Abstract
Mammalian target of rapamycin (mTOR) regulates various cellular functions, including tumorigenesis, and is inhibited by the tuberous sclerosis 1 (TSC1)–TSC2 complex. Here, we demonstrate that arrest-defective protein 1 (ARD1) physically interacts with, acetylates, and stabilizes TSC2, thereby repressing mTOR activity. The inhibition of mTOR by ARD1 inhibits cell proliferation and increases autophagy, thereby inhibiting tumorigenicity. Correlation between ARD1 and TSC2 abundance was apparent in multiple tumor types. Moreover, evaluation of loss of heterozygosity at Xq28 revealed allelic loss in 31% of tested breast cancer cell lines and tumor samples. Together, our findings suggest that ARD1 functions as an inhibitor of the mTOR pathway and that dysregulation of the ARD1-TSC2-mTOR axis may contribute to cancer development.
INTRODUCTION
Tumorigenesis is a complex, multistep process characterized by the dysregulation of many signaling cascades, including the mammalian target of rapamycin (mTOR) signaling pathway. Many of mTOR’s upstream regulators and downstream effectors are aberrantly activated in different types of human cancer, heightening interest in mTOR signaling. Because the malignant phenotype depends on these signaling proteins, it is not surprising that mTOR is viewed as a potential target for cancer therapy. Therefore, various approaches to inhibiting the mTOR signaling pathway are being pursued for clinical development (1–3).
mTOR is an evolutionarily conserved serine-threonine kinase (4, 5) that integrates signals from multiple inputs, including growth factors (6), amino acids (7), and intracellular energy supply (8, 9), to regulate diverse cellular functions, including transcription (10), ribosome biogenesis (11), translation initiation (12), and autophagic cell death (autophagy) (13). Autophagy is a process in which bulk cytoplasm and organelles are sequestered in double or multimembrane autophagic vesicles to be delivered to and degraded by the lysosome system. The recent implication of tumor suppressors [such as Bcl-2–interacting protein 1 (Beclin 1) and phosphatase and tensin homolog (PTEN)] in autophagic pathways indicates that deficiencies in autophagy may contribute to tumorigenesis (14, 15). The induction of autophagy by various anticancer therapies underlines its potential utility as a cancer treatment modality (16, 17).
Tuberous sclerosis 1 (TSC1) and TSC2 are upstream regulators of mTOR that form a functional complex and suppress cell growth by inhibiting mTOR activity (18, 19). Downstream targets of mTOR include two families of proteins involved in translational control, the ribosomal protein S6 kinases (S6Ks) and the eukaryotic initiation factor 4E binding proteins (4E-BPs). mTOR-dependent phosphorylation of S6K1 causes S6K1 activation (20), whereas mTOR-dependent phosphorylation of 4E-BP1 leads to its dissociation from the initiation factor eIF4E, thereby enabling eIF4E derepression (12). 4E-BPs and S6Ks have a central role in ribosomal biogenesis and cap-dependent translation, processes that are directly involved in translational control of cell-growth and cell-cycle regulators (21–25). In view of the importance of these proteins that are subject to mTOR-mediated translational control, it is not surprising that alterations in mTOR signaling should be implicated in cancer development.
Protein acetylation and deacetylation are posttranslational modifications that regulate normal cell functions and affect cancer development (26, 27). Of the mammalian protein acetyltransferases, arrest-defective protein 1 (ARD1) represents an atypical enzyme with both N-terminal a protein and ε protein acetylation activities (28, 29). Mouse ARD1 has been reported to acetylate Lys532 in hypoxia-inducible factor 1a (HIF-1α) and thereby enhance HIF-1α ubiquitination and degradation (29), although this observation is controversial (30, 31). In yeast, ARD1 has been implicated in cell fate specification, DNA repair, and maintenance of genomic stability (32, 33). In addition, several reports have implicated ARD1 in regulation of cell proliferation and apoptosis in mammalian cells (34–36).
Although a potential role for ARD1 in controlling cell proliferation and apoptosis has been identified (34–36), little is known about the relevance of ARD1 to cancer development. While searching for the relationship between ARD1 gene expression and clinical outcome in the database of Integrated Tumor Transcriptome Array and Clinical data Analysis (ITTACA) (http://bioinfo.curie.fr/ittaca), we found that increased ARD1 messenger RNA (mRNA) abundance correlated with better clinical outcome in patients with breast cancer. Further analysis revealed loss of heterozygosity (LOH) for ARD1 in a subset of breast cancers, suggesting that ARD1 may function as a tumor suppressor. We determined that ARD1 inhibited mTOR activity through acetylation and stabilization of TSC2. ARD1 suppressed cell proliferation, induced autophagy, and inhibited tumor growth. We thus conclude that ARD1 stabilizes TSC2, thereby inhibiting mTOR signaling and suppressing cancer development.
RESULTS
Association between ARD1 mRNA expression and clinical outcome
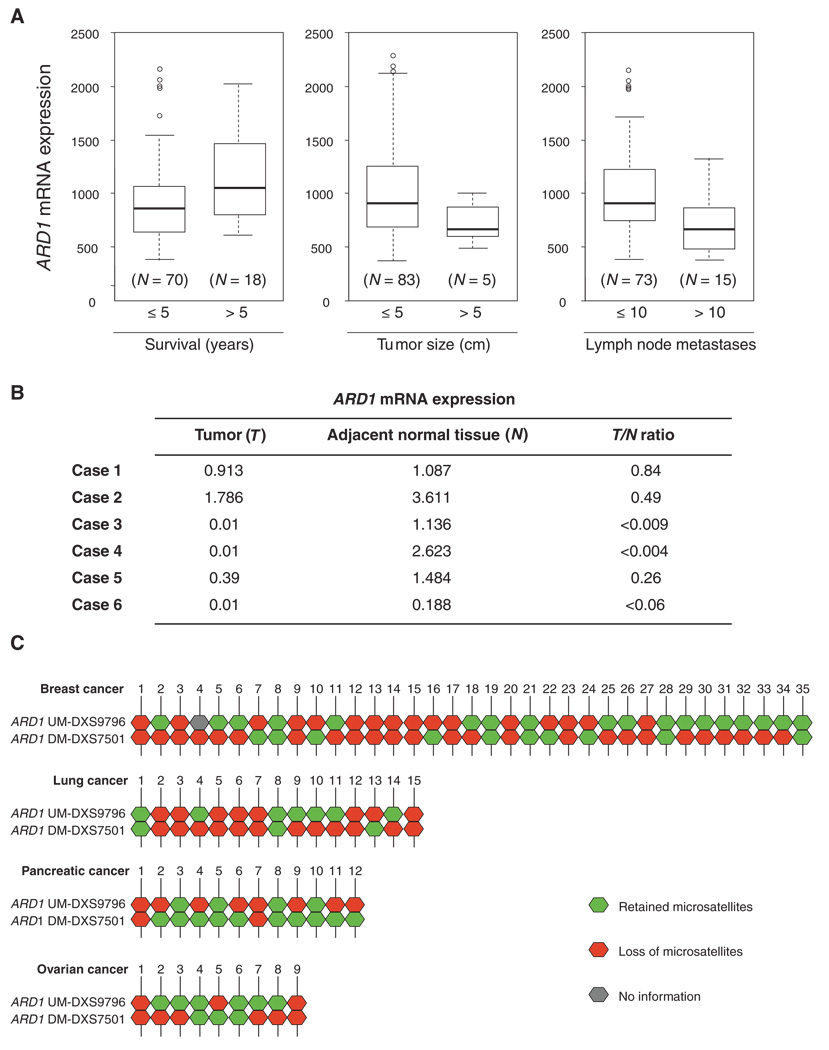
To determine the potential relevance of ARD1 to human breast cancer, we analyzed ARD1 mRNA expression in the ITTACA database. ITTACA was developed by Institut Curie Bioinformatics group and the Institut Curie, CNRS UMR144, to provide a central localization for public data sets containing both gene expression and clinical data (37). According to the database, and consistent with the results of a study by Huang and colleagues (38), we found that ARD1 mRNA expression was higher in samples from patients with longer relapse-free survival (>5 years, P < 0.05), smaller tumor size (≤5 cm, P < 0.05), and fewer lymph node metastases (≤10 metastases, P < 0.01) compared to that in samples from patients with shorter relapse-free survival, larger tumors, and more lymph node metastases (Fig. 1A). These data suggest that increased ARD1 expression is associated with better clinical outcome for patients with breast cancer.
Fig. 1.
Clinical significance of ARD1 mRNA expression and LOH at the ARD1 locus. (A) Analysis of gene expression data sets revealed that ARD1 mRNA expression positively correlates with relapse-free survival (P < 0.05) and negatively correlated with tumor size (P < 0.05) and number of lymph node metastases (P < 0.01). In each box plot, the upper and lower limits of the box indicate the 75th and 25th percentile, respectively, whereas the lines (whiskers) emerging above and below the box indicate the largest and smallest non-outlier observation. Open dots indicate the outliers within the data set. The sample sizes in each category are indicated in parentheses, and statistical significance was calculated with the Student’s t test. (B) Comparison of ARD1 mRNA expression in tumors and their adjacent normal tissues. ARD1 mRNA expression was examined with the Illumina humanRef-8 V2 expression Bead Chip containing six cases of non-small cell lung cancer. (C) Summary of LOH patterns of 35 breast cancer, 15 lung cancer, 12 pancreatic cancer, and 9 ovarian cancer samples. Retained microsatellites are indicated in green, markers demonstrating allelic loss in red, and noninformative markers in gray. ARD1 UM-DXS9796 represents upstream of the ARD1 locus, and ARD1 DM-DXS7501 represents downstream of the ARD1 locus.
To further study the role of ARD1 deficiency in tumorigenesis, we examined ARD1 mRNA abundance with the Illumina humanRef-8 V2 expression Bead Chip, which provides genome-wide expression patterns comparing tumor tissue to control tissue for six individuals with non–small cell lung cancer (39). ARD1 gene expression was 50% lower in tumor tissues compared to adjacent nontumor lung tissues in five out of six paired samples (Fig. 1B). Indeed, there was virtually no detectable ARD1 expression in three out of six tumor tissues, supporting the possibility that ARD1 acts as a tumor suppressor.
