from Schwarz/Mann. Other materials were obtained from more than one supplier with no apparent effect on the results.
Buffers and Solutions
A, lysis buffer: 9.5 m urea, 2% (w/v) NP-40, 2% Ampholines (comprised of 1.6% pH range 5 to 7 and 0.4% pH range 3 to 10) and 5% β-mercaptoethanol. (This lysis buffer is stored as frozen aliquots.) B, sonication buffer: 0.01 m Tris-HCl, pH 7.4, 5 mm MgCl2, 50 μg of pancreatic RNase per ml. C, stock solution of pancreatic DNase: 1 mg per ml in 0.01 m Tris-HCl pH 7.4, 1 mm MgCl2 (stored as frozen aliquots). D, 30% acrylamide stock for isoelectric focusing gels: 28.38% (w/v) acrylamide and 1.62% bisacrylamide. E, stock Nonidet P-40 solution: 10% (w/v) NP-40 in H2O. F, Ampholines were used as supplied: 40% (w/v) solutions. G, ammonium persulfate: a fresh 10% (w/v) solution was prepared every two weeks. H, gel overlay solution: 8 m urea (stored as frozen aliquots). I, anode electrode solution: 0.01 m H3PO4. J, cathode electrode solution: 0.02 m NaOH (degassed and stored under vacuum). K, sample overlay solution: 9 m urea, 1% Ampholines (comprised of 0.8% pH range 5 to 7 and 0.2% pH range 3 to 10, stored as frozen aliquots). L, lower gel buffer: 1.5 m Tris-HCl, pH 8.8, and 0.4% SDS. M, upper gel buffer: 0.5 m Tris-HCl, pH 6.8, and 0.4% SDS. N, 30% acrylamide stock for SDS gels: 29.2% (w/v) acrylamide and 0.8% bisacrylamide. O, SDS sample buffer: 10% (w/v) glycerol, 5% (v/v) β-mercaptoethanol, 2.3% (w/v) SDS, and 0.0625 m Tris-HCl, pH 6.8. P, agarose gel: 1 g of agarose was melted in 100 ml of Buffer O, immediately divided into aliquots, and stored at 4°. Q, running buffer: 0.025 m Tris base, 0.192 m glycine, and 0.1% SDS. R, running buffer with high SDS: 0.025 m Tris base, 0.192 m glycine, and 2% SDS. S, bromphenol blue: 0.1% bromphenol blue. T, staining solution: 50% (w/v) trichloroacetic acid, 0.1% Coomassie blue. U, destaining solution: 7% (v/v) acetic acid.
Solutions B, D, E, F, G, L, M, N, and O were stored at 4°. Solutions I, J, Q, R, S, T, and U were stored at room temperature and all other solutions were stored as indicated.
Radioactive Labeling of Proteins
The gel separations presented represent radioactively labeled proteins from Escherichia coli, bacteriophage T4 infected E. coli, or the nematode Caenorhabditis elegans. E. coli strains AS19 (17), 1100 (18), or 5333 (19) were grown at 30° in M9 media (20) plus the appropriate supplements to between 1 × 108 and 3 × 108 cells per ml. The cultures were labeled with mixed 14C-amino-acids from 5 μCi per ml to 20 μCi per ml. The labeling period varied from 5 to 30 min.
The T4 samples were prepared as follows. E. coli B AS19 (17) cultures were infected with the appropriate phage at a multiplicity of eight. These infections were labeled from 3 to 10 min postinfection with mixed 14C-amino-acids at an isotopic concentration of 5 μCi per ml. One- or two-milliliter cultures of either infected or uninfected cells were used for all labelings. All of these labelings were terminated by the addition of casamino acids (final concentration of 1%) and chilling the cultures on ice. These cultures were centrifuged at 10,000 × g for 5 min, and the pellets were treated as described under “Sample Preparation.”
In one case (Fig. 13), E. coli AS19 was labeled with [35S]sulfate. E. coli was grown to 109 cells per ml in M9 media, and 20 μl of this culture were added to 2 ml of sulfate-free M9 containing 200 μCi of carrier-free 35SO4−. This culture was incubated for 6 hours, at which time 50% of the radioactivity was incorporated.
Fig. 13.
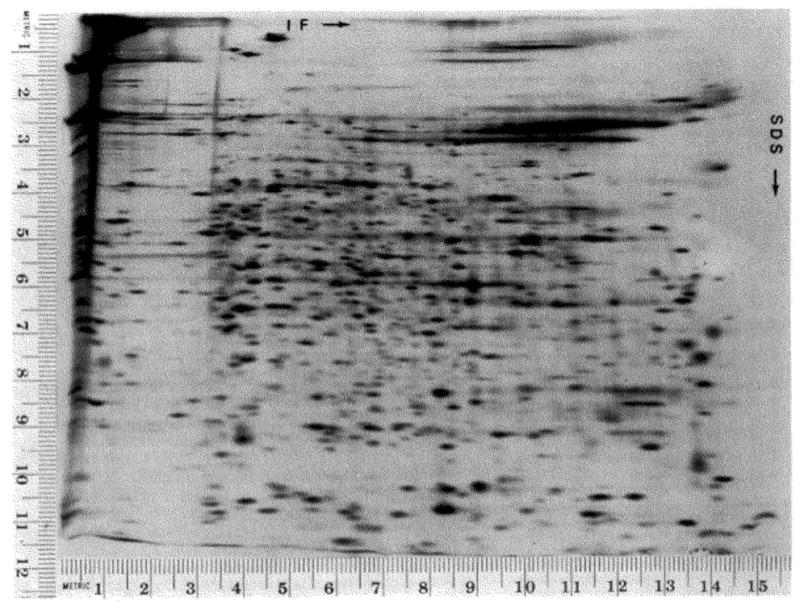
Separation of proteins from Caenorhabdilis elegans. C. elegans was labeled as described under “Materials and Methods,” and lysed by sonication. The lysate was treated with RNase and DNase, and urea and lysis buffer were added. The sample applied to the gel contained 400,000 cpm and 3 μg of protein. The autoradiogram shown was exposed to the gel for 515 hours.
Nematodes (C. elegans) were labeled by feeding them 35S-labeled E. coli. The labeling period was followed by a chase period of 30 min during which time the nematodes were fed unlabeled E. coli. After this time no labeled E. coli proteins could be detected.
Sample Preparation
All procedures for sample preparation must avoid conditions that result in the chemical modification of proteins. The gel system is sensitive to single charge changes and modifications frequently cause a single protein to give two or more spots. Nucleic acids in the sample interfere with the isoelectric focusing. Usually the effect of nucleic acids is not severe.
I have obtained satisfactory results by suspending a pellet of cells (E. coli) in lysis buffer, then freezing, and thawing (3 to 5 cycles). An improvement is to sonicate the cells (normally in 100 μl) in sonication buffer. After sonication, DNase is added to 50 μg per ml. This mixture is allowed to stand on ice for at least 5 min; then solid urea is added to bring the concentration to 9 m urea. This mixture is taken off ice, and 1 volume of lysis buffer (A) is added. The urea goes into solution at room temperature. If a sample is too dilute, the protein can be precipitated with saturated (NH4)2SO4 or lyophilized and then dissolved in lysis buffer.
Once samples are in lysis buffer, they should either be run immediately or frozen. Samples have been stored frozen in lysis buffer for up to 2 months without adverse effects. Under some other conditions of storage, artifactual spots are generated. Ideally, less than 40 μg of protein containing more than 100,000 cpm are loaded in 25 μl or less (for details, see “Results and Discussion”).
First Dimension
Isoelectric focusing gels are made in glass tubing (130 × 2.5 mm inside diameter) sealed at the bottom with Parafilm. The length of the gel tube and the length of the gel itself affect the reproducibility, and care should be taken to keep these measurements constant between samples that are to be compared. To make 10 ml of gel mixture (about 0.5 ml of gel mixture per gel tube), 5.5 g of urea are added to a 125-ml side arm flask, then 1.33 ml of acrylamide stock (D), 2 ml of stock NP-40 (E), 1.97 ml of H2O + 0.4 ml of Ampholines, pH range 5 to 7, + 0.1 ml of pH 3 to 10 (to make 2% Ampholines). The flask is swirled until the urea is completely dissolved, then 10 μl of 10% ammonium persulfate are added and the solution is degassed under vacuum for about 1 min. Immediately after addition of 7 μl of TEMED, the solution is loaded into the gel tubes. To avoid trapping bubbles, a syringe with a long narrow gauge hypodermic needle is used to fill the tubes with gel mixture. The tubes are filled to approximately 5 mm from the top. This can be varied to hold samples of different volumes. The gel is overlayed with gel overlay solution (H) and after 1 to 2 hours this overlay solution is removed and replaced with 20 μl of lysis buffer (A) overlayed with a small amount of water. The gels are allowed to set for 1 to 2 more hours. The Parafilm is removed, and the ends of the tubes are covered with dialysis membrane held in place by a 3-mm section of latex tubing. The gels are then placed in a standard tube gel electrophoresis chamber. The lysis buffer (A) and water are removed from the surface of the gel and 20 μl of fresh lysis buffer (A) are added. The tubes are then filled with 0.02 m NaOH. The lower reservoir is filled with 0.01 m H3PO4 and the upper reservoir is filled with 0.02 m NaOH which should be extensively degassed to remove CO2. The gels are then prerun according to the following schedule: (a) 200 volts for ¼ hour; (b) 300 volts for ½ hour; (c) 400 volts for ½ hour. The power is turned off, the upper reservoir is emptied, lysis buffer and NaOH are removed from the surface of the gels, and the samples are loaded. Samples are most easily loaded with an Eppendorf micropipette or equivalent. The samples are overlayed with 10 μl of sample overlay solution (K) then 0.02 m NaOH, and the chamber is refilled. After the samples are loaded, the gels are run at 400 volts for 12 hours and then at 800 volts for 1 hour. This schedule for the running of isoelectric focusing gels can be varied without effect on the quality of the gels; changes will affect the absolute positions of spots. The product of the voltage and the time (in hours) should total more than 5,000 and less than 10,000 volt hours for gels of this length, while the final voltage should be a minimum of 400. If gels are run at too high a voltage, bands become distorted.
