Abstract
Preserving mitochondrial mass, bioenergetic functions and ROS (reactive oxygen species) homoeostasis is key to neuronal differentiation and survival, as mitochondria produce most of the energy in the form of ATP to execute and maintain these cellular processes. In view of our previous studies showing that NeuroD6 promotes neuronal differentiation and survival on trophic factor withdrawal, combined with its ability to stimulate the mitochondrial biomass and to trigger comprehensive antiapoptotic and molecular chaperone responses, we investigated whether NeuroD6 could concomitantly modulate the mitochondrial biomass and ROS homoeostasis on oxidative stress mediated by serum deprivation. In the present study, we report a novel role of NeuroD6 as a regulator of ROS homoeostasis, resulting in enhanced tolerance to oxidative stress. Using a combination of flow cytometry, confocal fluorescence microscopy and mitochondrial fractionation, we found that NeuroD6 sustains mitochondrial mass, intracellular ATP levels and expression of specific subunits of respiratory complexes upon oxidative stress triggered by withdrawal of trophic factors. NeuroD6 also maintains the expression of nuclear-encoded transcription factors, known to regulate mitochondrial biogenesis, such as PGC-1α (peroxisome-proliferator-activated receptor γ co-activator-1α), Tfam (transcription factor A, mitochondrial) and NRF-1 (nuclear respiratory factor-1). Finally, NeuroD6 triggers a comprehensive antioxidant response to endow PC12-ND6 cells with intracellular ROS scavenging capacity. The NeuroD6 effect is not limited to the classic induction of the ROS-scavenging enzymes, such as SOD2 (superoxide dismutase 2), GPx1 (glutathione peroxidase 1) and PRDX5 (peroxiredoxin 5), but also to the recently identified powerful ROS suppressors PGC-1α, PINK1 (phosphatase and tensin homologue-induced kinase 1) and SIRT1. Thus our collective results support the concept that the NeuroD6–PGC-1α–SIRT1 neuroprotective axis may be critical in co-ordinating the mitochondrial biomass with the antioxidant reserve to confer tolerance to oxidative stress.
Keywords: mitochondria, NeuroD family, neuronal survival, reactive oxygen species (ROS), SIRT1, transcriptional co-regulator peroxisome-proliferator-activated receptor γ co-activator-1α (PGC-1α)
Abbreviations: AD, Alzheimer’s disease; AM, acetoxymethyl ester; bHLH, basic helix–loop–helix; COX, cytochrome c oxidase; DAPI, 4′,6-diamidino-2-phenylindole; DIC, differential interference contrast; Drp1, dynamin-related protein 1; ETC, electron transfer chain; GABP-α, GA-binding protein-α; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GFP, green fluorescent protein; GPx1, glutathione peroxidase 1; HSP, heat-shock protein; Mfn2, mitofusin 2; Mg-Gr, Magnesium Green; MMP, mitochondrial membrane potential; mtDNA, mitochondrial DNA; MTG, MitoTracker® Green; MTR, MitoTracker® Red; NRF, nuclear respiratory factor; NT-PGC-1α, N-terminal-truncated PGC-1α; OPA1, optic atrophy 1; OXPHOS, oxidative phosphorylation; PDL, poly-d-lysine; PGC-1α, peroxisome-proliferator-activated receptor γ co-activator-1α; PINK1, phosphatase and tensin homologue-induced kinase 1; PRDX5, peroxiredoxin 5; ROS, reactive oxygen species; SOD, superoxide dismutase; Tfam, transcription factor A, mitochondrial; WGA, wheatgerm agglutinin
INTRODUCTION
A wealth of studies have demonstrated that both mitochondrial dysfunction and oxidative stress are implicated in the pathogenesis of several neurodevelopmental disorders, such as spongiform encephalopathy (Melov et al., 2001; Golden et al., 2005), mitochondrial encephalopathy (Wallace, 1999; Patel, 2004; Khurana et al., 2008) and autism spectrum disorder (James et al., 2004, 2006; Pons et al., 2004; Chauhan and Chauhan, 2006; Rossignol and Bradstreet, 2008) as well as many neurodegenerative diseases, such as PD (Parkinson’s disease), AD (Alzheimer’s disease), HD (Huntington’s disease) and ALS (amyotrophic lateral sclerosis) (reviewed by Finkel and Holbrook, 2000; Fridovich, 2004; Wallace, 2005; Lin and Beal, 2006; Giorgio et al., 2007; Nicholls, 2008; Malkus et al., 2009). Thus preserving mitochondrial mass and function is key to neuronal differentiation and survival, as mitochondria produce most of the energy in the form of ATP through a series of oxidative reactions occurring in the ETC (electron transfer chain) necessary to execute and maintain neuronal differentiation in a developing or mature brain.
Mitochondria, being a key source of ROS (reactive oxygen species) as a result of electron transfer through the respiratory chain at the level of both complex I [COX1 (NADH: ubiquinone oxidoreductase)] and complex III (COX3; ubiquinone-cytochrome c reductase) (Sugioka et al., 1988; Trumpower, 1990; Demin et al., 1998; Han et al., 2001; St-Pierre et al., 2002; Chen et al., 2003), possess an intrinsic defence system to regulate ROS homoeostasis via the expression of an array of antioxidant regulators, such as non-enzymatic regulators (α-tocopherol, coenzyme Q10, cytochrome c and glutathione) and detoxifying enzymes [SOD (superoxide dismutase), glutathione peroxidase and peroxiredoxins] (reviewed by Finkel and Holbrook, 2000). Increased ROS production leads to oxidative damage of the mtDNA (mitochondrial DNA), potentially due to its limited repair system and location in the mitochondrial matrix near the released ROS (Esposito et al., 1999; Melov et al., 1999; Balaban et al., 2005), resulting in compromised mitochondrial function and integrity as well as further increased ROS levels.
Given the fact that mitochondria assume the dual role of regulating neuronal survival and controlling ROS levels, the degree of vulnerability of developing and mature neurons is most likely correlated to their functional mitochondrial mass and the extent of their antioxidant reserve. Thus it is of great interest to identify neurogenic transcription factors promoting interconnected transcriptional networks responsible for co-ordinating the mitochondrial biomass with a comprehensive antioxidant response, which can be tailored to developmental and cellular contexts. The neurogenic bHLH (basic helix–loop–helix) transcription factor NeuroD6 is an excellent candidate to assume such a dual function, based on our previous findings showing its role as a co-regulator of mitochondrial mass, cytoskeletal remodelling, anti-apoptotic and molecular chaperone networks (Uittenbogaard and Chiaramello, 2002, 2005; Baxter et al., 2009; Uittenbogaard et al., 2009, 2010).
NeuroD6 (previously known as Nex1/MATH-2) is a member of the neurogenic bHLH NeuroD family, which contributes to the specification of multipotential progenitors towards a glutamatergic pyramidal fate (Schwab et al., 1998; Wu et al., 2005). It remains expressed in the adult brain in areas associated with higher cognitive functions, such as the hippocampus, the neocortex and the entorhinal cortex, the first area to be affected in AD (Alzheimer’s disease) (Bartholomä and Nave, 1994; Shimizu et al., 1995). Due to functional redundancy between members of the NeuroD family, NeuroD6’s role during brain development has remained elusive (Schwab et al., 2000). Using our in vitro cellular paradigm, the PC12-ND6 cell line, which constitutively overexpresses NeuroD6 independently of NGF (nerve growth factor) signalling, we have demonstrated that NeuroD6 initiates neuronal differentiation, while promoting neuronal survival via the expression of anti-apoptotic regulators preserving mitochondrial integrity, such as Bcl-XL, Bcl-w, survivin and XIAP (X-linked inhibitor of apoptosis; Uittenbogaard and Chiaramello, 2002, 2004, 2005). Using a genome-wide microarray approach, we further identified NeuroD6-mediated transcriptional pathways linking neuronal differentiation to survival upon oxidative stress induced by serum deprivation. By applying a gene set enrichment strategy, we found that NeuroD6 regulates an extensive molecular chaperone network through the expression of a multitude of HSPs (heat-shock proteins), combined with increased expression of genes involved in mitochondrial biogenesis, bioenergetics and oxidative metabolism (Uittenbogaard et al., 2010). Most importantly, these genomic microarray-based findings are in keeping with our recently published study reporting a novel role for NeuroD6 in stimulating mitochondrial mass and ATP levels during the very early stages of neuronal differentiation (Baxter et al., 2009).
The main objective of the present study was to investigate whether NeuroD6 could maintain the mitochondrial biomass while triggering an antioxidant response upon oxidative stress induced by serum deprivation. Our original impetus was based on our previously reported NeuroD6-mediated long-term neuronal survival upon serum removal (Uittenbogaard and Chiaramello, 2005) combined with our recent microarray analysis suggestive of a co-ordinate expression between NeuroD6, mitochondria-related genes and a cluster of ROS-scavenging genes (Uittenbogaard et al., 2010). Using a combination of flow cytometry, confocal fluorescence microscopy and mitochondrial fractionation, we found that NeuroD6 sustained the mitochondrial content of serum-deprived PC12-ND6 cells, accompanied by maintained intracellular ATP levels and expression of key subunits of various respiratory complexes of the ETC. Furthermore, NeuroD6 triggered a comprehensive antioxidant response co-ordinated with the expression of key mitochondrial regulators, such as PINK1 (phosphatase and tensin homologue-induced kinase 1), PGC-1α (peroxisome-proliferator-activated receptor γ co-activator-1α) and SIRT1, resulting in low ROS levels upon oxidative stress mediated by serum withdrawal. Thus the present study provides the first evidence of a novel function of the neurogenic bHLH transcription factor NeuroD6 as a regulator of ROS homoeostasis. Combined with our previous studies, our results support the notion that NeuroD6 plays an integrative role in co-ordinating mitochondrial content and ROS metabolism necessary for both neuronal differentiation and survival.
MATERIALS AND METHODS
Cell culture
Control rat pheochromocytoma PC12 cells and PC12-ND6 cells (previously referred to as Nex1/MATH-2-overexpressing PC12-Nex1 cells) were grown on collagen I-coated plates (Becton Dickinson Labware, San Jose, CA, U.S.A.) in F12K modified medium (Kaighn’s modification) supplemented with 2.5% (v/v) fetal bovine serum and 15% (v/v) horse serum (Invitrogen, Carlsbad, CA, U.S.A.), as described in Uittenbogaard and Chiaramello (2002). For serum deprivation, cells were seeded at 70% confluency and then washed on the following day three times with serum-free medium, as described in Uittenbogaard and Chiaramello (2005).
Immunoblot analysis
Serum-grown control PC12 and PC12-ND6 cells as well as serum-deprived PC12-ND6 cells were lysed for specific time periods as indicated in the Figure legends in M-per mammalian protein extraction buffer (Pierce Biotechnology, Rockford, IL, U.S.A.) in the presence of a cocktail of protease inhibitors (Roche Applied Science, Indianapolis, IN, U.S.A.), as described in Uittenbogaard et al. (2010). Proteins (40 μg) were resolved on 10% NuPAGE Bis-Tris gels (Invitrogen) with either Mes/SDS or Mops/SDS running buffer and transferred to the nitrocellulose. Nitrocellulose membranes were stained with Ponceau-S (Sigma, St. Louis, MO, U.S.A.) to confirm uniform transfer of proteins and subsequently blocked using Superblock™ blocking buffer (Pierce). The membranes were probed with various antibodies described in Table 1 and corresponding secondary horseradish peroxidase-conjugated antibodies (Pierce). The antigen–antibody complex was detected using the Supersignal West Pico Chemiluminescent Substrate kit (Pierce). Blots were then stripped using Restore™ Western blot stripping buffer (Pierce) according to the manufacturer’s recommendations, and re-probed with a monoclonal antibody against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) to confirm equal protein loading. Protein levels visualized by immunoblot analyses were quantified using the NIH (National Institutes of Health) ImageJ software (http://rsb.info.nih.gov/ij/) and normalized against the GAPDH loading control as described in Uittenbogaard et al. (2010).
Table 1. List of antibodies used for immunoblot analyses and immunocytochemistry.