Genomic alterations of ARD1 in human breast cancer
Loss of genomic stability is believed to be an important step in the early stages of cancer development (40). The progressive accumulation of genetic alterations leading to oncogene activation and loss of tumor suppressor function results in the transformation of normal cells to malignant ones. Because we hypothesized that ARD1 might act as a tumor suppressor, we investigated the possibility that there was a genomic alteration in the ARD1 locus. To clarify the status of the ARD1 locus in breast cancer, we analyzed two microsatellite markers, ARD1 UM and ARD1 DM, which closely flank the ARD1 gene (within 50 kb on either side). Using these two markers, we genotyped 18 breast cancer cell lines and 17 primary breast tumors and demonstrated LOH for both markers in 11 out of 35 (31%) breast cancer specimens (Fig. 1C). Quantitative polymerase chain reaction (PCR) confirmed that the average copy number of genomic ARD1 was lower in ARD1 LOH samples compared to that in ARD1 wild-type samples (0.67 versus 1.19) (fig. S1). We further demonstrated LOH of ARD1 in 7 out of 15 (47%) lung cancer cell lines, 2 out of 12 (17%) pancreatic cancer cell lines, and 2 out of 9 (22%) ovarian cancer cell lines (Fig. 1C). The somatic alterations of ARD1 in different types of cancers suggest that ARD1 may act to suppress the development of multiple types of cancer.
ARD1 suppression of cell growth, clonogenicity, and tumor growth
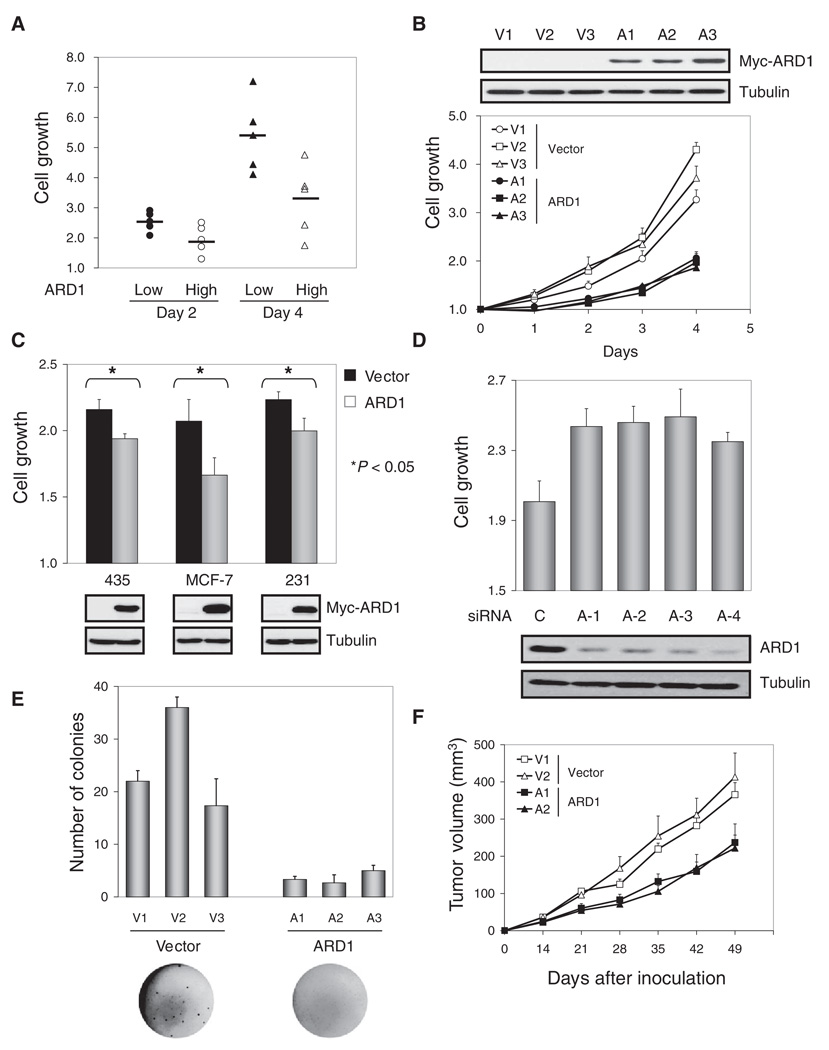
Analysis of 10 breast cancer cell lines revealed a reverse correlation between endogenous ARD1 abundance and the rate with which cell number increased over time (cell growth) (Fig. 2A and fig. S2). A stronger inhibitory effect of transient transfection with plasmids encoding ARD1 on cell growth was observed in tumor cells with less abundant endogenous ARD1 (fig. S3). To explore the possible roles of ARD1 in cancer development, we developed cell lines stably transfected with plasmids encoding ARD1 (ARD stable transfectants) or with empty vector controls (vector stable transfectants). All three stably transfected ARD1 clones grew more slowly than did those stably transfected with vector, as assessed with a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Fig. 2B). Transient transfection with plasmids encoding ARD1 inhibited cell growth in three different human breast cancer cell lines: MDA-MB-435, MCF-7, and MDA-MB-231 (Fig. 2C). Consistent with these results, ARD1 knockdown with small-interfering RNAs (siRNAs) increased cell growth in both MCF-10A and MDA-MB-435 cells (Fig. 2D and fig. S4). The ability of ARD1 stable transfectants to form clones in soft agar was impaired compared to that of vector control stable transfectants (Fig. 2E). Furthermore, the growth rate of tumors induced by inoculating ARD1 stable transfectants into the mammary fat pads of nude mice was also significantly lower than that of vector control stable transfectants (P < 0.001; t test; Fig. 2F). Together, these data indicate that ARD1 inhibits the growth, clonogenicity, and tumorigenicity of breast cancer cells.
Fig. 2.
Inhibition of cell growth, clonogenicity, and tumor development by ARD1. (A) An inverse correlation was found between endogenous ARD and cell growth (P < 0.05; t test). Cell growth was determined in five ARD1 low abundance-cells (MDA-MB-435, MDA-MB-468, BT549, HBL100, 11-9-1-4) and five ARD1 high abundance-cells (MDA-MB-436, MDA-MB-453, MDA-MB-361, ZR75-1, SKBr3) by an MTT assay. (B) The growth of ARD1 stable transfectants was slower than that of vector stable transfectants (P < 0.005; t test). V1, V2, V3 and A1, A2, A3 are three individual clones of vector control and ARD1 stable transfectants generated in MDA-MB-435 cells. Error bars represent SD (n = 3). (C) Transient transfection of Myc-ARD1 suppressed cell growth (*P < 0.05; t test). Error bars represent SD (n = 5). (D) Knockdown of ARD1 with any of four different siRNAs increased the growth of MCF-10A cells (P < 0.05; t test). Error bars represent SD (n = 5). (E) ARD1 stable transfectants showed impaired clonogenicity (P < 0.05; t test) in an anchorage-independent growth assay. The results shown are the average and SD of colony numbers in week 4 (n = 3). (F) The tumor growth rate was decreased with ARD1 stable transfectants (n = 10; P < 0.001; t test). Error bars represent SEM.
ARD1 suppression of cell proliferation and promotion of autophagy
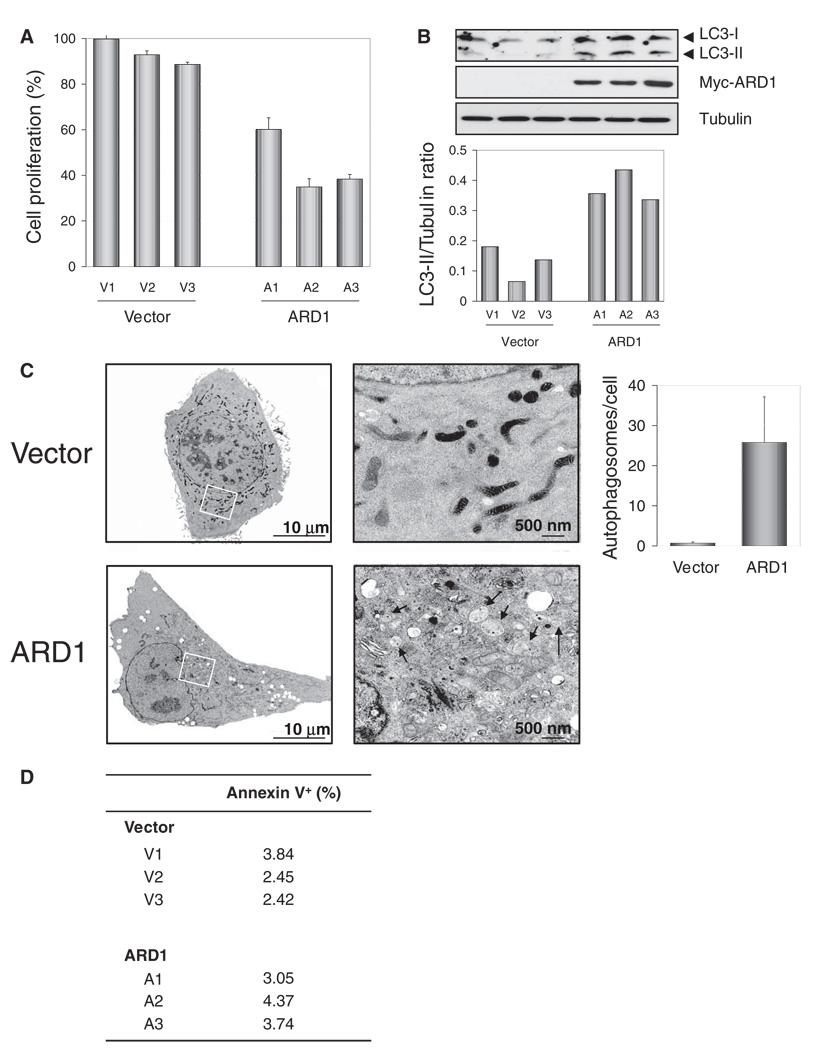
We assessed the effect of ARD1 on cell proliferation and cell death, both of which contribute to the overall cell growth rate. Using a bromodeoxyuridine (BrdU) incorporation assay, we found that ARD1 stable transfectants proliferated more slowly than did vector stable transfectants (Fig. 3A). We next investigated the possibility that ARD1 might induce autophagy or apoptosis. During autophagy, autophagosomes engulf cytoplasmic components, and a cytosolic form of microtubule-associated protein light chain 3 (LC3), LC3-I, is concomitantly conjugated to phosphatidyl-ethanolamine to form LC3-II. ARD1 stable transfectants showed enhanced conversion of LC3-I (18 kD) into LC3-II (16 kD) compared to vector stable transfectants, suggesting that ARD1 increased autophagic cell death (Fig. 3B). Additionally, transmission electron microscopy revealed more autophagosomes in ARD1 stable transfectants than in vector stable transfectants (Fig. 3C). In contrast, annexin V staining revealed no significant difference in apoptotic cell population between vector and ARD1 stable transfectants (Fig. 3D). These results suggest that ARD1 suppresses cell growth primarily by inhibiting cell proliferation and increasing autophagic cell death.