To remove the gels from the tubes, a 5-ml syringe is connected to the electrophoresis tube via a short piece of Tygon tubing, and the gel is slowly forced out by pressure on the syringe. The gel can be treated in any of the following four ways. It can be (a) equilibrated and immediately loaded on the second dimension gel, (b) equilibrated and frozen, (c) frozen without prior equilibration, and (d) loaded on an appropriate second dimension gel without any treatment. The first procedure will be described now and the others will be discussed later (see “Freezing and Storage of Isoelectric Focusing Gels” and “Nonequilibrated Gels”).
Equilibration
The isoelectric focusing gel is extruded (as described above) into 5 ml of SDS sample buffer and shaken at room temperature for at least 30 min. The best results have been obtained with a 2-hour equilibration time.
Second Dimension
The second dimension is the discontinuous SDS gel system which, as first described by Laemmli (1), consists of an Ornstein (21) and Davis (22) stacking system with SDS added.
Apparatus
The apparatus for running slabs is similar to that described by Reid and Bieleski (23) except the slot was replaced with a notch (24), and the height of the lower reservoir was increased to within 25 mm of the base of the notch so that the buffer in the lower chamber can provide cooling. A silicone rubber gasket was also added to the back of the apparatus for sealing the gel to the tank. The total height of the apparatus was increased to 185 mm to accommodate longer gels.
Glass plates used in making the slab gels are made as follows. Window glass is cut into squares 164 × 164 mm. Notches are cut 18 mm deep and 134 mm long on one edge. Rectangles of 164 × 33 mm are also cut. To form the back plate (Fig. 1C), a 164 × 33-mm rectangle is glued to a notched plate so that the notch is covered (Fig. 1, C and D). The corner formed between the base of the notch and the rectangular back is filled with epoxy to form a surface at a 45° angle (Fig. 1). The front plate is formed by gluing two notched plates together with one plate displaced by 6 mm (Fig. 1B). The corner at the base of the notch is again filled with epoxy (Fig. 1). Rubber strips (0.8 × approximately 5 mm) are used as spacers and a little silicone grease is used to make a water-tight seal. The beveled edges face each other when the plates are assembled and clamped together (Fig. 1A).
Fig. 1.
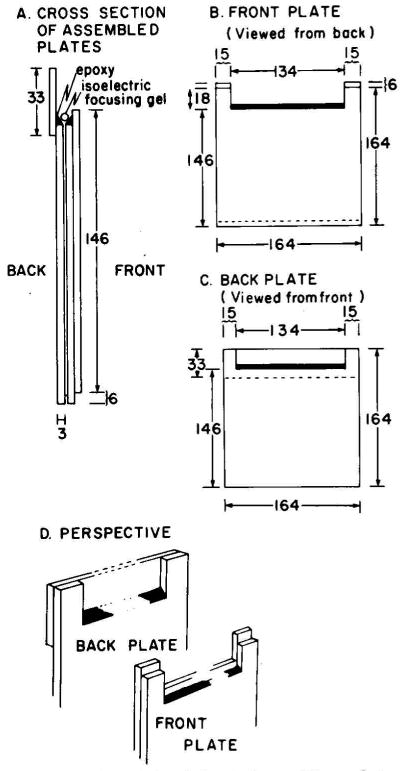
Illustration of the slab gel plates. These plates are prepared as described in the text. All measurements are given in mm.
Preparation of Slab Gels
Uniform gels of any acrylamide concentration can be poured as well as gradient gels. The best separation of proteins is achieved using an exponential acrylamide gradient gel (see “Distribution of Proteins” under “Results and Discussion”). These gradients can be poured from a standard two-chambered gradient mixer where the volume in the front chamber is held constant by plugging with a stopper (the pressure is equilibrated by momentary insertion of a hypodermic needle through the stopper). The front chamber is stirred with a magnetic stirrer and the gradient is poured at about 3 ml per min.
The stock solutions are mixed in the following way to make 1 volume of the running gel (5 to 22.5% acrylamide, 0.1% SDS, 0.375 m Tris-HCl, pH 8.8); 0.25 volume of lower gel buffer (L), 0.75 volume of acrylamide stock (N), and water (the proportions of acrylamide stock to water are varied to give different gel concentrations up to 22.5% acrylamide), 0.0033 volume of 10% ammonium persulfate (G). The above mixture is degassed, 0.0005 volume of TEMED is added, and it is poured between the glass plates. For gradient gels, 75% glycerol is used instead of water when the high concentration acrylamide solution is prepared and in both solutions the volume of 10% ammonium persulfate is reduced to 0.00145 volume. The polymerization mixture is added to a level 25 mm below the bottom of the notch and is gently overlayed with about 2 ml of H2O. This mixture is allowed to polymerize for 1 hour, then the water and the residual unpolymerized gel mixture are removed from the gel surface. The gel is covered with the lower gel buffer which has been diluted 4-fold. The gel is allowed to stand overnight. The following day, the stacking gel mixture (4.75% acrylamide, 0.1% SDS, and 0.125 m Tris-HCl, pH 6.8) is prepared as follows: 0.25 volume of upper gel buffer (M), 0.15 volume of the acrylamide stock for SDS gels (N), 0.6 volume of H2O, 0.003 volume of 10% ammonium persulfate, and 0.001 volume of TEMED. Prior to addition of the TEMED, this mixture is degassed and the solution which is covering the running gel is removed completely. The chamber is filled with the complete stacking gel mixture to the base of the notch. A Teflon strip (0.8 × 130 × 20 mm) is inserted 1 to 2 mm below the base of the notch to form a flat surface on top of the stacking gel. Care should be taken so that no bubbles are trapped under this Teflon strip. The stacking gel is allowed to polymerize for 30 to 60 min.
Loading Isoelectric Focusing Gel onto Slab Gel
A 1% agarose gel (P) is used to keep the cylindrical isoelectric focusing gel in place and to avoid mixing as the protein zones migrate out of the cylinder into the slab. The Teflon strip is removed from the gel and all unpolymerized gel solution is removed from the surface. The cylindrical gel is placed on a piece of Parafilm, straightened out, and placed close and parallel to one edge of the Parafilm. One milliliter of the melted agarose solution (held at about 80°) is put in the notch, then the Parafilm is used to transfer quickly the cylindrical gel into this solution.
Electrophoresis in Second Dimension
About 5 min are allowed for the agarose to set, then the bottom gasket is removed, and the gel is clamped to the tank. Running buffer is added and bubbles are removed from the bottom of the gel by a stream of running buffer from a cannula which is bent at the end and attached to a 10-ml syringe. Then 0.04 ml of 0.1% bromphenol blue is added to the upper reservoir. Gels are run at 20 ma constant current until the dye front reaches the bottom of the gel. The running time is about 5 hours.
Staining, Drying, and Autoradiography
The remaining rubber gaskets are removed and the gel plates are pried apart with a spatula. The gel is stained in 0.1% Coomassie blue in 50% trichloroacetic acid for 20 min and destained in several changes of 7% acetic acid. Destaining solutions are stirred by a magnetic stirrer with the magnet inside the bottom of an inverted plastic Petri dish with holes cut in the side; this protects the gel. Gels are dried as previously described (25) and exposed to No-Screen Kodak medical x-ray film which is processed according to standard procedures. Gels can also be dried without prior staining and destaining.
Nonequilibrated Gels
If it is important to avoid any losses of protein, gels can be run without equilibrating the isoelectric focusing gel. This requires the following modifications of the described procedures. The running gel is poured to 50 mm below the base of the notch. A 50-mm stacking gel is poured in the same way that the normal 25-mm stacking gel is formed. The isoelectric focusing gel is extruded directly onto a piece of Parafilm, and this gel is loaded on top of the slab in the same manner as described for equilibrated gels. These operations should be carried out rapidly to avoid crystalization of the urea which causes the isoelectric focusing gel to get rigid. The slab is mounted on the gel tank and high SDS running buffer (R) is put in the upper reservoir and regular running buffer (Q) is put in the lower reservoir. The gels are then run at 20 ma for 20 min. The power is turned off and as much of the high SDS running buffer is removed as is possible and is replaced with regular running buffer. Bromphenol blue is added and electrophoresis is continued until the dye front reaches the end of the gel.
Freezing and Storage of Isoelectric Focusing Gel
At anytime during the equilibration the gel can be frozen and equilibration continued when the gel is thawed. To freeze a gel, it is placed in 5 ml of SDS sample buffer in a stoppered tube (25 ml) and the tube is placed on its side in an ethanol-Dry Ice bath. Frozen gels are best stored at −70°, particularly if there is a large amount of residual urea, as in nonqeuilibrated gels. If gels are frozen immediately they can either be equilibrated when thawed or run as if unequilibrated. Frozen gels can be stored indefinitely without altering the separation of proteins eventually attained.
Cleaning Glass Plates and Tubes
Residual grease and polyacrylamide gel is wiped off the plates; they are soaked in Chem-Solve (Mallinckrodt Chemical Works), rinsed, and dried. The glass tubes used for isoelectric focusing are soaked in chromic acid, rinsed in H2O, soaked in alcoholic KOH, rinsed extensively, and air dried.