Abbreviations: Ab, antibody; IB, immunoblot; ICC, immunocytochemistry; mAb, monoclonal antibody; NA, not applicable; Rb, rabbit.
| Antibody | Type | Source (catalogue no.) | Dilution for IB | Dilution for ICC |
|---|---|---|---|---|
| Alexa Fluor® 488 anti-rabbit IgG | Goat Ab | Invitrogen (A11034) | NA | 1:2000 |
| Alexa Fluor® 568 anti-rabbit IgG | Goat Ab | Invitrogen (A11036) | NA | 1:2000 |
| ATP synthase α (COX5) | Mouse mAb | Mitosciences (MS502) | 1:1000 | 1:200 |
| COX1 (NDUFA9) | Mouse mAb | Mitosciences (MS111) | 1:1000 | 1:200 |
| COX3 (core2 protein) | Mouse mAb | Mitosciences (MS304) | 1:1000 | 1:1000 |
| COX4 (subunit IV) | Mouse mAb | Abcam (ab14744) | 1:500 | NA |
| Drp1 | Mouse mAb | Abcam (ab56788) | 1:400 | NA |
| GAPDH | Mouse mAb | Ambion (4300) | 1:10000 | NA |
| Gpx1 | Rb Ab | Abcam (ab59564) | 1:3000 | NA |
| Mfn2 | Rb Ab | Abcam (ab50838) | 1:1000 | NA |
| NRF1 | Rb Ab | Abcam (ab34682) | 1:5000 | NA |
| NRF2 (GABP-α) | Mouse mAb | Santa Cruz (sc-28312) | 1:200 | NA |
| OPA-1 | Rb Ab | Abcam (ab42364) | 1:500 | NA |
| Pink1 | Rb Ab | Abcam (ab23707) | 1:250 | NA |
| PGC-1α | Rb Ab | Santa Cruz (sc-13067) | 1:500 | NA |
| Prdx5 | Rb Ab | Abcam (ab16823) | 1:5000 | NA |
| SIRT1 | Goat Ab | Santa Cruz (sc-19857) | 1:1000 | NA |
| SOD1 | Rb Ab | Abcam (ab16831) | 1:10000 | NA |
| SOD2 | Rb Ab | Abcam (ab13534) | 1:2000 | 1:500 |
| Tfam | Goat Ab | Santa Cruz (sc-23588) | 1:1000 | NA |
Flow cytometry
Flow cytometry was performed as described in Baxter et al. (2009). Briefly, cells were grown on collagen I-coated plates to 80% confluency before staining with 70 nM MTG (MitoTracker® Green; Molecular Probes, Eugene, OR, U.S.A.) for 30 min at 37°C, washed with PBS and trypsizined before being centrifuged at 228 g for 5 min and analysed with a FACSCalibur™ flow cytometer (BD Bioscience, San Jose, CA, U.S.A.). Data were collected from 20000 cells for each sample and analysed using CellQuant software (BD Biosciences).
Mitochondrial area measurement
The mitochondrial area was measured using the histogram function of ImageJ software as described in Baxter et al. (2009). The cytoplasm was delineated using a combination of Alexa Fluor® 488-conjugated WGA (wheatgerm agglutinin; Molecular Probes) and TO-PRO®-3 staining (Molecular Probes).
Mitochondrial fractionation
Mitochondria-enriched fractions were isolated using a mitochondrial/cytosol fractionation kit (Sigma) as described in Baxter et al. (2009). Briefly, serum-grown and serum-deprived control PC12 and PC12-ND6 cells were trypsinized, centrifuged at 600 g for 5 min, washed twice with PBS and resuspended in an extraction buffer in the presence of protease inhibitors. Cells were gently homogenized using a Dounce homogenizer and the degree of homogenization was visualized by Trypan Blue staining. The homogenate was centrifuged at 1000 g for 10 min at 4°C. The supernatant was centrifuged again at 3500 g for 10 min at 4°C and the pelleted mitochondria were lysed with the lysis buffer provided in the presence of protease inhibitors. Protein concentration was determined by the Bradford assay (Bio-Rad Laboratories, Hercules, CA, U.S.A.).
Live cell imaging and mitochondrial morphology
Mitochondria of PC12-ND6 cells were labelled by electroporation (BTX, Harvard Apparatus, Holliston, MA, U.S.A.) using the GFP (green fluorescent protein)-mito vector (BD Pharmingen, San Jose, CA, U.S.A.), which encodes the GFP (green fluorescent protein) with the mitochondrial targeting sequence from subunit VIII of human COX. After 24 h of transfection, PC12-ND6 cells were grown on PDL (poly-d-lysine)-coated glass-bottom dishes (Warner Instruments, Hamden, CT, U.S.A.) in the presence or absence of serum for 48 h. Mitochondrial morphology was assessed using a ×100 Planapo Chromat (NA 1.4) objective and a Zeiss 510 LSM confocal system (Zeiss, Thornwood, NY, U.S.A.) equipped with TempModuleS and CO2 Module (Zeiss) to maintain the temperature and CO2 levels at 37°C and 5.0% respectively and the Zen navigation software (version 4.5; 2007) interface.
Measurement of intracellular ATP levels
Intracellular ATP levels were measured using the fluorescent dye Mg-Gr (Magnesium Green)-AM (acetoxymethyl ester) (Molecular Probes) as described in Bernstein and Bamburg (2003). Mg-Gr decreases emission intensity (λex = 475 nm; λem = 530 nm) with an increase in ATP levels due to its high binding affinity to ATP, as compared with that of ADP (Bernstein and Bamburg, 2003; Lee and Peng, 2008). Serum-grown and serum-deprived control PC12 and PC12-ND6 cells were labelled with 10 μM Mg-Gr resuspended in F12K serum- and Phenol Red-free medium (Invitrogen) for 30 min at 37°C. Cells were washed twice with PBS before being trypsinized and collected at 1300 g for 5 min and analysed with a FACSCalibur™ flow cytometer. Data were collected from 20000 cells and three independent experiments.
ROS quantification
Control PC12 and PC12-ND6 cells were grown on PDL-coated glass-bottom dishes, labelled with 0.5 μg/ml Hoechst 33342 (Invitrogen) for 30 min and 2.5 μM MitoSOX™ Red (Invitrogen) for 10 min at 37°C. Cells were imaged in Hoechst 33342 and MitoSOX™-free media with a ×63 Planapo Chromat (NA 1.4) objective using a Zeiss 510 LSM confocal system equipped with TempModuleS and CO2 Module. Using DIC (differential interference contrast) images to delineate the cells, the mean fluorescence intensity of MitoSOX™ labelling was calculated for each cell using NIH ImageJ software.
Immunocytochemistry and confocal fluorescence microscopy
For immunocytochemistry labelling, serum-grown PC12-ND6 cells were first seeded on to PDL-coated coverslips (Electron Microscopy Sciences, Hatfield, PA, U.S.A.) placed in a 6-well plate (Nunc, Rochester, NY, U.S.A.). For serum deprivation, cells were extensively washed with serum-free medium and grown in the absence of serum for 48 h. Immunocytochemistry and confocal fluorescence microscopy were carried out as described in Baxter et al. (2009). Mitochondria were labelled by incubating cells in the presence of 500 nM MTR (MitoTracker® Red; Molecular Probes) for 10 min at 37°C, as described in Baxter et al. (2009). Samples were fixed in 4.0% (w/v) paraformaldehyde (Electron Microscopy Sciences) for 5 min, and permeabilized in 0.2% Triton X-100 for 5 min. Cells were then blocked in 10% goat serum (Invitrogen) for 1 h at room temperature (25°C). All primary and secondary antibody incubations were performed at room temperature as follows: 1 h incubation with primary antibodies (Table 1), diluted in PBS, followed by 1 h incubation with the appropriate Alexa Fluor®-conjugated secondary antibodies diluted 1:1000 in PBS (Table 1). When indicated, cells were incubated with either nuclear counterstain TO-PRO®-3 or DAPI (4′,6-diamidino-2-phenylindole) (Molecular Probes). All samples were then mounted with Fluoromount G (Electron Microscopy Sciences). Images were acquired with a ×63 Planapo Chromat (NA 1.4) objective using a Zeiss LSM 710 confocal system.
Statistical analysis
Statistical analyses were performed using the unpaired Student’s t test. P values less than 0.05 were considered statistically significant. Data are expressed as means±S.D.
RESULTS
Constitutive expression of NeuroD6 in PC12-ND6 cells leads to maintained mitochondrial biomass upon oxidative stress mediated by serum deprivation
On the basis of NeuroD6-mediated increase in mitochondrial mass during the early stages of neuronal differentiation (Baxter et al., 2009) combined with NeuroD6 regulation of anti-apoptotic Bcl-2 members known to protect mitochondrial integrity (Uittenbogaard and Chiaramello, 2005; Uittenbogaard et al., 2009), we hypothesized that NeuroD6 could maintain the mitochondrial biomass of PC12-ND6 cells upon withdrawal of trophic factors. We used serum deprivation as an oxidative stress paradigm (Satoh et al., 1996), based on our previous studies showing that constitutive expression of NeuroD6 endows the PC12-ND6 cells with long-term survival properties while keeping them metabolically active (Uittenbogaard and Chiaramello, 2005). To test this hypothesis, we used three distinct but complementary experimental approaches: flow cytometry, confocal fluorescence microscopy and mitochondrial-enriched fractionation.
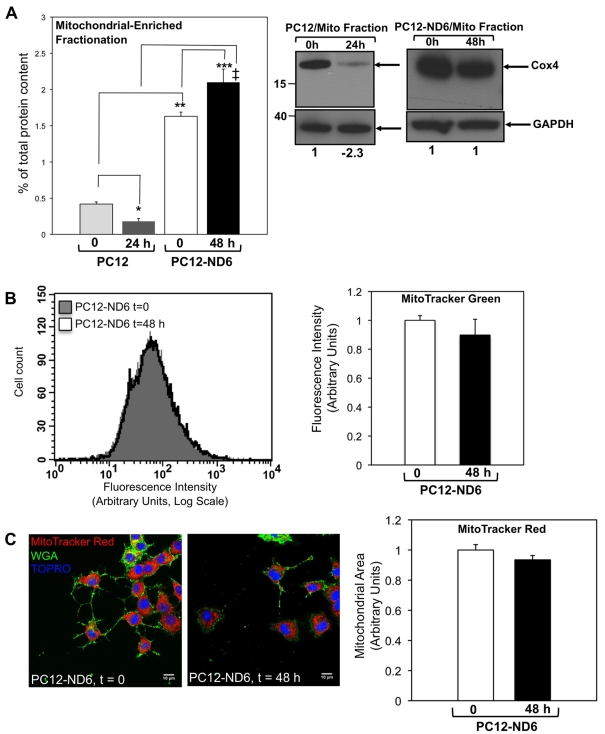
To initiate our studies, we isolated mitochondrial-enriched fractions from control PC12 and PC12-ND6 cells grown in the presence or absence of serum for 24 and 48 h respectively. Mitochondrial fractionation using control PC12 cells could not be performed beyond 24 h of serum deprivation due to loss of PC12 cells adherence and extensive apoptosis accompanied by caspase activation, cytochrome c leakage into the cytosol as a result of compromised mitochondrial membrane integrity, chromatin condensation and extensive membrane blebbing (Stefanis et al., 1996, 1998; Troy et al., 1997; François et al., 2001; Vyas et al., 2004; Uittenbogaard and Chiaramello, 2005). Results are expressed as a percentage of total protein content of the corresponding whole-cell extract, as described in Baxter et al. (2009). While PC12-ND6 cells displayed a statistically significant (P = 0.0153) 20% increase in their mitochondrial protein content after 48 h of serum deprivation, control PC12 cells lost more than 50% of their mitochondrial protein content after 24 h of serum deprivation (Figure 1A). Furthermore, serum-deprived PC12-ND6 cells (48 h) exhibit an 11-fold increase in mitochondrial protein content, as compared with that of serum-deprived control PC12 cells (24 h), in keeping with the reported increased survival capacity upon NeuroD6 expression (Uittenbogaard and Chiaramello, 2005; Uittenbogaard et al., 2010). Using a monoclonal antibody against COX4, we found a 2.3-fold decrease in COX4 expression to negligible levels in mitochondria-enriched fractions from serum-deprived (for 24 h) control PC12 cells, as compared with serum-grown control PC12 cells (Figure 1A, right panel). This stands in contrast with maintained COX4 expression levels in PC12-ND6 cells after 48 h of serum deprivation (Figure 1A, right panel). It bears mentioning that serum-grown PC12-ND6 cells exhibited higher levels of COX4 expression than serum-grown control PC12 cells, in keeping with the observed increase in mitochondrial protein content upon NeuroD6 expression (Figure 1A, left panel).