Fig. 3.
Suppression of cell proliferation and induction of autophagy by ARD1. (A) The cell proliferation rate, determined by BrdU incorporation, was decreased in ARD1 stable transfectants (P < 0.01; t test). Error bars represent SD (n = 3). (B) Increased conversion of LC3-I into LC3-II was found in ARD1 stable transfectants. (C) Transmission electron microscopy showed increased numbers of autophagosomes in ARD1 stable transfectants (P < 0.05; t test). Images on right represent rectangular areas in images on left. Arrowheads, autophagosomes. The number of autophagosomes per cell was assessed in 100 cells, and the results shown represent the mean ± 1 SD. (D) There was no significant difference in apoptotic cell population between the vector control and ARD1 stable transfectants (P = 0.248), which was determined by annexin V staining.
Requirement of TSC2 for mTOR inhibition by ARD1
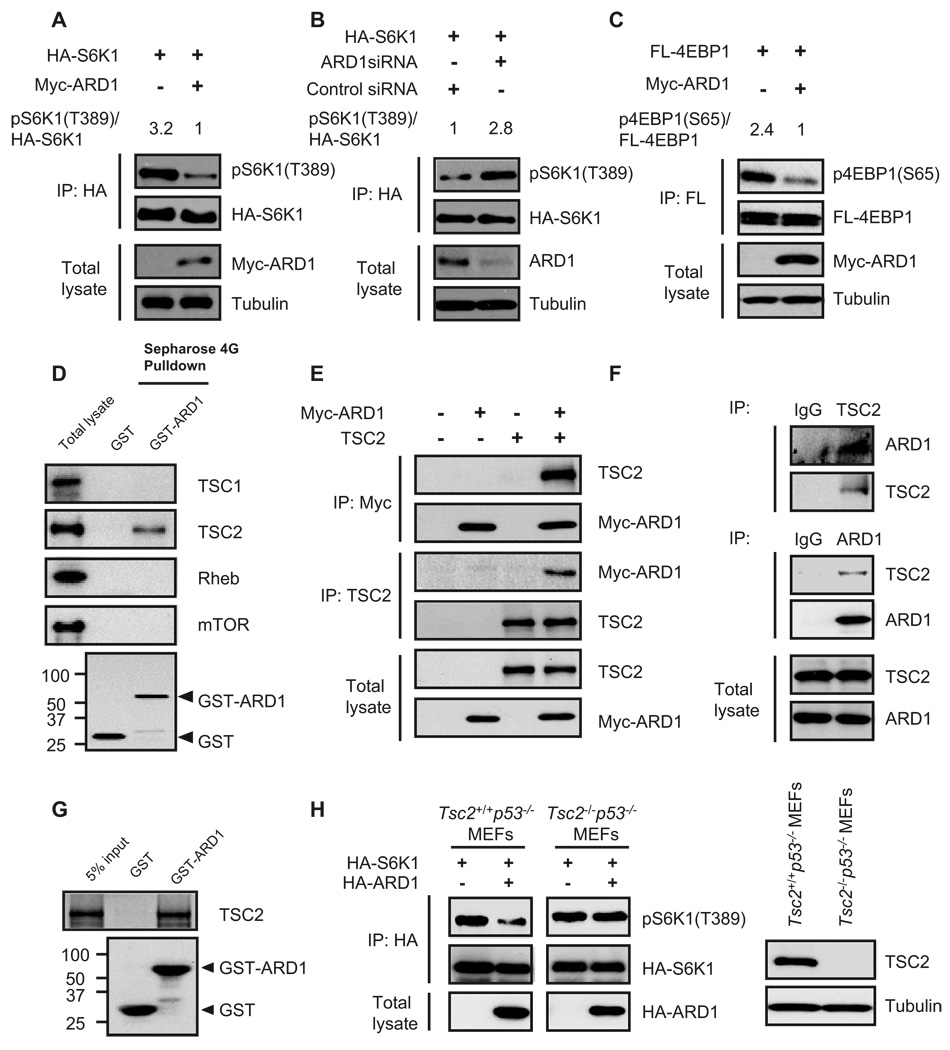
mTOR signaling has been implicated in regulating cell growth and autophagic cell death (13); therefore, we investigated the possibility that ARD1 affects mTOR activity. We transfected plasmids encoding Myc-tagged ARD1 or vector control into human embryonic kidney (HEK) 293T cells and assessed the phospho-rylation status of the mTOR substrate S6K1. To exclude signals from un-transfected cells and thereby increase the signal-to-noise ratio, we included a plasmid encoding hemagglutinin (HA)-tagged S6K1 in the transfection mixture. HA-tagged S6K1 was immunoprecipitated with an antibody against HA and immunoblotted with an antibody directed against phospho-S6K1. S6K1 phosphorylation at Thr389 [pS6K1(T389)] in ARD1-transfected cells was less than that in vector-transfected cells under conditions of normal growth (Fig. 4A), as well as during serum starvation and insulin-like growth factor (IGF) stimulation (fig. S5A). Depletion of ARD1 by siRNAs caused a marked increase in pS6K1(T389) abundance (Fig. 4B and fig. S5B). We also found that ARD1 inhibited 4EBP1 phosphorylation at Ser65 [p4EBP1(S65)] (Fig. 4C and fig. S6). These data indicate that ARD1 inhibits mTOR activity.
Fig. 4.
ARD1 inhibition of mTOR activity through TSC2. (A) Myc-ARD1 decreased S6K1 phosphorylation [pS6K1(T389)] in HEK293T cells. (B) ARD1 knockdown with siRNAs increased S6K1 phosphorylation [pS6K1(T389)] in HEK293T cells. (C) Myc-ARD1 decreased 4EBP1 phosphorylation [p4EBP1(S65)] in HEK293T cells. (D) TSC2, but not TSC1, Rheb, or mTOR, associated with GST-ARD1. GST or GST-ARD1 protein was pulled down with Sepharose 4G beads, and the associated proteins were analyzed by immunoblotting. (E) Interactions between exogenous ARD1 and TSC2 proteins. Lysates of HEK293T cells cotransfected with Myc-ARD1 and TSC2 were analyzed with antibodies directed against the Myc tag and TSC2 by reciprocal coimmunoprecipitation and immunoblotting. (F) Interaction between endogenous ARD1 and TSC2 proteins. Lysates of MDA-MB-435 cells were analyzed with antibodies directed against ARD1 and TSC2 by reciprocal coimmunoprecipitation and immunoblotting. (G) ARD1 interacted directly with TSC2. In vitro transcribed and translated TSC2 proteins were incubated with GST or GST-ARD1 proteins and then pulled down with Sepharose 4G beads. (H) Transient transfection of HA-ARD1 decreased pS6K1(T389) in Tsc2+/+ p53−/− MEFs but not in Tsc2−/− p53−/− MEFs.
The next question we addressed was which molecules upstream of mTOR might be associated with ARD1. We transfected HEK293T cells with plasmids encoding glutathione S-transferase–tagged ARD1 (GST-ARD1) and performed GST pull-down assays. We separated the GST pull-down lysate, containing ARD1-associated proteins, by SDS–polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting analysis with antibodies against mTOR, and several molecules known to participate in regulating mTOR function. We found that TSC2, but not TSC1, Rheb, or mTOR itself, associated with GST-ARD1 (Fig. 4D), suggesting that TSC2 may be involved in ARD1-mediated mTOR inhibition. Reciprocal coimmunoprecipitation assays of endogenous and exogenous proteins indicated that ARD1 physically interacted with TSC2 (Fig. 4, E and F). Moreover, in vitro pull-down assays revealed a direct association between ARD1 and TSC2 (Fig. 4G).
We transiently transfected plasmids encoding ARD1 into Tsc2+/+p53−/− and Tsc2−/−p53−/− mouse embryonic fibroblasts (MEFs) to verify that mTOR regulation by ARD1 was mediated through TSC2. pS6K1(T389) abundance was decreased in ARD1-bearing Tsc2+/+p53−/− MEFs but not in ARD1-bearing Tsc2−/−p53−/− MEFs (Fig. 4H), suggesting that TSC2 is indeed required for mTOR inactivation by ARD1.