Measurement of pH Gradient
The isoelectric focusing gel was cut into 5-mm sections which were either placed in individual vials with 2 ml of freshly prepared 9.2 m urea in degassed H2O or were placed in vials containing 2 ml of degassed H2O. These vials were capped and shaken for 5 to 10 min; then the pH was measured on a pH meter.
Determination of Spot Size and Standard Deviations
The spot areas were measured on photographic enlargements. The areas were measured by tracing the perimeters of the spots, cutting these out, and weighing them. The weights were corrected to give units of mm2 on the original autoradiogram.
The standard deviations (σ) of a spot were determined from tracings made using the Joyce Loebl microdensitometer. A tracing was made by scanning across the spot in the SDS dimension and measurements were made on the peak obtained. The peak width at one-half the maximal peak height was determined. This corresponds to σ in the SDS dimension. Similarly, the film was turned 90° and the spot was scanned in the isoelectric focusing dimension. Measurements made on the resulting tracing gave σ in the isoelectric focusing dimension.
Quantification of Autoradiograms
As a result of the high resolution of this system, a complete analysis of a separation by available procedures is very tedious. It is hoped that the analysis will eventually be automated through the use of a two-dimensional scanner and computer processing. In the present communication, measurements were made using the Joyce Loebl microdensitometer. Estimates of the amount of radioactivity in minor spots are made by determining the length of exposure required in order to detect a spot.
To measure the total radioactivity in a spot, the total optical density of the spot on the x-ray film is measured and related to radioactivity by means of a standard curve. The measurement of the optical density is made on the Joyce Loebl as follows. The width of the light beam must be sufficient to include the entire spot in one scan. The size of the recording beam can be changed by changing the objective in the condenser. Any selected spot can be positioned ahead of the light beam and scanned with the appropriate settings. The area under the peak (on the tracing obtained) is determined by cutting out the peak and weighing it. This area is assumed to correspond to the integrated optical density of the spot. Because the light beam averages transmittance rather than optical density, there is a small error in this assumption. Since transmittance does not decrease linearly as exposure is increased, this error should result in a nonlinear increase in the measured optical density of a spot as exposure is increased. However, all observed nonlinearities can be accounted for by the response of the film (see below). Since the error due to transmittance averaging was not detectable by these procedures, I have assumed that it can be neglected for determinations of integrated optical density.
To calibrate autoradiograms, standardization wedges were made as follows. Twenty 10% acrylamide gels (0.8 mm thick) were made. The first gel contained 2.75 × 104 cpm of 14C-labeled E. coli proteins per mm2 of surface area. Each successive gel contained half as much 14C-labeled protein as the preceding gel. These gels were fixed in 7% acetic acid, cut into small pieces (approximately 2.5 mm × 4 mm), arranged in sequences (1 through 20) on filter paper, and dried. Each of these sequences forms one standardization wedge. These wedges can be stored indefinitely. A wedge is exposed along with each film and provides an internal control for variations that can occur in autoradiography. Scans of these wedges were used to determine the sensitivity of a given exposure in cpm per mm2 and the range of optical densities across which the film response is linear, and to calibrate measurements of total optical density of a spot (described above).
The estimates of the amounts of radioactivity in the minor spots have been made using a series of different autoradiograms of known sensitivity. Since the area of a spot can be determined, the minimum detectable number of cpm in a protein can be estimated. Two autoradiograms of different sensitivity are compared. All spots which appear only on the more sensitive autoradiogram must contain less radioactivity than is required for detection on the less sensitive autoradiogram and more than is required for detection on the more sensitive autoradiogram. Autoradiograms of different sensitivities are prepared by exposing x-ray films to the same gel for different lengths of time. Since the areas of the major spots are greatly affected by the amount of protein present, these must be quantified by microdensitometry.
Subjective Quantification
The subjective impression that one spot is more or less intense than another is both sensitive and accurate. If two spots have a similar size, any intensity differences greater than 10% can be detected. However, subjective estimates of the degree of difference are extremely imprecise. The contrast range is compressed during photography of autoradiograms. Thus, photographic copies of x-ray films can enhance the apparent differences between some spots and greatly decrease the apparent differences between some others.
Results and Discussion
Fig. 2 demonstrates the capability of this gel electrophoresis system. It is clear from the number and the size of the spots that it represents a large improvement over any previously published one- or two-dimensional system for the separation of proteins. I will present a detailed description of the properties of this system and the effects of a number of variables.
Fig. 2.
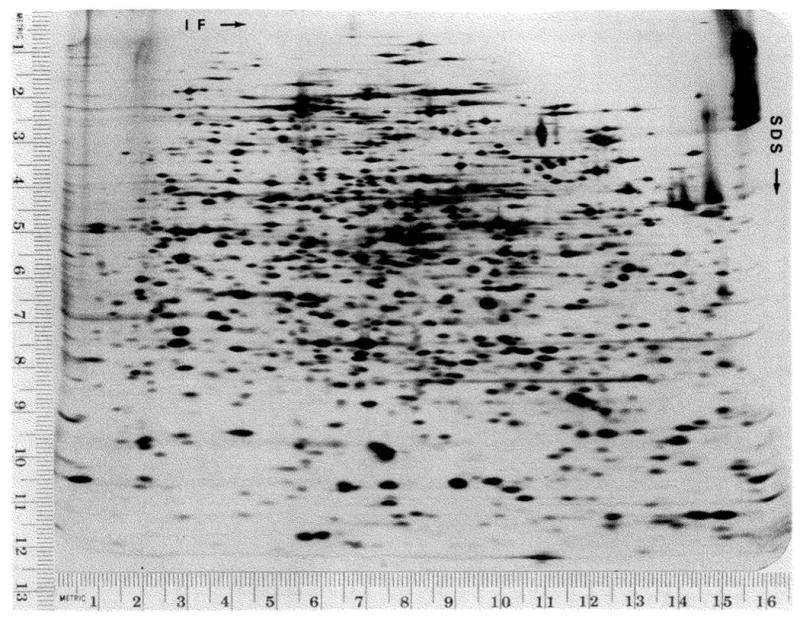
Separation of Escherichia coli proteins. E. coli (1100) was labeled with 14C-amino-acids as described under “Materials and Methods.” The cells were lysed by sonication, treated with DNase and RNase and dissolved in lysis buffer. Twenty-five microliters of sample containing 180,000 cpm and approximately 10 μg of protein were loaded on the gel. The isoelectric focusing gel was equilibrated for 30 min. The gel in the SDS dimension was a 9.25 to 14.4% exponential acrylamide gradient. A volume of 10 ml of 14.4% acrylamide was used in the front chamber of the gradient mixer. The total volume of the gel was 16 ml. At this exposure, 825 hours, it is possible to count 1000 spots on the original autoradiogram. All autoradiograms of two-dimensional gels were photographed with a metric ruler along two edges of the autoradiogram. These rulers establish a coordinate system which is used to give spot positions. The verticle scale is given in units from top to bottom. The horizontal scale is given in units from left to right. The coordinates are given as horizontal × vertical.
Resolving Power
Since isoelectric focusing resolves 70 Escherichia coli proteins and SDS electrophoresis on a gradient gel resolves 100, the use of these two procedures in a two-dimensional protocol should resolve 7000 components. However, I have never counted more than 1100 spots on a heavily exposed autoradiogram of a two-dimensional separation of E. coli proteins. This discrepancy, between the maximum calculated resolution and the observed, is primarily due to properties of the sample. As discussed under “Per cent Abundance,” in order to detect the minor components of the sample, the autoradiogram must be grossly overexposed with respect to the major components. This results in a loss of resolution due to a 2-fold increase in spot size for every 10-fold increase in exposure (autoradiographic spreading). In addition, E. coli, which has a genetic capacity to code for 4000 proteins (assuming that the E. coli chromosome is 4 × 106 base pairs and to code an average protein requires 1000 base pairs), is presumably incapable of saturating the resolving power of this gel system.
In order to determine whether there was a loss in resolution of one or both gel systems when they were combined as a two-dimensional system, the spot size on a two-dimensional gel was compared with the band widths on one-dimensional gels. Since the optical density across bands on both SDS gels and isoelectric focusing gels approximates a Gaussian distribution, and the distribution of densities in a spot on the two-dimensional gels approximates a two-dimensional Gaussian distribution, the widths will be given as the standard deviation (σ). The following abbreviations will be used in discussing the widths of bands and spots: σ 1D-SDS represents the standard deviation of bands on a one-dimensional SDS gel; σ 1D-IF represents the standard deviation of bands on a one-dimensional isoelectric focusing gel; σ 2D-SDS represents the standard deviation in the SDS dimension of a spot on a two-dimensional gel; and σ 2D-IF represents the standard deviation in the isoelectric focusing dimension of a spot on a two-dimensional gel. Since Gaussian distribution can be said to be resolved when their centers are separated by a distance 4 σ, resolution is inversely proportional to σ. Spots on a two-dimensional gel (Fig. 3) were selected and scanned in both dimensions. Spots were selected so that all optical densities were within the linear range of film response and so that spots from all areas of the film were scanned. Data from 25 spots were collected and averaged to give σ 2D-IF of 1.49 mm and σ 2D-SDS of 0.54 mm. Tracings of 16 bands on the SDS gel (Fig. 3) were analyzed to give a σ 1D-SDS of 0.65 mm. Similarly, tracings of 16 bands were used to determine a σ 1D-IF of 1.07 mm. The comparison of σ 1D-SDS to σ 2D-SDS shows no loss in resolution in the SDS dimension. The σ 2D-IF was 40% greater than σ 1D-IF. This was probably due to the diffusion of proteins during equilibration (see below) of the first dimension gel and running the second dimension gel. As a result of this increase in σ 2D-IF over σ 1D-IF, the resolution of the two-dimensional gel system is about 70% (σ 1D-IF/σ 2D-IF = 1/1.4) of the theoretical maximum. Thus, given an optimal sample it should be possible to resolve about 5000 components
Fig. 3.