Figure 1. Constitutive expression of NeuroD6 results in sustained mitochondrial biomass upon serum deprivation.
(A) Whereas PC12-ND6 cells retained their mitochondrial protein content after 48 h of serum deprivation, control PC12 cells lost more than half of their mitochondrial protein content after 24 h of serum deprivation. Mitochondrial-enriched fractions were isolated from serum-grown control PC12 and PC12-ND6 cells (t = 0) and serum-deprived control PC12 (t = 24 h) and PC12-ND6 (t = 48 h) cells. The protein content of mitochondrial fractions is expressed as a percentage of total protein content of the corresponding whole cell extract (left-hand panel). Data are expressed as the means±S.D. from three independent experiments (*P = 0.0002 as compared with serum-grown control PC12 cells; **P = 0.0001 as compared with serum-deprived control PC12 cells; ***P = 0.0153 as compared with serum-grown PC12-ND6 cells; ‡P = 0.0001 as compared with serum-deprived control PC12 cells). Expression levels of COX4 remained constant in mitochondrial-enriched fractions of PC12-ND6 cells after 48 h of serum deprivation (right-hand panel). Immunoblot analysis was performed using mitochondrial-enriched fractions isolated from serum-grown (0 h) and serum-deprived (24 h) control PC12 as well as serum-grown (0 h) and serum-deprived (48 h) PC12-ND6 cells using a monoclonal antibody against COX4. Equal loading was verified using an anti-GAPDH antibody. Molecular mass markers are indicated in kDa on the left side of each Western blot panel. Results shown are representative of three independent experiments. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. (B) Quantification of the fluorescence intensity of serum-grown and serum-deprived PC12-ND6 labelled with the mitochondrial dye MTG. PC12-ND6 cells were first grown in a serum-containing medium before being serum-deprived for 48 h. PC12-ND6 cells were then stained with MTG (70 nM) and analysed by flow cytometry. Flow cytometric profiles of serum-grown (t = 0) and serum-deprived (t = 48 h) PC12-ND6 labelled with MTG (left panel). Data are expressed as means±S.D. from five independent experiments (right panel). There was no statistical difference in the mitochondrial biomass of serum-deprived PC12-ND6 cells, as compared with serum-grown PC12 cells. (C) The mitochondrial area of PC12-ND6 cells remains constant after 2 days of serum deprivation. To stain the mitochondrial compartment, serum-grown (t = 0) and serum-deprived (t = 48 h) PC12-ND6 cells were labelled with MTR (red) and the plasma membrane marker Alexa Fluor® 488-conjugated WGA (green) and TO-PRO®-3® (blue) (left panel; scale bar, 10 μm). The right panel illustrates the quantification of mitochondrial area in serum-grown and serum-deprived PC12-ND6 cells. Data are expressed as the means±S.D. for three independent experiments with a minimum of n = 150 cells for each culture condition. No statistically significant variation of the mitochondrial area was observed on serum deprivation of PC12-ND6 cells.
We further quantified the mitochondrial biomass in serum-grown and serum-deprived PC12 ND6 cells using flow cytometry by staining PC12-ND6 cells with MTG, as fluorescence intensity emitted by MTG is directly proportional to mitochondrial mass as assessed by flow cytometry. We also opted to label the mitochondrial population with MTG, since this dye accumulates in the lipid compartment in an MMP-independent fashion (Pendergrass et al., 2004). Serum-deprived control PC12 cells could not be stained with this mitochondrial dye due to the compromised mitochondrial membrane integrity upon serum deprivation, therefore remaining non-fluorescent. Quantitative flow cytometry revealed that the mitochondrial mass of PC12-ND6 cells remained constant after 2 days of serum deprivation (Figure 1B).
To complement our quantification analysis, we measured another indicator of mitochondrial mass, mitochondrial area (Baxter et al., 2009), by labelling serum-grown and serum-deprived PC12-ND6 cells with the plasma membrane marker, Alexa Fluor® 488-conjugated WGA, the mitochondrial dye MTR, and the nuclear counterstain TO-PRO®-3 followed by confocal fluorescence analysis. Serum-deprived control PC12 cells could not be stained with MTR, as this dye cannot be retained in the mitochondrial matrix when the mitochondrial membrane is compromised (Deshmukh et al., 2000; Buckman et al., 2001). Mitochondrial area was calculated using the plasma membrane and nuclear stains to delineate the cytoplasm and MTR to stain the mitochondrial population. We found no statistically significant difference in the mitochondrial area of serum-grown and serum-deprived PC12-ND6 cells (Figure 1C). Taken together, these results reveal that NeuroD6 maintains the mitochondrial mass of PC12-ND6 cells upon removal of trophic factors, in keeping with the previously reported NeuroD6 neuro-protective properties using our PC12-ND6 cellular paradigm and withdrawal of trophic factors (Uittenbogaard and Chiaramello, 2005; Uittenbogaard et al., 2010).
Serum-deprived PC12-ND6 cells display sustained intracellular ATP levels and expression levels of specific subunits of respiratory complexes, preventing mitochondrial fractionation
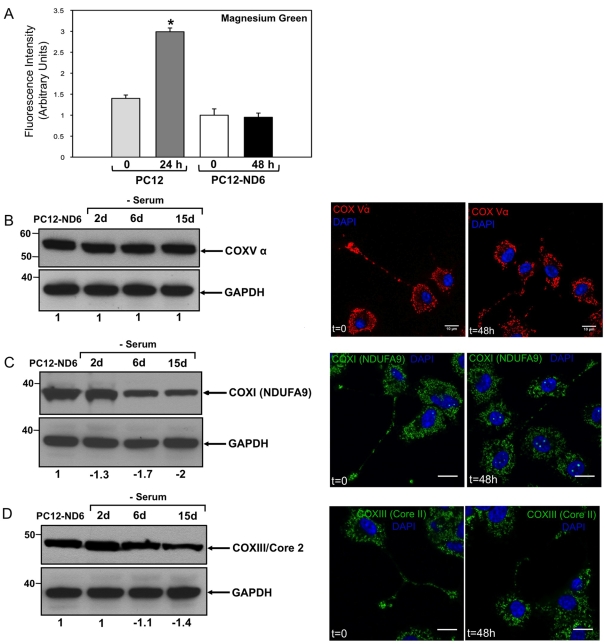
The lack of variation in MTR labelling coupled with sustained mitochondrial mass and COX4 expression levels in serum-deprived PC12-ND6 cells (Figure 1C) is suggestive of maintained bioenergetic functions, as accumulation and retention of MTR have been shown to be MMP (mitochondrial membrane potential)-dependent (Pendergrass et al., 2004). Thus we first assessed the intracellular ATP levels in control PC12 and PC12-ND6 cells in the presence or absence of serum for 24 and 48 h respectively. We opted for a flow cytometry approach with the fluorescent dye Mg-Gr-AM, as previous studies have established a correlation between MTR labelling and ATP levels estimated by Mg-Gr-AM in Xenopus spinal neurons (Lee and Peng, 2008). It has been well documented that the fluorescence intensity of this probe decreases with ATP content due to its higher binding affinity to ATP than that to ADP (Leyssens et al., 1996; Bernstein and Bamburg, 2003). Quantitative flow cytometry revealed that serum deprivation of PC12-ND6 cells did not alter intracellular ATP levels (Figure 2A), consistent with the sustained fluorescence intensity emitted by the mitochondrial dye MTR (Figure 1C). As expected, serum-deprived control PC12 cells lost more than 50% of intracellular ATP, resulting in increased fluorescence emission from Mg-Gr-AM (Figure 2A). Such decreased ATP levels correlate with the collapsed COX4 expression levels in mitochondria-enriched fractions isolated from serum-deprived (24 h) control PC12 cells and an overall decrease in mitochondrial content (Figure 1A). Moreover, they are in full concordance with the well-documented catastrophic PC12 cell death after 24 h of serum deprivation (Stefanis et al., 1996, 1998; Troy et al., 1997; François et al., 2001; Vyas et al., 2004; Uittenbogaard and Chiaramello, 2005) and the recently reported 40% loss of mitochondrial membrane depolarization of PC12 cells after 24 h of serum deprivation (Krzyzanski et al., 2007).
Figure 2. Serum-deprived PC12-ND6 cells sustain intracellular ATP levels and expression levels of specific subunits of the respiratory complexes I, III and V.
(A) Intracellular ATP levels of PC12-ND6 cells remained constant after 2 days of serum deprivation, while serum-deprived control PC12 cells (for 24 h) lost half of their original ATP levels. Serum-grown and serum-deprived control PC12 and PC12-ND6 cells were labelled with Mg-Gr-AM (10 μM) for 30 min at 37°C before being analysed with a FACSCalibur™ flow cytometer. Data were collected from 20000 cells and three independent experiments. Mg-Gr decreases emission intensity (λex = 475 nm; λem = 530 nm) with an increase in ATP levels due to its high binding affinity to ATP, as compared with that of ADP. Results are expressed as relative fluorescence intensity. No statistically significant difference in intracellular ATP levels was observed after 2 days of serum deprivation (P = 0.7992). (B) The α-subunit of the ATP synthase (COXV) remains steadily expressed throughout the duration of serum deprivation in PC12-ND6 cells. Immunoblot analysis was performed using whole-cell extracts from serum-grown PC12-ND6 cells and PC12-ND6 cells serum-deprived for the indicated periods of time. Equal loading was verified using an anti-GAPDH antibody. Molecular masses in kDa are indicated on the left side of each Western blot panel. The left-hand panel shows immunoblot results representative of three independent experiments. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. The right-hand panel illustrates the representative immunocytochemistry showing the retained mitochondrial localization of the α-subunit of the ATP synthase (COX5) on serum deprivation (scale bar, 10 μm). (C) The subunit NDUFA9 of complex I remains expressed throughout the duration of serum deprivation in PC12-ND6 cells, albeit at lower levels after 6 days of serum removal. The left-hand panel shows immunoblot results representative of at least three independent experiments. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. The right-hand panel illustrates representative immunocytochemistry showing the retained mitochondrial localization of the subunit NDUFA9 upon serum deprivation. Cells were labelled with an anti-NDUFA9 monoclonal antibody and the nuclear counterstain DAPI before being analysed by confocal fluorescence microscopy (scale bar, 10 μm). (D) The core2 protein of COX3 remains expressed throughout the duration of serum deprivation in PC12-ND6 cells. The left-hand panel shows immunoblot results representative of three independent experiments. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. The right-hand panel illustrates the representative immunocytochemistry showing the retained mitochondrial localization of the core2 protein of COX3 on serum deprivation (scale bar, 10 μm).
To understand how PC12-ND6 cells maintain their intracellular ATP levels upon serum deprivation, we examined the expression levels of the α-subunit of the ATP synthase (complex V), as our genome-wide microarray analysis revealed a 1.52-fold increase in serum-deprived PC12-ND6 cells after 48 h (Uittenbogaard et al., 2010). By immunoblot analysis, we found that the α-subunit of the ATP synthase (complex V) remained expressed at constant levels throughout serum deprivation (Figure 2B). We complemented our immunoblot analysis by examining the expression levels of the subunit NDUFA9 of complex I and the core2 protein of complex III. Although their expression levels remained unaltered during the first 2 days of serum deprivation, they decreased 2- and 1.4-fold respectively after 15 days of serum deprivation (Figures 2C and 2D). Next, we assessed their localization by immunocytochemistry, which confirmed their sustained expression levels as well as their localization in growth cones of serum-deprived PC12-ND6 cells (Figures 2B–2D).
