Summary
Hepatitis B viral infection remains a serious global health problem despite the availability of a highly effective vaccine. Approximately 5% of HBV-infected adults develop chronic hepatitis B, which may result in liver cirrhosis or hepatocellular carcinoma. Variants of interleukin-10 (IL10) have been previously associated with chronic hepatitis B infection and progression to hepatocellular carcinoma. Single nucleotide polymorphisms (SNPs, n = 42) from the IL10, IL19, and IL20 gene regions were examined for an association with HBV infection outcome, either chronic or recovered, in a nested case-control study of African Americans and European Americans. Among African Americans, three nominally statistically significant SNP associations in IL10, two in IL20, and one haplotype association were observed with different HBV infection outcomes (P = 0.005–0.04). The SNP, rs1518108, in IL20 nominally deviated significantly from Hardy-Weinberg equilibrium in African Americans, with a large excess of heterozygotes in chronic HBV-infected cases (P = 0.0006), which suggests a strong genetic effect. Among European Americans, a nominally statistically significant SNP association in IL20, as well as an IL20 haplotype were associated with HBV recovery (P = 0.01–0.04). These results suggest that IL10 and IL20 gene variants influence HBV infection outcome and encourage the pursuit of further studies of these cytokines in HBV pathogenesis.
Keywords: Interleukin-10, Inflammation, African American, Immunogenetics, Hepatitis b, HIV co-infection
Introduction
Worldwide, 350 million people have chronic hepatitis B virus (HBV) infection; approximately 1.25 million of these people live in the United States of America (WHO, 2000). Approximately 5% of all hepatitis B infected adults will develop chronic hepatitis B (WHO, 2000). Nearly 5,000 people in the United States die each year from hepatitis B-related complications like liver cirrhosis and hepatocellular carcinoma (HCC). Treatment of hepatitis B-related liver disease results in annual health care costs and lost wages equaling $700 million (CDC, 2002).
HBV is transmitted via contact with infected body fluids, including blood, saliva, and semen. In high endemicity areas, including much of Asia and most of sub-Saharan Africa, HBV is usually acquired perinatally or during childhood, however in low endemicity regions, such as the United States and Europe, the virus is usually contracted in adulthood via high-risk sexual behaviors or percutaneously through injection drug use. Co-infection of HBV and the human immunodeficiency virus (HIV) is common given both viruses share similar routes of transmission in adults (Rodriguez-Mendez et al., 2000; Salmon-Ceron et al., 2005).
The natural history of chronic HBV infection is altered by HIV co-infection, with significantly more liver-related deaths among HIV-infected individuals (Thio et al., 2002; Konopnicki et al., 2005; Yachimski & Chung, 2005). While highly active anti-retroviral therapy (HAART) has improved the survival of HIV-infected people, unfortunately, many of those co-infected with HBV now go on to develop end-stage liver disease (Palella et al., 1998).
The normal cellular immune response of an HBV infection leads to liver damage that may result in cirrhosis and HCC (Lai et al., 2003). The pro-inflammatory T-helper 1 (Th1) and anti-inflammatory T-helper 2 (Th2) cells regulate this cellular immune response. Interleukin-10 (IL-10) is one of the critical modulators of this balance by suppressing the host Th1 immune response. IL-10 is produced by Th2 cells and inhibits expression of other pro-inflammatory cytokines such as IFN-gamma, IL-2 and TNF-alpha in Th1 cells (Pestka et al., 2004). IL-10 influences the natural history of HBV infection and other viral diseases such as Epstein-Barr, herpes zoster, HIV, and hepatitis C (Shin et al., 2000; Vicari & Trinchieri, 2004; Oleksyk et al., 2005).
Polymorphisms in the IL10 proximal promoter region affect production of IL-10 (Crawley et al., 1999; Edwards-Smith et al., 1999) and have been examined extensively for associations with HBV infection and progression. Among populations throughout the world there are three classic promoter haplotypes, GCC, ACC, and ATA (Turner et al., 1997). The GCC haplotype produces high levels of IL-10, ACC medium, and ATA low (Crawley et al., 1999; Edwards-Smith et al., 1999). The three proximal promoter SNPs and haplotypes have been associated with hepatitis B infection outcomes in Chinese (Zhu et al., 2005; Peng et al., 2006), Korean (Shin et al., 2003; Cheong et al., 2006), and Japanese (Miyazoe et al., 2002; Migita et al., 2005) populations. These reports suggest an important role for IL10 variation in HBV infection outcome, which we sought to examine further. Additionally, the gene paralogs, IL19 and IL20, located in the 100 kb 1q31-q32 region telomeric to IL10 (Pestka et al., 2004), have been recently identified as having early pro-inflammatory cytokine and immunoregulatory activities (Liao et al., 2002; Parrish-Novak et al., 2002; Wolk et al., 2002).
We examined 42 single nucleotide polymorphisms (SNPs) located in the IL10, IL19, and IL20 genes in European Americans and African Americans for host genetic differences between individuals with chronic hepatitis B infection or serologic evidence of recovery. Host genetic variation was evaluated by differences in allele, genotype, and haplotype frequencies using a nested case-control study design.
Materials and Methods
Patients
The null hypothesis that host genetic differences did not exist between chronic hepatitis B infected and recovery individuals was tested in a nested case-control study among African Americans (AA) and European Americans (EA). Individuals selected for this study were participants in several HIV infection and progression cohorts: AIDS Link to the Intravenous Experience (ALIVE, number of AA (nAA) = 93, number of EA (nEA) = 3), Multicenter Hemophilia Cohort Study (MHCS, nAA = 7, nEA = 36), Hemophilia Growth and Development Study (HGDS, nAA = 7, nEA = 55), and the Multicenter AIDS Cohort Study (MACS, nAA = 14, nEA = 398) for HIV-1/AIDS (demograpic characteristics are described in Karacki et al., 2004). Cases had chronic hepatitis B, which was defined by testing positive for HBsAg at two visits separated by a minimum of six months. Controls were individuals who spontaneously recovered without treatment and had serologic evidence of prior infection (antibodies against hepatitis B core antigen anti-HBc, and against HBsAg, anti-HBs) with HBsAg undetectable at two time points separated by a minimum of six months (Lok & McMahon, 2004). Each chronic hepatitis B case was matched with at least one, and when possible, two recovery controls for age, gender, race, and HIV status, the demographic factors known to be associated with HBV infection outcome (Karacki et al., 2004). A total of 121 African Americans (45 chronic cases, 76 recovery controls) and 492 European Americans (179 chronic cases, 313 recovery controls) were studied. The majority of subjects were HIV infected (68.5% in European Americans, 76.9% in African Americans), which was one of the very significant factors matched between cases and controls.