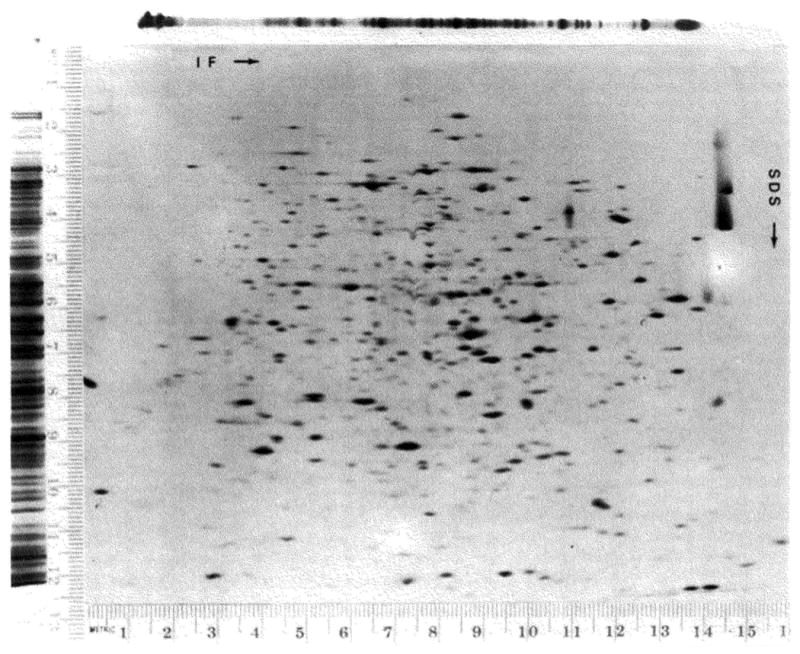
Comparison of spot dimensions to band widths. The one-dimensional SDS gel was run at the same time as the second dimension of the two-dimensional gel. Both SDS gels are exponential gradient gels (10 to 14% with 8 ml in the front chamber). The isoelectric focusing gel shown above the two-dimensional gel was run at the same time as the isoelectric focusing gel which was loaded on the second dimension. The one-dimensional isoelectric focusing gel shown was fixed in 50% trichloroacetic acid, rinsed in 7% acetic acid (overnight), and the whole gel was dried without any slicing. Lower exposures of the one-dimensional gels were scanned to obtain tracings from which the standard deviation was measured.
Loading Capacity
The size of a spot increases as the amount of protein increases as shown in Fig. 4. As seen from this figure, in order to run gels in which the spot size is not detectably influenced by the amount of material loaded, less than 0.1 μg of an individual protein can be loaded. For mixtures of proteins, loading is strongly influenced by the relative amounts of the component proteins. Since the major component in E. coli comprises about 10% of the total protein (see “Per cent Abundance”), 1 μg of total E. coli proteins can be loaded on the gels without any loading effect. However, since spot size can be considered for each spot independently, and since in a complex mixture of proteins only a small number of species of protein are present as major components, the gels can be severely overloaded for major species with only a small decrease in total resolution. In general the less protein loaded on the gel, the better the resolution. However, if detection (see below) requires the addition of a large amount of protein, up to 100 μg can be added.
Fig. 4.
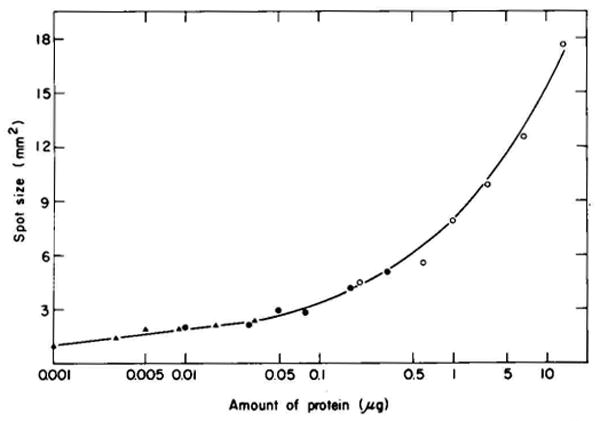
The effect of the amount of protein on spot size. The data were obtained from an analysis of six gels containing known amounts of Escherichia coli protein. Four of the gels used are shown in Fig. 5. The area of each spot and the amount of protein in each spot were determined as described under “Materials and Methods.” A different symbol is used to record the data collected for different spots. The coordinates given refer to Fig. 5A; ○ = 8.8 × 3.4, ● = 9.5 × 3.1, ▲ = 7.0 × 3.3. The size of these spots was measured from autoradiograms of much lower exposure than those shown in Fig. 5. Low exposure autoradiograms were used to avoid possible increases in spot size due to overexposure of the film.
In addition to the described effect on spot size, an increase in the amount of protein can cause distortion of spot shape, changes in spot position, and even selective loss of some spots. These effects become more severe as the amount of protein is increased (see Fig. 5). When less than 20 μg of E. coli protein was loaded, no significant loading effects were observed other than the effect on spot size (Fig. 5, compare A and B). Thus, when samples are to be compared, either less than 20 μg should be subjected to electrophoresis or the same amount of total protein must be loaded.
Fig. 5.
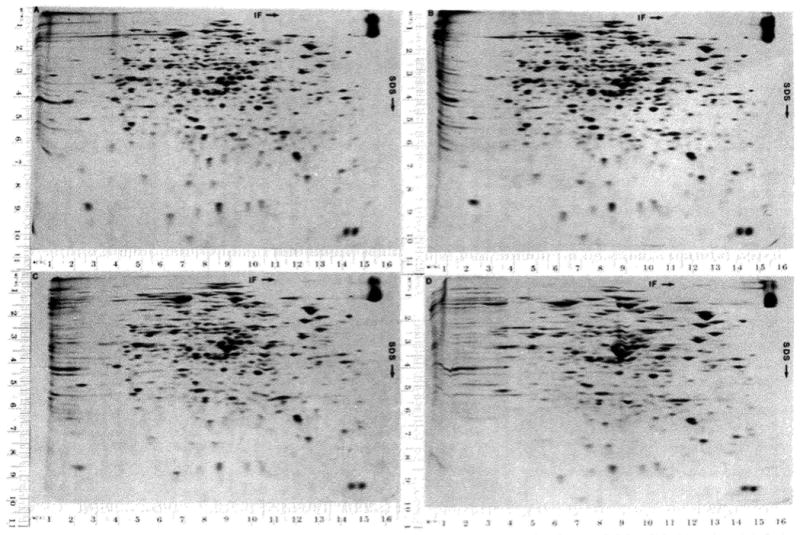
Effect of increasing the amount of protein on the resolution is illustrated by the four two-dimensional separations shown. In all cases 350,000 cpm in 4μ g of 14C-labeled Escherichia coli protein were loaded on the gel. In addition, 3, 16, 32, and 128 μg of unlabeled E. coli protein were loaded on Gels A, B, C, and D, respectively. These preparations of E. coli protein were not treated with RNase and DNase. The slab gel is a uniform gel of 12.5% acrylamide. The autoradiograms illustrated were exposed for 72 hours.
As the amount of protein is increased, the spots change in shape. Although this has little effect on the separation, it can be difficult to decide where the true position is. The true position is required for accurate matching of gels and for the accurate determination of molecular weight. As illustrated schematically in Fig. 6, and as can be seen in Fig. 5, the spot expands symmetrically in the isoelectric focusing dimension. However, in the SDS dimension the spot enlarges primarily on the lower molecular weight side. Thus, the spot center always corresponds to the true position in the isoelectric focusing dimension. A line drawn through the extremes of a spot as indicated in Fig. 6 gives the true position of a spot in the SDS dimension.
Fig. 6.
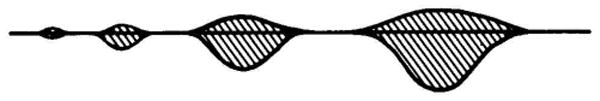
Effect of amount of protein on spot shapes. The asymmetric expansion of a spot is illustrated. The spots toward the right represent larger amounts of protein. The horizontal line indicates the position which should be used for determination of molecular weight. The spots are symmetric in the isoelectric focusing dimension.
One protein can exclude another from its normal position. This effect is partially responsible for the change in spot positions as the amount of protein loaded is increased as illustrated in Fig. 5 C and D (coordinates2 8.8 × 3.2). As a result, when a major spot is close to a minor spot and the gel is overloaded, the minor spot is shifted rather than being engulfed by the expanded major spot.
Reproducibility
When a sample is run on two different two-dimensional gels, each spot on one gel can be seen unambiguously to correspond to a single spot on the other gel. This is demonstrated in Fig. 7 and Fig. 5, A and B. The intensities of corresponding spots are indistinguishable and all artifacts such as trailing are reproduced. When the isoelectric focusing gels are run at different times, occasionally, there is a slight variation at the top (basic end) of the isoelectric focusing gel. This variation can cause ambiguities in the matching of about 1 % of the spots. The SDS gel can be run at different times without affecting reproducibility.
Fig. 7.