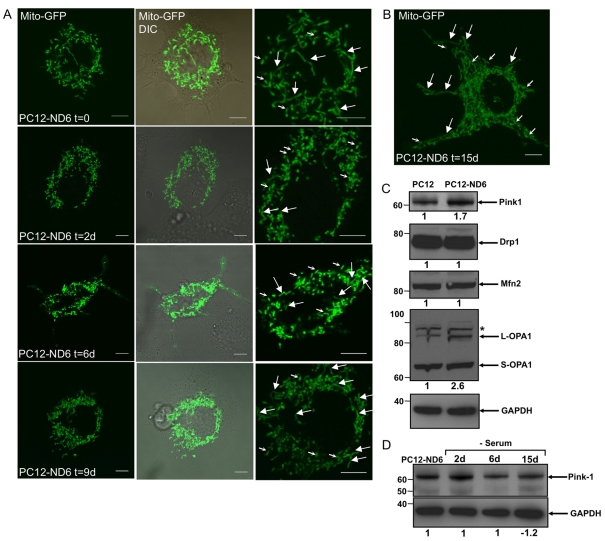
Since a strong link exists between mitochondrial morphology and bioenergetic functions, with an elongated morphology linked to the normal metabolic state and cell survival and a fragmented morphology (spheroid-like) associated with compromised respiratory activities and programmed cell death (reviewed by Karbowski and Youle, 2003; Okamoto and Shaw, 2005; Chan 2006), we evaluated by confocal live cell imaging whether PC12-ND6 cells maintained a healthy mitochondrial population at distinct time points of serum deprivation. PC12-ND6 cells were first transfected by electroporation using the mito-GFP vector, which encodes the GFP with the mitochondrial targeting sequence from subunit VIII of human COX, and were then serum-deprived after 24 h of transfection for defined periods of time. Figure 3(A) shows that serum-grown PC12-ND6 cells displayed a preponderance of elongated mitochondria, as previously reported by immunocytochemistry using fixed PC12-ND6 cells (Baxter et al., 2009). After 48 h of serum deprivation, most mitochondria showed a short rod-like morphology and a few showed an elongated morphology, while spheroid-like fragmented mitochondria were rare (Figure 3A). A similar overall mitochondrial morphology was sustained after 6–9 days of serum deprivation (Figure 3A). PC12-ND6 cells, which were serum-deprived for 15 days, displayed elongated mitochondria (Figure 3B), suggestive of an ongoing dynamic mitochondrial fusion–fission process, associated with proper mitochondrial functions. Collectively, our flow cytometry analysis and confocal live cell imaging have revealed that NeuroD6 not only maintains the mitochondrial biomass, but also a properly regulated mitochondrial fusion–fission dynamics throughout the different phases of long-term serum deprivation.
Figure 3. Analysis of mitochondrial morphology by live cell imaging and expression levels of proteins involved in mitochondrial fusion and fission prior to and after serum removal.
(A) Live cell confocal fluorescence imaging of serum-grown and serum-deprived PC12-ND6 cells. PC12-ND6 cells were first transfected with the mito-GFP vector to label the mitochondrial mass and then switched to a serum-free medium for the indicated periods of time. Mitochondrial morphology was assessed by confocal fluorescence microscopy using a ×100 oil objective and an environmental chamber to keep the CO2 level and temperature constant. The left panels show GFP-labelled mitochondria, while the middle panels show the merge with the corresponding DIC pictures. The right panels show high magnification of the labelled mitochondria (large arrows for elongated mitochondria and small for rod-like mitochondria). Scale bar, 5 μm. (B) High magnification of serum-deprived PC12-ND6 cells transfected with the mito-GFP vector. This image illustrates the range of mitochondrial length, suggestive of a dynamic fusion–fission activity even after 15 days of serum deprivation. Scale bar, 5 μm. (C) Expression profile of protein involved in the fusion–fission process of mitochondria. Immunoblot analyses were performed using whole-cell extracts from serum-grown control PC12 and PC12-ND6 cells and equal loading was verified using an anti-GAPDH antibody. Relevant molecular masses (in kDa) are indicated on the left side of each Western blot panel. The immunoblots are representative of at least three independent experiments. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. (D) Full-length PINK1 protein remains expressed in serum-deprived PC12-ND6 cells, even after 15 days of serum deprivation. Immunoblot analysis was performed using whole-cell extracts from PC12-ND6 cells grown in a serum-free medium for the indicated periods of time. Equal loading was verified using an anti-GAPDH antibody. Relevant molecular masses (in kDa) are indicated on the left side of the Western blot panels. Results shown are representative of three independent experiments. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates.
On the basis of the fact that PINK1 is critical for mitochondrial integrity in several neuronal paradigms, with PINK1 deficiency inducing mitochondrial fragmentation and increased sensitivity to oxidative stress (Clark et al., 2006; Park et al., 2006; Gautier et al., 2008; Wood-Kaczmar et al., 2008; Dagda et al., 2009), we examined PINK1 expression levels upon constitutive NeuroD6 expression and serum deprivation. We consistently observed the expression level of the full-length PINK1 protein to increase 1.7-fold in serum-grown PC12-ND6 cells, as compared with that of control PC12 cells by immunoblot analysis (Figure 3B). We supplemented our analysis by examining the expression levels of proteins known to regulate mitochondrial morphology, such as the mitochondrial fusion Mfn2 (mitofusin 2) and OPA1 (optic atrophy 1) proteins and the mitochondrial fission factor Drp1 (dynamin-related protein 1) (reviewed by Lackner and Nunnari, 2009; Thomas and Cookson, 2009). Among those proteins, only the long OPA1 isoform showed a 2.6-fold increase in expression level in PC12-ND6 cells (Figure 3B), which is the fusion-competent form of OPA1 in the rat species (Ishihara et al., 2006). Finally, the PINK1 protein remained uncleaved and expressed throughout the whole duration of serum deprivation (Figure 3C), an observation that is in keeping with the overall mitochondrial morphology observed in serum-deprived PC12-ND6 cells. Thus our collective results support the notion that constitutive expression of NeuroD6 not only allows serum-deprived PC12-ND6 cells to retain a steady healthy mitochondrial population, but also to sustain intracellular ATP levels and expression levels of key subunits of respiratory complexes I, III, IV and V.
PGC-1α, Tfam (transcription factor A, mitochondrial) and NRF-1 (nuclear respiratory factor-1) expression levels are sustained in serum-deprived PC12-ND6 cells
To initiate our studies on the NeuroD6-mediated transcriptional pathways responsible for maintenance of the mitochondrial biomass throughout serum deprivation, we examined the expression levels of key transcriptional regulators, known to stimulate mitochondrial biogenesis, such as the master transcriptional co-activator PGC-1α and the nuclear-encoded transcriptional regulators, Tfam and NRFs NRF-1 and -2 (reviewed by Kelly and Scarpulla, 2004; Scarpulla 2008).
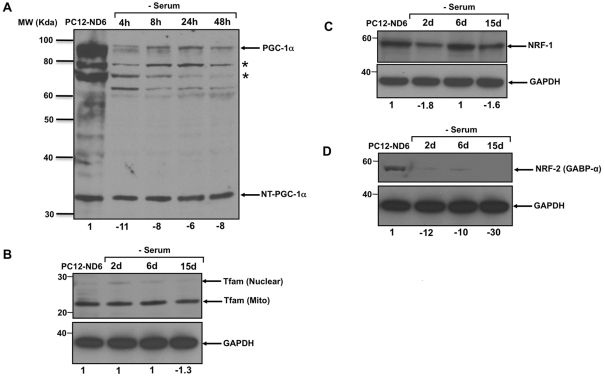
We initially focused on PGC-1α expression levels using a polyclonal antibody raised against the N-terminal domain of PGC-1α (Table 1), as PGC-1α not only regulates mitochondrial biogenesis, but also confers protection from oxidative stress in the brain (St-Pierre et al., 2006; Wareski et al., 2009). We detected two predominant isoforms of full-length PGC-1α as well as the recently identified truncated form of PGC-1α (35 kDa), NT-PGC-1α (N-terminal-truncated PGC-1α; Figure 4A). This truncated form has been shown to be transcriptionally competent to activate a subset of known PGC-1α target genes in brown adipocyte tissue (Zhang et al., 2009). Interestingly, NT-PGC-1α remained expressed at constant levels throughout the whole duration of serum deprivation, whereas the full-length PGC-1α expression levels rapidly decreased 11-fold after 4 h of serum removal, failing to display a recovery after 48 h of serum deprivation (Figure 4A). Thus the dynamics of PGC-1α expression pattern suggests that the truncated form of NT-PGC-1α rather than the full-length PGC-1α may be involved in maintaining the mitochondrial mass upon serum deprivation.
Figure 4. Expression pattern of key regulators involved in mitochondrial biogenesis upon serum deprivation.
The immunoblots are representative of three independent experiments. Equal loading was verified using an anti-GAPDH antibody and the relevant molecular masses (in kDa) are indicated on the left side of each Western blot panel. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. (A) Only the expression levels of the truncated form of PGC-1α, NT-PGC-1α, remain unaltered throughout serum deprivation of PC12-ND6 cells. The asterisks indicate isoforms of the full-length PGC-1α. Quantification pertains to the full length of PGC-1α. (B) Tfam expression levels in PC12-ND6 cells were unaffected by serum deprivation. The mitochondrial-specific form of Tfam was the main form detected in both serum-grown and serum-deprived PC12-ND6 cells. (C) Levels of NRF-1 expression remained steady throughout serum deprivation of PC12-ND6 cells. (D) Levels of NRF2 expression collapsed 2 days after serum deprivation of PC12-ND6 cells.
We supplemented our analysis by examining the expression levels of Tfam in serum-deprived PC12-ND6 cells, since Tfam regulates mitochondrial transcription and maintenance of mtDNA copy number (Ekstrand et al., 2004; Kanki et al., 2004; Ikeuchi et al., 2005; Cotney et al., 2007). We found that serum deprivation of PC12-ND6 cells triggered only a negligible decrease (1.3-fold) of the mitochondrial form of Tfam protein (Figure 4B). These Tfam levels are in keeping with the maintained mitochondrial mass in serum-deprived PC12-ND6 cells assessed by flow cytometry, confocal microscopy and mitochondrial-enriched fractionation (Figure 1).
Since recent studies have shown that the transcriptional co-activator PGC-1α interacts with NRF-1 to stimulate mitochondrial biogenesis by activating the transcription of all ten subunits of COX in neurons (Dhar et al., 2008), we investigated whether NRF-1 expression levels paralleled PGC-1α and Tfam levels in serum-deprived PC12-ND6 cells. By immunoblot analysis, we showed a tight correlation between PGC-1α, Tfam and NRF-1 expression levels throughout serum deprivation (Figure 4C). Finally, we also examined the expression levels of NRF-2 [GABP-α (GA-binding protein-α)] in serum-deprived PC12-ND6 cells, as NRF-2 in conjunction with the transcriptional co-regulator PGC-1α also regulates the expression of all ten nuclear-encoded subunits of COX in neurons (Ongwijitwat and Wong-Riley, 2005; Ongwijitwat et al., 2006) and Tfam (Virbasius et al., 1993). We used a mouse monoclonal antibody against the α-subunit of NRF-2 (GABP-α), as this subunit is required for NRF-2 DNA-binding activity (Batchelor et al., 1998). GABP-α expression levels were barely detectable, with a 12-fold decrease after 2 days of serum deprivation (Figure 4D), suggesting that maintenance of the mitochondrial mass and mtDNA copy number via Tfam expression upon serum deprivation does not require NRF-2-mediated transcriptional regulation. Collectively, our results support the notion that NeuroD6 triggers a specific transcriptional programme to maintain the mitochondrial biomass of serum-deprived PC12-ND6 cells in order to mediate neuronal survival and mitochondrial integrity as previously reported (Uittenbogaard and Chiaramello, 2005; Uittenbogaard et al., 2009).