This study was conducted following the principles of the Helsinki Declaration of 1975 (1996). Study participants provided written informed consent to be included in the parent cohorts, which were approved by their respective Institutional Review Boards.
Study design
We examined SNPs located in and around the IL10, IL19, and IL20 genes for differences in allele, genotype, and haplotype frequencies for African Americans and European Americans. A total of 42 SNPs were chosen for genotyping, 25 from IL10, 10 from IL19, and seven from IL20. The 25 SNPs genotyped in IL10 resulted in saturated coverage of the gene. For the IL19 and IL20 genes, haplotype tagging or functionally important SNPs were chosen. SNPs were genotyped using multiplexed length-modified single-base extension and 5′ exonuclease assays (Oleksyk et al., 2005). The forward and reverse primers, as well as the probe sequences for each SNP are reported in Supplementary Table 1. SDS v2.2 software (Applied Biosystems, Foster City, CA) was used to call genotypes generated with the 5’ exonuclease assays. GeneScan v3.5 and Genotyper v3.6 software programs (Applied Biosystems, Foster City, CA) were used to call genotypes from the single-base extension assays. Both standard Qiagen (Hilden, Germany) and phenol-chloroform extraction methods were used to isolate DNA from EBV transformed B-cell lines established from each individual (Dean et al., 1994).
Given the departure from Hardy-Weinberg equilibrium (HWE) observed at rs1518108 in the IL20 gene (Fig. 1), genotypes were confirmed by sequencing all 45 African American cases and 10 controls. The forward primer TCAAGTCCTTATCTTTGTGTCCAA and reverse primer TGTTGGGCAGCTGTTACTTG were used for sequencing with Big Dye v.1.1 terminator kit and the AB3730 sequencer (Applied Biosystems, Foster City, CA). Sequencher v.4.5 software (Gene Codes, Ann Arbor, MI) was used to align the sequences and to validate the genotyping results.
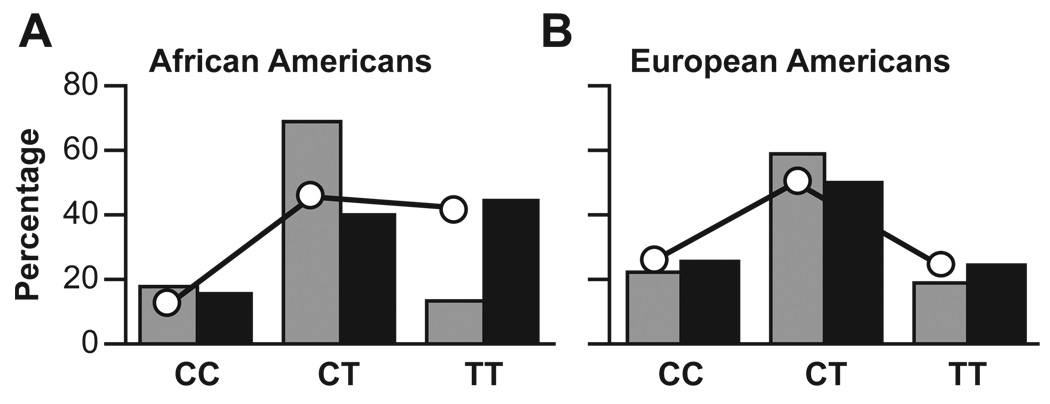
Fig. 1.
Deviations from Hardy-Weinberg equilibrium (HWE) for rs1518108. Chronic HBV-infected cases are associated with a SNP in IL20, rs1518108, which deviates from HWE in African Americans. The genotypes CC, CT, and TT are shown for chronic cases (grey bar) and recovery controls (black bars) in A) African Americans and B) European Americans. The open circles represent genotypic frequencies expected by HWE based on recovery control allele frequencies. The observed genotypic frequencies for African American chronic cases deviate from HWE, with less TT and more CT genotypes (χ2 = 14.8, d.f. = 2, P = 0.0006). Deviations from HWE remained in African Americans even after utilizing a different set of control allele frequencies (C = 44%, T = 56%) from 1,480 African Americans (χ2 = 8.148, d.f. = 2, P = 0.017). Support for a similar association in Europeans is suggestive but not statistically significant (χ2 = 5.83, d.f. = 2, P = 0.054).
Statistical analyses
The statistical package SAS v9.1 and SAS/Genetics (SAS Institute Inc., Cary, NC) was utilized for the statistical analysis. The P values reported throughout this paper are nominal (uncorrected for multiple testing). African Americans and European Americans were analyzed separately for allele, genotype, and haplotype associations. Seven loci in IL10 (rs3024510, rs3024506, rs5743625, rs3024489, rs1800895, rs5743623, and rs7349077) had a minor allele frequency of less than 5% in cases and controls among both racial populations and were not considered in further SNP or haplotype analyses. Chi-square tests (χ2) were performed to determine whether an association existed for alleles and genotypes. If a nominally significant allelic or genotypic association was present (P < 0.05) or suggestive (P < 0.10) at a locus, then conditional logistic regression (CLR) was performed. The most frequent homozygote allele in the control group was chosen as the referent, which allowed for separate evaluation of heterozygotes and rare allele homozygotes. A sub-analysis of individuals stratified by HIV status was also performed to examine whether the effect of any associations between the two groups differed.
Haplotypes were characterized based on SNPs in the two regions of linkage disequilibrium present around IL10, IL19, and IL20. One LD block contains the IL10 gene and the other both IL19 and IL20 (Oleksyk et al., 2005). A haplotype trend regression (HTR) model was used to assess associations between haplotypes and HBV recovery (Shrestha et al., 2006). Briefly, haplotype frequencies were calculated first, using the expectation-maximum (EM) algorithm methods (Excoffier & Slatkin, 1995) modeled after the SNPHAP software program (Clayton). Statistical methods were then used to estimate a posterior probability matrix of values based on all possible haplotypes for each individual, given their genotypes at each locus (Schaid et al., 2002; Zaykin et al., 2002) that were then included in a conditional logistic regression analysis (Breslow & Day, 1980). A stepwise regression approach identified the most parsimonious model. Rare haplotypes, with frequencies < 3% were combined into one haplotype. Odds ratios (OR), 95% confidence intervals (CI), and probabilities were calculated for haplotypes with P < 0.10.
Results
Allele and genotype frequencies as well as p-values for the allelic and genotypic χ2 tests for chronic HBV-infected cases and recovery controls for all 35 SNP loci are shown for African Americans in Supplementary Table 2A and for European Americans in Supplementary Table 2B. The CLR results for SNPs with P < 0.05 from either the allelic or genotypic χ2 analysis are shown in Table 1.
Table 1.