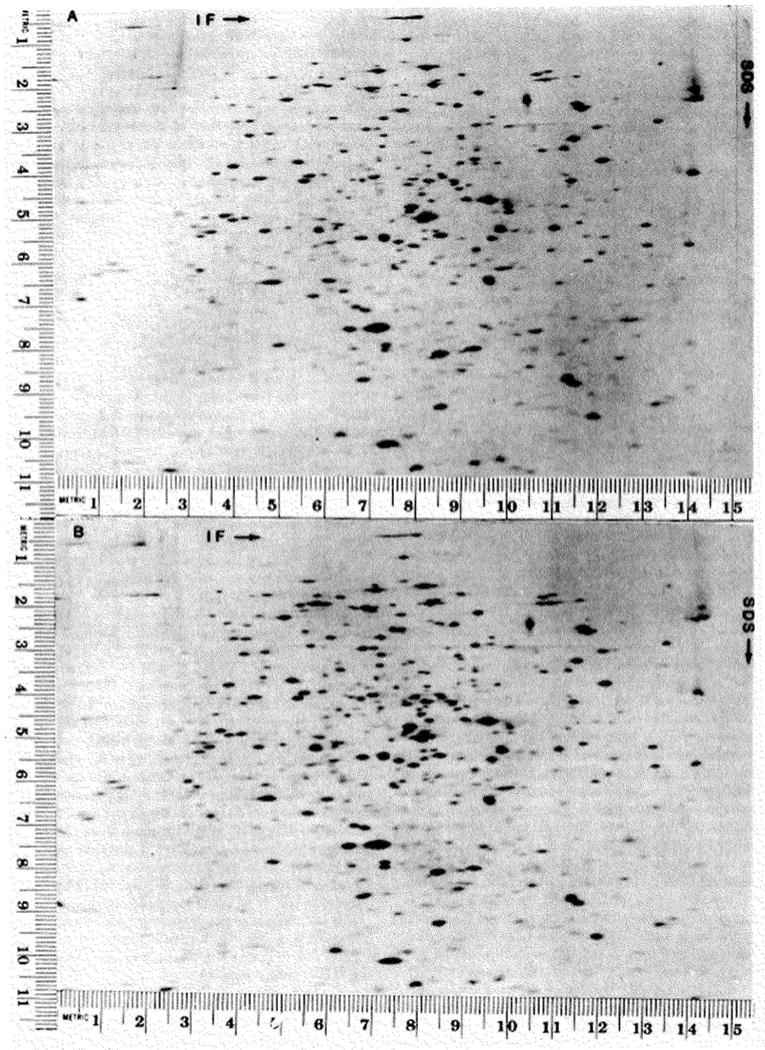
Positional reproducibility is illustrated in this figure as well as Fig. 5, A and B. The samples subjected to electrophoresis in these gels were prepared from Escherichia coli 5333 (19) (cyclic adenosine 3′:5′-monophosphate binding protein mutant) grown in the presence of adenosine 5′-monophosphate (A) or cyclic guanosine 3′:5′-monophosphate (B). These samples contained 300,000 cpm and approximately 15 μg of protein. The isoelectric focusing gels were run at the same time, equilibrated for 30 min, and run on 9 to 15% exponential, acrylamide gradient, slab gels (6 ml in the front chamber). The gels were exposed for 30 hours. The slight difference in growth conditions for the bacteria used to prepare the two samples has resulted in several quantitative differences which are evident on close inspection.
Two criteria are used to match separations. One is the absolute spot position which is particularly useful when the isoelectric focusing gels are run at the same time and there is no change in electrophoresis conditions. When these gels are compared the absolute spot positions vary less than 2 mm (after aligning the films by eye). The other information used for matching is relative spot positions and intensities. These parameters are much less sensitive to the conditions of electrophoresis and are sufficiently distinctive to permit matching of patterns even if the electrophoresis conditions have been altered substantially.
Sensitivity of Detection
Because of the small spot size, the detection of proteins by both stain and autoradiography is very sensitive. Staining with Coomassie blue can detect about 0.01 μg of protein. The sensitivity varies with spot size and the sensitivity of detection of low-molecular weight proteins, which form larger spots, is 4-fold less than optimum (calculated from the measurements made on the gel shown in Fig. 3). Calibration curves for autoradiography show that a spot containing 1 cpm can be detected (the minimum optical density of the film required for detection is 0.01) after a 20-day exposure and that the film response is linear to an optical density of 0.5. Since the linear range is small in comparison to the range of intensities (Fig. 8) found in most sources of protein, several exposure times are required in order to quantify proteins across the entire range.
Fig. 8.
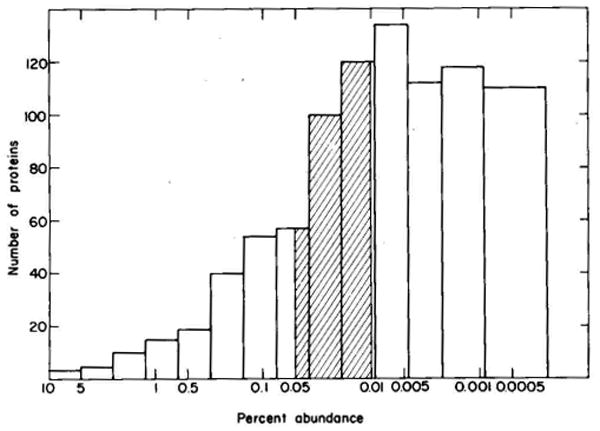
A histogram representing the number of proteins versus per cent abundance. A sample prepared from Escherichia coli 1100 and containing 300,000 cpm and 25 μg of protein was subjected to electrophoresis. The isoelectric focusing gel was equilibrated for 2 hours and run on a 12.8% acrylamide gel. Films were exposed to this gel (and a standardization wedge) for various times from 30 min to 50 days. All of the spots selected on these autoradiograms were quantified and the number of spots within an interval was plotted. The quantity interval used was a factor of 2 and these invervals are plotted on a logarithmic scale. The per cent abundance is the amount of an individual protein expressed as a per cent of the total amount of protein. The shaded area represents the number of spots summed in order to determine the number of proteins whose per cent abundance is between 0.05 and 0.01% of the total (used for the calculation presented under “Results and Discussion”).
The detection of proteins which constitute a very small percentage of a complex sample is dependent on the sensitivity and the total amount of material which is loaded on the gel. For example, a protein (MW 40,000) present at a level of one molecule per cell in E. coli would constitute 2 × 10−5% of the total protein. To detect it on a 10-day exposure 5 × 106 cpm would have to be loaded on the gel. Since the amount of protein which can be loaded on the gel is limited (see “Loading Capacity”) the protein must be labeled to high specific activity to detect rare proteins. Due to autoradiographic spreading some proteins constituting less than 10−4% of the total will be masked by the major species. Proteins which migrate to relatively lightly exposed areas of the film can be detected at levels of 10−5% of the total protein.
The fraction of the total protein detectable by staining with Coomassie blue is dependent on the amount of protein loaded. For a heavily loaded gel (100 μg of E. coli proteins) staining can detect a spot that constitutes 10−2% of the total protein. Staining can detect a maximum of 400 E. coli proteins.
Per cent Abundance
Fig. 8 is a frequency histogram of the number of E. coli proteins that have a particular per cent abundance. There is no per cent abundance at which there is a large number of proteins. The major component was overexposed by a factor of 3 × 104 in order to detect the least intense spots. Thus, the area of the major spot was some 20 times larger (autoradiographic spreading) than its minimal size. If all spots increased by this factor the resolving power of the gels would be decreased 20-fold (i.e. roughly 250 components could be resolved). The ability of this technique to detect trace components is limited more by this phenomenon than by the true sensitivity (see “Sensitivity of Detection”).
The wide distribution of relative amounts has two advantages. It is easy to recognize and match even complex patterns without any possibility of mismatch because of the large differences in the intensities of different spots. Also, the probability that any detected spot is contaminated to a significant extent is small because the number of protein species within a range of relative amounts is small. The probability that a given spot is at least 90% pure can be estimated as follows. If a spot constitutes 0.1% of the total protein and if a contaminant were present at a level greater than 10% of the given spot, then such a contaminant must constitute between 0.01% and 0.05% (since a contaminant cannot be greater than 50% of the total) of the total protein. The approximate number of proteins in this abundance interval is determined from the per cent abundance frequency histogram (22 + 100 + 108 = 230, see shaded area in Fig. 8). If any of these were to cause contamination, they must be in a small area around the given spot. If this area is 4 σ 2D-SDS × 4 σ 2D-1F, then there would be contamination. However, the spots might be resolved and any contamination could be corrected for. If, however, a secondary spot was in the area 2 σ 2D-SDS × 2 σ 2D-IF centering around the given spot, it would not be resolved and therefore would contaminate the given spot. The probability that this would occur is given by (2 σ 2D-SDS × 2 σ 2D-IF × # of spots)/(total area of the gel) or (2 × 0.55 mm × 2 × 1.5 mm × 230)/(15,000 mm2) = 0.05. Therefore, the probability that a spot representing 0.1 % of the total protein is more than 90% pure is 95%.
Streaking
Streaking can result in a large increase in spot size and thus, a decrease in resolution. Streaking occurs primarily during the first dimension (streaking in the second dimension is discussed under “Equilibration”) and is due to low solubility of some proteins in the gels. The presence of nucleic acids in a sample greatly increases streaking. I have found that nucleic acids are precipitated by basic Ampholines, that high molecular weight 32P-labeled nucleic acid forms a smear at the top of the isoelectric focusing gel, and that this effect is not due to sieving of nucleic acids. It appears as if the nucleic acids form a highly ionic precipitate that binds many proteins. Streaking is minimized by treatment with DNase and RNase as described under “Materials and Methods.”
Streaking increases if the urea concentration decreases significantly in the sample or at the top interface of the gel. The urea concentration should always be above 8 m.
