Abstract
Objectives
Biochemical (PSA) recurrence of prostate cancer following radical prostatectomy remains a major problem. Better biomarkers are needed to identify high-risk patients. DNA methylation of promoter regions leads to gene silencing in many cancers. In this study, we assessed the impact of DNA methylation on the identification of recurrent prostate cancer.
Methods
We studied the methylation status of fifteen pre-specified genes using MSPCR (Methylation Specific PCR) on tissue samples from 151 patients with localized prostate cancer with at least five years of follow-up after prostatectomy.
Results
In a multivariable logistic regression analysis, high Gleason score and involvement of the capsule, lymph nodes, seminal vesicles, or surgical margin were associated with an increased risk of biochemical recurrence. Methylation of CDH13 by itself (OR=5.50; 95% CI=1.34–22.67; P=0.02) or combined with methylation of ASC (OR=5.64 (95% CI=1.47–21.7; P=0.01) was also associated with an increased risk of biochemical recurrence. The presence of methylation of ASC and/or CDH13 yielded a sensitivity of 72.3% (95% CI=57–84.4%) and negative predictive value of 79% (95% CI=66.8–88.3%), which was similar to Rw′, a powerful clinico-pathologic prognostic score. However, 34% (95% CI=21–49%) of the patients who recurred were identified by the methylation profile of ASC and CDH13 rather than Rw′.
Conclusions
Methylation of CDH13 alone or combined with methylation of ASC is independently associated with an increased risk of biochemical recurrence after radical prostatectomy even when one considers the Rw′ score. These findings should be validated in an independent, larger cohort of prostate cancer patients who have undergone radical prostatectomies.
Keywords: DNA methylation, prostate cancer, biochemical recurrence, CDH13, ASC
INTRODUCTION
Prostate cancer is the most common and second most lethal malignancy affecting men in the United States.1 Despite PSA testing and increased primary treatment, recurrences remain problematic.2,3 Several clinico-pathologic scoring systems have been developed to identify patients at greatest risk of recurrence post-surgery including the RW′ and the Kattan nomograms.4,5 However, many patients defined as low-risk still recur. Identification of biomarkers could aid in risk stratification and give a clearer understanding of the biologic basis for recurrence.
Prostate cancer is a disease whose complexity continues to unfold. Genetic events such as deletions of PTEN and NKX3.1 and gene fusions involving TMPRSS and ETS transcription factors are well-characterized events in prostate tumorigenesis.6–8 There is also strong evidence for the role of DNA methylation-induced gene silencing in the pathogenesis of prostate cancer.9–11 We studied the methylation status of fifteen pre-specified genes whose loss of expression is involved in the progression of cancer.6,9–18 In most cases, DNA methylation had previously been shown to silence these genes.
MATERIALS AND METHODS
Study Population
The rate of biochemical recurrence at 5 years post-radical prostatectomy ranges from 16 to 22 percent.4,19 Using a retrospective, nested case-control design, we identified 151 patients with at least 5 years of follow-up after surgery for whom tissue specimens were available. One hundred and four patients, or two-thirds, were without biochemical recurrence while 47, or one-third, had recurred. All patients underwent a radical prostatectomy at Johns Hopkins in the PSA screening era, and none received adjuvant therapy. The patient characteristics are shown in Table 1.
Table 1.