Allelic and genotypic associations of single IL10 and IL20 SNPs with chronic hepatitis B
| SNPs | Frequency n (%) | χ2 | CLR | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sample | Gene | NCBI ID | Position | Genotype | Cases | Controls | Allelic P | Genotypic P | OR (95% CI) | P |
| African American | ||||||||||
| IL10 | rs1800893 | Promoter −1353 |
GG | 12(27) | 35(46) | 0.04 | 0.10 | 1+ | - | |
| AG | 26(58) | 34(45) | 2.03 (0.78–5.31) | 0.15 | ||||||
| AA | 7(15) | 7(9) | 3.02 (0.64–14.15) | 0.16 | ||||||
| rs1800896 | Promoter −1082 |
AA | 15(33) | 38(50) | 0.01 | 0.02 | 1 | - | ||
| AG | 24(53) | 32(42) | 1.56 (0.69–3.55) | 0.29 | ||||||
| GG | 6(13) | 6(8) | 2.35 (0.59–9.41) | 0.23 | ||||||
| rs1518110 | Intronic +954 |
GG | 16(48) | 22(35) | 0.04 | 0.08 | 1 | - | ||
| GT | 16(48) | 29(46) | 0.81 (0.32–2.06) | 0.66 | ||||||
| TT | 1(3) | 12(19) | 0.11 (0.01–0.93) | 0.04 | ||||||
| IL20 | rs1400986 | Intergenic 1807 |
CC | 8(18) | 25(33) | 0.76 | 0.03 | 1 | - | |
| CT | 16(36) | 45(59) | 0.34 (0.10–1.13) | 0.08 | ||||||
| TT | 21(47) | 6(8) | 0.63 (0.19–2.15) | 0.46 | ||||||
| rs1518108 | Intergenic 6296 |
TT | 6(13) | 34(45) | 0.01 | 0.001 0.03 | 1 | - | ||
| CT | 31(69) | 30(39) | 7.58 (2.14–26.89) | 0.0017 | ||||||
| CC | 8(18) | 12(16) | 5.45 (1.31–22.77) | 0.02 | ||||||
| European American | ||||||||||
| IL20 | rs1400986 | Intergenic 1807 |
TT | 124(69) | 196(63) | 0.04 | 1 | - | ||
| CT | 52(29) | 97(31) | 0.87 (0.57–1.31) | 0.49 | ||||||
| CC | 3(2) | 20(6) | 0.23 (0.07–0.80) | 0.02 | ||||||
| rs3024517 | Intronic 3374 |
GG | 125(70) | 200(64) | 0.03 | 0.02 | 1 | - | ||
| AG | 52(29) | 93(30) | 0.90 (0.60–1.35) | 0.60 | ||||||
| AA | 2(1) | 20(6) | 0.16 (0.04–0.70) | 0.01 | ||||||
The referent group is the most common homozygote among the recovery controls. SNP locations given for IL10 are derived from the start position of the first exon and match previous reports (Turner et al., 1997; Gibson et al., 2001). SNP locations for IL20 are from AF402002.1 at NCBI, which contains the complete gene sequence.
Among African Americans, a χ2 analysis revealed three nominally statistically significant allelic associations with HBV infection outcome in the IL10 region (rs1800893, P = 0.04; rs1800896, P = 0.01; and rs1518110, P = 0.04) and one in the IL20 region (rs1518108, P = 0.01). There were three nominally statistically significant genotypic associations for African Americans; one at IL10 (rs1800896, P = 0.02) and two at IL20 (rs1400986, P = 0.03, and rs1518108, P = 0.002). Among European Americans, rs3024517 in IL20 was nominally statistically significantly associated for alleles and genotypes (P = 0.03, P = 0.02, respectively). This SNP was found to be in strong LD (r2= 0.974, D′= 0.993) with rs1400986, which also had nominally statistically significant allelic and genotypic associations, (P = 0.04 and P = 0.03, respectively).
Odds ratios (OR) and 95% confidence intervals (CI) were computed using CLR for SNPs with P ≤ 0.05 for either the allelic or genotypic χ2 tests (Table 1). Three genotypic associations were observed in African Americans and two genotypic associations in European Americans with HBV infection outcome. African American recovery controls were more likely to have the TT genotype compared to the GG referent genotype at rs1518110 with a protective ORTT = 0.11 (0.01–0.93), P = 0.04. Chronic HBV infected cases were more likely to have the CT or CC genotypes at rs1518108 in IL20 compared to the referent TT with ORCT = 7.58 (2.14–26.89), P = 0.0017 and ORCC = 5.45 (1.31–22.77), P = 0.02. Among European Americans, recovery controls were more likely to have the CC genotype at rs1400986 and the AA genotype at rs3024517 due to strong LD, compared to the referent genotypes, TT and GG. The protective odds ratios were ORCC = 0.23 (0.07–0.80), P = 0.02 for rs1400986 and ORAA = 0.16 (0.04–0.70), P = 0.01 for rs3024517. These associations were examined further in an analysis stratified by HIV status. The direction of the associations reported above among HIV positive and negative European Americans, as well as HIV positive African Americans were consistent with those reported above (analysis not shown, and the African American HIV negative sample had insufficient numbers for examination).
All 35 loci genotyped were tested for HWE. African American cases deviated from HWE at one locus, rs1518108 in IL20, based on allele frequencies from African American controls (Fig. 1, P=0.0006). As noted above, genotypes at this locus have a strong association with susceptibility to chronic hepatitis B infection in African Americans (P = 0.0017). Genotypes for rs1518108 were confirmed by two independent methods, both genotyping and sequencing, and using multiple replications.
Haplotypes were analyzed in the two blocks of LD that encompass IL10 and the IL19/IL20 combination (Oleksyk et al., 2005) for an association with hepatitis B infection outcome. The LD block containing IL10 was analyzed using the proximal promoter SNPs (Turner et al., 1997), the distal promoter SNPs (Gibson et al., 2001), and the SNPs that covered the length of the gene (Oleksyk et al., 2005). In the IL19/IL20 block, haplotypes for each individual gene as well as both genes combined were tested separately.
Associations were evident in the IL20 gene among African Americans and European Americans (Table 2). Haplotype results for all three genes are shown in Tables 2 and 3. Differences in haplotype frequencies existed between cases and controls in the IL20 region, which were reflected with a global P = 0.005 for African Americans and P = 0.036 for European Americans. In African Americans, the IL20 haplotype GTGTC was susceptible with OR = 14.41 (2.23 – 93.28), P = 0.005. In European Americans, the IL20 haplotype GCATT was protective with OR = 0.38 (.15 – .97), P = 0.04.
Table 2.