Distribution of Proteins
The distribution of proteins across the two-dimensional gel also affects the resolution of the gel system. If the proteins are clustered in small areas of the gel, the total resolving power might be poor even if the spots are very small. Although proteins can differ substantially in both their isoelectric points and molecular weights, the majority cluster around the means for both of these parameters. A series of different mixtures of Ampholines were tested to obtain a pH gradient that gave uniform distribution of E. coli proteins. The approximate shape of a pH gradient achieved with the described conditions is given in Fig. 9. It is difficult to make a precise determination of the pH gradient because the high concentration of urea causes large changes in pH as well as the isoelectric points of proteins. Despite the inaccuracies, it is clear that the pH gradient does not extend significantly above pH 7. Extension of this pH gradient to pH 10 results in a large loss in resolution of the majority of the proteins and few new spots appear on the two-dimensional gel. The measurement of the pH gradient at the acid end of the isoelectric focusing gel is least accurate because of diffusion of the urea out of the gel and because of the strong influence of urea on the pKa values of carboxyl groups. The E. coli ribosomal proteins L7 and L12 can be used as pH markers for this end of the isoelectric focusing gel (see “Resolution of Single Charge Differences”). The pI of L12 is 4.85 and that of L7 is 4.7 (26). Their positions on the gels (Fig. 5A, coordinates 14.2 × 9.7 and 14.6 × 9.7) suggest that the end of the gel must be about pH 4.5. When more acidic Ampholines are used, there is an increase in the separation of the acidic proteins, few (less than 1% of total number of proteins) new acidic proteins are detected and there is a large loss of resolution of the majority of the proteins. The shape of the pH gradient can be altered in a predictable manner, since increasing the relative concentration of Ampholines of a certain nominal pH range causes a decrease in the slope of the pH curve in that pH range. Although the described pH gradient has given excellent resolution of proteins from rabbit embryos, hepatoma cells, nematodes, and E. coli, there can be noticeable differences in the mean pI (compare Figs. 7 and 13). In such cases, it may be desirable to modify the shape of the pH gradient.
Fig. 9.
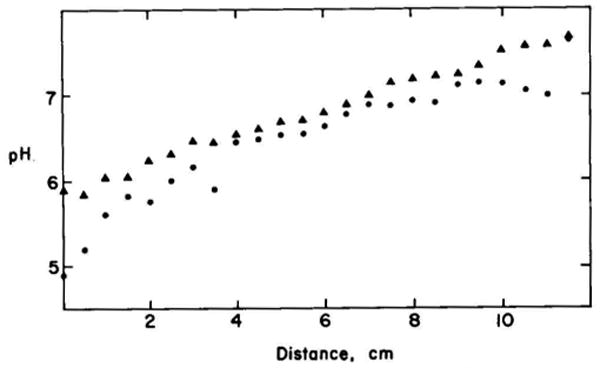
The pH gradient measured with and without urea. The pH gradient was measured as described under “Materials and Methods.” In one case the gel selections were placed in degassed water (●) for measurement, whereas in the other case the sections were placed in 9.2 m urea (▲). The top of the gel is on the right.
There is a group of proteins with isoelectric points beyond the range of commercially available Ampholines, and in this gel system these proteins are either lost or remain at the top of the isoelectric focusing gel. A second technique for the separation of all basic proteins would be a useful complement to the present technique. Commercial Ampholines were reacted with ethyl acetimidate (generously provided by Norman M. Whiteley) to produce carrier ampholytes with isoelectric points in a pH range of 11 to 13. These failed to give a stable pH gradient at pH values above 10. Nonequilibrium isoelectric focusing (isotachophoresis) (27, 28) has in recent attempts given very good separation of basic ribosomal proteins.3
Although the distribution of proteins in the SDS dimension can be drastically altered by changing the porosity of the lower (running) gel, no single acrylamide concentration can give a uniform distribution of the proteins. The best resolution was achieved using an exponential gradient gel. A uniform gel of 10% acrylamide provides a fairly uniform distribution of proteins across the gel, but a number of proteins are lost in the dye front. In 12.5% acrylamide gels, almost no proteins migrate at the dye front, but the distribution of proteins is not as uniform.
Bacteriophage T4 early proteins run on a one-dimensional SDS gel and a two-dimensional system demonstrated that the distances the proteins migrated in the SDS dimensions were the same. Thus, the mobilities of the proteins in the SDS dimension are proportional to the log of their molecular weights, and it is possible to use a standard curve produced by running markers in the SDS dimension only.
Equilibration
Two methods for transfer of the first dimension gel onto the second were given. The method involving equilibration of the first dimension gel results in a loss of about 5 to 25% of the total protein loaded (actual loss depends on the protein sample and duration of equilibration). There is a selective loss of protein which has not adequately entered the isoelectric focusing gel. The advantage of this method is that there is almost no streaking of proteins in the SDS dimension.
When gels are not equilibrated, there is no loss of protein and slightly less spreading due to diffusion. However, prolonged exposure reveals streaking of several high molecular weight components in the SDS dimension. The intensity of these streaks will not mask spots that constitute more than 10−3% of the total protein. Streaking is diminished if the time during which 2% SDS is subjected to electrophoresis through the isoelectric focusing gel is increased. However, as described below, this causes an increase in the streaking of low molecular weight proteins.
The low molecular weight proteins occasionally show a smear below the spot (see Fig. 5A, coordinates 8.5 × 8.6). This is an artifact due to incomplete stacking of small proteins. This problem is overcome if a gradient gel is used since small proteins can stack in the upper part of the gradient. Otherwise, this effect can be eliminated by increasing the length of the stacking gel or by decreasing the amount of SDS on the top of the slab gel (i.e. decreasing the volume of the agar embedding gel) or in the case of nonequilibrated gels, decreasing the time during which 2% SDS is subjected to electrophoresis through the isoelectric focusing gel.
Sample
At urea concentrations below 8 m, a number of proteins aggregate and never enter the gel. Salts in the sample, even in high concentrations, have very little effect on the gels. The only observed effect was a small change in the shape of the pH gradient. If samples of different salt content are to be compared, it would be advantageous to normalize the salt content. The effect of changes in the sample volume has been tested for volumes from 5 to 50 μl. No effects have been detected, although again it is recommended that samples to be compared be applied in equal volumes.
Surprisingly, the sample applied to the isoelectric focusing gel can contain SDS without destroying the separation. Although SDS binds to protein with a high binding constant (29), it comes off the protein and forms mixed micelles with the NP-40 and these micelles migrate to the acidic end of the gel. When fairly high concentrations of SDS are used, virtually all of the detergent (nonionic as well as anionic) is in a short region (1 or 2 cm) at the acidic end of the gel. The pH in this region of the gel is much lower than in normal isoelectric focusing gels and there do not appear to be any proteins in this region. Although the effective pH gradient is shorter, patterns can still be compared to patterns obtained in the absence of SDS. SDS can cause changes by dissociating aggregates. For example, SDS treatment results in the disappearance of an aggregate seen on all gels of E. coli when SDS is not used. This aggregate consists of ribosomal RNA and some protein which is not removed by 9.5 m urea and 2% NP-40, and it produces a series of spots and a smear on the second dimension (Fig. 5A, coordinates 5.3 × 1). The plug of detergent at the acidic end of the gel results in a distortion of the dye front on the second dimension, but this distortion does not extend into the region where the proteins migrate.
Behavior of Ampholines and NP-40 in Second Dimension Gel
The Ampholines behave like very small proteins. They bind SDS and the vast majority migrate at the dye front. In the presence of SDS, Ampholines are acid precipitable and remain in the gel under the described procedure. Ampholines also stain with common protein stains. As a result, the gels will always have intensely staining material at the dye front which can obscure the staining of small proteins migrating at or very near the dye front. SDS can be extracted from the Ampholines, and the Ampholines removed from the gel if the gel is soaked in acidic alcohol solutions. I have soaked gels in 50% alcohol, 7% acetic acid, and 0.005% Coomassie blue for 36 hours, followed by rehydration in 7% acetic acid-0.005% Coomassie blue (the Coomassie blue prevents the destaining of proteins in the alcohol solution). This procedure removes all Ampholines except for a fine line at the dye front which stains faintly.
The mixed micelles of SDS and NP-40 migrate into the second dimension gel. As long as there is an excess of SDS, the NP-40 has no detrimental effects.
Resolution of Single Charge Differences
In order to use the gel system in combination with genetics, it is important that single charge changes produce a detectable change in spot position so that some missense mutations can be detected. From the genetic code, it can be calculated that 30% of all of the base substitutions will give a charge change in a protein. In one-dimensional isoelectric focusing, single charge changes are known to produce a significant change in band position. To test the capability of this gel system, four T4 temperature-sensitive (ts) mutants in gene 32 were analyzed on the gels. One of these mutants produced an obvious alteration in the position of the spot corresponding to gene 32 protein (Fig. 10). These ts mutations are presumably missense mutations. One of these mutations must have produced an amino acid substitution resulting in a charge change, consequently changing the pI.
Fig. 10.