ARD1 suppression of tumor growth through TSC2
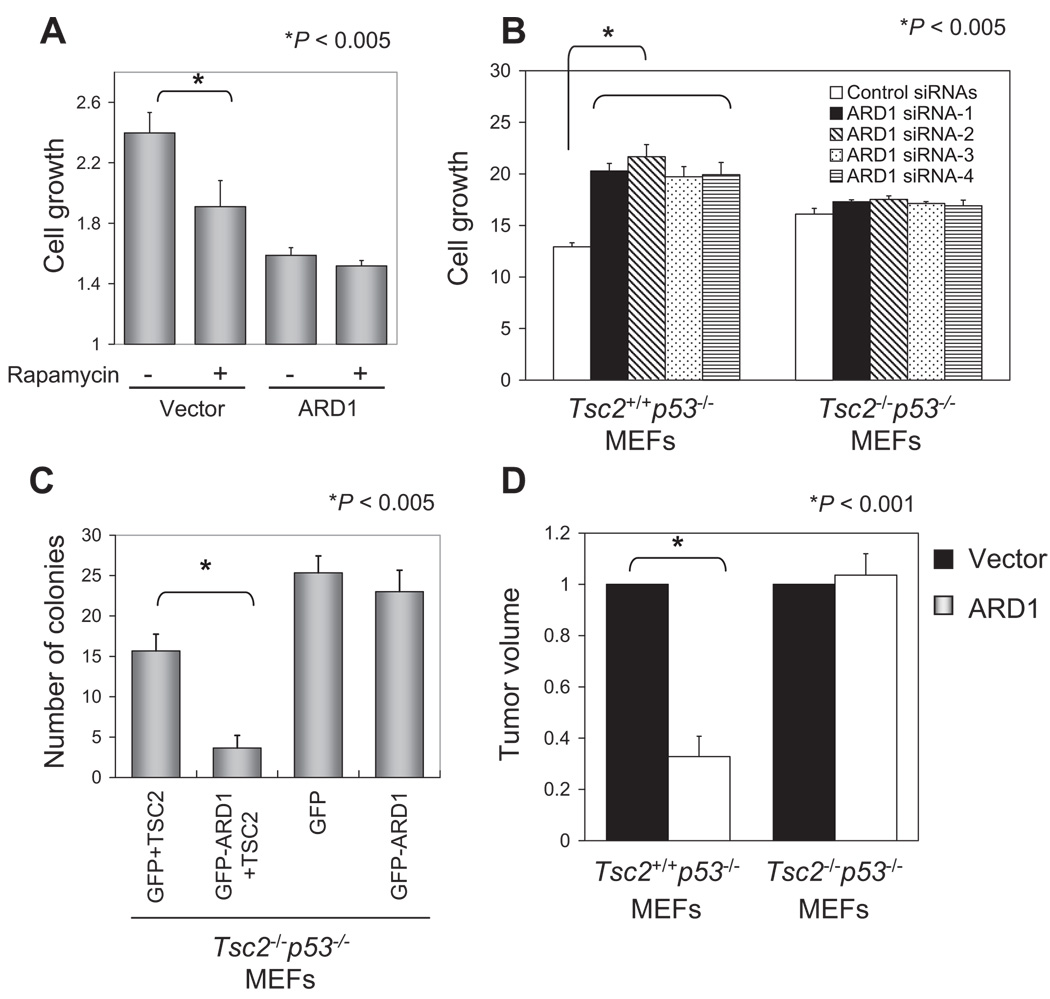
To determine whether inhibition of the mTOR pathway is involved in ARD1-induced suppression of cell growth, we treated stable transfectants with the mTOR inhibitor rapamycin. Rapamycin inhibited growth of vector stable transfectants but not that of ARD1 stable transfectants (Fig. 5A). Moreover, ARD1 knockdown with any of four different siRNAs significantly increased Tsc2+/+p53−/− MEF growth but had no effect on that of Tsc2−/−p53−/− MEFs (Fig. 5B). These data support the notion that the growth-suppressive effect of ARD1 is mediated through the TSC2-mTOR pathway. To determine whether ARD1 regulates clonogenicity in soft agar through TSC2, we transfected plasmids encoding green fluorescent protein (GFP) vector or GFP-ARD1 with or without TSC2 into TSC2−/−p53−/− MEFs, sorted the GFP-positive cells by flow cytometry, and assessed anchorage-independent growth. ARD1 only inhibited colony formation when it was cotransfected with TSC2 in TSC2−/−p53−/− MEFs (Fig. 5C), indicating that TSC2 was required for ARD1-dependent suppression of cell transformation. To investigate the role of TSC2 in ARD1-mediated suppression of tumor growth, we injected ARD1 plasmid DNA or vector controls complexed with N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methyl sulfate DOTAP-cholesterol (Chol) liposome using a DNA delivery system (41) intratumorally into nude mice bearing subcutaneous Tsc2+/+p53−/− or TSC2−/−p53−/− MEF xenografts. ARD1 significantly suppressed tumor growth compared to the vector control in TSC2+/+p53−/− but not in TSC2−/−p53−/− MEFs (Fig. 5D), supporting the idea that ARD1 inhibits tumor growth through TSC2.
Fig. 5.
ARD1 suppression of tumor growth through TSC2. (A) The mTOR inhibitor rapamycin inhibited growth of the vector but not ARD1 stable transfectants (*P < 0.005; t test). Cells were treated with 100 nM rapamycin for 24 hours, and cell growth rate was determined by an MTT assay. Error bars represent SD (n = 5). (B) ARD1 knockdown with siRNAs increased cell growth in Tsc2+/+p53−/− MEFs (*P < 0.005; t test). Four different ARD1 siRNAs were transfected into Tsc2+/+p53−/− or Tsc2−/− p53−/− MEFs; cell growth rate was determined by an MTT assay after 4 days. Error bars represent SD (n = 5). (C) ARD1 decreased the clonogenicity of Tsc2−/− p53−/− MEFs only when TSC2 was reintroduced (*P < 0.005; t test) in an anchorage-independent growth assay. The results shown are the average and SD of colony numbers in week 4 (n = 3). (D) Intratumoral injection of the ARD1 plasmid DNA: Liposome mixture inhibited tumor growth of Tsc2+/+ p53−/− MEFs (*P < 0.001; t test) but not Tsc2−/− p53−/− MEFs. Graph shows relative tumor volume normalized to 1 for the vector control-injected tumor. Five mice were used in each group. Error bars represent SD.
ARD1 stabilization of TSC2 through interaction and acetylation
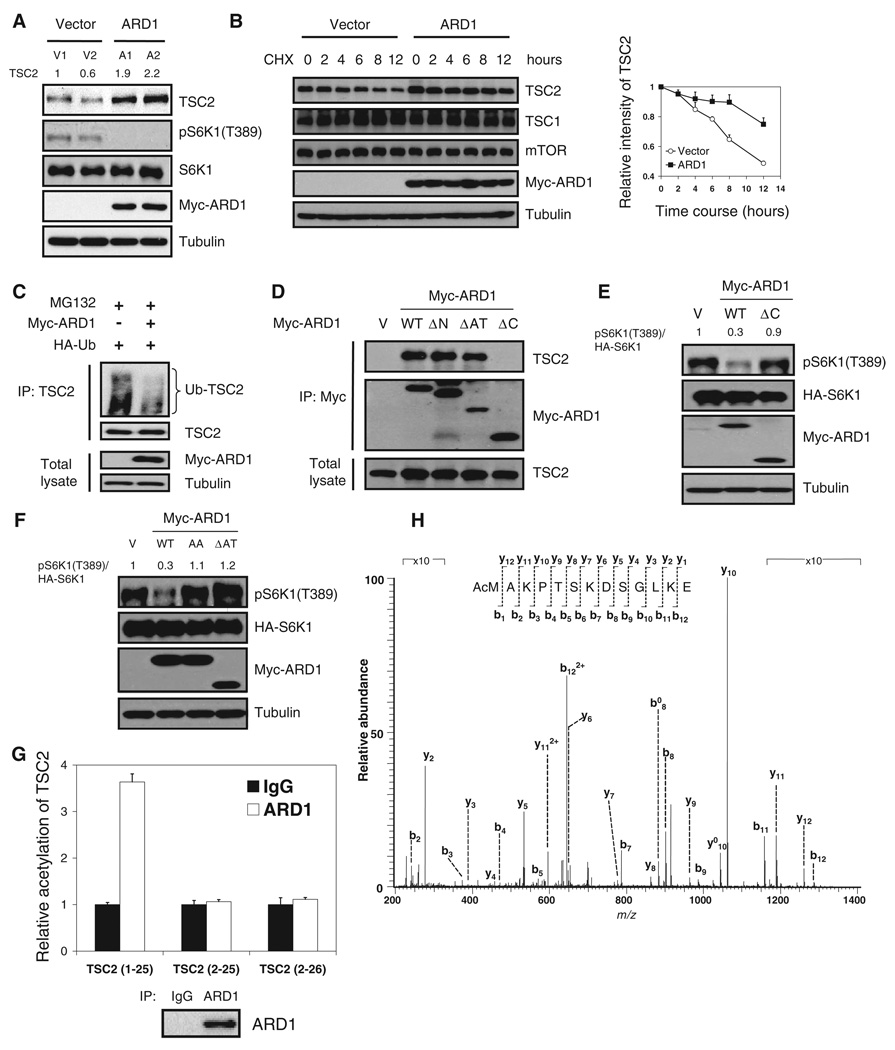
We observed that TSC2 abundance was higher in ARD1 stable transfectants than in vector stable transfectants (Fig. 6A). We obtained similar results with transient ARD1 expression in HEK293T cells (fig. S7). The implication that ARD1 physically interacts with TSC2 and thereby increases TSC2 abundance led us to investigate whether ARD1 enhances TSC2 stability. Treatment with cycloheximide to inhibit protein synthesis indicated that TSC2 stability increased in cells cotransfected with ARD1 (Fig. 6B) and decreased in ARD1-depleted cells (fig. S8). To determine TSC2 half-life, we graphed the data of Fig. 6B on a semilog plot (fig. S9) and determined that ARD1 significantly increased TSC2 half-life from 12.7 to 33.0 hours. Treatment with MG132, which inhibits the proteasome degradation pathway, also increased TSC2 abundance (fig. S10). Moreover, ARD1 decreased TSC2 ubiquitination (Fig. 6C). Together, these data suggest that ARD1 increases TSC2 stability and inhibits TSC2 degradation through the ubiquitin proteasome pathway, thereby increasing TSC2 abundance.
Fig. 6.
ARD1 stabilization of TSC2 through interaction and acetylation. (A) Abundance of TSC2 was increased in ARD1 stable transfectants. (B) ARD1 increased TSC2 stability, measured at 4, 6, 8, and 12 hours after cycloheximide (100 µg/ml) treatment (P < 0.05; mixed-effects model). Graph shows relative intensity of TSC2 standardized to 1 for the cycloheximide-pretreated (CHX 0 hours) sample. Error bars represent SD (n = 3). (C) ARD1 decreased ubiquitination of TSC2. HEK293T cells co-transfected with Myc-ARD1 or vector control with HA-ubiquitin were treated with MG132 for 6 hours, and the lysates were immunoprecipitated (IP) by specific antibody to TSC2 and then immunoblotted with antibody to HA tag. (D) ARD1 C-terminal domain is required for its association with TSC2. Interaction between ARD1 constructs and TSC2 was examined by coimmunoprecipitation and immunoblotting. (E) Wild-type (WT) ARD1 but not ARD1 ΔC reduced phosphorylated S6K1 [pS6K1 (T389)] in HEK293T cells. (F) WT ARD1 but not ARD1 AA or ARD1 ΔAT decreased pS6K1 (T389). (G) ARD1 acetylated TSC2 at the first amino acid, methionine. Immunoprecipitated ARD1 was used to acetylate TSC2 peptides TSC2 (1 to 25), TSC2 (2 to 25), or TSC2 (2 to 26) in the N-α-acetylation assay. (H) In vitro acetylation of TSC2 at the first methionine was confirmed by mass spectrometry analysis.