Haplotype Trend Regression analysis for SNPs in IL19 and IL20
| African Americans |
European Americans |
|||||
|---|---|---|---|---|---|---|
| Gene | Frequency (%) | Frequency (%) | ||||
| Haplotype | Cases | Controls | Global P | Cases | Controls | Global P |
| IL20 | 0.005 | 0.036 | ||||
| GTGCT | 12.8 | 24.3 | ||||
| GTGTC1 | 35.0 | 18.1 | 35.7 | 37.0 | ||
| GTGTT | 9.5 | 16.3 | 34.9 | 28.6 | ||
| GCGTT | 4.3 | 15.9 | ||||
| GCGTC | 8.1 | 9.8 | ||||
| GCATT2 | 7.8 | 6.2 | 8.3 | 13.6 | ||
| GCATC | 7.2 | 4.6 | 7.0 | 7.6 | ||
| CTGTT | 5.8 | 0.0 | 5.3 | 6.4 | ||
| GCGCT3 | 6.1 | 0.0 | ||||
| GTGCC | 3.1 | 0.0 | ||||
| CTGTC | 8.6 | 6.8 | ||||
| IL19 | > 0.10 | > 0.10 | ||||
| ACC | 57.1 | 57.5 | 75.9 | 75.8 | ||
| TCT | 10.8 | 15.6 | 7.8 | 8.1 | ||
| TCC | 13.6 | 15.3 | ||||
| ATC | 9.2 | 6.8 | ||||
| ATT | 6.4 | 3.1 | 13.5 | 12.4 | ||
| IL19/IL20 | 0.053 | 0.055 | ||||
| ACCGTGCT | 9.3 | 21.2 | ||||
| ACCGTGTT | 12.4 | 15.2 | 31.0 | 26.5 | ||
| ACCGTGTC4 | 19.9 | 12.6 | 31.8 | 30.9 | ||
| TCCGCGTT | 0.0 | 11.5 | ||||
| TCTGTGTC | 6.9 | 7.5 | 3.3 | 6.7 | ||
| TCTGTGCT | 4.1 | 7.2 | ||||
| TCCGCGTC | 13.0 | 5.5 | ||||
| ACCGCGTC | 0.0 | 5.4 | ||||
| ACCGCATT5 | 5.5 | 5.2 | 8.5 | 14.0 | ||
| ACCGCATC | 5.2 | 4.6 | 7.7 | 7.7 | ||
| ACCGCACT | 4.7 | 0.0 | ||||
| ATCGCGTT | 3.8 | 0.0 | ||||
| ATTCTGTT | 3.1 | 0.0 | 5.1 | 5.9 | ||
| TCTGTGTT | 4.9 | 0.0 | ||||
| ATTCTGTC | 0.0 | 6.6 | ||||
GTGTC : OR = 14.41 [2.23 – 93.28], P = 0.005 for African Americans
GCATT : OR = .38 [.15 – .97], P = 0.04 for European Americans
GCGCT : OR = 51.16 [0.88 – 2977.64], P = 0.06 for African Americans
ACCGTGTC : OR = 6.13 [.90 – 41.83], P = 0.06 for African Americans
ACCGCATT : OR = .41 [.16 – 1.05], P = 0.06 for European Americans
The SNPs typed in the IL19 and IL20 genes were analyzed for haplotype associations using the haplotype trend regression method (Shrestha et al., 2006). For each haplotype with P (χ2) < .10, an odds ratio (OR), 95% confidence ratio (CI), and CLR P value was determined. The global P value is based on the likelihood ratio test between cases and controls. IL20 haplotypes include the ordered SNPs: rs1713239, rs1400986, rs3024517, rs1109461, and rs1518108. Two SNPs in IL20, rs2981573 and rs2232360, were not included because of some missing genotypes between essential cases and matched controls. IL19 haplotypes include the ordered SNPs: rs2243168, rs2243176, and rs2243191. The SNPs comprising the IL19 haplotypes were chosen because they contained the most complete set of data for the most individuals and may not fully represent the biological impact IL19 given the presence of other variants which were not included in this analysis. The combined IL19/IL20 extended haplotype analysis includes the ordered SNPs: rs2243168, rs2243176, rs2243191, rs1713239, rs1400986, rs3024517, rs1109461, and rs1518108. Nucleotide substitutions and positions for SNP loci are presented in Supplemental Table 1.
Table 3.
Haplotype Trend Regression analysis for SNPs in IL10
| African Americans |
European Americans |
|||||
|---|---|---|---|---|---|---|
| Gene | Frequency (%) | Frequency (%) | ||||
| Haplotype | Cases | Controls | Global P | Cases | Controls | Global P |
| IL10 Proximal Promoter | > 0.10 | > 0.10 | ||||
| ATA | 34.4 | 44.0 | 22.3 | 24.0 | ||
| GCC | 40.0 | 28.9 | 45.6 | 45.0 | ||
| ACC | 25.5 | 27.0 | 32.0 | 31.0 | ||
| IL10 Distal Promoter | 0.057 | > 0.10 | ||||
| AGC1 | 48.8 | 62.4 | 60.8 | 59.3 | ||
| TAA | 20.9 | 13.7 | 18.1 | 18.2 | ||
| AGA | 5.0 | 8.1 | ||||
| AAA | 14.2 | 7.3 | ||||
| TGA | 11.0 | 6.5 | 14.0 | 12.5 | ||
| TAC | 3.1 | 0.0 | ||||
| TGC | 3.9 | 3.4 | ||||
| IL10 Gene | > 0.10 | > 0.10 | ||||
| AGCCGAATTGGTTIAGC | 24.2 | 39.1 | 18.6 | 19.6 | ||
| TAACAGCGCCGTCIGGC | 12.1 | 10.9 | 18.1 | 19.6 | ||
| AGCCGAATTGATTIAGC | 7.6 | 8.7 | ||||
| AGCCGACGCGGTCDAAT | 6.1 | 8.7 | ||||
| AAACGACGCGGTTIAGC | 4.5 | 6.5 | ||||
| AGCCAGCGCCGTCIAGC | 7.6 | 6.5 | ||||
| AGATAGCGCGGTTIAGC | 6.1 | 5.4 | ||||
| TAACAGCGCCGTCIAGC | 7.6 | 5.4 | ||||
| AGCCGACGCGGTTIAGC | 9.1 | 4.3 | 34.3 | 30.8 | ||
| TGACAGCGCCGTCIAGC | 6.1 | 4.3 | 13.7 | 14.0 | ||
| AAACAACGCGGTCIAGC | 6.1 | 0.0 | ||||
| TGACGACGCGGTCDAAT | 3.0 | 0.0 | ||||
| AGCCAGCGCCGCCIAGC | 6.4 | 8.9 | ||||
| TGCCAGCGCCGTCIAGC | 4.9 | 3.8 | ||||
| AGCCGAATCGGTTIAGC | 0.0 | 3.3 | ||||
| TACCAGCGCCGTCIGGC | 4.1 | 0.0 | ||||
AGC : OR = .28 [.07 – 1.12] P = 0.07 in African Americans
The distal and proximal promoter SNPs, a subset of the total variants typed in IL10, were analyzed for haplotype associations using the haplotype trend regression method (Shrestha et al., 2006). For each haplotype with P (χ2 ) < .10, an odds ratio (OR), 95% confidence ratio (CI), and CLR P value was determined. The global P value is based on the likelihood ratio test between cases and controls. Distal promoter haplotypes include the ordered SNPs: rs1800890, rs6703630, and rs6693899. Proximal promoter haplotypes include the ordered SNPs: −1082 (rs1800896), −819 (rs1800871), and −592 (rs1800872). The IL10 gene includes the ordered SNPs: rs1800890, rs6703630, rs6693899, rs5743624, rs1800893, rs1800896, rs1800872, rs1518110, rs1554286, rs1878672, rs3024494, rs3024509, rs3024496, rs9282739, rs3024498, rs6697497, and rs6687786. Nucleotide substitutions and positions for SNP loci are presented in Supplemental Table 1.