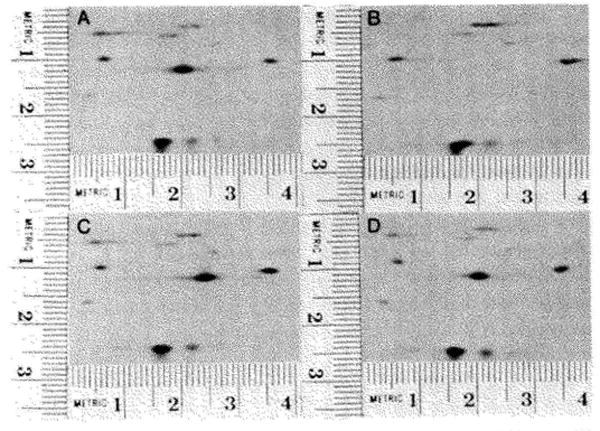
The detection of a missense mutation in T4 gene 32. Samples prepared from T4 infected Escherichia coli (AS19) were subjected to electrophoresis and autoradiograms prepared. Sections of these autoradiograms are shown. The patterns shown were produced by infections with wild type T4 (A) an amber mutant in gene 32 (B), a ts mutant (tsP7) in gene 32 (C), and another ts mutant (tsL170) in gene 32 (D). The spot corresponding to the gene 32 protein was identified by comparing Patterns A and B. A major spot (A, coordinates 2.05 × 1.15) is missing in Pattern B. This spot corresponds to the gene 32 protein. In the pattern shown in Panel C the spot corresponding to the gene 32 protein can still be seen; however, its position (coordinates 2.45 × 1.2) has shifted in comparison to the wild type gene 32. The gene 32 proteins seen in Panel D appears to be in the same position as wild type gene 32 protein. The pattern obtained from the gene 32 amber mutant is missing one minor spot in addition to the gene 32 protein. This minor spot is seen in Panels A, C, and D (coordinates 0.5 × 0.6). It is concluded that this amber mutant (HL618) contains two amber mutations, one in gene 32 and the other in a nonessential gene.
To test further the sensitivity of this system to single charge changes, E. coli ribosomes were subjected to electrophoresis on the two-dimensional gels. It was possible to identify the ribosomal proteins L7 and L12 on these gels because their isoelectric points and molecular weights are known and these parameters are very distinctive. These two proteins differ only in that L7 is acetylated on the α-amino group while L12 is not (30). The clear separation of these proteins again demonstrates that single charge differences can be resolved. The separation of ribosomal proteins is not shown. However, L7 and L12 can be seen on separations of total E. coli protein (see Fig. 5A, coordinates 14.2 × 9.7, and 14.6 × 9.7).
The degree to which a single charge change influences the pI of a protein (i.e. the distance between the original spot and the spot produced by the modified protein), is dependent on the pI of the protein as well as its molecular weight and amino acid composition. If the pI of the protein is close to the pKa of a major dissociating group in the protein (i.e. the carboxyl groups of glutamic and aspartic acid), then a small change in pH, near the pI, will result in the titration of many groups, and therefore a large change in charge. A single charge change will cause the isoelectric point of a protein with a pI of 5.5 (native conditions) to shift about 10 times as much as a protein with an original pI of 4.5 (31). It should be noted that the pKa values of carboxyl groups are about 1 pH unit higher in 10 m urea than in water.
Since pI is the point at which the net charge is zero, the influence of a single charge difference will decrease as the total number of titratable charged groups increases. As a result, the effect of a charge change decreases as the molecular weight increases. The amino acid compositions will also have an influence. If a protein is rich in histidine and has a pI near 7, a charge alteration would cause a small change, whereas a protein with no histidine would be greatly affected. If the ratio of charged amino acids to uncharged amino acids varied greatly, it would be possible for a large protein to have less titratable groups than a small protein. However, amino acid compositions seldom differ widely enough to dominate the molecular weight and isoelectric point effects which cause the resolution of single charge differences to vary widely from one area of the two-dimensional gel to another (Fig. 11). Since resolution of single charge differences depends on molecular weight and isoelectric point, and since this technique separates proteins according to these parameters, the degree of resolution of single charge differences can be predicted from the position on the gel.
Fig. 11.
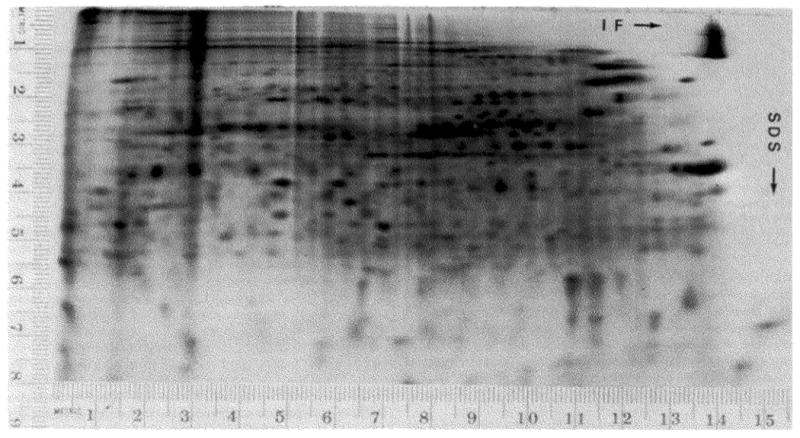
Artifactual multiple spots due to induced charge heterogeneity. Escherichia coli (AS19) was labeled with 35SO4 as described under “Materials and Methods” and the labeled cells were stored frozen for 1 month. The cells were lysed in lysis buffer without RNase or DNase treatment. The SDS slab gel was a uniform concentration (13%) of acrylamide. Since there were almost no spots on the lower part of this gel, the lower one-third of the gel is not shown.
Charge Heterogeneity
Charge heterogeneity could be due to in vivo modifications, such as phosphorylation, acetylation, or addition of charged carbohydrate groups, or it could be due to artifactual modification. It is unavoidable and perhaps desirable that in vivo induced charge modifications are detected as satellite spots. However, artifactual induction of charge heterogeneity would reduce the usefulness of this technique.
There are a large number of reports of charge heterogeneity of purified proteins, as detected by isoelectric focusing. This has lead to considerable skepticism, particularly since heterogeneity has been reported for a number of well characterized proteins for which there are no known in vivo charge modifications (33–34). In most cases, the cause of the heterogeneity is not reported.
Artifactual charge heterogeneity of a single protein produces a distinctive pattern which can be recognized even in a complex mixture of proteins. The spots produced by a protein possessing charge heterogeneity form a series of spots with the same molecular weight (mobility in the SDS dimension); the spacing of the spots is consistent with single charge differences between consecutive spots; the more acidic spots are less intense (Fig. 11). In this pattern almost all of the spots are present as multiple spots. Single charge differences affect the position of smaller, more basic proteins (pH 5 to 7) so drastically it is difficult to recognize multiples (see “Resolution of Single Charge Differences”). Since smaller proteins have a smaller number of targets for modification, they are less sensitive to random modifications, and therefore smaller proteins are less heterogeneous than larger proteins. The severe heterogeneity seen in Fig. 11 is artifactual, and must involve the modification of at least one common constituent of proteins.
When the samples are prepared as described under “Materials and Methods” almost no multiple spots are detected. It is clear that the described method of treatment does not induce modification of any common protein constituent since there are a large number of high molecular weight proteins which form single spots (for example, Fig. 7, A and B, coordinates 12.2 × 3.6, 9.4 × 2.1, and 5.1 × 2.2). Investigators should, however, be aware of this problem since it is not yet clear what type of treatments (other than those described) are permissible and what conditions give rise to these multiple spots.
The pattern illustrated in Fig. 11 was obtained from an E. coli sample which had been stored as frozen cells (−20°) for 1 month prior to lysing in the urea sample buffer. When a cell lysate was lyophilized and stored as a powder at −70° for one month prior to dissolving in the urea sample buffer, artifactual heterogeneity resulted. When the lyophilized powder was not stored, no heterogeneity was observed. Thus, drastic modifications can occur under mild conditions. The modifications decrease the pI; the number of modifications is correlated with molecular weight; the distances between consecutive spots are consistent with single charge changes; and the distribution of intensities of the spots is consistent with random modification. Because of their reactivity, it is felt that the most likely targets for this modification are cysteine, asparagine, and glutamine. Cysteine can be readily oxidized to form cysteic acid while asparagine and glutamine are known to undergo spontaneous deamidation (35). Many of the reported cases of protein heterogeneity share the features of this observed artifactual heterogeneity.
A few multiple spots are always obtained using the described technique (Fig. 2 coordinates 3.1 × 1.8). These may be due to in vivo modifications or to selective artifactual modification.
Possible Artifacts
It is also possible to form multiple spots due to solubility effects. These artifactual spots can usually be detected because there is normally some streaking associated with these spots. Some ambiguity can arise when other proteins migrate to a position within the streak. These multiple spots and associated streaks occur only in the isoelectric focusing dimension. High concentrations of urea and NP-40 greatly increase solubility and thereby decrease artifacts due to insolubility.
Isocyanate formed by decomposition of urea might result in carbamylation of protein. The following precautions are taken to prevent carbamylation: all urea solutions are prepared fresh or are stored as frozen aliquots; Ampholines are present in all urea solutions which contact the protein (Ampholines contain reactive amines); once the protein is dissolved in a urea solution, the time which it is not frozen is kept to a minimum; and finally, the isoelectric focusing gels are prerun to remove isocyanate (pKa 3.75). The absence of generalized modifications suggests that no carbamylation has occurred. Tests in which the sample was prepared in SDS and no urea, and tests in which the gels were either prerun or not, showed no differences and suggest that carbamylation could not account for any of the multiple spots.
I have observed artifactual heterogeneity in the SDS dimension on only five two-dimensional gels out of more than 200. I do not know the source of this artifact. Fig. 12 illustrates the appearance of these artifactual spots. The pattern produced is sufficiently distinctive that it should always be possible to identify this artifactual heterogeneity.
Fig. 12.