Demographic, Clinical and Pathologic Characteristics for the 151 Patients
| Variable | Total (n=151) | Relapse (n=47) | Non-relapse (n=104) | P value |
|---|---|---|---|---|
| Age (yrs) | ||||
| Mean (± SD) | 58.5 (± 6.1) | 58.8 (± 5.5) | 58.3 (± 6.4) | 0.703* |
| Median (range) | 59.0 (41 – 71) | 59.0 (47 – 69) | 60.0 (41 – 71) | |
|
| ||||
| Race | ||||
| Caucasian | 141 (93.4) | 43 (91.5) | 98 (94.2) | 0.503 |
| Non Caucasian | 10 (6.6) | 4 (8.5) | 6 (5.8) | |
|
| ||||
| Clinical stage | ||||
| T1a | 1 (0.7) | 0 (0) | 1 (1.0) | 0.326 |
| T1b | 4 (2.7) | 1 (2.1) | 3 (2.9) | |
| T1c | 66 (43.7) | 18 (38.3) | 48 (46.1) | |
| T2a | 44 (29.1) | 16 (34.0) | 28 (26.9) | |
| T2b | 23 (15.2) | 7 (14.9) | 16 (15.4) | |
| T2c | 11 (7.3) | 4 (8.5) | 7 (6.7) | |
| T3a | 2 (1.3) | 1 (2.1) | 1 (1.0) | |
|
| ||||
| Preoperative PSA | ||||
| ≤ 4.01 | 27 (17.9) | 3 (6.4) | 24 (23.1) | < 0.0001* |
| 4.0 – 10.0 | 80 (53.0) | 15 (31.9) | 65 (62.5) | |
| 10.0 – 20.0 | 35 (23.1) | 24 (51.1) | 11 (10.6) | |
| > 20.0 | 9 (6.0) | 5 (10.6) | 4 (3.8) | |
|
| ||||
| Postoperative Gleason score | ||||
| 0 - Gleason sum 5 | 5 (3.3) | 0 (0) | 5 (4.8) | < 0.0001* |
| 1 - Gleason pattern 3+3 | 61 (40.4) | 4 (8.5) | 57 (54.8) | |
| 2 - Gleason pattern 3+4 | 49 (32.4) | 15 (31.9) | 34 (32.7) | |
| 3 - Gleason patter 4+3 | 16 (10.6) | 13 (27.7) | 3 (2.9) | |
| 4 - Gleason sum 8–10 | 20 (13.3) | 15 (31.9) | 5 (4.8) | |
|
| ||||
| Extra capsular penetration | ||||
| No | 82 (54.3) | 9 (19.1) | 73 (70.2) | < 0.0001 |
| Yes | 69 (45.7) | 38 (80.9) | 31 (29.8) | |
|
| ||||
| Surgical margin involvement | ||||
| No | 117 (77.5) | 27 (57.5) | 90 (86.5) | < 0.0001 |
| Yes | 34 (22.5) | 20 (42.5) | 14 (13.5) | |
|
| ||||
| Lymph node involvement | ||||
| No | 133 (88.1) | 31 (66.0) | 102 (98.1) | < 0.0001 |
| Yes | 18 (11.9) | 16 (34.0) | 2 (1.9) | |
|
| ||||
| Seminal vesicle involvement | ||||
| No | 131 (86.7) | 29 (61.7) | 102 (98.1) | < 0.0001 |
| Yes | 20 (13.3) | 18 (38.3) | 2 (1.9) | |
Analyzed using t test or nonparametric Wilcoxon rank sum test for continuous data;
Numbers shown are frequencies with percentages in parentheses except for age.
Our endpoint was biochemical recurrence-free survival as defined by a PSA≤ 0.2ng/mL at five years post-surgery. Since most patients had not yet experienced overt, measurable disease recurrences, we did not examine this endpoint. The investigators carrying out and interpreting all methylation assays (JJA and JGH) were blinded to the pathological and outcome data.