Discussion
Our study provides new insights into the pathogenesis of chronic hepatitis B in regards to the IL10, IL19 and IL20 candidate genes. There was evidence that HBV infection outcome in African Americans was influenced by SNP variants of the IL10 and IL20 genes, whereas recovery in European Americans was influenced solely by SNP variants in IL20. Our strongest findings were among variants of IL20, especially the SNP rs1518108, which had a very strong association in African Americans. Additionally, an IL20 haplotype in African Americans and a different IL20 haplotype in European Americans were associated with differences in HBV infection outcome. This is an important finding since IL20 variants have not been implicated with HBV pathogenesis before, although they have been associated with hepatitis C recovery in African Americans (Oleksyk et al., 2005).
The IL20 gene is a paralog of IL10 and belongs to the IL10 family of cytokine genes (Xu, 2004). Two highly correlated SNPs, rs1400986 and rs3024517 (r2 = 0.974), were associated with HBV recovery in European Americans (ORCC = 0.23 [0.07–0.80], P = 0.02 and ORAA = 0.16 [0.04–0.70], P = 0.01 respectively). In African Americans the association observed with rs1518108 is the most striking and significant (P = 0.0017). This intergenic SNP that lacks a known functional role and creates no obvious deleterious genetic change is positioned nearly 1Kb downstream from the IL20 polyadenylation site and 28 Kb upstream from IL24 in an area of moderate conservation. In European Americans, the minor allele (T) had a frequency of 49% in both cases and controls. However, in African Americans, the T allele was more common in recovery controls (64%) than chronic HBV-infected cases (48%, allelic P = 0.01). We saw a departure from HWE among African American cases (more CT and less TT genotypes than expected at rs1518108, P = 0.0006, Fig. 1). At this locus, chronic HBV-infected African American cases were more likely to have the CT genotype than were those who had recovered (who were more likely to have TT). The heterozygous disadvantage observed in African American cases could be due to very recent admixture between stratified populations. However, this is very unlikely because excess heterozygotes are not observed in controls at rs1518108, or at the other 34 loci examined in this study, or at additional loci examined among African Americans from the same cohort (Duggal et al., 2005; Oleksyk et al., 2005; Javanbakht et al., 2006; Shrestha et al., 2006). The TC genotype of rs1518108 has also been associated with reduced susceptibility to psoriasis in Estonians (Kingo et al., 2004). Taken together, these results suggest that heterozygotes at rs1518108 (or a haplotype it tags) are more susceptible to HBV infection in African Americans, and that polymorphisms at the locus have a role in other inflammatory disorders.
Several studies indicate that the role of IL10 variants, especially those in the promoter region, are important in HBV infection and progression to liver disease (Miyazoe et al., 2002; Shin et al., 2003; Zhu et al., 2005). We tested these promoter region variants of IL10 for an association with HBV infection outcome. A positive association was observed between the proximal promoter SNP IL10-1082, known for its effects on IL-10 production (Turner et al., 1997; Eskdale et al., 1998), among allelic (P = 0.01) and genotypic tests (P = 0.02) with chronic hepatitis B infection in African American cases. Associations with the other proximal promoter SNP, IL10-592, also involved in IL-10 production, were not observed in either population. Two additional SNPs in IL10, rs1800893 and rs1518110, were associated with HBV infection outcome in allelic tests (P = 0.04) among African Americans. The effect of IL10 polymorphisms on HBV infection outcome is consistent with the role of IL-10 as an immunomodulatory cytokine.
SNP associations alone often cannot represent the complexity of the variation among hosts. Rather, haplotype analysis offers more power and further insight when one studies multifactorial diseases such as HBV infection (de Bakker et al., 2005). Two blocks of LD are present in the genomic region examined: the IL10 gene and the IL19/IL20 genes. These LD blocks have been reported in our previous study using the same SNP markers (Oleksyk et al., 2005). The SNP, rs1518108 was found to be in weak LD (|D’| between 0.07 and .25) with other SNPs of IL20 in an Estonian population (Koks et al., 2004), and is located in an area with elevated rates of recombination based on HapMap data (Altshuler et al., 2005). We have included rs1518108 in the IL20 haplotype analysis based on the |D’| value of 0.72 we observed in African Americans with one of its neighboring SNPs in IL20, rs1109461.
Our study is the first to demonstrate an association between haplotypes of IL20 and susceptibility to chronic hepatitis B infection among African Americans and European Americans (Table 2, P = 0.005 – 0.04). Haplotypes of IL19 and IL20 have been associated with hepatitis C clearance (Oleksyk et al., 2005) and susceptibility to psoriasis (Kingo et al., 2004; Koks et al., 2004; Koks et al., 2005). IL20 may be a common link between these diseases. In transgenic mice, IL-20 over-expression causes a shiny skin appearance and neonatal lethality, possibly due to skin-barrier defects (Otkjaer et al., 2005). In humans, IL-20 stimulates immune cells in the skin effecting keratinocyte proliferation that can result in psoriasis (Stenderup et al., 2007). The observed altered epidermal differentiation and hyper-proliferation resembles human psoriatic abnormalities. In humans, IL-20 is known to induce production of IL-6 and TNF-alpha in monocytes; stimulate expression of keratinocyte growth factor, IL-6, TNF-alpha, and proto-oncogene tyrosine-protein kinase ROS in CD8+ T cells, and likely functions as an early pro-inflammatory cytokine (Xu, 2004). Recently, IL-19 and IL-20 were identified as modulators of the Th1/Th2 balance, likely acting through receptor subunits shared with IL-10 (Oral et al., 2006). Lastly, the emergence or reappearance of psoriasis is a common side effect of interferon-alpha treatment, a cornerstone of therapy for patients with chronic hepatitis infection (Seckin et al., 2004; Kartal et al., 2005; Ketikoglou et al., 2005).
