Abstract
Carbohydrate Response Element Binding Protein (ChREBP) is a Mondo family transcription factor that activates a number of glycolytic and lipogenic genes in response to glucose stimulation. We have previously reported that high glucose can activate the transcriptional activity of ChREBP independent of the protein phosphatase 2A (PP2A)-mediated increase in nuclear entry and DNA binding. Here we found that formation of glucose-6-phosphate (G-6-P) is essential for glucose activation of ChREBP. The glucose response of GAL4-ChREBP is attenuated by D-mannoheptulose, a potent hexokinase inhibitor, as well as over-expression of glucose-6-phosphatase (G6Pase); kinetics of activation of GAL4-ChREBP can be modified by exogenously expressed GCK. Further metabolism of G-6-P through the two major glucose metabolic pathways, glycolysis and pentose phosphate pathway, is not required for activation of ChREBP; over-expression of glucose-6-phosphate dehydrogenase (G6PD) diminishes, whereas RNAi knockdown of the enzyme enhances, the glucose response of GAL4-ChREBP, respectively. Moreover, the glucose analogue 2-deoxyglucose (2-DG), which is phosphorylated by hexokinase, but not further metabolized, effectively upregulates the transcription activity of ChREBP. In addition, over-expression of phosphofructokinase (PFK) 1 and 2, synergistically diminishes the glucose response of GAL4-ChREBP. These multiple lines of evidence support the conclusion that G-6-P mediates the activation of ChREBP.
Keywords: carbohydrate response element binding protein (ChREBP), Glucose-6-phosphate (G-6-P), Transcriptional activation
Introduction
It is well known that glucose stimulates the expression of a number of lipogenic and glycolytic genes independently of its secondary humoral effects such as insulin secretion [1–3]. Many glucose-regulated genes share a conserved consensus sequence, called a carbohydrate response element (ChoRE), which is required for their glucose-responsive expression [4, 5]. ChREBP, a member of Mondo family of bHLH/ZIP transcription factors, has been shown to bind to ChoRE and mediate the glucose-responsive transcription of these genes [6]. ChREBP has been implicated in a number of metabolic disease processes, such as diabetes, obesity, hepatosteatosis and hyperlipidemia [7–10].
To date two mechanisms have been described to underlie different aspects of glucose responsiveness of ChREBP, one dependent on protein phosphatase 2A (PP2A), and one independent. Xylulose-5-phosphate, a metabolite in the pentose-phosphate pathway, was reported to be responsible for activation of PP2A-dependent DNA binding and nuclear entry of ChREBP in hepatocytes [11–13]. This pathway is particularly important in lipogenic organs such as liver, and appears to be much less active in other glucose-sensitive tissues such as pancreatic islets [14] and an insulinoma cell line 832/13 [11, 15].
The high glucose-induced ChREBP activation is mediated by the release of an intramolecular inhibition that occurs under low glucose [11, 15, 16]. Unlike the PP2A-dependent pathway, this PPA2-independent pathway is active in both 832/13 cells and hepatocytes [11, 16]. The domain that mediates this function, called glucose-sensing module (GSM), is highly conserved across species in ChREBP as well as its paralogue MondoA [15]. Importantly, the two residues crucial for PP2A-dependent activation, i.e., S196 and T666, are not conserved in ChREBP or MondoA from nonmammalian species such as Drosophila and C. elegans [15], in which glucose responsiveness also occurs, suggesting that PP2A-independent mechanisms may play a more fundamental role in activation of the Mondo family of transcription factors across species and tissue types.
While the detailed mechanism whereby the intramolecular inhibition and its release are effected is being examined by structure-function analysis of ChREBP [11, 17], the pathway responsible for PP2A-independent transactivation of ChREBP remains to be elucidated. It stands to reason that an intermediate metabolite of glucose metabolism may mediate the activation of ChREBP. A plausible candidate molecule is glucose 6-phosphate (G-6-P), which serves as the nodal point for major glucose metabolic pathways, such as the glycolytic and pentose phosphate pathways. Indeed, in adipocytes, the level of G-6-P correlates strongly with the expression level of several lipogenic genes [18, 19]. In addition, 2-deoxyglucose (2-DG), a glucose analogue that is phosphorylated by hexokinase but cannot be further metabolized downstream of hexokinase, is capable of stimulating the expression of ChoRE-containing genes in fat and insulinoma cells [19, 20].