To see whether the association between ARD1 and TSC2 is necessary for TSC2 stabilization and ARD1-dependent mTOR inhibition, we generated three truncated forms of ARD1, ARD1 ΔN, ARD1 ΔAT, and ARD1 ΔC, with deletion of ARD1 N-terminal domain (amino acids 1 to 44), acetyltransferase domain (amino acids 45 to 130), and C-terminal domain (amino acids 131 to 235), respectively (fig. S11). Coimmunoprecipitation assays indicated that the C-terminal domain of ARD1 is required for its association with TSC2 (Fig. 6D). Increased TSC2 stability was apparent in cells transfected with wild-type ARD1 but not in cells transfected with ARD1 ΔC (fig. S12). Furthermore, expression of wild-type ARD1 but not that of ARD1 ΔC decreased pS6K1(T389) (Fig. 6E). Together, these results suggest that interaction between ARD1 and TSC2 is required for TSC2 stabilization and inhibition of mTOR signaling.
The next question we addressed was whether ARD1 acetyltransferase activity was required for its regulation of TSC2-mTOR signaling and its suppression of cell growth. It has previously been shown that the consensus sequence R82-x-x-G85-x-A is critical for ARD1 enzyme activity, and the acetyltransferase activity of mutant ARD1 R82A or G85A, in which Ala is substituted for Arg82 or Gly85, is much lower than that of wild-type (WT) ARD1 (42). Here, we generated two catalytically inactive ARD1 mutants. In the first (ARD1 AA), we substituted Ala for both Arg82 and Gly85. In the second, we truncated the ARD1 acetyltransferase motif (amino acids 45 to 130) to generate ARD1 ΔAT. We found that wild-type ARD1, but not ARD1 AA or ARD1 ΔAT, increased TSC2 stability (fig. S13) and decreased TSC2 ubiquitination (fig. S14), suggesting that ARD1 enzyme activity is required for it to stabilize TSC2 and prevent its ubiquitination. wild-type ARD1, but not ARD1 AA or ARD1 ΔAT, reduced pS6K1(T389), indicating that ARD1 enzyme activity is necessary for its inhibition of mTOR signaling (Fig. 6F). Wild-type ARD1 significantly suppressed growth of MDA-MB-435 and MCF-7 cells compared to ARD1 AA or ARD1 ΔAT (fig. S15), indicating that the growth-suppressing effect of ARD1 also depends on its enzymatic activity.
To determine whether ARD1 caused the acetylation of TSC2, we knocked down ARD1 in HEK293T cells and measured acetylation of endogenous TSC2. The ARD1-depleted cells showed decreased acetylation of endogenous TSC2 (fig. S16). α-Protein acetylation is believed to protect proteins from N-terminal degradation, and ARD1 is a well-known α-acetyltransferase in yeast (32, 33). ARD1 has also been reported to mediate ε-acetylation in mammalian cells (29, 36). Thus, we investigated whether either of these two modifications was involved in TSC2 regulation by ARD1. To examine ARD1 α-protein acetylation activity, we performed an N-terminal acetyltransferase assay on peptides derived from TSC2. ARD1-induced α-protein acetylation was only found on TSC2 peptide 1 to 25 (MAKPTSKDSGLKEKFKILLGLGTPR), not on TSC2 peptide 2 to 25 (AKPTSKDSGLKEKFKILLGLGTPR) or TSC2 peptide 2 to 26 (AKPTSKDSGLKEKFKILLGLGTPRP) (43) (Fig. 6G), suggesting that ARD1 acetylates TSC2 on the first methionine (Met1). This was confirmed by the results of a mass spectrometry analysis that demonstrated α-acetylation on TSC2 Met1 (Fig. 6H). We next assessed whether ARD1 mediates ε-acetylation of TSC2. MDA-MB-435 cells were cotransfected with ARD1 and TSC2 and treated with sodium butyrate to prevent protein deacetylation, and the acetylated TSC2 was detected by antibodies against acetyl lysine. We were unable to see any ε-acetylation of TSC2, even after performing analyses with seven different antibodies (fig. S17), indicating that ARD1 does not induce ε-acetylation of TSC2. We conclude that ARD1 acetylates TSC2 at Met1, thereby stabilizing TSC2 and increasing its abundance.
Correlation between ARD1 and TSC2 abundance
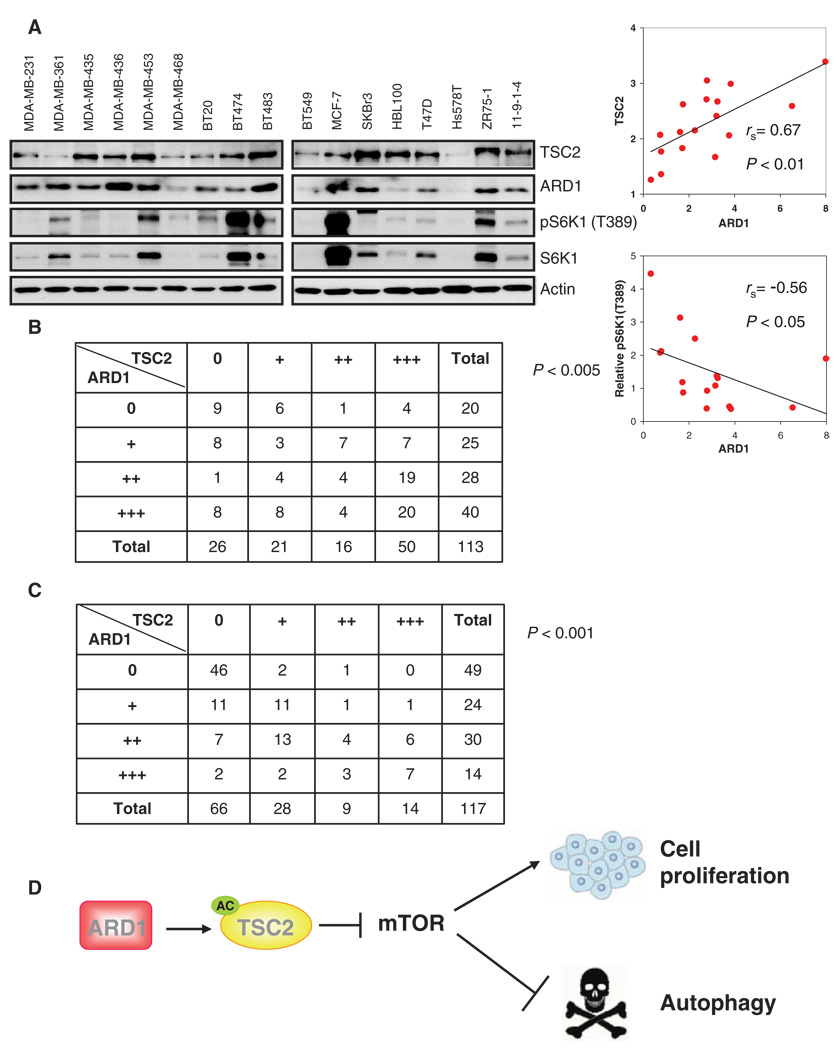
To determine whether the relationship between ARD1 and TSC2 was generally applicable in breast cancer cells, we measured ARD1 and TSC2 abundance in 17 breast cancer cell lines and found a significantly positive correlation between the abundance of ARD1 and that of TSC2 (P < 0.01; Fig. 7A). We also found a significantly negative correlation between ARD1 abundance and the degree of S6K1 phosphorylation [pS6K1(T389)] substantiating the inhibitory role of ARD1 in mTOR signaling (P < 0.05; Fig. 7A). ARD1 abundance, detected by immunohistochemical staining, was also positively correlated with that of TSC2 (P < 0.005; Fig. 7B and fig. S18) in a previously described cohort of breast tumors containing 113 human primary breast tumor specimens (44). To explore whether the association between ARD1 and TSC2 occurred in other tumor types, we examined 117 tumor tissue array samples (representing cancers of the oral cavity, nasopharynx, salivary gland, esophagus, stomach, small intestine, colorectum, liver, gallbladder, pancreas, larynx, and lung). The results showed that the abundance of ARD1 was associated with TSC2 not only in breast cancer, but in many other types of cancer as well (P < 0.001; Fig. 7C, fig. S19, and table S1). Together, these results strengthen the notion of TSC2 regulation by ARD1 and the physiological importance of ARD1 in maintaining TSC2 stability.
Fig. 7.
The association between ARD1 and TSC2. (A) ARD1 abundance was positively associated with that of TSC2 (P < 0.01) and negatively associated with pS6K1(T389) (normalized to total S6K1 expression) (P < 0.05) in a panel of human breast cancer cell lines. Correlation analyses were performed with the Spearman rank correlation test. (B) ARD1 abundance was associated with that of TSC2 in 113 primary human breast cancer specimens. Correlation analyses were performed with the Pearson chi-square test (P < 0.005). (C) ARD1 abundance was associated with that of TSC2 in 117 human tumor tissue specimens. Correlation analyses were performed by the Pearson chi-square test (P < 0.001). (D) A model in which stabilization of TSC2 by ARD1 inhibits mTOR signaling and thereby suppresses cell proliferation and induces autophagy.
DISCUSSION
There are conflicting data in the literature regarding the role of ARD1 in tumorigenesis. Early studies by Arnesen et al. reported that ARD1 knockdown induces apoptosis, an effect that depended on cell type (34). They found that HeLa and GaMg cells underwent apoptosis in response to treatment with siRNA directed against ARD1, whereas the HeLaS3 subclone showed only a weak apoptotic response. In contrast, Yi et al. found that ARD1 knockdown with any of six different ARD1 siRNAs protected fly cells and HeLa cells from apoptosis after DNA damage (35). Yi et al. hypothesized that the discrepancy between their data and those of Arnesen et al. may have arisen from differences among the siRNAs used and the ensuing off-target effects. Specifically, the siRNAs used by Arnesen et al. decreased cellular ATP concentrations by about 20% and therefore may decrease cell survival, whereas the siRNAs used by Yi et al. did not affect ATP concentration. Using the same siRNAs used in the latter study, we showed that ARD1 depletion increased the growth of MCF-10A and MDA-MB-435 cells. We also showed that ARD1 overexpression consistently decreased cell growth in breast cancer cell lines. In contrast, Fisher et al. found that ARD1 depletion suppressed proliferation of HepG2 cells (45). This discrepancy may result from the different siRNAs and cell types used in the two studies. ARD1 may associate with and acetylate various substrates or bind to diverse protein partners and, therefore, play different roles in different cell types. Thus, determination of the precise role of ARD1 in biological functions and identification of the spectrum of ARD1 substrates remain challenges for the future.