Constitutive expression of NeuroD6 prevents generation of the ROS superoxide anions upon serum deprivation
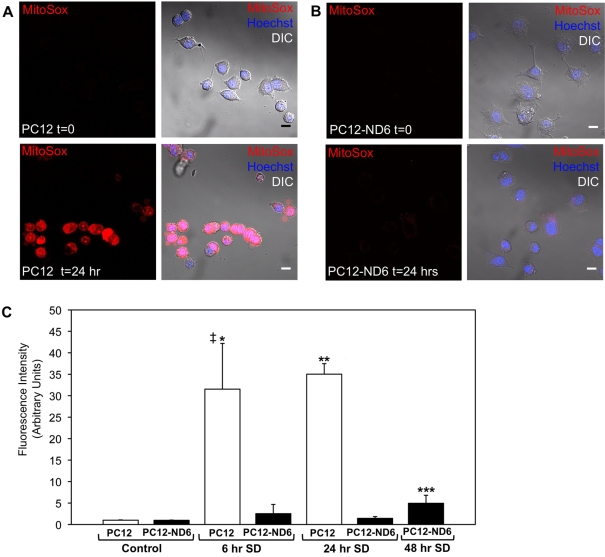
Based on our recent finding that serum-grown PC12-ND6 cells produce more ATP than the serum-grown control PC12 cells (Baxter et al., 2009), combined with the fact that the main source of electron leakage originates from the mitochondrial ETC to reduce O2 to the predominant form of ROS superoxide anions (Turrens, 1997; Demin et al., 1998), we investigated whether such an increased energy production would result in higher levels of superoxide anions (O2−) than that in control PC12 cells. To address this question, we measured relative levels of O2− using the redox-sensitive dye, MitoSOX™, which is a fluorogenic dye for highly selective detection of superoxide anions. Since the MitoSOX™ dye is live-cell-permeant and selectively targeted to the mitochondria, we labelled live control PC12 and PC12-ND6 cells with the MitoSOX™ dye and nuclear counterstain Hoechst 33342 and we measured by confocal microscopy the amount of red fluorescence from oxidized MitoSOX™ dye, as an indicator of mitochondrial O2− levels (Johnson-Cadwell et al., 2007). Figure 5 shows that serum-grown PC12-ND6 cells did not display higher levels of O2− than serum-grown control PC12 cells.
Figure 5. Constitutive expression of NeuroD6 prevents ROS production upon serum deprivation.
(A, B) Live cell confocal images of control PC12 and PC12-ND6 cells grown in the presence or absence of serum for 24 h before being labelled with the MitoSOX™ dye (red) and the nuclear counterstain Hoechst 33342 (blue). Images are representative of three independent experiments. The left panels illustrate MitoSOX™-labelled cells before (t = 0) and after serum deprivation (t = 24 h), while the right panels show the merge with the corresponding DIC pictures (scale bar, 10 μm). (C) Quantification of ROS production at different time points of serum deprivation for control PC12 and PC12-ND6 cells. The mean fluorescence intensity of MitoSOX™ labelling was measured in the presence or absence of serum for 6 and 24 h in control PC12 and PC12-ND6 cells and for 48 h in PC12-ND6 cells. At the outset of serum deprivation, we did not observe any statistical difference of ROS production between control PC12 and PC12-ND6 cells (P = 0.6074). The graph represents the averages from 150 cells from three independent experiments. ‡P = 0.0077 as compared with serum-grown control PC12 cells; *P = 0.0098 as compared with serum-deprived control PC12 cells for 6 h; **P = 0.0004 as compared with serum-deprived control PC12 cells for 24 h; ***P = 0.0207 as compared with untreated PC12-ND6 cells.
Previous studies have demonstrated that serum deprivation simulates oxidative stress in the context of the PC12 cellular paradigm, resulting in ROS generation within 8 h of treatment and ultimately inducing apoptosis within 24 h of serum withdrawal (Rukenstein et al., 1991; Troy and Shelanski, 1994; Satoh et al., 1996). Thus we investigated whether constitutive expression of NeuroD6 would prevent ROS generation on serum withdrawal. As expected, we observed a 32-fold increase in superoxide anion levels in control PC12 cells after 6 h of serum deprivation, which was slightly more exacerbated after 24 h of serum deprivation to reach a final 35-fold increase (Figures 5A and 5C). In contrast, serum-deprived PC12-ND6 cells failed to display increased levels of ROS within the first 24 h of serum deprivation (Figures 5B and 5C). However, we observed a slight increase by a factor of 5, after 48 h of serum deprivation, levels that remained significantly lower than those of serum-deprived PC12 cells (Figure 5). Collectively, these results indicate that NeuroD6 protects PC12-ND6 cells from generating significant levels of superoxide anions in mitochondria, which is in keeping with our previous finding of NeuroD6-mediated neuronal survival upon trophic factor withdrawal via the mitochondrial pathway (Uittenbogaard and Chiaramello, 2005; Uittenbogaard et al., 2009).
NeuroD6 sustains the expression of ROS detoxifying enzymes during oxidative stress mediated by serum deprivation
Because our recent genome-wide microarray analysis supplemented with a GSEA (gene set enrichment analysis) has revealed a correlated expression between NeuroD6 and genes involved in oxidative stress, ROS metabolism and the molecular chaperone response prior to and during serum deprivation (Uittenbogaard et al., 2010), we examined the expression profile of the ROS detoxifying enzymes copper/zinc SOD (SOD1), manganese SOD (SOD2), GPx1 (glutathione peroxidase 1) and PRDX5 (peroxiredoxin 5) throughout 15 days of serum deprivation by immunoblot analysis to acquire a perspective on the dynamic aspect of NeuroD6-mediated tolerance to prolonged stress upon removal of trophic factors.
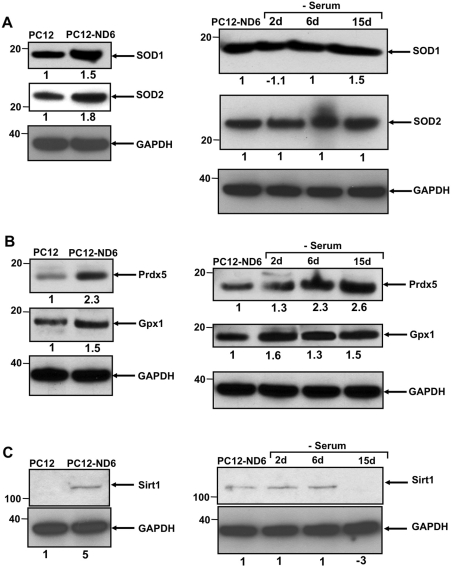
We initially focused on the ROS detoxifying enzymes, SOD1 and SOD2, as they are critical for survival of PC12 cells upon serum deprivation (Troy and Shelanski, 1994; Troy et al., 1996) and, more importantly, as SOD2 protein is localized in the mitochondrial matrix. We found that constitutive expression of NeuroD6 not only increased the expression levels of both SOD1 and SOD2 by 1.5- and 1.8-fold respectively but also maintained their expression levels throughout the whole period of serum deprivation (Figure 6A). We confirmed by immunocytochemistry that SOD2 protein remained present in the mitochondria of serum-deprived PC12-ND6 cells (results not shown). In contrast, serum-deprived control PC12 cells showed decreased expression levels of SOD1 and SOD2 (see Supplementary Figure S1 at http://www.asnneuro.org/an/002/an002e034add.htm).
Figure 6. NeuroD6 triggers an antioxidant response in the absence of stress, which remains expressed throughout serum deprivation.
The immunoblots are representative of three independent experiments. Equal loading was verified using an anti-GAPDH antibody. Indicated quantification values are specific to the blot shown, with an S.D. less than 10% compared with the corresponding replicates. (A) Constitutive expression of NeuroD6 results in increased expression of SOD1 and SOD2 in the absence of stress stimulus, which remains unaltered during oxidative stress mediated by serum deprivation. (B) Expression levels of the antioxidant regulators, Gpx1 and Prdx5, are increased upon constitutive expression of NeuroD6 and throughout the duration of serum deprivation treatment. (C) SIRT1 expression increases in the absence of stress stimulus in serum-grown PC12-ND6 cells, while remaining constant for up to 6 days of serum deprivation. Relevant molecular masses in kDa are indicated on the left side of each Western blot panel.
Given the fact that our microarray analysis reflected a 1.81-fold increase in expression levels of PRDX5 mRNA upon constitutive expression of NeuroD6 combined with its known antioxidant and anti-apoptotic properties (Zhou et al., 2000; Banmeyer et al., 2004, 2005; Dubuisson et al., 2004; Wong et al., 2004; Kropotov et al., 2006; De Simoni et al., 2008; Radyuk et al., 2009), we determined whether such a direct correlation remained at the protein level prior to and upon serum deprivation. By immunoblot analysis, we validated that the protein PRDX5 was indeed significantly up-regulated 2.3-fold upon constitutive expression of NeuroD6 (Figure 6B). Most importantly, PRDX5 expression levels steadily increased throughout the whole serum deprivation treatment, to a final 2.6-fold after 15 days of serum deprivation, suggesting a potential pivotal role in the long-term NeuroD6-mediated tolerance to oxidative stress (Figure 6B). In contrast, control PC12 cells, which expressed very low levels of PRDX5 even in the presence of serum, maintained negligible levels of PRDX5 expression after 24 h of serum deprivation (see Supplementary Figure S1).
We next examined the expression levels of another ROS detoxifying enzyme, GPx1, as our genome-wide microarray analysis revealed a 1.54-fold increase at the mRNA level (Uittenbogaard et al., 2010) and it plays a well-established role in the O2 reduction pathway associated with mitochondrial respiratory functions (reviewed by D’Autréaux and Toledano, 2007; Giorgio et al., 2007). We found that NeuroD6 by itself was sufficient to stimulate levels of the GPx1 protein 1.5-fold, which further increased by up to 1.6-fold throughout oxidative stress mediated by serum deprivation (Figure 6B). Again, serum-deprived control PC12 cells failed to maintain GPx1 expression levels (see Supplementary Figure S1). We supplemented our immunoblot analysis by examining the expression levels of another detoxifying enzyme, catalase, which revealed substantially decreased levels of catalase in serum-deprived PC12-ND6 cells (results not shown), implying its lack of involvement in NeuroD6-mediated tolerance to oxidative stress and control of ROS production.
Given that PC12 cells transiently express the mammalian NAD-dependent deacetylase orthologue of the yeast silent information regulator 2, SIRT1, after 16 h of serum deprivation (Nemoto et al, 2004; Wu et al., 2006), combined with its role in cellular energy and redox homoeostasis (Fulco et al., 2003; Brunet et al., 2004; Nemoto et al., 2005), embryonic brain development (Cheng et al., 2003; McBurney et al., 2003; Sakamoto et al., 2004; Prozorovski et al., 2008) and neuroprotection in the context of neurodegenerative diseases (Parker et al., 2005; Sinclair 2005), we investigated whether NeuroD6 could up-regulate SIRT1 expression independently of a stress stimulus or only upon serum deprivation. In the presence of serum, the SIRT1 protein was expressed in PC12-ND6 cells, while it was absent in control PC12 cells (Figure 6C), in keeping with the reported studies (Nemoto et al., 2004). Interestingly, serum deprivation did not result in a further increase in SIRT1 expression levels in PC12-ND6 cells, but rather resulted in sustained expression levels for up to 6 days of treatment before collapsing after 15 days of serum removal (Figure 6C). Collectively, these results indicate that NeuroD6 triggers a comprehensive antioxidant response in the absence of stress, which generates a reserve of antioxidant ready to inactivate generated ROS upon serum deprivation thereby maintaining ROS homoeostasis.
DISCUSSION
Given the ability of NeuroD6 to increase the mitochondrial biomass in the early stages of neuronal differentiation (Baxter et al., 2009) and to trigger a comprehensive anti-apoptotic response and a molecular chaperone network through the expression of multiple HSPs (Uittenbogaard and Chiaramello, 2005; Uittenbogaard et al., 2009, 2010), we extended our study on the neuroprotective properties of NeuroD6 by investigating whether constitutive expression of NeuroD6 could concomitantly regulate the mitochondrial biomass and ROS homoeostasis upon oxidative stress mediated by serum deprivation, as suggested by our recent genome-wide microarray analysis (Uittenbogaard et al., 2010). In the present study, we report a novel role for NeuroD6 as a regulator of ROS homoeostasis, resulting in enhanced tolerance to oxidative stress. First, constitutive expression of NeuroD6 results in sustained mitochondrial mass upon oxidative stress triggered by withdrawal of trophic factors as shown by distinct but complementary experimental approaches, such as flow cytometry, confocal fluorescence microscopy and mitochondrial fractionation. Secondly, NeuroD6-mediated maintenance of mitochondrial mass is accompanied by sustained intracellular ATP levels and expression of specific subunits of distinct respiratory complexes located in the mitochondrial inner membrane. Thirdly, serum-deprived PC12-ND6 cells bear mitochondria displaying an elongated morphology compatible with bioenergetic capacity to ensure cell survival upon withdrawal of trophic factors. Fourthly, NeuroD6 sustains the expression of the transcriptional co-activator PGC-1α and the nuclear-encoded transcription factors Tfam and NRF-1, all of them known to regulate mitochondrial biogenesis. Lastly, NeuroD6 triggers a comprehensive antioxidant response in the absence of stress, which endows PC12-ND6 cells with intracellular ROS scavenging capacity, resulting in low ROS levels upon oxidative stress.