In this study, we developed multiple lines of evidence using pharmacologic and genetic manipulations to determine the glucose metabolite that mediates the release of self-inhibition of ChREBP in response to glucose. We found that glucose (or its analog 2-deoxyglucose) has to be phosphorylated by hexokinase to G-6-P for activation of ChREBP, but neither the subsequent flux through glycolysis nor the pentose phosphate pathway is required for the process. We conclude that G-6-P mediates the high glucose-induced activation of ChREBP.
Materials and Methods
Chemicals
D-mannoheptulose was from Glycoteam GmbH (Hamburg, Germany). Cloning enzymes and lipofectamine 2000 were from Invitrogen (Carlsbad, CA). All other chemicals were from Sigma-Aldrich (St. Louis, MO).
Plasmid Construction
Construction of pGAMPAC-ChREBP and pGAMPAC-CA-ChREBP (Δ1-196) has been reported earlier [15]. Mouse GCK (genbank ID: 15029831), mouse PFK1 (genebank ID: BC020097) cDNA and rat G6PD cDNA (genbank ID: NM_017006) were cloned into mammalian expression vector pCMX [21] and/or retroviral vector pSRQT [17]. The previously described plasmids expressing canine G6Pase (genbank ID: U91844) and pcDNA3.1-cG6Pase were generously provided by Dr. D. Koeberl (Duke University, Durham, NC) [22]. The expression plasmid for inducible PFK2 (pIRESneo-iPFK2) was a generous gift of Dr. J. Chesney (University of Louisville, KY). The cDNA for tetracycline controlled transactivator (tTA) was cloned into PMSCVhygro (Clontech). The shRNA for G6PD or its scrambled version were cloned into the lentiviral vector pLL3.7 [23]. UAS-driven luciferase reporter pG5-luc and the internal control TK promoter driven Renilla luciferase expression plasmid pRL-TK were from Promega (Madison, WI). UAS-driven rapid response luciferase reporter pG5-R2.2luc was reported earlier [15]. All cloning primers, shRNA sequences and real-time PCR primers are listed in Supplementary Table 1.
Cell Culture, Transfection and Luciferase reporter assay
832/13 cells (gift of Dr. C. Newgard, Duke University, Durham, NC) were cultured as described [24]. BOSC23 and 293T cells were cultured in DMEM media containing 10% FBS. Cells were transfected with Lipofectamine 2000 (Invitrogen) with indicated plasmids. Dual-luciferase kit (Promega) was used for the luciferase assay according to the manufacturer's instructions. Results were shown as arbitrary units normalized to the TK luciferase activity.
Retrovirus, Lentivirus Production and Infection
Retroviruses expressing pSRQT-GCK and PMSCVhygro-tTA were generated using enveloping plasmid pCL-eco (Imgenex, Sorrento Valley, CA) and packaging cells BOSC23 (American Type Culture Collection, Manassas, VA), as described earlier [17]. 832/13 cells were sequentially infected with these two viruses, and selected with both 1 μg/ml puromycin and 200 μg/ml hygromycin to realize strong expression of GCK in these cells. Lentiviral constructs expressing shRNAs were co-transfected with the packaging plasmid psPAX and the envelope plasmid pMD2.G into HEK293T cells. The media were collected and used to infect 832/13 cells with polybrene added at 8 μg/ml. The procedure was repeated once to ensure 100% infection as examined with fluorescent microscopy as described [23].
RNA Extraction and Real-time PCR
We extracted RNAs using RNeasy miniprep kit (Qiagen, Valencia, CA), synthesized cDNA using Ominscript RT (Qiagen), and performed real-time quantitative RT-PCR on the Strategene MX3000 real time detection system using iQ™ SYBR Green PCR reagent kit (Biorad).
Glucose-6-phosphate Measurement
Cultured cells were washed twice with ice-cold PBS and scraped with 30% ice-cold perchloric acid. The cell extracts were neutralized with KIK (6 N KOH, 0.4 M imidazole, and 0.4 M KCl) buffer. We measured glucose-6-phosphate by the coupled reaction of glucose-6-phosphate dehydrogenase to NADPH production [25]. Increase in absorbance was monitored at 340 nm. Sample quantification was based on a standard curve realized with known concentrations of G-6-P.