We found that ARD1 induces autophagy in MDA-MB-435 cells. Previous studies have shown that ARD1 knockdown decreased the mRNA abundance of Beclin 1 (45), a mammalian gene that is required for autophagy (14). Here, we provide evidence that ARD1 suppresses the mTOR signaling pathway through TSC2. It has been reported that TOR controls autophagy in yeast (46) and suppression of mTOR activity is also associated with increased autophagy in mammals (47). Further studies are required to investigate the relationship between ARD1-mediated TSC2-mTOR signaling and Beclin 1 expression.
A previous report showed that ARD1 abundance is decreased in most neoplastic thyroid tissue compared to that in nonneoplastic tissue (48), suggesting that ARD1 may be a tumor suppressor candidate. Gene expression and clinical data from the ITTACA database revealed that ARD1 mRNA expression correlates with better clinical outcome for patients with breast cancer (38), supporting a role for ARD1 as a tumor suppressor. However, Ren et al. found that ARD1 expression is higher in colorectal cancer tissues than in normal tissues (49). It is not yet clear why there is distinct expression of ARD1 in various tumors compared to their adjacent normal tissues, raising the question of whether ARD1 plays different roles in different tissues.
The analysis of LOH in tumors is a powerful tool for mapping the sites of tumor suppressor genes in the human genome. High frequencies of LOH observed at sites on chromosome Xq in human cancers of the breast (50), uterine cervix (51), and lung (52) suggest the presence of important tumor suppressor genes in this region. Moreover, there are reports of frequent LOH at chromosome Xq28, where ARD1 is located (53), in ovarian cancer (54) and cervical carcinoma (55). Here, we showed that LOH of ARD1 occurs in human breast, lung, pancreatic, and ovarian cancer samples, further supporting a role for ARD1 in cancer development.
We also demonstrated that ARD1 stabilizes TSC2 and prevents its degradation through a ubiquitin proteasome pathway, thereby inhibiting mTOR activity and cell growth (Fig. 7D). The identification of ARD1 as an upstream regulator of the mTOR signaling pathway provides an important basis for understanding the molecular basis of ARD1-mediated tumor suppression. Future investigation should address the physiological functions of ARD1 and its relevance to tumorigenesis.
MATERIALS AND METHODS
Plasmids, antibodies, and chemicals
We constructed Myc-ARD1– and HA-ARD1–expressing plasmids by inserting hARD1 complementary DNA (cDNA) into pcDNA6 and pCMV5 vectors containing the Myc and HA tags, respectively. We generated the GST-ARD1–expressing plasmid by subcloning the ARD1 fragment into the pGEX6P-1 GST vector. Different truncated forms of ARD1 constructs were generated by subcloning the indicated ARD1 fragment into pcDNA6 vector.
We used antibodies to FLAG (F3165, Sigma), Myc (11667203001, Roche), HA (11666606001, Roche), ARD1 (15-288-22667, GenWay; SC-33256, Santa Cruz Biotechnology), TSC1 (37-0400 Zymed; 4906, Cell Signaling Technology), TSC2 (SC-893, Santa Cruz Biotechnology), S6K1 (SC-230, Santa Cruz Biotechnology), pS6K1(T389) (9205, Cell Signaling Technology), 4EBP1 (9452L, Cell Signaling Technology), p4EBP1(S65) (9451s, Cell Signaling Technology), Rheb (SC-6341, Santa Cruz Biotechnology), mTOR (2972, Cell Signaling Technology), MAP LC3 (SC-28266, Santa Cruz Biotechnology; NB100-2331, Novus Biologicals), acetyl lysine (623402, Biolegend; MA1-2021, Affinity BioReagents; ST1027, Calbiochem; 05-515, Upstate; 9441s, Cell Signaling Technology; 9681s, Cell Signaling Technology; 06-933, Upstate), α-tubulin (T-5168, Sigma), and actin (A2066, Sigma).
Rapamycin, cycloheximide, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide, and MG132 were purchased from Sigma. Blasticidin S was purchased from InvivoGen. G418 was purchased from Cellgro.
siRNAs
HEK293T, MDA-MB-435, and MCF-10A cells were transfected with ARD1 ON-TARGETplus SMARTpool siRNAs (L-009606-00, Dharmacon RNA Technologies), ARD1 ON-TARGETplus SMARTpool duplex siRNAs (J-009606-05, J-009606-06, J-009606-07 and J-009606-08, Dharmacon RNA Technologies), or ON-TARGETplus siCONTROL not-targeting pool siRNAs (D-001810-10-20) by using DharmaFECT Transfection Reagents (T-2001-02) and the cells were harvested for analysis 72 hours after transfection. Tsc2−/−p53−/− and Tsc2+/+p53−/− MEFs were transfected with ARD1 ON-TARGETplus set of four duplex siRNAs (J-049547-09, J-049547-10, J-049547-11 and J-049547-12, Dharmacon RNA Technologies) or ON-TARGETplus siCONTROL not-targeting pool siRNAs by using DharmaFECT Transfection Reagents and harvested for analysis.
Cell culture, stable transfectants, and transient transfection
MDA-MB-435, MDA-MB-231, MCF-7, and HEK293T cells and Tsc2−/−p53−/− and Tsc2+/+p53−/− MEFs were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium supplemented with 10% fetal bovine serum (FBS). MCF-10A cells were cultured in DMEM/F12 medium supplemented with 5% equine serum, insulin (10 µg/ml), epidermal growth factor (EGF; 20 ng/ml), choleratoxin (100 ng/ml), and hydrocortisone (0.5 µg/ml). Vector control or ARD1 stable transfectants were generated in MDA-MB-435 cells (56, 57) and selected with blasticidin S (12 µg/ml). For transient transfection, cells were transfected with DNA either by SN liposome (58), Lipofectamine 2000 (11668-019, Invitrogen), or Lipofectamine with plus reagent (18324-012 and 11514-015, Invitrogen).
MTT assays
Cells were plated at a density of 5000 cells per well for MDA-MB-435 cells, or 500 to 1000 cells per well for MEFs in 96-well microplates. At different time points, 50 µl of MTT (0.5% in phosphate-buffered saline) were added to each well, and incubation was continued for 2 hours. The formazan crystals resulting from mitochondrial enzymatic activity on the MTT substrate were solubilized with 100 µl of dimethyl sulfoxide and the light absorbance was measured at 570 nm with a multiwell spectrophotometer (Labsystems). The light absorbance at day 0 was set to 1 and the y axes shown in the figures indicate the relative units to day 0.
Anchorage-independent growth assays
Anchorage-independent growth of stable transfectants was determined by a previously described method (59). Briefly, the cell-growth matrix consisted of base agar and top agarose in six-well culture plates. The base layer (1.5 ml) contained DMEM/F12 medium, 10% FBS, and 0.5% agar. The top layer (1.5 ml) contained DMEM/F12 medium, 10% FBS, 0.35% agarose, and the suspension of cells (5 × 103). The number of foci was counted after 4 weeks. The modifications used for TSC2−/−p53−/− MEFs are as follows: GFP-positive cells (5 × 104) were sorted, mixed with 0.5% agarose, and poured onto a bed of 1% agar. Both the top and the bottom layers were prepared in DMEM/F12 medium with 10% FBS.
Mouse model for tumorigenesis
We performed the tumorigenesis assay for stable transfectants with a breast cancer orthotopic mouse model that has been described elsewhere (60). Cells (1 × 106) were injected into the mammary fat pads of female nude mice (Jackson Laboratory), 10 mice per group, and the length (L) and width (W) of each tumor mass were measured by calipers once a week. Tumor volume (TV) was calculated according to the formula described by Yaguchi et al. (61): TV = 0.5 × L × W2.
For mouse model of MEF xenograft, TSC2+/+p53−/− MEFs consistently produced tumors in nude mice as previously described (62). Tumorigenic TSC2−/−p53−/− MEFs were generated with a previously described method (62). Briefly, 1 × 107 parental TSC2−/−p53−/− MEFs were subcutaneously injected into nude mice. Two weeks later, the tumor cells were harvested and recultured for tumor inoculation. Intratumoral injection of plasmid DNA complexed with DOTAP-Chol liposome was performed twice per week when the tumor reached 5 to 6 mm in diameter. The tumor volume was measured after three times of treatments.
BrdU incorporation assays
Cell proliferation was assessed by the Cell Proliferation enzyme-linked im-munosorbent assay (ELISA, Roche) per the manufacturer’s instructions.
Transmission electron microscopy
To demonstrate the induction of autophagy in ARD1 stable transfectants, we performed an electron microscopy analysis. Cells were fixed with a solution containing 3% glutaraldehyde plus 2% paraformaldehyde in 0.1 M cacodylate (pH 7.3) for 1 hour. After fixation, the cells were treated with tannic acid (0.1% in cacodylate buffer), postfixed with 1% OsO4 for 30 min, and stained with 1% uranyl acetate. Then, the samples were dehydrated in increasing concentrations of ethanol, infiltrated and embedded in Poly-bed 812 medium, and subjected to polymerization in a 60°C oven for 2 days. Representative areas were chosen for ultrathin sectioning, stained with uranyl acetate/lead citrate, and examined with a JEM 1010 transmission electron microscope (JEOL) at an accelerating voltage of 80 kV. Digital images were obtained with the AMT Imaging System (Advanced Microscopy Techniques).
Apoptosis assays
Annexin V–PE (BD Biosciences) was used to recognize the phospholipid phosphatidylserine (PS) exposed on the outer membrane of apoptotic cells, and flow cytometry was used to quantitatively determine the percentage of apoptotic cells within a population.
Immunoprecipitation and immunoblotting assays
Immunoprecipitation and immunoblotting assays were performed as described previously (63).
In vitro pull-down assays
GST-ARD1 protein was first purified with glutathione Sepharose 4B beads and then incubated with in vitro transcription and translation lysate of TSC2, which was produced with a TNT-coupled reticulocyte lysate system (Promega) in binding buffer [0.5 mM dithiothreitol, 0.5 mM EDTA, 0.1% NP-40, 50 mM NaCl, 10 mM Hepes (pH 7.5)] at 4°C for 3 hours. The resulting pull-down product was washed extensively with binding buffer, and the bound proteins were eluted with SDS-PAGE sampling buffer and analyzed.