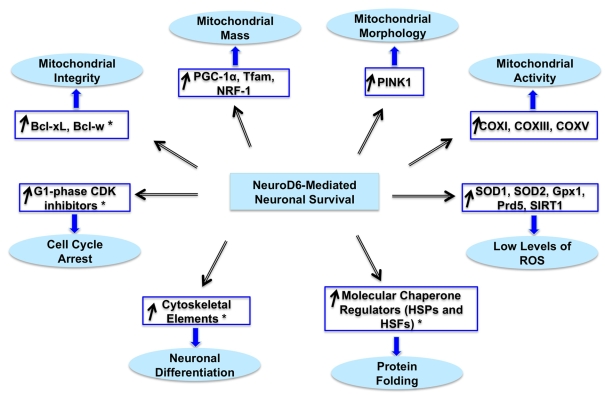
In view of the prevailing theory that oxidative stress, mitochondrial dysfunction and impaired protein folding/degradation are common underlying causes for many neurodevelopmental disorders, neurodegenerative diseases and brain aging (reviewed by Wallace, 2005; Lin and Beal, 2006; Giorgio et al., 2007; Nicholls, 2008), the ability of NeuroD6 to regulate these three interconnected pathways may provide therapeutic opportunities to mediate neuroprotection. This is particularly relevant in the context of NeuroD6 exclusive expression in the entorhinal cortex and subiculum (Schwab et al., 1998), two areas of the brain to undergo degeneration at the very early stages of AD (Braak and Braak, 1991; deToledo-Morrell et al., 2004) and to functionally connect the hippocampus to the neocortex (de la Prida et al., 2006; Yasuda and Mayford, 2006), where NeuroD6 is also highly expressed. To our knowledge, our collective studies provide the first demonstration of a neurogenic bHLH transcription factor able to concomitantly co-ordinate the regulation of cell cycle, neuronal differentiation, neuronal survival, molecular chaperone network, mitochondrial biogenesis and ROS homoeostasis (Figure 7), pathways known to be dysregulated in many neurodegenerative diseases, resulting in compromised mitochondrial integrity and aberrant mitochondrial homoeostasis (reviewed by Nicholls and Budd, 2000; Greene et al., 2004; Langley and Ratan, 2004; Herrup and Yang, 2007; Wyttenbach and Arrigo, 2007; Mattson et al., 2008; Nicholls, 2008; Gibson et al., 2010).
Figure 7. Working model of NeuroD6-mediated tolerance to oxidative stress.
This comprehensive working model integrates newly identified transcriptional modules with modules identified in our published studies (Uittenbogaard and Chiaramello, 2004, 2005; Baxter et al., 2009; Uittenbogaard et al., 2010), as indicated with asterisks. Whereas representative markers for each module are indicated in blue-framed boxes, the corresponding cellular processes are specified in blue elliptical objects.
Given the central importance of the transcriptional co-activator PGC-1α to control mitochondrial biogenesis and bioenergetic functions in various cellular differentiation paradigms, such as brown adipocyte and muscle cell differentiation (reviewed by Scarpulla, 2008; Hock and Kralli, 2009), and more recently in neuronal paradigms (Cowell et al., 2007; Meng et al., 2007; Wareski et al., 2009), we examined the expression of the transcriptional co-activator PGC-1α in serum-deprived PC12-ND6 cells throughout the entire period of serum deprivation. To our surprise, expression levels of the full-length PGC-1α protein rapidly collapsed after 4 h of serum deprivation, while the NT-PGC-1α isoforms remained expressed at constant levels throughout the whole serum deprivation treatment. NT-PGC-1α, a recently discovered isoform of PGC-1α, contains an in-frame stop codon as a result of alternative 3′ splicing, which gives rise to a truncated PGC-1α form carrying only the transactivation and nuclear receptor interaction domains (Zhang et al., 2009). More importantly, NT-PGC-1α is not only expressed at the highest level in rat brain, when compared with that of liver and brown adipose tissue, but also its spliced form is also favoured over the full-length form (Zhang et al., 2009). The notion that the full-length and truncated forms of PGC-1α have unique functions has been articulated based on the retention of a subset of shared functional domains (Zhang et al., 2009), which is further strengthened by their non-overlapping expression profiles in PC12-ND6 cells throughout serum deprivation. Future studies addressing NT-PGC-1α’s regulatory role in modulating mitochondrial mass within the context of neuronal differentiation and survival using our cellular paradigm are relevant based on the recent evidence of NT-PGC-1α enhancing mitochondrial biogenesis in brown adipocytes (Zhang et al., 2009).
In view of the integrative role of PGC-1α in co-ordinating a network of nuclear-encoded transcription factors responsible for mitochondrial gene expression, we examined the expression levels of the transcription factors NRF-1 and NRF-2 (GABP-α), whose transcriptional activities are known to be regulated upon protein–protein interactions with PGC-1α (reviewed by Scarpulla, 2008; Hock and Kralli, 2009). Interestingly, NRF-1 and NRF-2 (GABP-α) did not display a similar expression profile upon serum deprivation. While a tight correlation between expression levels of NT-PGC-1α and NRF-1 was detected during the whole duration of serum deprivation, it did not apply to the DNA-binding subunit GABP-α. Such expression profiles suggest that GABP-α is not necessary to maintain the mitochondrial biomass as well as the expression of genes encoding mitochondrial proteins upon stress in neuronal PC12-ND6 cells. This notion is further reinforced by a previous study using conditional gabp-α mutant mice demonstrating that GABP-α is not essential for mitochondrial biogenesis and corresponding gene expression in skeletal muscle cells (Jaworski et al., 2007). The fact that a direct correlation has been observed between expression of GABP-α and several respiratory enzymes upon neural activity but in the absence of a stress stimulus (Zhang and Wong-Riley, 2000; Yang et al., 2004; Wong-Riley et al., 2005) highlights the influence of the cellular context in favouring NRF-1- over NRF-2-mediated transcriptional pathways. This may be further explained by the notion that each transcription factor has broad developmental functions beyond mitochondrial biogenesis and respiratory functions, as NRF-1- and NRF-2-null mice display pre-implantation lethality (Huo and Scarpulla, 2001; Ristevski et al., 2004). Thus it appears that NRF-1 and NRF-2 have the ability to distinctly co-ordinate mitochondrial functions by tailoring nucleo-mitochondrial interactions to a specific cellular context, such as differentiation, stress stimulus and cell lineage.
Given the functional interplay between mitochondrial morphology and functions (reviewed by Karbowski and Youle, 2003; Chen and Chan, 2009; Lackner and Nunnari, 2009), we confirmed the absence of mitochondrial fragmentation throughout serum deprivation by live cell confocal fluorescence microscopy. This is in keeping with sustained levels of the serine/threonine kinase PINK1 protein in PC12-ND6 cells throughout the whole duration of serum deprivation. PINK1, which is located in both the cytosol and mitochondria via an N-terminal mitochondrial targeting sequence (Valente et al., 2004; Zhou et al., 2008), regulates mitochondrial morphology in several neuronal paradigms (reviewed by Thomas and Cookson, 2009). In addition, our observations of sustained expression of mitochondrial OXPHOS (oxidative phosphorylation) complex subunits, intracellular ATP levels, fluorescence intensity emitted by the MTR dye, whose accumulation and retention are MMP dependent, are congruent with the notion that NeuroD6 confers tolerance to oxidative stress by sustaining the content of energized mitochondria.
Finally, the present study has revealed a novel role for NeuroD6 in mediating a comprehensive antioxidant response in the absence of stress, thereby generating an antioxidant reserve to enhance tolerance to oxidative stress. The NeuroD6 effect is not limited to the classic induction of the ROS-scavenging enzymes, such as SOD1, SOD2, GPx1 and PRDX5, but also to recently identified powerful suppressors of ROS, such as PGC-1α, PINK1 and SIRT1. PINK1 possesses neuroprotective properties via its catalytic properties (Haque et al., 2008) and confers tolerance to oxidative stress by preventing mitochondrial fragmentation and subsequent increased ROS production in neuronal cells (Benard and Rossignol, 2008; Wood-Kaczmar et al., 2008; Dagda et al., 2009; Thomas and Cookson, 2009). Also key is the role of PGC-1α as a broad and robust regulator of ROS metabolism in addition to its well-established role as a regulator of mitochondrial biogenesis (St-Pierre et al., 2006; Wareski et al., 2009), which is consistent with the reported neurodegenerative lesions in the brains of PGC-1α-null mice (Lin et al., 2004; Leone et al., 2005). The co-expression of PGC-1α and two major ROS-detoxifying mitochondrial enzymes, SOD2 and GPx1, in PC12-ND6 cells prior to and during oxidative stress bears mentioning in the context that PGC-1α has been reported to induce their expression in neuronal cells (St-Pierre et al., 2003, 2006; Valle et al., 2005; Kukidome et al., 2006). Similarly important is the co-ordinate expression of SOD2 and the NAD-dependent class III protein deacetylase, SIRT1, in PC12-ND6 cells prior to and during serum deprivation, in keeping with its well-documented transcriptional control of the SOD2 gene (Nemoto and Finkel, 2002; Brunet et al., 2004; Daitoku et al., 2004). Moreover, expression of the endogenous SIRT1 protein in serum-grown PC12-ND6 cells may be a result of the constitutive expression of NeuroD6, as SIRT1 protein is known to be transiently expressed upon short-term serum starvation in the parental PC12 cell line (Nemoto et al., 2004). Of interest is the co-expression of PGC-1α and SIRT1 in serum-grown PC12-ND6 cells in light of the fact that SIRT1 deacetylates PGC-1α to stimulate its transcriptional activity (Nemoto et al., 2005; Wareski et al., 2009). Such a PGC-1α–SIRT1 functional interaction may contribute to the NeuroD6-mediated increase in mitochondrial mass in the absence of stress. Further studies need to be carried out to investigate such interactions and their functional impact in regulating mitochondria density within the context of NeuroD6-mediated transcriptional pathways and oxidative stress.
In summary, our collective results support the concept that the NeuroD6–PGC–1α–SIRT1 neuroprotective axis may be critical in achieving a balance between the mitochondrial biomass and antioxidant reserve to confer tolerance to oxidative stress. Thus NeuroD6 may constitute an instructive target towards the design of novel therapeutic strategies for the treatment of neurodegenerative diseases characterized by a mitochondrial deficit and oxidative damage.
ACKNOWLEDGEMENTS
We thank Dr Vittorio Gallo (Children's National Medical Center, Center for Neuroscience Research, Washington, DC, U.S.A.) for a critical review of the paper prior to acceptance. We also thank Dr A Popratiloff (Center for Microscopy and Image Analysis, George Washington University School of Medicine, Washington, DC, U.S.A.) for his help and expertise in confocal microscopy analysis, and Teresa Hawley for her help and expertise in flow cytometry. This work will be part of a dissertation presented to the graduate programme of Molecular Medicine, The George Washington University Institute for Biomedical Sciences, in partial fulfilment of the requirements for the award of Ph.D. degree.
Footnotes
This work was supported in part by the National Institutes of Health [grant number R01-NS041391; to A.C.]; by the National Institute of Child Health and Development [P30HD40677]; and by the National Center for Research Resources [S10RR025565].