Tissue Samples
After IRB permission was granted, sections from 151 paraffin-embedded prostate cancer samples from radical prostatectomies were mounted on H&E slides. Adjacent sections representing the highest Gleason score corresponding to the H&E slides were then taken. Specimens were deparaffinized with xylene, washed twice with 100% ethanol, and digested with Proteinase K. Then, DNA was extracted with phenol-chloroform, and 1 microgram was treated with sodium bisulfite.20
Nested Methylation Specific PCR
Nested MSP was performed using in vitro methylated DNA and peripheral blood lymphocytes from a normal volunteer as methylated and unmethylated controls.20 All reactions also had negative H20 controls. The PCR products were loaded onto 2.5% agarose gels stained with GelStar (Cambrex, E. Rutherford, NJ) and subjected to electrophoresis. The products were subsequently diluted 1:500 in H20 and served as the template for the second round PCR reaction with both unmethylated and methylated primers, respectively. All samples were run on 2.5% agarose gels. Samples with a methylated band were scored as a methylated. Samples with only an unmethylated band were scored as unmethylated. Samples with neither band were scored as non-evaluable. Primer sequences and PCR conditions are available on request.
Statistical Analysis
The absence of a PSA> 0.2ng/mL at 5 years post-surgery was used to define a dichotomous outcome of biochemical recurrence. DNA methylation was treated as a binary variable (presence versus absence of methylation). Student’s t-test or its nonparametric alternative was used to analyze continuous data, and Fisher’s exact test was used for categorical data. A logistic regression approach was used to determine the associations of the factors with biochemical recurrence, including the best known clinico-pathological risk factors and the methylation of the genes. The model building followed two steps: 1) univariate analysis was used to identify important covariates and all variables whose p-values were < 0.25; 2) the variables selected in step 1 were simultaneously included to fit a multiple logistic regression model to verify the importance of each variable in the multivariable setting. The lack of fit of the final model was examined using the Hosmer and Lemeshow goodness-of-fit test. Odds ratios and 95% confidence intervals were reported. Operating characteristics of selected gene methylation and high Rw′ score were summarized using sensitivity, specificity, positive and negative predictive values, and likelihood ratios.4 McNemar’s test was used to compare paired proportions. All statistical tests were two-sided with p values< 0.05 considered statistically significant. Genes included in hypothesis testing were pre-specified and an adjustment for multiple comparisons was not made. Analyses were performed with SAS software (Version 9.1, Cary, NC).
RESULTS
Methylation was not detected in CDH1, PTEN, CHFR, and AR genes in the first 50 prostate cancers examined suggesting low frequencies of methylation which did not merit further study on the remaining tumors. For the other 11 genes, all 151 samples were evaluated. We found the following methylation frequencies: GSTP1 60%, MGMT 30%, ASC 37%, CDKN2A 30%, EDNRB 15%, CDH13 45%, CD44 19%, TIMP3 4%, RUNX3 44%, APC 71%, and WIF-1 28%. Overall, 99% of the PCR reactions were successful and informative. Representative gels for ASC and CDH13 are shown in Figure 1.