In vitro N-α-acetylation assays
N-α-acetylation assays were performed as described (28). Briefly, ARD1 was immunoprecipitated and purified by protein G–agarose. The pellet of protein G–agarose–bound ARD1 was added to a mixture of 10 µl of TSC2 peptides (0.5 mM, QCB), 4 µl of [3H]acetyl coenzyme A (1 µCi, ICN MP), and 136 µl of 0.2 M K2HPO4 (pH 8.1). The mixture was then incubated at 37°C for 1 hour. After centrifugation, the supernatant was added to 150 µl of SP Sepharose (50% slurry in 0.5 M acetic acid; Sigma) and incubated for 5 min. The pellet was further washed three times with 0.5 M acetic acid and finally with methanol. Radioactivity was determined by scintillation counting. The activity of ARD1 immunoprecipitates was compared to that of control immunoglobulin G immunoprecipitates.
Immunohistochemical staining
Immunohistochemical staining was performed as described (63, 64). Briefly, human HistoArrays of multiple cancerous tissues (IMH-365, Imgenex) were incubated with antibodies directed against ARD1 or TSC2, detected with biotin-conjugated secondary antibody and avidinperoxidase, and visualized with aminoethylcarbazole chromogen. According to the histologic scoring, the intensity of staining was ranked into four groups: negative (−), low (+), medium (++), and high (+++). The Pearson chi-square test was used for statistical analysis, and P < 0.05 was considered statistically significant.
Microsatellite-based LOH analysis
Fluorescent LOH analysis with genomic DNA was performed as described (65). Contig U52112.2.1.181343, containing ARD1 flanking sequences, was downloaded from the Ensembl database (http://www.ensembl.org). Two pairs of fluorescence-labeled microsatellite primers flanking ARD1 [ARD1 UM-DXS9796 (GCTTCTTTCACACTCACGCAGC and CCCTGATCCAACCAAACAATGG), representing upstream of the ARD1 locus, and ARD1 DM-DXS7501 (TTTCCAGCCCTCCCCTAC and AAACGTGACATTTTCCACAGC), representing downstream of the ARD1 locus] were designed according to the Ensembl database and synthesized by Sigma Genosys. Briefly, each PCR reaction was performed in a total volume of 20 µl containing 100 ng DNA, 0.2 mM deoxynucleotide triphosphates, 1.5 mM MgCl2, 2 pmol of each primer, and 0.25 U of Ampli Taq Gold DNA polymerase (Applied Biosystems). Cycling conditions were 94°C (10 min) for one cycle, 94°C (30 s), 61°C to 65°C (30 s), and 72°C (30 s) for 25 cycles and a final extension of 72°C (30 min). The reactions were carried out in a T3 Thermocycler PCR System (Biometra). We then analyzed the data with the ABI Genescan and Genotyper software packages (PerkinElmer) and scored the allelic loss. The alleles were defined as the two highest peaks within the expected size range, using the normal breast cell line MCF-10A, normal lung cell line HBE4, normal pancreatic cell line HPDE6, and normal ovarian cell line IOSE as the reference. In our system, a relative allele ratio of less than 0.6 or more than 1.5 was defined as LOH.
Statistical analyses
Statistical analyses were performed with the Student’s t test, Spearman rank correlation test, mixed-effects model, or the Pearson chi-square test as indicated. A P value of < 0.05 was considered statistically significant.
Supplementary Material
Footnotes
Author contributions: H.-P.K., D.-F.L., C.-T.C., M.L., C.-K.C., H.-J.L., Y.D., X.X., Y.W., W.X., Z.W., J.-Y.Y., C-J.Y., T.-H.H., M.T., G.X., and C.-H.L. participated in the experimental design; H.-P.K., Y.Z., S.-F.T., I.J.F., and M.-C.H. participated in data acquisition and analysis; H.-P.K. and M.-C.H. wrote the manuscript. In memoriam, Tiong Loi Ang for his courageous fight against cancer.
REFERENCES AND NOTES
- 1.Peralba JM, DeGraffenried L, Friedrichs W, Fulcher L, Grünwald V, Weiss G, Hidalgo M. Pharmacodynamic evaluation of CCI-779, an inhibitor of mTOR, in cancer patients. Clin. Cancer Res. 2003;9:2887–2892. [PubMed] [Google Scholar]
- 2.Boulay A, Zumstein-Mecker S, Stephan C, Beuvink I, Zilbermann F, Haller R, Tobler S, Heusser C, O’Reilly T, Stolz B, Marti A, Thomas G, Lane HA. Antitumor efficacy of intermittent treatment schedules with the rapamycin derivative RAD001 correlates with prolonged inactivation of ribosomal protein S6 kinase 1 in peripheral blood mononuclear cells. Cancer Res. 2004;64:252–261. doi: 10.1158/0008-5472.can-3554-2. [DOI] [PubMed] [Google Scholar]
- 3.Basso AD, Mirza A, Liu G, Long BJ, Bishop WR, Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J. Biol. Chem. 2005;280:31101–31108. doi: 10.1074/jbc.M503763200. [DOI] [PubMed] [Google Scholar]
- 4.Brown EJ, Beal PA, Keith CT, Chen J, Shin TB, Schreiber SL. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature. 1995;377:441–446. doi: 10.1038/377441a0. [DOI] [PubMed] [Google Scholar]
- 5.Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- 6.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 7.Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- 8.Dennis PB, Jaeschke A, Saitoh M, Fowler B, Kozma SC, Thomas G. Mammalian TOR: A homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 9.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 10.Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD. mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol. Cell. Biol. 2003;23:8862–8877. doi: 10.1128/MCB.23.23.8862-8877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Terada N, Patel HR, Takase K, Kohno K, Nairn AC, Gelfand EW. Rapamycin selectively inhibits translation of mRNAs encoding elongation factors and ribosomal proteins. Proc. Natl. Acad. Sci. U.S.A. 1994;91:11477–11481. doi: 10.1073/pnas.91.24.11477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–826. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 13.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell. 2004;7:167–178. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 15.Arico S, Petiot A, Bauvy C, Dubbelhuis PF, Meijer AJ, Codogno P, Ogier-Denis E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001;276:35243–35246. doi: 10.1074/jbc.C100319200. [DOI] [PubMed] [Google Scholar]
- 16.Bursch W, Ellinger A, Kienzl H, Török L, Pandey S, Sikorska M, Walker R, Hermann RS. Active cell death induced by the anti-estrogens tamoxifen and ICI 164 384 in human mammary carcinoma cells (MCF-7) in culture: The role of autophagy. Carcinogenesis. 1996;17:1595–1607. doi: 10.1093/carcin/17.8.1595. [DOI] [PubMed] [Google Scholar]
- 17.Shao Y, Gao Z, Marks PA, Jiang X. Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc. Natl. Acad. Sci. U.S.A. 2004;101:18030–18035. doi: 10.1073/pnas.0408345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tee AR, Fingar DC, Manning BD, Kwiatkowski DJ, Cantley LC, Blenis J. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U.S.A. 2002;99:13571–13576. doi: 10.1073/pnas.202476899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 20.Jefferies HB, Fumagalli S, Dennis PB, Reinhard C, Pearson RB, Thomas G. Rapamycin suppresses 5’TOP mRNA translation through inhibition of p70s6k. EMBO J. 1997;16:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montagne J, Stewart MJ, Stocker H, Hafen E, Kozma SC, Thomas G. Drosophila S6 kinase: A regulator of cell size. Science. 1999;285:2126–2129. doi: 10.1126/science.285.5436.2126. [DOI] [PubMed] [Google Scholar]
- 22.Radimerski T, Montagne J, Rintelen F, Stocker H, van der Kaay J, Downes CP, Hafen E, Thomas G. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 2002;4:251–255. doi: 10.1038/ncb763. [DOI] [PubMed] [Google Scholar]
- 23.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 24.Fingar DC, Richardson CJ, Tee AR, Cheatham L, Tsou C, Blenis J. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 2004;24:200–216. doi: 10.1128/MCB.24.1.200-216.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen HY, Lavu S, Bitterman KJ, Hekking B, Imahiyerobo TA, Miller C, Frye R, Ploegh H, Kessler BM, Sinclair DA. Acetylation of the C terminus of Ku70 by CBP and PCAF controls Bax-mediated apoptosis. Mol. Cell. 2004;13:627–638. doi: 10.1016/s1097-2765(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 27.Mahlknecht U, Hoelzer D. Histone acetylation modifiers in the pathogenesis of malignant disease. Mol. Med. 2000;6:623–644. [PMC free article] [PubMed] [Google Scholar]
- 28.Arnesen T, Anderson D, Baldersheim C, Lanotte M, Varhaug JE, Lillehaug JR. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem. J. 2005;386:433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong JW, Bae MK, Ahn MY, Kim SH, Sohn TK, Bae MH, Yoo MA, Song EJ, Lee KJ, Kim KW. Regulation and destabilization of HIF-1a by ARD1-mediated acetylation. Cell. 2002;111:709–720. doi: 10.1016/s0092-8674(02)01085-1. [DOI] [PubMed] [Google Scholar]
- 30.Arnesen T, Kong X, Evjenth R, Gromyko D, Varhaug JE, Lin Z, Sang N, Caro J, Lillehaug JR. Interaction between HIF-1α (ODD) and hARD1 does not induce acetylation and destabilization of HIF-1α. FEBS Lett. 2005;579:6428–6432. doi: 10.1016/j.febslet.2005.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murray-Rust TA, Oldham NJ, Hewitson KS, Schofield CJ. Purified recombinant hARD1 does not catalyse acetylation of Lys532 of HIF-1α fragments in vitro. FEBS Lett. 2006;580:1911–1918. doi: 10.1016/j.febslet.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 32.Whiteway M, Szostak JW. The ARD1 gene of yeast functions in the switch between the mitotic cell cycle and alternative developmental pathways. Cell. 1985;43:483–492. doi: 10.1016/0092-8674(85)90178-3. [DOI] [PubMed] [Google Scholar]
- 33.Lee FJ, Lin LW, Smith JA. Nα acetylation is required for normal growth and mating of Saccharomyces cerevisiae. J. Bacteriol. 1989;171:5795–5802. doi: 10.1128/jb.171.11.5795-5802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arnesen T, Gromyko D, Pendino F, Ryningen A, Varhaug JE, Lillehaug JR. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-α-acetyltransferase complex. Oncogene. 2006;25:4350–4360. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- 35.Yi CH, Sogah DK, Boyce M, Degterev A, Christofferson DE, Yuan J. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J. Cell Biol. 2007;179:619–626. doi: 10.1083/jcb.200708090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim JH, Park JW, Chun YS. Human arrest defective 1 acetylates and activates β-catenin, promoting lung cancer cell proliferation. Cancer Res. 2006;66:10677–10682. doi: 10.1158/0008-5472.CAN-06-3171. [DOI] [PubMed] [Google Scholar]
- 37.Elfilali A, Lair S, Verbeke C, La Rosa P, Radvanyi F, Barillot E. ITTACA: A new database for integrated tumor transcriptome array and clinical data analysis. Nucleic Acids Res. 2006;34:D613–D616. doi: 10.1093/nar/gkj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, Nevins JR, Huang AT. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 39.Shiao YM, Chang YH, Liu YM, Li JC, Su JS, Liu KJ, Liu YF, Lin MW, Tsai SF. Dysregulation of GIMAP genes in non-small cell lung cancer. Lung Cancer. 2008;62:287–294. doi: 10.1016/j.lungcan.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 40.Lee DF, Kuo HP, Liu M, Chou CK, Xia W, Du Y, Shen J, Chen CT, Huo L, Hsu MC, Li CW, Ding Q, Liao TL, Lai CC, Lin AC, Chang YH, Tsai SF, Li LY, Hung MC. KEAP1 E3 ligase-mediated downregulation of NF-κB signaling by targeting IKKβ. Mol. Cell. 2009;36:131–140. doi: 10.1016/j.molcel.2009.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie X, Xia W, Li Z, Kuo HP, Liu Y, Li Z, Ding Q, Zhang S, Spohn B, Yang Y, Wei Y, Lang JY, Evans DB, Chiao PJ, Abbruzzese JL, Hung MC. Targeted expression of BikDD eradicates pancreatic tumors in noninvasive imaging models. Cancer Cell. 2007;12:52–65. doi: 10.1016/j.ccr.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Asaumi M, Iijima K, Sumioka A, Iijima-Ando K, Kirino Y, Nakaya T, Suzuki T. Interaction of N-terminal acetyltransferase with the cytoplasmic domain of β-amyloid precursor protein and its effect on Aβ secretion. J. Biochem. 2005;137:147–155. doi: 10.1093/jb/mvi014. [DOI] [PubMed] [Google Scholar]
- 43.Abbreviations for the amino acids are as follows: A, Ala; D, Asp; E, Glu; F, Phe; G, Gly; I, Ile; K, Lys; L, Leu; M, Met; P, Pro; R, Arg; S, Ser; and T, Thr.