REFERENCES
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Banmeyer I, Marchand C, Clippe A, Knoops B. Human mitochondrial peroxiredoxin 5 protects from mitochondrial DNA damages induced by hydrogen peroxide. FEBS Lett. 2005;579:2327–2333. doi: 10.1016/j.febslet.2005.03.027. [DOI] [PubMed] [Google Scholar]
- Banmeyer I, Marchand C, Verhaeghe C, Vucic B, Rees JF, Knoops B. Overexpression of human peroxiredoxin 5 in subcellular compartments of chinese hamster ovary cells: Effects on cytotoxicity and DNA damage caused by peroxides. Free Radical Biol Med. 2004;36:65–77. doi: 10.1016/j.freeradbiomed.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Bartholomä A, Nave KA. NEX-1: a novel brain-specific helix–loop–helix protein with autoregulation and sustained expression in mature cortical neurons. Mech Dev. 1994;48:217–228. doi: 10.1016/0925-4773(94)90061-2. [DOI] [PubMed] [Google Scholar]
- Batchelor AH, Piper DE, de la Brousse FC, McKnight SL, Wolberger C. The structure of GABPalpha/beta: an ETS domain–ankyrin repeat heterodimer bound to DNA. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- Baxter KK, Uittenbogaard M, Yoon J, Chiaramello A. The neurogenic basic helix–loop–helix transcription factor neuroD6 concomitantly increases mitochondrial mass and regulates cytoskeletal organization in the early stages of neuronal differentiation. ASN NEURO 1(4) 2009:art:e00016. doi: 10.1042/AN20090036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard G, Rossignol R. Ultrastructure of the mitochondrion and its bearing on function and bioenergetics. Antioxid Redox Signal. 2008;10:1313–1342. doi: 10.1089/ars.2007.2000. [DOI] [PubMed] [Google Scholar]
- Bernstein BW, Bamburg JR. Actin-ATP hydrolysis is a major energy drain for neurons. J. Neurosci. 2003;23:1–6. doi: 10.1523/JNEUROSCI.23-01-00002.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Buckman JF, Hernàndez H, Kress GJ, Votyakova TV, Pal S, Reynolds IJ. MitoTracker labeling in primary neuronal and astrocytic cultures: influence of mitochondrial membrane potential and oxidants. J Neurosci Res. 2001:165–176. doi: 10.1016/s0165-0270(00)00340-x. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- Chauhan A, Chauhan V. Oxidative stress in autism. Pathophysiology. 2006;13:171–181. doi: 10.1016/j.pathophys.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics-fusion, fission, movement, and mitophagy in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–R176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria: Central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homologue (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, Yoo SJ, Hay BA, Guo M. Drosophila pink 1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441:1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell RM, Blake KR, Russell JW. Localization of the transcriptional coactivator PGC-1alpha to GABAergic neurons during maturation of the rat brain. J Comp Neurol. 2007;502:1–18. doi: 10.1002/cne.21211. [DOI] [PubMed] [Google Scholar]
- Dagda RK, Cherra SJ, III, Kulich SM, Tandon A, Park D, Chu CT. Loss of PINK1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J Biol Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci USA. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Autréaux B, Toledano MB. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- de la Prida LM, Totterdell S, Gigg J, Miles R. The subiculum comes of age. Hippocampus. 2006;16:916–923. doi: 10.1002/hipo.20220. [DOI] [PubMed] [Google Scholar]
- De Simoni S, Goemaere J, Knoops B. Silencing of peroxiredoxin 3 and peroxiredoxin 5 reveals the role of mitochondrial peroxiredoxins in the protection of human neuroblastoma SH-SY5Y cells toward MPP+. Neurosci Lett. 2008;433:219–224. doi: 10.1016/j.neulet.2007.12.068. [DOI] [PubMed] [Google Scholar]
- Demin OV, Kholodenko BN, Skulachev VP. A model of O2•− generation in the complex III of the electron transport chain. Mol Cell Biochem. 1998;184:21–33. [PubMed] [Google Scholar]
- Deshmukh M, Kuida K, Johnson EM., Jr Caspase inhibition extends the commitment to neuronal death beyond cytochrome c release to the point of mitochondrial depolarization. J Cell Biol. 2000;150:131–143. doi: 10.1083/jcb.150.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA. MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging. 2004;25:1197–1203. doi: 10.1016/j.neurobiolaging.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Dhar SS, Ongwijitwat S, Wong-Riley MTT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubuisson M, Vander Stricht D, Clippe A, Etienne F, Nauser T, Kissner R, Koppenol WH, Rees JF, Knoops B. Human peroxiredoxin 5 is a peroxynitrite reductase. FEBS Lett. 2004;571:161–165. doi: 10.1016/j.febslet.2004.06.080. [DOI] [PubMed] [Google Scholar]
- Ekstrand MI, Falkenberg M, Rantanen A, Park CB, Gaspari M, Hultenby K, Rustin P, Gustafsson CM, Larsson NG. Mitochondrial transcription factor A regulates mtDNA copy number in mammals. Hum Mol Genet. 2004;13:935–944. doi: 10.1093/hmg/ddh109. [DOI] [PubMed] [Google Scholar]
- Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820–4825. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- François F, Godinho MJ, Dragunow M, Grimes ML. A population of PC12 cells that is initiating apoptosis can be rescued by nerve growth factor. Mol Cell Neurosci. 2001;18:347–362. doi: 10.1006/mcne.2001.1035. [DOI] [PubMed] [Google Scholar]
- Fridovich I. Mitochondria: are they the seat of senescence? Aging Cell. 2004;3:13–16. doi: 10.1046/j.1474-9728.2003.00075.x. [DOI] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- Gautier CA, Kitada T, Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc Natl Acad Sci USA. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson GE, Starkov A, Blass JP, Ratan RR, Beal MF. Cause and consequence: Mitochondrial dysfunction initiates and propagates neuronal dysfunction, neuronal death and behavioral abnormalities in age-associated neurodegenerative diseases. Biochim Biophys Acta. 2010;1802:122–134. doi: 10.1016/j.bbadis.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- Golden TR, Hubbard A, Morten KJ, Hinerfeld D, Melov S. Pharmacogenomic profiling of an oxidative stress-mediated spongiform encephalopathy. Free Radical Biol Med. 2005;39:152–163. doi: 10.1016/j.freeradbiomed.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Greene LA, Biswas SC, Liu DX. Cell cycle molecules and vertebrate neuron death: E2F at the hub. Cell Death Differ. 2004;11:49–60. doi: 10.1038/sj.cdd.4401341. [DOI] [PubMed] [Google Scholar]
- Han D, Williams E, Cadenas E. Mitochondrial respiratory chain-dependent generation of superoxide anion and its release into the intermembrane space. Biochem J. 2001;353:411–416. doi: 10.1042/0264-6021:3530411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque ME, Thomas KJ, D’Souza C, Callaghan S, Kitada T, Slack RS, Fraser P, Cookson MR, Tandon A, Park DS. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc Natl Acad Sci USA. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8:368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- Hock MB, Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71:177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M, Matsusaka H, Kang D, Matsushima S, Ide T, Kubota T, Fujiwara T, Hamasaki N, Takeshita A, Sunagawa K, Tsutsui H. Overexpression of mitochondrial transcription factor a ameliorates mitochondrial deficiencies and cardiac failure after myocardial infarction. Circulation. 2005;112:683–690. doi: 10.1161/CIRCULATIONAHA.104.524835. [DOI] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SJ, Cutler P, Melnyk S, Jernigan S, Janak L, Gaylor DW, Neubrander JA. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr. 2004;80:1611–1617. doi: 10.1093/ajcn/80.6.1611. [DOI] [PubMed] [Google Scholar]
- James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, Cutler P, Bock K, Boris M, Bradstreet JJ, Baker SM, Gaylor DW. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:947–956. doi: 10.1002/ajmg.b.30366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski A, Smith CL, Burden SJ. GA-binding protein is dispensable for neuromuscular synapse formation and synapse-specific gene expression. Mol Cell Biol. 2007;27:5040–5046. doi: 10.1128/MCB.02228-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson-Cadwell LI, Jekabsons MB, Wang A, Polster BM, Nicholls DG. ‘Mild uncoupling’ does not decrease mitochondrial superoxide levels in cultured cerebellar granule neurons but decreases spare respiratory capacity and increases toxicity to glutamate and oxidative stress. J Neurochem. 2007;101:1619–1631. doi: 10.1111/j.1471-4159.2007.04516.x. [DOI] [PubMed] [Google Scholar]
- Kanki T, Ohgaki K, Gaspari M, Gustafsson CM, Fukuoh A, Sasaki N, Hamasaki N, Kang D. Architectural role of mitochondrial transcription factor A in maintenance of human mitochondrial DNA. Mol Cell Biol. 2004;24:9823–9834. doi: 10.1128/MCB.24.22.9823-9834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karbowski M, Youle RJ. Dynamics of mitochondrial morphology in healthy cells and during apoptosis. Cell Death Differ. 2003;10:870–880. doi: 10.1038/sj.cdd.4401260. [DOI] [PubMed] [Google Scholar]
- Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- Khurana D, Salganicoff L, Melvin J, Hobdell E, Valencia I, Hardison H, Marks H, Grover W, Legido A. Epilepsy and respiratory chain defects in children with mitochondrial encephalopathies. Epilepsia. 2008;49:1972. doi: 10.1111/j.1528-1167.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- Kropotov A, Gogvadze V, Shupliakov O, Tomilin N, Serikov VB, Tomilin NV, Zhivotovsky B. Peroxiredoxin V is essential for protection against apoptosis in human lung carcinoma cells. Exp Cell Res. 2006;312:2806–2815. doi: 10.1016/j.yexcr.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Krzyzanski W, Oberdoester J, Rabin RA. Mechanism of ethanol enhancement of apoptosis and caspase activation in serum-deprived PC12 cells. Life Sci. 2007;81:756–764. doi: 10.1016/j.lfs.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Kukidome D, Nishikawa T, Sonoda K, Imoto K, Fujisawa K, Yano M, Motoshima H, Taguchi T, Matsumura T, Araki E. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes. 2006;55:120–127. [PubMed] [Google Scholar]
- Lackner LL, Nunnari JM. The molecular mechanism and cellular functions of mitochondrial division. Biochim Biophys Acta. 2009;1792:1138–1144. doi: 10.1016/j.bbadis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley B, Ratan RR. Oxidative stress-induced death in the nervous system: cell cycle dependent or independent? J Neurosci Res. 2004;77:621–629. doi: 10.1002/jnr.20210. [DOI] [PubMed] [Google Scholar]
- Lee CW, Peng BH. The function of mitochondria in presynaptic development at the neuromuscular junction. Mol Biol Cell. 2008;19:150–158. doi: 10.1091/mbc.E07-05-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone TC, Lehman JJ, Finck BN, Schaeffer PJ, Wende AR, Boudina S, Courtois M, Wozniak DF, Sambandam N, Bernal-Mizrachi C, Chen Z, Holloszy JO, Medeiros DM, Schmidt RE, Saffitz JE, Abel ED, Semenkovich CF, Kelly DP. PGC-1alpha deficiency causes multi-system energy metabolic derangements: muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyssens A, Nowicky AV, Patterson L, Crompton M, Duchen MR. The relationship between mitochondrial state, ATP hydrolysis, [Mg2+]i and [Ca2+]i studied in isolated rat cardiomyocytes. J Physiol. 1996;496:111–128. doi: 10.1113/jphysiol.1996.sp021669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, Sr-Pierre J, Zhang CY, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Malkus KA, Tsika E, Ischiropoulos H. Oxidative modifications, mitochondrial dysfunction, and impaired protein degradation in Parkinson’s disease: how neurons are lost in the bermuda triangle. Mol Neurodegener. 2009;4:24. doi: 10.1186/1750-1326-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60:748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K, Hixon M, Boekelheide K, Webb JR, Lansdorp PM, Lemieux M. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Doctrow SR, Schneider JA, Haberson J, Patel M, Coskun PE, Huffman K, Wallace DC, Malfroy B. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J Neurosci. 2001;21:8348–8353. doi: 10.1523/JNEUROSCI.21-21-08348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melov S, Coskun P, Patel M, Tuinstra R, Cottrell B, Jun AS, Zastawny TH, Dizdaroglu M, Goodman SI, Huang TT, Miziorko H, Epstein CJ, Wallace DC. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc Natl Acad Sci USA. 1999;96:846–851. doi: 10.1073/pnas.96.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Liang HL, Wong-Riley M. Quantitative immuno-electron microscopic analysis of depolarization-induced expression of PGC-1alpha in cultured rat visual cortical neurons. Brain Res. 2007;1175:10–16. doi: 10.1016/j.brainres.2007.07.063. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Fergusson MM, Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- Nicholls DG. Oxidative stress and energy crises in neuronal dysfunction. Ann NY Acad Sci. 2008;1147:53–60. doi: 10.1196/annals.1427.002. [DOI] [PubMed] [Google Scholar]