Statistic analysis
Results were shown as representative of data from ≥3 independent experiments. Statistical analyses of the data were performed using a two-tailed unpaired t test with unequal variance. All data are presented as the means ± SD from ≥3 replicates with statistical significance set at a P value of <0.05.
Results
In searching for the signaling metabolite and analyzing the effect of glucose metabolism on PP2A-independent activation of ChREBP transcriptional activity, we used the reporter system, GAL4-ChREBP fusion protein and UAS-driven luciferase reporter in 832/13 insulinoma cells, as the PP2A-dependent pathway has only minimal effect on nuclear entry and no effect at all on the transcriptional activity of GAL4-ChREBP in this system [15].
Formation of G-6-P is required for glucose activation of ChREBP
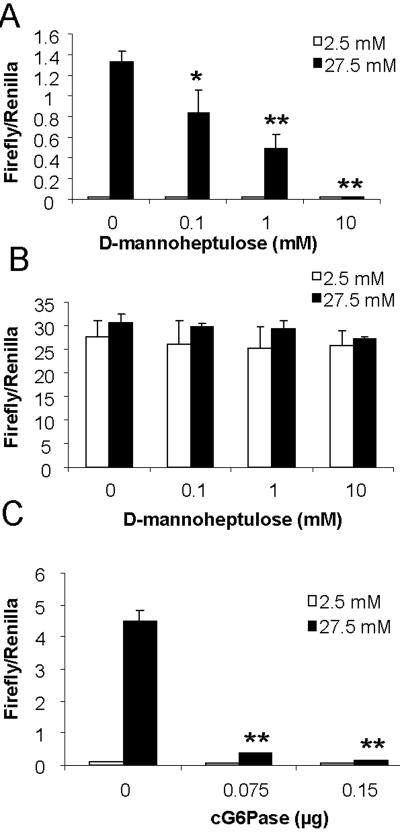
After entry of glucose into the cell, phosphorylation by hexokinase is the first step of glucose catabolism to trap glucose inside the cell and commit it to the downstream metabolic pathways [26]. To test whether G-6-P itself or one of its derivatives activates ChREBP, we assessed the effect of a specific inhibitor of hexokinase, D-mannoheptulose, on the glucose responsiveness of ChREBP using luciferase reporter assay in 832/13 cells. We found that D-mannoheptulose dose-dependently shut down the glucose response of GAL4-ChREBP (Fig. 1A), suggesting that formation of G-6-P is essential for the glucose response of ChREBP. In contrast, D-mannoheptulose had no effects on the transactivtion activity of the constitutively active mutant (Δ1-196) ChREBP (Fig. 1B), indicating that D-mannoheptulose affects the sensing of glucose by ChREBP without directly inhibiting its transactivation activity.
Fig. 1. Metabolism of glucose is required for activation of GAL4-ChREBP.
Luciferase assay results under following conditions: (A) pGAMPAC-ChREBP, pG5-luc and pRL-TK, (B) pGAMPAC-CA-ChREBP, pG5-luc and pRL-TK, (C) pGAMPAC-ChREBP, pG5-luc, pRL-TK and indicated amount of pcDNA3.1-cG6Pase or pcDNA3.1 were cotransfected into 832/13 cells. Transfected cells were incubated in media with low glucose concentration (2.5 mM) overnight, and then treated with 2.5 (open bar) or 27.5 (solid bar) mM glucose, and for (A) and (B), also indicated concentrations of D-mannoheptulose, for 6 hrs. * p< 0.05; ** p<0.005 vs vehicle treated at 27.5 mM glucose (A) or empty vector transfected at 27.5 mM glucose (C).
We also measured the glucose responsiveness of GAL4-ChREBP in the presence of G6Pase, which converts G-6-P into glucose, thus reducing the intracellular concentration of G-6-P. As shown in Fig. 1C, over-expression of G6Pase potently and dose-dependently suppressed the glucose activation of GAL4-ChREBP, further corroborating that formation of G-6-P via hexokinase is essential for glucose response of ChREBP in 832/13 cells.
GCK acts as the glucose sensor for activation of ChREBP in 832/13 cells
In glucose-responsive tissues such as liver and pancreatic islet β-cells, one particular hexokinase, i.e., GCK, is highly expressed to convert glucose into G-6-P. The distinguishing feature of GCK is its low affinity for glucose, which allows it to respond to physiological changes in glucose concentration in the portal vein [27]. Interestingly, ChREBP was also found to be most sensitive to a similar range of glucose variation (5–20 mM) in the insulinoma cell line 832/13 [15]. The similar kinetic behavior of ChREBP and GCK suggests that hexokinase, and in the case of liver and islet β-cells, GCK in particular, may serve as the glucose sensor for ChREBP.