Figure 1. Gel Illustrating MSPCR Reaction.
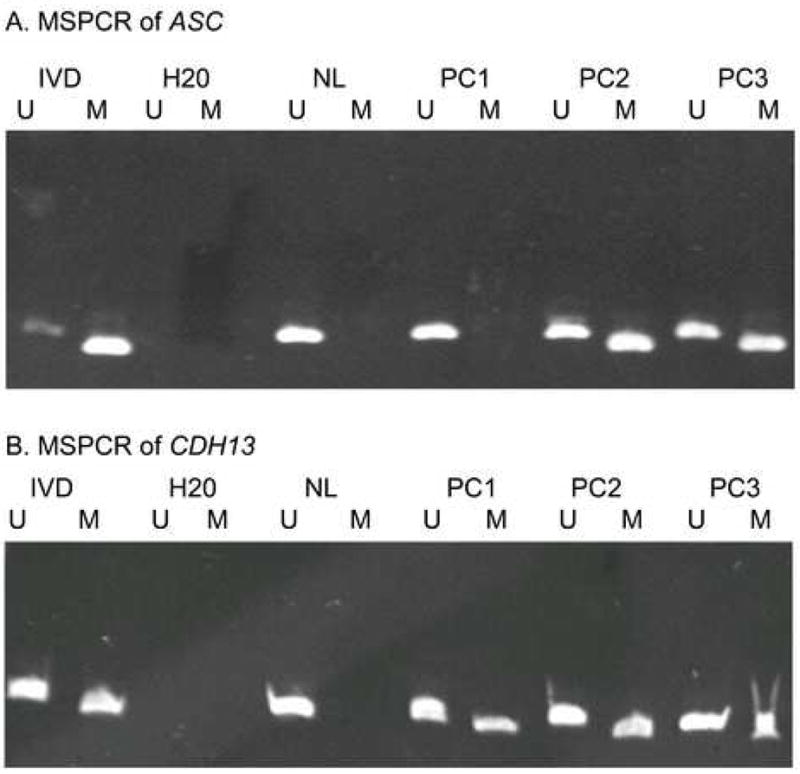
IVD=In vitro methylated DNA, the methylated control; NL=Normal lymphocytes, the unmethylated control; PC1, 2, 3=Prostate cancer samples; U=Unmethylated reaction, M=Methylated reaction
We then compared methylation of each gene as a variable for biochemical recurrence. Methylation of CDKN2A was associated with a decreased risk of biochemical recurrence with borderline significance (OR=0.43; 95% CI=0.19–0.98; P=0.05) in univariate analysis. While no individual gene’s methylation was associated with a statistically significant increased risk of recurrence in univariate analysis (Table 2A), two genes showed strong trends: ASC (OR=1.64; 95% CI=0.81–3.32; P=0.17) and CDH13 (OR=1.80; 95% CI=0.90–3.61; P=0.10). ASC was methylated in 37% of cases, while CDH13 was methylated in 45% of cases. Following the step 1 selection with the univariate analysis, the clinico-pathologic variables and methylation of CDKN2A, ASC, CDH13, RUNX3, MGMT, and GSTP1 were considered in a step 2 multivariable analysis (Table 2B). We included GSTP1 in the multivariable analysis despite its p-value >0.25 due to its importance in prostate cancer.
Table 2.
DNA Methylation is Associated with an Increased Risk of PSA Recurrence
| A. Univariate Analysis of the Risk of Biochemical Recurrence | |||
|---|---|---|---|
| Variable | Odds ratio | 95% CI# | P value |
| Preoperative PSA* | 1.15 | 1.07 – 1.24 | 0.0001 |
| Postoperative Gleason score* | 4.05 | 2.55 – 6.44 | < 0.0001 |
| Extra capsular penetration | 9.67 | 4.17 – 22.4 | < 0.0001 |
| Lymph node involvement | 25.8 | 5.62 – 118.5 | < 0.0001 |
| Seminal vesicle involvement | 31.0 | 6.80 – 141.6 | < 0.0001 |
| Surgical margin involvement | 4.66 | 2.08 – 10.4 | 0.0002 |
| CDKN2A methylation | 0.43 | 0.19 – 0.98 | 0.05 |
| CD44 methylation | 0.98 | 0.41 – 2.36 | 0.97 |
| WIF1 methylation | 0.82 | 0.38 – 1.80 | 0.63 |
| GSTP1 methylation | 1.07 | 0.53 – 2.18 | 0.85 |
| EDNRB methylation | 1.49 | 0.59 – 3.74 | 0.40 |
| ASC methylation | 1.64 | 0.81 – 3.32 | 0.17 |
| CDH13 methylation | 1.80 | 0.90 – 3.61 | 0.10 |
| RUNX3 methylation | 0.65 | 0.32 – 1.32 | 0.23 |
| MGMT methylation | 0.62 | 0.28 – 1.37 | 0.24 |
| TIMP3 methylation | 0.43 | 0.05 – 3.75 | 0.44 |
| APC methylation | 1.26 | 0.58 – 2.74 | 0.57 |
| B. Multivariable Analysis of the Risk of Biochemical Recurrence | |||
|---|---|---|---|
| Covariate | Odds ratio | 95% CI* | P value |
| Preoperative PSA | 1.00 | 0.91 – 1.09 | 0.99 |
| Postoperative Gleason score | 3.77 | 1.87 – 7.62 | 0.0002 |
| Extra capsular penetration | 4.92 | 1.31 – 18.52 | 0.02 |
| Lymph node involvement | 7.26 | 0.92 – 57.59 | 0.06 |
| Seminal vesicle involvement | 13.41 | 1.76 – 101.84 | 0.01 |
| Surgical margin involvement | 7.73 | 1.90 – 31.46 | 0.004 |
| CDKN2A methylation | 0.43 | 0.10 – 1.90 | 0.27 |
| GSTP1 methylation | 0.30 | 0.07 – 1.24 | 0.10 |