There are some limitations to this study. We have examined a subset of the variation known to be present in the IL10, IL19 and IL20 gene regions. Although genotyping additional SNPs and other genetic variants could yield additional associations, our selection of SNPs was designed to encompass the major known haplotypes in the gene region. Another limitation is the lack of functional studies of the association between serum levels of cytokines and the genetic polymorphisms assayed in this specific group of patients. Also, about half of the patients examined are co-infected with HIV, which was taken into account in two ways. First, this is a nested case-control study where those with and without HBV recovery were matched based on HIV infection status. Second, stratified analyses that were undertaken based on HIV infection and race showed no difference in associations for rs1518110, rs1518180, rs3024517 and rs1400986. Third, HBV infection generally occurs before HIV infection; thus, HBV recovery or persistence is usually determined prior to HIV infection (Kingsley et al., 1990; Levine et al., 1996). The inclusion of samples from HIV positive individuals complicates interpretation of the study, but we believe the correction for the complication of HIV infection makes results reported valid albeit more difficult to interpret..
In our study we applied association tests to 35 different SNPs. Using the Bonferroni correction, our significance levels would have to be lowered from P < 0.05 to P < 0.0014 to identify positive associations with certainty when analyzing the two racial groups separately for each of the allelic, genotypic, and HWE tests (Bonferroni, 1935). Nominally the most statistically significant associations we observed were P = 0.0006 for genotypic deviations from HWE (Fig. 1) and P = 0.0017 for CLR of genotypes for the locus rs1518108 in African Americans (Table 1), essentially meeting these Bonferroni criteria. None of the nominally significant P-values meets the strictest criteria of Bonferroni correction for 6 times 35 tests (P < 0.00024), but this cut-off is too strict because the comparisons are not independent due to haplotype structure and the statistical tests are related by common data and hypotheses under evaluation. Some of the allelic and genotypic χ2 tests in Table 1 (0.05 < P < 0.0017) among both racial populations are also likely to be true statistically significant associations, but are confounded by a number of false discoveries (Supplementary Table 2A and 2B footnote) (Jung et al., 2005). Given the importance of IL-10 in regulating the immune response and its impact on the natural history of many diseases (Moore et al., 2001), along with the newly discovered roles of IL-19 and IL-20 in regulating the Th1/Th2 balance (Oral et al., 2006), exploring the associations we observed with rs1518108 as well as with SNPs with a P < 0.05 provides a reasonable approach to choosing targets for future research in samples from the same and different human racial and ethnic groups.
In summary, we found evidence to support the hypothesis that host genetic variability in IL10 and IL20 influence the outcome of acute HBV infection. Populations at higher risk of acquiring HBV should be the focus for replication of these results and furthering our understanding of the complex relationship between host genetics and HBV infection outcome.
Supplementary Material
Acknowledgements
We thank Yvette Berthier-Schaad, Randy Johnson, Bailey Kessing, Michael Malasky, Mary McNally, Kai Zhou, Andrea Smith, Nicole Crumpler, Melissa Levasseur, Andrea Atkinson, and Shanise Hill from the Laboratory of Genomic Diversity of the National Cancer Institute at Frederick for help with carrying out this project. For help in preparing this manuscript for publication we also thank Allen Kane, Carolyn Whistler, and Marrita Grau of the Scientific Publications, Graphics and Media at SAIC-Frederick, Inc. We would also like to thank Abbott Laboratories for supplying kits to perform serologic HBV testing. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. CT is supported in part by the Investigators in Pathogenesis of Infectious Disease Award from the Burroughs-Wellcome Fund. DT is supported in part by R01 DA13324.
Contributor Information
Ann L. Truelove, Email: truelovea@ncifcrf.gov.
Taras K. Oleksyk, Email: oleksyk@ncifcrf.gov.
Sadeep Shrestha, Email: sshresth@uab.edu.
Chloe L. Thio, Email: cthio@jhmi.edu.
James J. Goedert, Email: goedertj@mail.nih.gov.
Sharyne M. Donfield, Email: sdonfield@rhoworld.com.
Gregory D. Kirk, Email: gkirk@jhsph.edu.
David L. Thomas, Email: dthomas@jhmi.edu.
Stephen J. O’Brien, Email: obrien@ncifcrf.gov.
Michael W. Smith, Email: smithm@ncifcrf.gov.
References
- Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonferroni CE. Il calcolo delle assicurazioni su gruppi di teste". Studi in Onore del Professore Salvatore Ortu Carboni. 1935:13–60. [Google Scholar]
- Breslow NE, Day NE. Statistical methods in cancer research. Volume I - The analysis of case-control studies. IARC Sci Publ. 1980:5–338. [PubMed] [Google Scholar]
- CDC. Viral Hepatitis and Injection Drug Users; Fact Sheet Series. 2002 [Google Scholar]
- Cheong JY, Cho SW, Hwang IL, Yoon SK, Lee JH, Park CS, Lee JE, Hahm KB, Kim JH. Association between chronic hepatitis B virus infection and interleukin-10, tumor necrosis factor-alpha gene promoter polymorphisms. J. Gastroenterol. Hepatol. 2006;21:1163–1169. doi: 10.1111/j.1440-1746.2006.04304.x. [DOI] [PubMed] [Google Scholar]
- Clayton D. SNPHAP. ( http://www-gene.cimr.cam.ac.uk/clayton/software/)