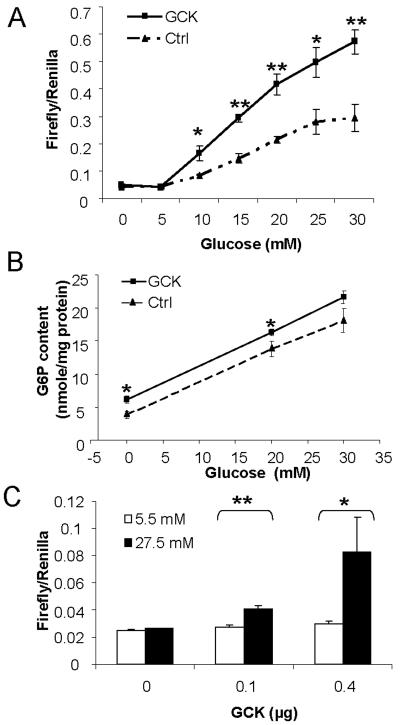
To test this hypothesis, we assessed the effects of GCK over-expression in 832/13 on the kinetics of the glucose responsiveness of GAL4-ChREBP. As was done in our earlier publication on the kinetic behavior of ChREBP [15], the rapid response luciferase reporter was used in this experiment to better indicate the real-time transcriptional activity of GAL4-ChREBP in response to changes in glucose concentration. Indeed, in the GCK-overexpressing cells, the glucose dose-response curve of the transcriptional activity of GAL4-ChREBP was significantly shifted upwards and leftwards (Fig. 2A) when compared with control cells. We also measured the G-6-P content in response to different glucose concentrations with or without GCK overexpression. As depicted in Fig. 2B, the G-6-P level was increased almost linearly between 0 and 30 mM glucose in both control and GCK overexpressing 832/13 cells. Overexpression of GCK resulted in a higher amount of cellular G-6-P compared with control (Fig 2B), which correlates with its higher transcriptional activity (Fig. 2A).
Fig. 2. GCK acts as glucose sensor for ChREBP activation.
(A) Over-expression of GCK in 832/13 changed kinetic behavior of GAL4-ChREBP. 832/13 cells were infected with retrovirus which over-expresses GCK as described in Materials and Methods. Infected cells or uninfected control cells were transfected with pGAMPAC-ChREBP, pG5-R2.2luc and pRL-TK, and treated with low glucose over night, and then treated with indicated concentrations of glucose for 3 hrs before luciferase assay. (B) 832/13 cells overexpressing GCK and the control cells were cultured under low glucose (2.5 mM) over night and then treated with 0, 20 and 30 mM glucose for 1 hr followed by G-6-P measurement. (C) pGAMPAC-ChREBP, pG5-luc, pRL-TK and indicated amount of pCMX-GCK were transfected into HEK293T cells. The total amount of transfected DNA was held the same by adding appropriate amount of vector DNA. The transfected cells were then treated with low (5.5 mM) or high (27.5 mM) glucose concentration for 24 hrs before luciferase assay. * p < 0.05; ** p<0.005 vs vector transfected control cells at the same glucose concentration or as indicated (C).
We next overexpressed GCK in a normally non-glucose-responsive cell line, HEK293T, and tested if this maneuver would render GAL4-ChREBP responsive to high glucose. Indeed, compared to vector control, the transfection-induced GCK expression dose-dependently rendered GAL4-ChREBP responsive to high glucose concentration in this normally glucose unresponsive cell line (Fig. 2C).
Overall, the above data suggest that the response of ChREBP to glucose is determined by glucose availability and the hexokinase expressed, especially in hepatocytes and islet β-cells, where the kinetic attributes of GCK makes it an ideal glucose sensor for ChREBP activation.