| ASC methylation | 2.08 | 0.57 – 7.60 | 0.27 |
| CDH13 methylation | 5.51 | 1.34 – 22.67 | 0.02 |
| RUNX3 methylation | 0.60 | 0.16 – 2.28 | 0.45 |
| MGMT methylation | 0.33 | 0.07 – 1.63 | 0.17 |
Preoperative PSA and postoperative Gleason score were treated as continuous variables
CI: Confidence interval
CDH13 was the only gene whose methylation was found to be significantly associated with recurrence (OR=5.50; 95% CI=1.34–22.67; P=0.02) after adjusting for all of the other factors. Results of the Hosmer and Lemeshow test showed no evidence of a lack of fit in the final model (P=0.60). We further evaluated a profile combining the two genes (CDH13 and ASC) associated with an increased odds ratio of recurrence in another multivariable analysis with a similar multivariable logistic regression model (data not shown). Tumors with methylation of ASC and/or CDH13 were independently associated with an increased risk of recurrence compared to tumors without methylation of both of these genes (OR=5.64; 95% CI=1.47–21.7; P=0.01).
The operating characteristics of combining the two genes were further evaluated and compared with the previously defined Rw′ score.4 A high Rw′ score was more specific and was associated with a higher positive predictive value and higher likelihood ratio for recurrence than the methylation status of ASC and CDH13 (Table 3).4 However, a trend toward higher sensitivity for the methylation of ASC in combination with CDH13 (72.3%; 95% CI=57.4–84.4%) compared to the Rw′ score (55.3%; 95% CI=40.1–69.8%) was observed, although it did not reach statistical significance (P=0.10). Additionally, the methylation status of these 2 genes had a negative predictive value and a negative likelihood ratio not statistically different from the Rw′ score (Table 3).
Table 3.
Performance of # Rw′ Score and Gene Methylation in Predicting Biochemical Recurrence
| Variable | * Senstivity (%) (95% CI£) | § Specificity (%) (95% CI) | ¥ PPV (%) (95% CI) | † NPV (%) (95% CI) | € LR+ (95% CI) | ‡ LR− (95% CI) |
|---|---|---|---|---|---|---|
| # Rw′ (>2.84) | 55.3 (40.1 – 69.8) | 95.1 (88.9 – 98.4) | 83.9 (66.3 – 94.5) | 82.2 (74.1 – 88.6) | 11.3 (4.6 – 27.5) | 0.5 (0.3 – 0.6) |
| ASC, CDH13 (at least 1 methylated) | 72.3 (57.4 – 84.4) | 48.0 (38.0 – 58.2) | 39.1 (28.8 – 50.1) | 79.0 (66.8 – 88.3) | 1.4 (1.1 – 1.8) | 0.6 (0.3 – 0.9) |
Rw′, weighted risk of recurrence, based on lymph node status, seminal vesicle status, surgical margin status, and postoperative Gleason score
Sensitivity is calculated as the number of true positives divided by the number of true positives plus false negatives
Specificity is calculated as the number of true negatives divided by the number of true negatives plus false positives
PPV (positive predictive value) is calculated as the number of true positives divided by the number of true positives plus false positives
NPV (negative predictive value) is calculated as the number of true negatives divided by the number of true negatives plus false negatives
LR+ (likelihood ratio of a positive test result) is calculated as sensitivity/(1-specificity)
LR- (likelihood ratio of a negative test result) is calculated as (1-sensitivity)/specificity.