- Crawley E, Kay R, Sillibourne J, Patel P, Hutchinson I, Woo P. Polymorphic haplotypes of the interleukin-10 5' flanking region determine variable interleukin-10 transcription and are associated with particular phenotypes of juvenile rheumatoid arthritis. Arthritis Rheum. 1999;42:1101–1108. doi: 10.1002/1529-0131(199906)42:6<1101::AID-ANR6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Dean M, Stephens JC, Winkler C, Lomb DA, Ramsburg M, Boaze R, Stewart C, Charbonneau L, Goldman D, Albaugh BJ, et al. Polymorphic admixture typing in human ethnic populations. Am J Hum Genet. 1994;55:788–808. [PMC free article] [PubMed] [Google Scholar]
- Duggal P, Winkler CA, An P, Yu XF, Farzadegan H, O'Brien SJ, Beaty TH, Vlahov D. The effect of RANTES chemokine genetic variants on early HIV-1 plasma RNA among African American injection drug users. J. Acquir. Immune Defic. Syndr. 2005;38:584–589. doi: 10.1097/01.qai.0000134741.49208.03. [DOI] [PubMed] [Google Scholar]
- Edwards-Smith CJ, Jonsson JR, Purdie DM, Bansal A, Shorthouse C, Powell EE. Interleukin-10 promoter polymorphism predicts initial response of chronic hepatitis C to interferon alfa. Hepatology. 1999;30:526–530. doi: 10.1002/hep.510300207. [DOI] [PubMed] [Google Scholar]
- Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW. Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci U S A. 1998;95:9465–9470. doi: 10.1073/pnas.95.16.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Slatkin M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol Biol Evol. 1995;12:921–927. doi: 10.1093/oxfordjournals.molbev.a040269. [DOI] [PubMed] [Google Scholar]
- Gibson AW, Edberg JC, Wu J, Westendorp RG, Huizinga TW, Kimberly RP. Novel single nucleotide polymorphisms in the distal IL-10 promoter affect IL-10 production and enhance the risk of systemic lupus erythematosus. J Immunol. 2001;166:3915–3922. doi: 10.4049/jimmunol.166.6.3915. [DOI] [PubMed] [Google Scholar]
- Javanbakht H, An P, Gold B, Petersen DC, O'Huigin C, Nelson GW, O'Brien SJ, Kirk GD, Detels R, Buchbinder S, Donfield S, Shulenin S, Song B, Perron MJ, Stremlau M, Sodroski J, Dean M, Winkler C. Effects of human TRIM5alpha polymorphisms on antiretroviral function and susceptibility to human immunodeficiency virus infection. Virology. 2006;354:15–27. doi: 10.1016/j.virol.2006.06.031. [DOI] [PubMed] [Google Scholar]
- Jung SH, Bang H, Young S. Sample size calculation for multiple testing in microarray data analysis. Biostatistics. 2005;6:157–169. doi: 10.1093/biostatistics/kxh026. [DOI] [PubMed] [Google Scholar]
- Karacki PS, Gao X, Thio CL, Thomas DL, Goedert JJ, Vlahov D, Kaslow RA, Strathdee S, Hilgartner MW, O'Brien SJ, Carrington M. MICA and recovery from hepatitis C virus and hepatitis B virus infections. Genes Immun. 2004;5:261–266. doi: 10.1038/sj.gene.6364065. [DOI] [PubMed] [Google Scholar]
- Kartal ED, Colak H, Ozgunes I, Usluer G. Exacerbation of psoriasis due to peginterferon alpha-2b plus ribavirin treatment of chronic active hepatitis C. Chemotherapy. 2005;51:167–169. doi: 10.1159/000085626. [DOI] [PubMed] [Google Scholar]
- Ketikoglou I, Karatapanis S, Elefsiniotis I, Kafiri G, Moulakakis A. Extensive psoriasis induced by pegylated interferon alpha-2b treatment for chronic hepatitis B. Eur J Dermatol. 2005;15:107–109. [PubMed] [Google Scholar]
- Kingo K, Koks S, Nikopensius T, Silm H, Vasar E. Polymorphisms in the interleukin-20 gene: relationships to plaque-type psoriasis. Genes Immun. 2004;5:117–121. doi: 10.1038/sj.gene.6364046. [DOI] [PubMed] [Google Scholar]
- Kingsley LA, Rinaldo CR, Jr, Lyter DW, Valdiserri RO, Belle SH, Ho M. Sexual transmission efficiency of hepatitis B virus and human immunodeficiency virus among homosexual men. JAMA. 1990;264:230–234. [PubMed] [Google Scholar]
- Koks S, Kingo K, Ratsep R, Karelson M, Silm H, Vasar E. Combined haplotype analysis of the interleukin-19 and −20 genes: relationship to plaque-type psoriasis. Genes Immun. 2004;5:662–667. doi: 10.1038/sj.gene.6364141. [DOI] [PubMed] [Google Scholar]
- Koks S, Kingo K, Vabrit K, Ratsep R, Karelson M, Silm H, Vasar E. Possible relations between the polymorphisms of the cytokines IL-19, IL-20 and IL-24 and plaque-type psoriasis. Genes Immun. 2005;6:407–415. doi: 10.1038/sj.gene.6364216. [DOI] [PubMed] [Google Scholar]
- Konopnicki D, Mocroft A, de Wit S, Antunes F, Ledergerber B, Katlama C, Zilmer K, Vella S, Kirk O, Lundgren JD. Hepatitis B and HIV: prevalence, AIDS progression, response to highly active antiretroviral therapy and increased mortality in the EuroSIDA cohort. Aids. 2005;19:593–601. doi: 10.1097/01.aids.0000163936.99401.fe. [DOI] [PubMed] [Google Scholar]
- Lai CL, Ratziu V, Yuen MF, Poynard T. Viral hepatitis B. Lancet. 2003;362:2089–2094. doi: 10.1016/S0140-6736(03)15108-2. [DOI] [PubMed] [Google Scholar]
- Levine OS, Vlahov D, Brookmeyer R, Cohn S, Nelson KE. Differences in the incidence of hepatitis B and human immunodeficiency virus infections among injecting drug users. J Infect Dis. 1996;173:579–583. doi: 10.1093/infdis/173.3.579. [DOI] [PubMed] [Google Scholar]
- Liao YC, Liang WG, Chen FW, Hsu JH, Yang JJ, Chang MS. IL-19 induces production of IL-6 and TNF-alpha and results in cell apoptosis through TNF-alpha. J Immunol. 2002;169:4288–4297. doi: 10.4049/jimmunol.169.8.4288. [DOI] [PubMed] [Google Scholar]
- Lok AS, McMahon BJ. Chronic hepatitis B: update of recommendations. Hepatology. 2004;39:857–861. doi: 10.1002/hep.20110. [DOI] [PubMed] [Google Scholar]
- Migita K, Miyazoe S, Maeda Y, Daikoku M, Abiru S, Ueki T, Yano K, Nagaoka S, Matsumoto T, Nakao K, Hamasaki K, Yatsuhashi H, Ishibashi H, Eguchi K. Cytokine gene polymorphisms in Japanese patients with hepatitis B virus infection--association between TGF-beta1 polymorphisms and hepatocellular carcinoma. J Hepatol. 2005;42:505–510. doi: 10.1016/j.jhep.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Miyazoe S, Hamasaki K, Nakata K, Kajiya Y, Kitajima K, Nakao K, Daikoku M, Yatsuhashi H, Koga M, Yano M, Eguchi K. Influence of interleukin-10 gene promoter polymorphisms on disease progression in patients chronically infected with hepatitis B virus. Am J Gastroenterol. 2002;97:2086–2092. doi: 10.1111/j.1572-0241.2002.05926.x. [DOI] [PubMed] [Google Scholar]
- Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- Oleksyk TK, Thio CL, Truelove AL, Goedert JJ, Donfield SM, Kirk GD, Thomas DL, O'Brien SJ, Smith MW. Single nucleotide polymorphisms and haplotypes in the IL10 region associated with HCV clearance. Genes Immun. 2005;6:347–357. doi: 10.1038/sj.gene.6364188. [DOI] [PubMed] [Google Scholar]
- Oral HB, Kotenko SV, Yilmaz M, Mani O, Zumkehr J, Blaser K, Akdis CA, Akdis M. Regulation of T cells and cytokines by the interleukin-10 (IL-10)-family cytokines IL-19, IL-20, IL-22, IL-24 and IL-26. Eur. J. Immunol. 2006;36:380–388. doi: 10.1002/eji.200425523. [DOI] [PubMed] [Google Scholar]
- Organization WM. Declaration of Helsinki. British Medical Journal. 1996:1448–1449. [Google Scholar]
- Otkjaer K, Kragballe K, Funding AT, Clausen JT, Noerby PL, Steiniche T, Iversen L. The dynamics of gene expression of interleukin-19 and interleukin-20 and their receptors in psoriasis. Br. J. Dermatol. 2005;153:911–918. doi: 10.1111/j.1365-2133.2005.06800.x. [DOI] [PubMed] [Google Scholar]
- Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- Parrish-Novak J, Xu W, Brender T, Yao L, Jones C, West J, Brandt C, Jelinek L, Madden K, McKernan PA, Foster DC, Jaspers S, Chandrasekher YA. Interleukins 19, 20, and 24 signal through two distinct receptor complexes. Differences in receptor-ligand interactions mediate unique biological functions. J Biol Chem. 2002;277:47517–47523. doi: 10.1074/jbc.M205114200. [DOI] [PubMed] [Google Scholar]
- Peng XM, Huang YS, Ma HH, Gu L, Xie QF, Gao ZL. Interleukin-10 promoter polymorphisms are associated with the mode and sequel of HBeAg seroconversion in patients with chronic hepatitis B virus infection. Liver Int. 2006;26:326–333. doi: 10.1111/j.1478-3231.2005.01241.x. [DOI] [PubMed] [Google Scholar]
- Pestka S, Krause CD, Sarkar D, Walter MR, Shi Y, Fisher PB. Interleukin-10 and related cytokines and receptors. Annu Rev Immunol. 2004;22:929–979. doi: 10.1146/annurev.immunol.22.012703.104622. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mendez ML, Gonzalez-Quintela A, Aguilera A, Barrio E. Prevalence, patterns, and course of past hepatitis B virus infection in intravenous drug users with HIV-1 infection. Am J Gastroenterol. 2000;95:1316–1322. doi: 10.1111/j.1572-0241.2000.01981.x. [DOI] [PubMed] [Google Scholar]
- Salmon-Ceron D, Lewden C, Morlat P, Bevilacqua S, Jougla E, Bonnet F, Heripret L, Costagliola D, May T, Chene G. Liver disease as a major cause of death among HIV infected patients: role of hepatitis C and B viruses and alcohol. J Hepatol. 2005;42:799–805. doi: 10.1016/j.jhep.2005.01.022. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckin D, Durusoy C, Sahin S. Concomitant vitiligo and psoriasis in a patient treated with interferon alfa-2a for chronic hepatitis B infection. Pediatr Dermatol. 2004;21:577–579. doi: 10.1111/j.0736-8046.2004.21512.x. [DOI] [PubMed] [Google Scholar]
- Shin HD, Park BL, Kim LH, Jung JH, Kim JY, Yoon JH, Kim YJ, Lee HS. Interleukin 10 haplotype associated with increased risk of hepatocellular carcinoma. Hum Mol Genet. 2003;12:901–906. doi: 10.1093/hmg/ddg104. [DOI] [PubMed] [Google Scholar]
- Shin HD, Winkler C, Stephens JC, Bream J, Young H, Goedert JJ, O'Brien TR, Vlahov D, Buchbinder S, Giorgi J, Rinaldo C, Donfield S, Willoughby A, O'Brien SJ, Smith MW. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc Natl Acad Sci U S A. 2000;97:14467–14472. doi: 10.1073/pnas.97.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha S, Strathdee SA, Galai N, Oleksyk T, Fallin MD, Mehta S, Schaid D, Vlahov D, O'Brien SJ, Smith MW. Behavioral risk exposure and host genetics of susceptibility to HIV-1 infection. J Infect Dis. 2006;193:16–26. doi: 10.1086/498532. [DOI] [PubMed] [Google Scholar]
- Stenderup K, Rosada C, Worsaae A, Clausen JT, Norman Dam T. Interleukin-20 as a target in psoriasis treatment. Ann. N. Y. Acad. Sci. 2007;1110:368–381. doi: 10.1196/annals.1423.039. [DOI] [PubMed] [Google Scholar]
- Thio CL, Seaberg EC, Skolasky R, Jr, Phair J, Visscher B, Munoz A, Thomas DL. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS) Lancet. 2002;360:1921–1926. doi: 10.1016/s0140-6736(02)11913-1. [DOI] [PubMed] [Google Scholar]
- Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Vicari AP, Trinchieri G. Interleukin-10 in viral diseases and cancer: exiting the labyrinth? Immunol Rev. 2004;202:223–236. doi: 10.1111/j.0105-2896.2004.00216.x. [DOI] [PubMed] [Google Scholar]
- WHO. Hepatitis B Fact Sheet. World Health Organization; 2000
- Wolk K, Kunz S, Asadullah K, Sabat R. Cutting edge: immune cells as sources and targets of the IL-10 family members? J Immunol. 2002;168:5397–5402. doi: 10.4049/jimmunol.168.11.5397. [DOI] [PubMed] [Google Scholar]
- Xu W. Interleukin-20. Int Immunopharmacol. 2004;4:627–633. doi: 10.1016/j.intimp.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Yachimski P, Chung RT. Update on Hepatitis B and C Coinfection in HIV. Curr Infect Dis Rep. 2005;7:299–308. doi: 10.1007/s11908-005-0063-4. [DOI] [PubMed] [Google Scholar]
- Zaykin DV, Westfall PH, Young SS, Karnoub MA, Wagner MJ, Ehm MG. Testing association of statistically inferred haplotypes with discrete and continuous traits in samples of unrelated individuals. Hum Hered. 2002;53:79–91. doi: 10.1159/000057986. [DOI] [PubMed] [Google Scholar]
- Zhu QR, Ge YL, Gu SQ, Yu H, Wang JS, Gu XH, Fei LE, Dong ZQ. Relationship between cytokines gene polymorphism and susceptibility to hepatitis B virus intrauterine infection. Chin Med J (Engl) 2005;118:1604–1609. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.