Pentose-phosphate pathway is not involved in PP2A-independent activation of ChREBP
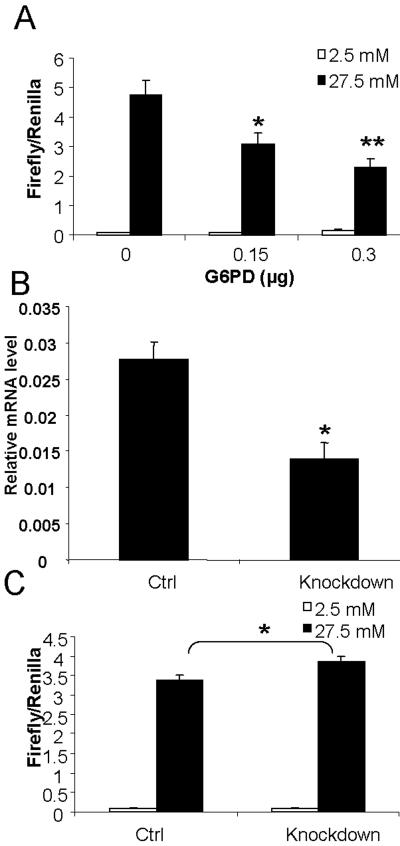
The pentose phosphate pathway is active mainly in lipogenic tissues such as liver and fat [26], but contributes to not more than 3–4% of glucose oxidation in islet β-cells [14], making it much less likely to play a significant role in insulinoma 832/13 cells. To further examine this notion, we tested if perturbation of this metabolic pathway will affect PP2A-independent activation of ChREBP in 832/13 cells. We used genetic approaches to increase or decrease the expression of Glucose 6 phosphate dehydrogenase (G6PD), a rate-limiting enzyme of the pentose phosphate pathway, to manipulate the flux in this pathway in 832/13 cells. As shown in Fig. 3A, not only did over-expression of G6PD fail to increase, instead, it actually dose-dependently decreased the glucose response of GAL4-ChREBP, suggesting that at a minimum, metabolites generated in the pentose phosphate pathway do not mediate transactivation of ChREBP; furthermore, G6PD may shunt G-6-P away from other metabolic pathways. In contrast, when we knocked down 50% of the expression of endogenous G6PD mRNA with shRNA in 832/13 cells (Fig. 3B); the glucose transactivation of ChREBP was actually increased, although only to a modest degree (Fig. 3C). This limited effect can be explained by an already very low endogenous expression of G6PD in 832/13 cells (our observation). Taken together, the above data suggest that the pentose phosphate pathway is not directly involved in the generation of the metabolite responsible for activation of ChREBP in 832/13 cells.
Fig. 3. Pentose phosphate pathway is not involved in PP2A-independent activation of ChREBP.
(A) pGAMPAC-ChREBP, pG5-luc, pRL-TK and indicated amount of pCMX-G6PD were infected into 832/13 cells. The total amount of transfected DNA was held the same by adding appropriate amount of vector DNA. Transfected cells were subsequently cultured under 2.5 mM low glucose over night, and then treated with either low (2.5 mM) or high (27.5 mM) glucose for 6 hrs before luciferase assay. * p< 0.05; ** p< 0.005 vs vector only transfected at 27.5 mM glucose. (B) Real-time PCR measurement of mRNA content of G6PD in 832/13 cells infected with lentivirus expressing shRNA against G6PD (knockdown) or its scrambled counterpart (ctrl). Data are normalized to mRNA level of the Eukaryotic Elongation factor 1 gamma (EEF1g). * < 0.05 vs ctrl. (C) Luciferase assay: pGAMPAC-ChREBP, pG5-luc and pRL-TK were transfected into 832/13 cells infected with lentivirus expressing either shRNA against G6PD (knockdown) or scrambled shRNA (ctrl). The transfected cells were then cultured in low glucose overnight, and treated with either 2.5 mM or 27.5 mM glucose for 6 hrs before luciferase assay. * < 0.05 vs ctrl at 27.5 mM glucose.