- Nicholls DG, Budd SL. Mitochondria and neuronal survival. Physiol Rev. 2000;80:315–360. doi: 10.1152/physrev.2000.80.1.315. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, Bae E, Kim J, Shong M, Kim JM, Chung J. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by Parkin. Nature. 2006;441:1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Neri C. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. 2005;37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- Patel M. Mitochondrial dysfunction and oxidative stress: Cause and consequence of epileptic seizures. Free Radical Biol Med. 2004;37:1951–1962. doi: 10.1016/j.freeradbiomed.2004.08.021. [DOI] [PubMed] [Google Scholar]
- Pendergrass W, Wolf N, Poot M. Efficacy of MitoTracker green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 2004;61:162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- Pons R, Andreu AL, Checcarelli N, Vila MR, Engelstad K, Sue CM, Shungu D, Haggerty R, de Vivo DC, DiMauro S. Mitochondrial DNA abnormalities and autistic spectrum disorders. J Pediatr. 2004;144:81–85. doi: 10.1016/j.jpeds.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, Zipp F, Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- Radyuk SN, Michalak K, Klichko VI, Benes J, Rebrin I, Sohal RS, Orr WC. Peroxiredoxin 5 confers protection against oxidative stress and apoptosis and also promotes longevity in Drosophila. Biochem J. 2009;419:437–445. doi: 10.1042/BJ20082003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol DA, Bradstreet JJ. Evidence of mitochondrial dysfunction in autism and implications for treatment. Am J Bioc Biotech. 2008;4:208–217. [Google Scholar]
- Ristevski S, O’Leary DA, Thornell AP, Owen MJ, Kola I, Hertzog PJ. The ETS transcription factor GABPalpha is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rukenstein A, Rydel RE, Greene LA. Multiple agents rescue PC12 cells from serum-free cell death by translation- and transcription-independent mechanisms. J Neurosci. 1991;11:2552–2563. doi: 10.1523/JNEUROSCI.11-08-02552.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto J, Miura T, Shimamoto K, Horio Y. Predominant expression of Sir2alpha, an NAD-dependent histone deacetylase, in the embryonic mouse heart and brain. FEBS Lett. 2004;556:281–286. doi: 10.1016/s0014-5793(03)01444-3. [DOI] [PubMed] [Google Scholar]
- Satoh T, Sakai N, Enokido Y, Uchiyama Y, Hatanaka H. Survival factor-insensitive generation of reactive oxygene species induced by serum deprivation in neuronal cells. Brain Res. 1996;733:9–14. doi: 10.1016/0006-8993(96)00527-6. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Schwab MH, Druffel-Augustin S, Gass P, Jung M, Klugmann M, Bartholomä A, Rossner MJ, Nave KA. Neuronal basic helix–loop–helix proteins (NEX, neuroD, NDRF): spatiotemporal expression and targeted disruption of the NEX gene in transgenic mice. J Neurosci. 1998;18:1408–1418. doi: 10.1523/JNEUROSCI.18-04-01408.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab MH, Bartholomä A, Heimrich B, Feldmeyer D, Druffel-Augustin S, Goebbels S, Naya FJ, Zhao S, Frotscher M, Tsai MJ, Nave KA. Neuronal basic helix–loop–helix proteins (NEX and BETA2/Neuro D) regulate terminal granule cell differentiation in the hippocampus. J Neurosci. 2000;20:3714–3724. doi: 10.1523/JNEUROSCI.20-10-03714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu C, Akazawa C, Nakanishi S, Kageyama R. MATH-2, a mammalian helix–loop–helix factor structurally related to the product of Drosophila proneural gene atonal, is specifically expressed in the nervous system. Eur J Biochem. 1995;229:239–248. doi: 10.1111/j.1432-1033.1995.tb20461.x. [DOI] [PubMed] [Google Scholar]
- Sinclair D. Sirtuins for healthy neurons. Nat Genet. 2005;37:339–340. doi: 10.1038/ng0405-339. [DOI] [PubMed] [Google Scholar]
- Stefanis L, Park DS, Yun C, Yan I, Farinelli SE, Troy CM, Shelanski ML, Greene LA. Induction of CPP32-like activity in PC12 cells by withdrawal of trophic support. J Neurosci. 1996;271:30663–30671. doi: 10.1074/jbc.271.48.30663. [DOI] [PubMed] [Google Scholar]
- Stefanis L, Troy CM, Qi H, Shelanski ML, Greene LA. Caspase-2 (Nedd-2) processing and death of trophic factor-deprived PC12 cells and sympathetic neurons occur independently of caspase-3 (cpp32)-like activity. J Neurosci. 1998;18:9204–9215. doi: 10.1523/JNEUROSCI.18-22-09204.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Lin J, Krauss S, Tarr PT, Yang R, Newgard CB, Spiegelman BM. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Sugioka K, Nakano M, Naito I, Tero-Kubota S, Ikegami Y. Properties of a coenzyme, pyrroloquinoline quinone: Generation of an active oxygen species during a reduction-oxidation cycle in the presence of NAD(P)H and O2. Biochim Biophys Acta. 1988;964:175–182. doi: 10.1016/0304-4165(88)90164-x. [DOI] [PubMed] [Google Scholar]
- Thomas KJ, Cookson MR. The role of PTEN-induced kinase 1 in mitochondrial dysfunction and dynamics. Int J Biochem Cell Biol. 2009;41:2025–2035. doi: 10.1016/j.biocel.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Shelanski ML. Down-regulation of copper/zinc superoxide dismutase causes apoptotic death in PC12 neuronal cells. Proc Natl Acad Sci USA. 1994;91:6384–6387. doi: 10.1073/pnas.91.14.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Derossi D, Prochiantz A, Greene LA, Shelanski ML. Downregulation of Cu/Zn superoxide dismutase leads to cell death via the nitric oxide–peroxynitrite pathway. J Neurosci. 1996;16:253–261. doi: 10.1523/JNEUROSCI.16-01-00253.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troy CM, Stefanis L, Greene LA, Shelanski ML. Nedd2 is required for apoptosis after trophies factor withdrawal, but not superoxide dismutase (SOD1 downregulation, in sympathetic neurons and PC12 cells. J Neurosci. 1997;17:1911–1918. doi: 10.1523/JNEUROSCI.17-06-01911.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpower BL. The protonmotive Q cycle. Energy transduction by coupling of proton translocation to electron transfer by the cytochrome bc1 complex. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- Turrens JF. Superoxide production by the mitochondrial respiratory chain. Biosci Rep. 1997;17:3–8. doi: 10.1023/a:1027374931887. [DOI] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. The basic helix–loop–helix transcription factor nex-1/Math-2 promotes neuronal survival of PC12 cells by modulating the dynamic expression of anti-apoptotic and cell cycle regulators. J Neurochem. 2005;92:585–596. doi: 10.1111/j.1471-4159.2004.02886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. Expression profiling upon Nex1/MATH-2-mediated neuritogenesis in PC12 cells and its implication in regeneration. J. Neurochem. 2004;91:1332–1343. doi: 10.1111/j.1471-4159.2004.02814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Chiaramello A. Constitutive overexpression of the basic helix–loop–helix Nex1/MATH-2 transcription factor promotes neuronal differentiation of PC12 cells and neurite regeneration. J Neurosci Res. 2002;67:235–245. doi: 10.1002/jnr.10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Baxter KK, Chiaramello A. NeuroD6 genomic signature bridging neuronal differentiation to survival via the molecular chaperone network. J Neurosci Res. 2010;88:33–54. doi: 10.1002/jnr.22182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uittenbogaard M, Baxter KK, Chiaramello A. Cloning and characterization of the 5′UTR of the rat anti-apoptotic bcl-w gene. Biochem Biophys Res Commun. 2009;389:657–662. doi: 10.1016/j.bbrc.2009.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Dei Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auberger G, Wood NW. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- Valle I, Alvarez-Barrientos A, Arza E, Lamas S, Monsalve M. PGC-1alpha regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- Vyas S, Juin P, Hancock D, Suzuki Y, Takahashi R, Triller A, Evan G. Differentiation-dependent sensitivity to apoptogenic factors in PC12 cells. J. Biol. Chem. 2004;279:30983–30993. doi: 10.1074/jbc.M400692200. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Wareski P, Vaarmann A, Choubey V, Safiulina D, Liiv J, Kuum M, Kaasik A. PGC-1{alpha} and PGC-1{beta} regulate mitochondrial density in neurons. J Biol Chem. 2009;284:21379–21385. doi: 10.1074/jbc.M109.018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CM, Siu KL, Jin DY. Peroxiredoxin-null yeast cells are hypersensitive to oxidative stress and are genomically unstable. J Biol Chem. 2004;279:23207–23213. doi: 10.1074/jbc.M402095200. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT, Yang SJ, Liang HL, Ning G, Jacobs P. Quantitative immuno-electron microscopic analysis of nuclear respiratory factor 2 alpha and beta subunits: normal distribution and activity-dependent regulation in mammalian visual cortex. Vis Neurosci. 2005;22:1–18. doi: 10.1017/S0952523805221016. [DOI] [PubMed] [Google Scholar]
- Wood-Kaczmar A, Gandhi S, Yao Z, Abramov AY, Miljan EA, Keen G, Stanyer L, Hargreaves I, Klupsch K, Deas E, Downward J, Mansfield L, Jat P, Taylor J, Heales S, Duchen MR, Latchman D, Tabrizi SJ, Wood NW. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS One. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Ying Z, Gomez-Pinilla F. Oxidative stress modulates Sir2alpha in rat hippocampus and cerebral cortex. Eur J Neurosci. 2006;23:2573–2580. doi: 10.1111/j.1460-9568.2006.04807.x. [DOI] [PubMed] [Google Scholar]
- Wu SX, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave KA, Tamamaki N. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci USA. 2005;102:17172–17177. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach A, Arrigo AP. The role of heat shock proteins during neurodegeneration in Alzheimer’s, Parkinson’s, and Huntington’s diseases. In: Richter-Landsberg C, editor. In Heat Shock Proteins in Neural Cells. Springer Landes Bioscience, New York: Neuroscience Intelligent Unit; 2007. pp. 81–99. [Google Scholar]
- Yang SJ, Liang HL, Ning G, Wong-Riley MT. Ultrastructural study of depolarization-induced translocation of NRF-2 transcription factor in cultured rat visual cortical neurons. Eur J Neurosci. 2004;19:1153–1162. doi: 10.1111/j.1460-9568.2004.03250.x. [DOI] [PubMed] [Google Scholar]
- Yasuda M, Mayford MR. CaMKII activation in the entorhinal cortex disrupts previously encoded spatial memory. Neuron. 2006;50:309–318. doi: 10.1016/j.neuron.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wong-Riley MT. Depolarizing stimulation upregulates GA-binding protein in neurons: a transcription factor involved in the bigenomic expression of cytochrome oxidase subunits. Eur J Neurosci. 2000;12:1013–1023. doi: 10.1046/j.1460-9568.2000.00997.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Huypens P, Adamson AW, Chang JS, Henagan TM, Boudreau A, Lenard NR, Burk D, Klein J, Perwitz N, Shin J, Fasshauer M, Kralli A, Gettys TW. Alternative mRNA splicing produces a novel biologically active short isoform of PGC-1alpha. J Biol Chem. 2009;284:32813–32826. doi: 10.1074/jbc.M109.037556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Shao Y, May J, Prou D, Perier C, Dauer W, Schon EA, Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Kok KH, Chun AC, Wong CM, Wu HW, Lin MC, Fung PC, Kung H, Jin DY. Mouse peroxiredoxin V is a thioredoxin peroxidase that inhibits p53-induced apoptosis. Biochem Biophys Res Commun. 2000;268:921–927. doi: 10.1006/bbrc.2000.2231. [DOI] [PubMed] [Google Scholar]