- 44.Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- 45.Fisher TS, Etages SD, Hayes L, Crimin K, Li B. Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J. Biol. Chem. 2005;280:17749–17757. doi: 10.1074/jbc.M412055200. [DOI] [PubMed] [Google Scholar]
- 46.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 47.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Arnesen T, Gromyko D, Horvli O, Fluge O, Lillehaug J, Varhaug JE. Expression of N-acetyl transferase human and human Arrest defective 1 proteins in thyroid neoplasms. Thyroid. 2005;15:1131–1136. doi: 10.1089/thy.2005.15.1131. [DOI] [PubMed] [Google Scholar]
- 49.Ren T, Jiang B, Jin G, Li J, Dong B, Zhang J, Meng L, Wu J, Shou C. Generation of novel monoclonal antibodies and their application for detecting ARD1 expression in colorectal cancer. Cancer Lett. 2008;264:83–92. doi: 10.1016/j.canlet.2008.01.028. [DOI] [PubMed] [Google Scholar]
- 50.Loupart ML, Adams S, Armour JA, Walker R, Brammar W, Varley J. Loss of heterozygosity on the X chromosome in human breast cancer. Genes Chromosomes Cancer. 1995;13:229–238. doi: 10.1002/gcc.2870130402. [DOI] [PubMed] [Google Scholar]
- 51.Kersemaekers AM, Kenter GG, Hermans J, Fleuren GJ, van de Vijver MJ. Allelic loss and prognosis in carcinoma of the uterine cervix. Int. J. Cancer. 1998;79:411–417. doi: 10.1002/(sici)1097-0215(19980821)79:4<411::aid-ijc17>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 52.Girard L, Zöchbauer-Muller S, Virmani AK, Gazdar AF, Minna JD. Genome-wide allelotyping of lung cancer identifies new regions of allelic loss, differences between small cell lung cancer and non-small cell lung cancer, and loci clustering. Cancer Res. 2000;60:4894–4906. [PubMed] [Google Scholar]
- 53.Tribioli C, Mancini M, Plassart E, Bione S, Rivella S, Sala C, Torri G, Toniolo D. Isolation of new genes in distal Xq28: Transcriptional map and identification of a human homologue of the ARD1 N-acetyl transferase of Saccharomyces cerevisiae. Hum. Mol. Genet. 1994;3:1061–1067. doi: 10.1093/hmg/3.7.1061. [DOI] [PubMed] [Google Scholar]
- 54.Shen XJ, Ali-Fehmi R, Weng CR, Sarkar FH, Grignon D, Liao DJ. Loss of heterozygosity and microsatellite instability at the Xq28 and the A/G heterozygosity of the QM gene are associated with ovarian cancer. Cancer Biol. Ther. 2006;5:523–528. doi: 10.4161/cbt.5.5.2610. [DOI] [PubMed] [Google Scholar]
- 55.Edelmann J, Richter K, Hänel C, Hering S, Horn LC. X chromosomal and autosomal loss of heterozygosity and microsatellite instability in human cervical carcinoma. Int. J. Gynecol. Cancer. 2006;16:1248–1253. doi: 10.1111/j.1525-1438.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- 56.Chambers AF. MDA-MB-435 and M14 cell lines: Identical but not M14 melanoma? Cancer Res. 2009;69:5292–5293. doi: 10.1158/0008-5472.CAN-09-1528. [DOI] [PubMed] [Google Scholar]
- 57.Sellappan S, Grijalva R, Zhou X, Yang W, Eli MB, Mills GB, Yu D. Lineage infidelity of MDA-MB-435 cells: Expression of melanocyte proteins in a breast cancer cell line. Cancer Res. 2004;64:3479–3485. doi: 10.1158/0008-5472.CAN-3299-2. [DOI] [PubMed] [Google Scholar]
- 58.Zou Y, Peng H, Zhou B, Wen Y, Wang SC, Tsai EM, Hung MC. Systemic tumor suppression by the proapoptotic gene bik. Cancer Res. 2002;62:8–12. [PubMed] [Google Scholar]
- 59.Ma C, Wang J, Luo J. Exposure to asphalt fumes activates activator protein-1 through the phosphatidylinositol 3-kinase/Akt signaling pathway in mouse epidermal cells. J. Biol. Chem. 2003;278:44265–44272. doi: 10.1074/jbc.M309023200. [DOI] [PubMed] [Google Scholar]
- 60.Chang JY, Xia W, Shao R, Sorgi F, Hortobagyi GN, Huang L, Hung MC. The tumor suppression activity of E1A in HER-2/neu-overexpressing breast cancer. Oncogene. 1997;14:561–568. doi: 10.1038/sj.onc.1200861. [DOI] [PubMed] [Google Scholar]
- 61.Yaguchi S, Fukui Y, Koshimizu I, Yoshimi H, Matsuno T, Gouda H, Hirono S, Yamazaki K, Yamori T. Antitumor activity of ZSTK474, a new phosphatidylinositol 3-kinase inhibitor. J. Natl. Cancer Inst. 2006;98:545–556. doi: 10.1093/jnci/djj133. [DOI] [PubMed] [Google Scholar]
- 62.Lee L, Sudentas P, Donohue B, Asrican K, Worku A, Walker V, Sun Y, Schmidt K, Albert MS, El-Hashemite N, Lader AS, Onda H, Zhang H, Kwiatkowski DJ, Dabora SL. Efficacy of a rapamycin analog (CCI-779) and IFN-γ in tuberous sclerosis mouse models. Genes Chromosomes Cancer. 2005;42:213–227. doi: 10.1002/gcc.20118. [DOI] [PubMed] [Google Scholar]
- 63.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, Kobayashi R, Hung MC. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 64.Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, He X, Hung JY, Lai CC, Ding Q, Su JL, Yang JY, Sahin AA, Hortobagyi GN, Tsai FJ, Tsai CH, Hung MC. IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell. 2007;130:440–455. doi: 10.1016/j.cell.2007.05.058. [DOI] [PubMed] [Google Scholar]
- 65.Singh A, Misra V, Thimmulappa RK, Lee H, Ames S, Hoque MO, Herman JG, Baylin SB, Sidransky D, Gabrielson E, Brock MV, Biswal S. Dysfunctional KEAP1–NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:e420. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Acknowledgments: We thank D. J. Kwiatkowski, J. Blenis, and K.-L. Guan for providing the cells and constructs; S. Zhang, J. Shi, B. Spohn, J.-F. Lee, Z. Han, H.-W. Yeh, T.-L. Liao, Y.-H. Chang, and K. Dunner Jr., for their technical support; The University of Texas M. D. Anderson Cancer Center (MDACC) Institutional Core Grant CA16672 High Resolution Electron Microscopy facility for electron microscopy analysis; The University of Texas MDACC DNA Core Facility for fluorescent DNA fragment analysis; and the Department of Scientific Publications at M. D. Anderson for editing this manuscript. This work was partially supported by NIH grants R01 CA109311, CCSG CA16672, and P01 CA099031; M. D. Anderson Specialized Programs of Research Excellence grants (P50 CA116199 for breast Cancer and P50 CA83639 for ovarian cancer), MDACC–China Medical University and Hospital Sister Institution Fund, Patel Memorial Breast Cancer Endowment Fund, and grants from the Kadoorie Charitable Foundations, the National Breast Cancer Foundation, Inc., and Taiwan National Science Council (NSC-96-3111-B) to M.-C.H.; a grant R01 CA126832 to Y.Z.; a predoctoral fellowship from the U. S. Army Breast Cancer Research Program (grant W81XWH-08-1-0397), and the Andrew Sowell–Wade Huggins Scholarship from the University of Texas Graduate School of Biomedical Sciences at Houston to H.-P.K.; a predoctoral fellowship from the U.S. Army Breast Cancer Research Program (grant W81XWH-05-1-0252) and the T. C. Hsu Endowed Memorial Scholarship, Andrew Sowell-Wade Huggins Scholarship and Presidents’ Research Scholarship from the University of Texas Graduate School of Biomedical Sciences at Houston to D.-F.L.; a predoctoral fellowship from the U. S. Army Breast Cancer Research Program (grant W81XWH-06-1-0709) to C.-K.C.; and a research assistant scholarship from the University of Texas Graduate School of Biomedical Sciences at Houston to M.L.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.