The glycolytic pathway downstream of G-6-P is not involved in PP2A-independent activation of ChREBP
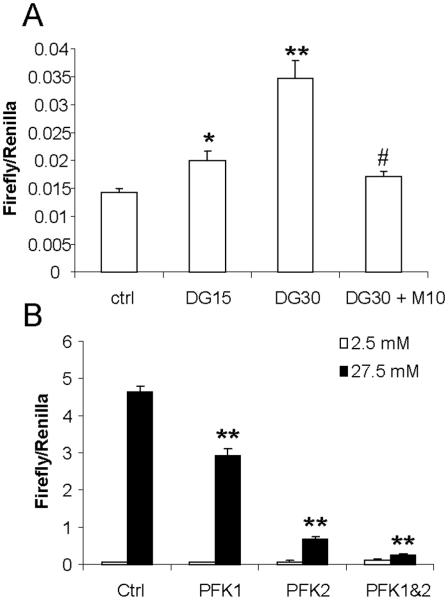
The glycolytic pathway controls the bulk of glucose flux. It feeds directly into other metabolic pathways, such as the Krebs cycle, the non-oxidative branch of the pentose phosphate pathway, and the hexosamine biosynthetic pathway. Therefore, it is possible that this pathway may generate the signaling compound for activation of ChREBP. To test this hypothesis, we took advantage of the glucose analogue, 2-deoxyglucose (2-DG), which can be phosphorylated by hexokinase, but not further metabolized. We found that 2-DG directly stimulates the activity of GAL4-ChREBP although with a lower efficiency (Fig. 4A), indicating that further metabolism of G-6-P is not required for high glucose-induced transactivation of ChREBP. Interestingly, the effect of 2-DG on transactivation of ChREBP is abrogated by cotreatment with D-mannoheptulose, the hexokinase inhibitor (Fig. 4A); again indicating that phosphorylation of glucose (or its nonmetabolizable analogue, 2-DG) is an essential step in transactivation of ChREBP.
Fig. 4. Glycolytic pathway is not involved in PP2A-independent activation of ChREBP.
(A) pGAMPAC-ChREBP, pG5-luc, and pRL-TK were transfected into 832/13 cells, which were subsequently cultured under low glucose overnight, and then treated with indicated concentrations of 2-DG and/or D-mannoheptulose for 6 hrs before luciferase assay. DG15, 15 mM 2-DG; DG30, 30 mM 2-DG; M10, 10 mM D-mannoheptulose:* p < 0.05; ** p < 0.005 vs ctrl; # p< 0.05 vs DG30. (B) pGAMPAC-ChREBP, pG5-luc, and pRL-TK were cotransfected with control vector, pCMX-PFK1 and/or pIRESpuro-iPFK2 (0.15 μg each) into 832/13 cells. The total amount of transfected DNA was held the same by adding appropriate amount of vector DNA. Transfected cells were subsequently cultured under low glucose overnight, and then treated with either 2.5 mM or 27.5 mM glucose for 6 hrs before luciferase assay. ** p < 0.005 vs ctrl at high glucose.
Using a complementary approach, we modulated the flux in the glycolytic pathway by modulating the next rate-limiting enzyme in this pathway, phosphofructokinase 1 (PFK1). We (i) overexpressed PFK1 directly, or (ii) stimulated the synthesis of fructose-2, 6-bisphosphate, a potent allosteric agonist of PFK1, by overexpressing phosphofructokinase 2 (PFK2), or (iii) overexpressed both together simultaneously. We found that overexpression of PFK1 and PFK2 individually reduced the glucose response of GAL4-ChREBP, and, when overexpressed together, they synergistically shut down, almost completely, the high glucose-induced transactivation activity of GAL4-ChREBP (Fig. 4B).
Taken together, these data indicate that glucose metabolizing pathways downstream of G-6-P are not involved in high glucose-stimulated activation of ChREBP.
Discussion
Previous work has addressed one aspect of activation of ChREBP, i.e., xylulose-5-phosphate mediated nuclear entry and DNA binding [12, 13]. However, the signaling pathway that leads to the other aspect of ChREBP activation, i.e., turning on its transcriptional activity, remains unknown. We have previously shown that high glucose-induced activation exists in cells and tissues where the pentose phosphate pathway is almost nonexisent [10], implicating the involvement of signaling molecules other than xylulose-5-phosphate. In this study, we focused our attention on the transcription activation of ChREBP using the GAL4-ChREBP system. The system is extremely sensitive, and is particularly suited for this purpose, because it excludes the effects of glucose on nuclear entry and DNA binding activity of ChREBP and focuses on the transcriptional activation itself [15]. We found that inhibition of hexokinase by D-mannoheptulose completely shuts down transactivation of ChREBP (Fig. 1A), suggesting that this second pathway is also dependent on the metabolism of glucose. Overexpression of G6Pase, which reverses the reaction of hexokinase by dephosphorylating G-6-P, also dose-dependently abolishes glucose responsiveness of GAL4-ChREBP (Fig. 1C), implicating the formation of G-6-P by hexokinase is an essential step for ChREBP activation. We further showed that the hexokinase expressed in the cell partly determined the glucose response of ChREBP, as predicted from the kinetic behavior of ChREBP activation when GCK is overexpressed, consistent with hexokinase acting as the glucose sensor for ChREBP in our experimental system.