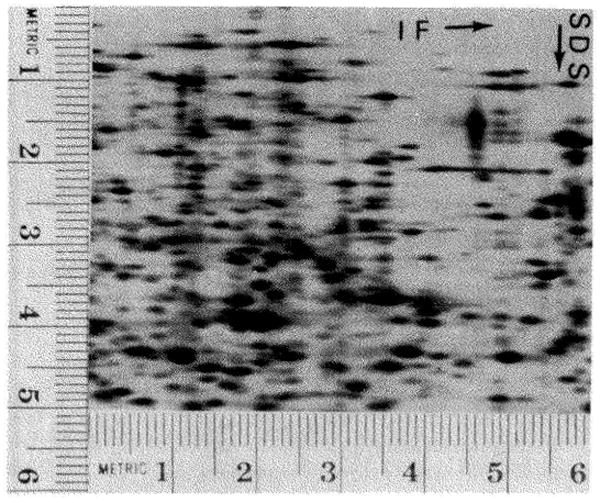
Artifactual multiple spots in the SDS dimension. Escherichia coli 5333 was grown in the presence of guanosine 5′-monophosphate and labeled with 14C-amino-acids. The cells were lysed by sonication, the lysate was treated with RNase and DNase, and urea and lysis buffer were added. This sample was subjected to electrophoresis under conditions similar to those described in Fig. 7. The film was exposed for 26 days and a section of the resulting autoradiogram is shown. Although this is a longer exposure it can be compared to the autoradiograms shown in Fig. 7. The section shown here corresponds to an area included between two vertical lines at 5.8 and 11.8 and two horizontal lines at 0.8 and 5.8 on the autoradiogram shown in Fig. 7A. The artifactual spots appear as a series of spots at the same position in the isoelectric focusing dimension and with different positions in the SDS dimension. These spots appear below many of the more abundant proteins, particularly below major high molecular weight proteins. A series of artifactual multiple spots can be seen along the vertical line at position 1.1.
If the upper surface of the stacking gel (second dimension) is not uniform, a slight distortion of the spots is produced (Fig. 3, coordinates 7.4 × 2.8 to 9). These effects do not hamper matching and spot identification unless they are very severe.
Possible artifacts caused by overloading the gel are discussed under “Loading Capacity.”
Separation of Proteins from Other Sources
This separation system was designed as a general technique; it separates proteins from almost any source. This technique separates all types of proteins except basic proteins such as ribosomal proteins and histones. I have separated the proteins of rabbit embryos (blastocyst stage (36)), hepatoma cells, and the nematode Caenorhabditis elegans. In all cases, the resolution and reproducibility are comparable to those obtained with proteins from E. coli. The separation of total nematode proteins is shown in Fig. 13. The isoelectric focusing dimension was run at the same time as those shown in Fig. 7. The mean isoelectric point is slightly higher for nematode proteins than for E. coli proteins (also see “Distribution of Proteins”). There are a number of proteins from nematodes which do not form as distinct spots as all the others (Fig. 13, coordinates 13.4 × 7.2).
High resolution, sensitivity, and reproducibility make this technique a powerful analytical tool which could potentially find use in a wide range of investigations.
Acknowledgments
I am grateful to Dr. David Hirsh and Dr. Larry Gold for helpful advice and criticism throughout the course of this work, for use of laboratory facilities and for comments on this manuscript. In addition, I would like to thank Dr. Jacques Pène for laboratory facilities and for his comments on this manuscript.
Footnotes
This work was supported by National Science Foundation Grant GB-37949.
The abbreviations used are: SDS, sodium dodecyl sulfate; TEMED, N,N,N′,N′-tetramethylethylenediamine; IF, isoelectric focusing.
All autoradiograms of two-dimensional gels were photographed with a metric ruler along two edges of the autoradiogram. These rulers establish a coordinate system which is used to give spot positions. The verticle scale is given in units from top to bottom. The horizontal scale is given in units from left to right. The coordinates are given as horizontal × vertical.
P. H. O'Farrell, in preparation.
References
- 1.Laemmli UK. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 2.O'Farrell PZ, Gold LM, Huang WM. J Biol Chem. 1973;248:5499–5501. [PubMed] [Google Scholar]
- 3.O'Farrell PZ, Gold LM. J Biol Chem. 1973;248:5502–5511. [PubMed] [Google Scholar]
- 4.O'Farrell PZ, Gold LM. J Biol Chem. 1973;248:5512–5519. [PubMed] [Google Scholar]
- 5.Studier FW. Science. 1972;176:367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 6.Raymond S. Ann N Y Acad Sci. 1964;121:350–365. doi: 10.1111/j.1749-6632.1964.tb14208.x. [DOI] [PubMed] [Google Scholar]
- 7.Macko V, Stegemann H. Z Physiol Chem. 1969;350:917–919. [PubMed] [Google Scholar]
- 8.Margolis J, Kenrick KG. Nature. 1969;221:1056–1057. doi: 10.1038/2211056a0. [DOI] [PubMed] [Google Scholar]
- 9.Mets LJ, Bogorad L. Anal Biochem. 1974;57:200–210. doi: 10.1016/0003-2697(74)90065-7. [DOI] [PubMed] [Google Scholar]
- 10.Barrett T, Gould HJ. Biochim Biophys Acta. 1973;294:165–170. [Google Scholar]
- 11.Orrick LR, Olson MOJ, Busch H. Proc Natl Acad Sci U S A. 1973;70:1316–1320. doi: 10.1073/pnas.70.5.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaltschmidt E, Wittmann HG. Anal Biochem. 1970;36:401–412. doi: 10.1016/0003-2697(70)90376-3. [DOI] [PubMed] [Google Scholar]
- 13.Kaltschmidt E, Wittmann HG. Proc Natl Acad Sci U S A. 1970;67:1276–1282. doi: 10.1073/pnas.67.3.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittmann HG, Stoffler G, Hindennach I, Kurland CG, Randall-Hazelbauer L, Birge EA, Nomura M, Kaltschmidt E, Mizushima S, Traut RR, Bickle TA. Mol Gen Genet. 1971;111:327–333. doi: 10.1007/BF00569784. [DOI] [PubMed] [Google Scholar]
- 15.Fahnestock S, Erdmann V, Nomura M. Biochemistry. 1973;12:220–224. doi: 10.1021/bi00726a007. [DOI] [PubMed] [Google Scholar]
- 16.Held WA, Nomura M. Biochemistry. 1973;12:3273–3281. doi: 10.1021/bi00741a020. [DOI] [PubMed] [Google Scholar]
- 17.Sekiguchi M, Iida S. Proc Natl Acad Sci U S A. 1967;58:2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox CF, Wilson G. Proc Natl Acad Sci U S A. 1968;59:988–995. doi: 10.1073/pnas.59.3.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emmer M, de Crombrugghe B, Pastan I, Perlman R. Proc Natl Acad Sci U S A. 1970;66:480–487. doi: 10.1073/pnas.66.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolle A, Epstein RH, Salser W, Geiduschek EP. J Mol Biol. 1968;33:339–362. doi: 10.1016/0022-2836(68)90193-9. [DOI] [PubMed] [Google Scholar]
- 21.Ornstein L. Ann N Y Acad Sci. 1964;121:321–349. doi: 10.1111/j.1749-6632.1964.tb14207.x. [DOI] [PubMed] [Google Scholar]
- 22.Davis BJ. Ann N Y Acad Sci. 1964;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- 23.Reid MS, Bieleski RL. Anal Biochem. 1968;22:374–381. doi: 10.1016/0003-2697(68)90278-9. [DOI] [PubMed] [Google Scholar]
- 24.Studier FW. J Mol Biol. 1973;79:237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- 25.Fairbanks G, Jr, Levinthal C, Reeder RH. Biochem Biophys Res Commun. 1965;20:393–399. doi: 10.1016/0006-291x(65)90589-9. [DOI] [PubMed] [Google Scholar]
- 26.Möller W, Groene A, Terhorst C, Amons R. Eur J Biochem. 1972;25:5–12. doi: 10.1111/j.1432-1033.1972.tb01660.x. [DOI] [PubMed] [Google Scholar]
- 27.Haglund H. Sci Tools LKB Instrum J. 1970;17:1–13. [Google Scholar]
- 28.Griffith A, Catsimpoolas N, Kenney J. Ann N Y Acad Sci. 1973;209:457–468. doi: 10.1111/j.1749-6632.1973.tb47548.x. [DOI] [PubMed] [Google Scholar]
- 29.Ray A, Reynolds JA, Polet H, Steinhardt J. Biochemistry. 1966;5:2606–2616. doi: 10.1021/bi00872a019. [DOI] [PubMed] [Google Scholar]
- 30.Terhorst C, Wittmann-Liebold B, Möller W. Eur J Biochem. 1972;25:13–19. doi: 10.1111/j.1432-1033.1972.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 31.Bobb D, Hofster BHJ. Anal Biochem. 1971;40:209–217. doi: 10.1016/0003-2697(71)90094-7. [DOI] [PubMed] [Google Scholar]
- 32.Williamson AR, Salaman MR, Kreth HW. Ann N Y Acad Sci. 1973;209:210–224. doi: 10.1111/j.1749-6632.1973.tb47530.x. [DOI] [PubMed] [Google Scholar]
- 33.Righetti PG, Drysdale JW. Ann N Y Acad Sci. 1973;209:163–186. doi: 10.1111/j.1749-6632.1973.tb47527.x. [DOI] [PubMed] [Google Scholar]
- 34.Lai CY, Chen C, Horecker BL. Biochem Biophys Res Commun. 1970;40:461–468. doi: 10.1016/0006-291x(70)91031-4. [DOI] [PubMed] [Google Scholar]
- 35.Robinson AB. Proc Natl Acad Sci U S A. 1974;71:885–888. doi: 10.1073/pnas.71.3.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Blerkom J, Manes C. Dev Biol. 1974;40:40–51. doi: 10.1016/0012-1606(74)90105-5. [DOI] [PubMed] [Google Scholar]