The observation that over-expression of G6PD leads to down-regulation of glucose response in GAL4-ChREBP activity, and knockdown of G6PD, a modest increase (Fig. 3), ruled out the involvement of pentose phosphate pathway in PP2A-independent activation of ChREBP in 832/13 cells. Additionally, the pentose phosphate pathway was known to be inactive in pancreatic islet β-cells [14]. This might have contributed to the susceptibility of these cells to oxidative stress for lack of supply of reducing equivalent, i.e., NADPH. In fact, the survival of INS-1 cells, from which 832/13 cells were derived, depends on the reducing agent 2-mercaptoethanol in the media [28]. Therefore, it is unlikely that the pentose phosphate pathway plays a significant role in activation of ChREBP in 832/13 cells and pancreatic islet β-cells.
Since glycolysis is the major pathway of glucose metabolism, it is important to determine if metabolites downstream of G-6-P are involved in transactivation of ChREBP in 832/13 cells. However, in our study, a glucose analogue, 2-DG, which can be directly phosphorylated by hexokinase but not further metabolized through the glycolytic pathway, was found to dose-dependently activate the transcriptional activity of GAL4-ChREBP, an effect that can be negated by treatment with D-mannoheptulose (Fig. 4A). This finding, which is consistent with a previous report [20], strongly suggests that phosphorylation by hexokinase is sufficient for activation of ChREBP. Meanwhile, artificially increasing the glycolytic flux by over-expression of PFK1 and PFK2 actually decreased the glucose response of ChREBP (Fig. 4B); again, corroborating the conclusion that further metabolism through glycolytic pathway is not required for high glucose-induced ChREBP activation.
We have probed the major metabolic pathways downstream of G-6-P and found that they were not required for activating ChREBP in 832/13 cells. Although the other fates of G-6-P (glycogen and hexosamine synthesis) were examined only indirectly, G-6-P itself remains the most likely candidate especially with the evidence that there is a correlation between intracellular G-6-P levels with the ChREBP transcriptional activity under different glucose concentrations (Fig. 2B). This interpretation is compatible with all existing data as well as those described in this report.
Conclusion
We have used multiple pharmacologic and genetic manipulations in 832/13 cells to dissect the glucose metabolic pathways to identify the metabolite that mediates glucose-responsive transactivation of ChREBP via a PP2A-independent pathway. All experimental evidence presented is consistent with G-6-P being the metabolite that mediates the high glucose-stimulated activation of ChREBP in 832/13 cells.
Supplementary Material
Acknowledgements
This work was supported by NIH grants DK68037, HL51586 and HL-037162, the Diabetes and Endocrinology Research Center (P30DK079638) at Baylor College of Medicine, the Rutherford Chair for Diabetes Research from St. Luke's Episcopal Hospital, and the T.T. & W.F. Chao Foundation. We thank Dr. Jason Chesney for pIRESneo3-iPFK2.
Abbreviations
- bHLH/ZIP
basic helix-loop-helix/leucine zipper
- ChoRE
carbohydrate response element
- ChREBP
carbohydrate response element binding protein
- liver pyruvate kinase
- PP2A
protein phosphatase 2A
- G-6-P
Glucose 6 phosphate
- 2-DG
2-deoxyglucose
- G6PD
Glucose 6 phosphate dehydrogenase
- PFK
phosphofructokinase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Goodridge AG. Annu Rev Nutr. 1987;7:157–85. doi: 10.1146/annurev.nu.07.070187.001105. [DOI] [PubMed] [Google Scholar]
- [2].Towle HC, Kaytor EN, Shih HM. Annu Rev Nutr. 1997;17:405–33. doi: 10.1146/annurev.nutr.17.1.405. [DOI] [PubMed] [Google Scholar]
- [3].Mitanchez D, Doiron B, Chen R, Kahn A. Endocr Rev. 1997;18:520–40. doi: 10.1210/edrv.18.4.0307. [DOI] [PubMed] [Google Scholar]
- [4].Thompson KS, Towle HC. J Biol Chem. 1991;266:8679–82. [PubMed] [Google Scholar]
- [5].Bergot MO, Diaz-Guerra MJ, Puzenat N, Raymondjean M, Kahn A. Nucleic Acids Res. 1992;20:1871–7. doi: 10.1093/nar/20.8.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, Uyeda K. Proc Natl Acad Sci U S A. 2001;98:9116–21. doi: 10.1073/pnas.161284298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Iizuka K, Miller B, Uyeda K. Am J Physiol Endocrinol Metab. 2006;291:E358–64. doi: 10.1152/ajpendo.00027.2006. [DOI] [PubMed] [Google Scholar]
- [8].Kooner JS, Chambers JC, Aguilar-Salinas CA, Hinds DA, Hyde CL, Warnes GR, Perez F.J. Gomez, Frazer KA, Elliott P, Scott J, Milos PM, Cox DR, Thompson JF. Nat Genet. 2008;40:149–51. doi: 10.1038/ng.2007.61. [DOI] [PubMed] [Google Scholar]
- [9].Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JR, Girard J, Postic C. Diabetes. 2006;55:2159–70. doi: 10.2337/db06-0200. [DOI] [PubMed] [Google Scholar]
- [10].da Silva Xavier G, Rutter GA, Diraison F, Andreolas C, Leclerc I. J Lipid Res. 2006;47:2482–91. doi: 10.1194/jlr.M600289-JLR200. [DOI] [PubMed] [Google Scholar]
- [11].Davies MN, O'Callaghan BL, Towle HC. J Biol Chem. 2008;283:24029–38. doi: 10.1074/jbc.M801539200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kabashima T, Kawaguchi T, Wadzinski BE, Uyeda K. Proc Natl Acad Sci U S A. 2003;100:5107–12. doi: 10.1073/pnas.0730817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kawaguchi T, Takenoshita M, Kabashima T, Uyeda K. Proc Natl Acad Sci U S A. 2001;98:13710–5. doi: 10.1073/pnas.231370798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hedeskov CJ, Capito K. Biochem J. 1975;152:571–6. doi: 10.1042/bj1520571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li MV, Chang B, Imamura M, Poungvarin N, Chan L. Diabetes. 2006;55:1179–89. doi: 10.2337/db05-0822. [DOI] [PubMed] [Google Scholar]
- [16].Tsatsos NG, Towle HC. Biochem Biophys Res Commun. 2006;340:449–56. doi: 10.1016/j.bbrc.2005.12.029. [DOI] [PubMed] [Google Scholar]
- [17].Li MV, Chen W, Poungvarin N, Imamura M, Chan L. Mol Endocrinol. 2008;22:1658–72. doi: 10.1210/me.2007-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Girard J, Ferre P, Foufelle F. Annu Rev Nutr. 1997;17:325–52. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- [19].Foufelle F, Gouhot B, Pegorier JP, Perdereau D, Girard J, Ferre P. J Biol Chem. 1992;267:20543–6. [PubMed] [Google Scholar]
- [20].Marie S, Diaz-Guerra MJ, Miquerol L, Kahn A, Iynedjian PB. J Biol Chem. 1993;268:23881–90. [PubMed] [Google Scholar]
- [21].Forman BM, Umesono K, Chen J, Evans RM. Cell. 1995;81:541–50. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- [22].Kishnani PS, Bao Y, Wu JY, Brix AE, Lin JL, Chen YT. Biochem Mol Med. 1997;61:168–77. doi: 10.1006/bmme.1997.2600. [DOI] [PubMed] [Google Scholar]
- [23].Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Zhang M, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. Nat Genet. 2003;33:401–6. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- [24].Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. Diabetes. 2000;49:424–30. doi: 10.2337/diabetes.49.3.424. [DOI] [PubMed] [Google Scholar]
- [25].Lang G, Michal G. D-Glucose-6-phosphate and D-fructose-6-phosphate in Methods of Enzymatic Analysis. Verlag Chemie International, Deerfield Beach; Florida: 1974. [Google Scholar]
- [26].D.a.C. Nelson M. Lehninger's Principles of Biochemistry. W. H. Freeman; New York: 2004. [Google Scholar]
- [27].Hillgartner FB, Salati LM, Goodridge AG. Physiol Rev. 1995;75:47–76. doi: 10.1152/physrev.1995.75.1.47. [DOI] [PubMed] [Google Scholar]
- [28].Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. Endocrinology. 1992;130:167–78. doi: 10.1210/endo.130.1.1370150. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.