CI: Confidence interval.
COMMENT
While a pathology report after radical prostatectomy gives valuable information about recurrence risk, there are limitations. For the growing group of patients with early stage, low-grade prostate cancer, outcomes can be quite variable with some patients experiencing recurrences, some of which will be lethal. In addition, there can be significant inter-pathologist variability in interpretation of the Gleason score.21
There are only two reports, both using quantitative MSP, that describe gene methylation associated with an increased risk of biochemical recurrence or a reduced time to biochemical recurrence, respectively.13,18 In the first report, PTGS2 methylation was found in over 90% of the primary tumor samples examined.13 In the other report, the frequency of APC methylation was not stated, but we and others have found that the APC methylation frequency in prostate cancer exceeds 71%.13,18 Hence, these reports do not separate outcomes according to the presence or absence of gene methylation. Since quantitative MSPCR approaches do not adjust for the percentage of tumors cells which comprise a sample, increased methylation may simply reflect a sample with more transformed cells. Higher tumor burden, as measured by involvement of multiple cores or a high percentage of tumor cells within each core, is associated with prostate cancer aggressiveness.22 Finally, pre-defined, binary cut-offs make one’s findings most applicable to future study populations.
Of note, we found that CDH13 methylation was independently associated with a statistically significant increased risk of biochemical recurrence (OR=5.50; 95% CI=1.34–22.67; P=0.02). Cadherins are calcium-dependent cell-cell adhesion molecules whose loss in solid tumors may be important for epithelial to mesenchymal transition and increased metastatic potential.23–25 CDH13 methylation is associated with high Gleason tumors, which increases one’s risk of recurrence.11 CDH13 expression is diminished in many human prostate cancers, and knockdown of CDH13 in normal prostate cells resulted in enhanced tumorigenicity, providing an explanation for our findings.26
ASC methylation was also associated with an increased risk of recurrence in multivariable analysis. ASC was first identified in human leukemia and is involved in apoptosis, providing an explanation for the risk of recurrence.27 The selection of ASC and CDH13 as a combination for the step 2 multivariable model was based on their individual trends predicting an increased risk of biochemical recurrence in univariate analysis and their increased odds ratios of recurrence in multivariable analysis. Methylation of these 2 genes (ASC and CDH13), while not associated with a greater odds ratio of recurrence than CDH13 alone, was associated with improved sensitivity (72.3%, 95% CI=57–84.4%) for detecting recurrences versus CDH13 alone (sensitivity= 55.3% (95% CI=40.1 – 69.8%). Methylation of ASC and/or CDH13 was independently associated with an increased risk of recurrence (OR=5.64; 95% CI 1.39–18.2; P=0.01) in multivariable analysis adjusting for all the covariates present in Table 2.
As opposed to the methylation of ASC and CDH13, we observed a decrease in odds ratios of recurrence for the individual clinico-pathological variables in multivariable analysis versus univariate analysis, which suggests that many recurring patients have multiple high-risk clinico-pathological features present simultaneously (Table 2). This highlights the importance of our DNA methylation findings, which appear to correctly identify patients misclassified as low-risk (due to absence of involvement of these clinico-pathological variables) who still recur. Of note, 34% percent (95% CI=21–49%) of the men who recurred were classified as low risk due to Rw′ scores <2.84, but were appropriately identified as recurrences by methylation of ASC or CDH13. Our findings suggests that determination of methylation of these genes may have the greatest utility for those patients treated with surgery whose tumors lack adverse clinico-pathological features or for patients treated with radiotherapy, for whom pathological staging is not available.
This study of DNA methylation in 151 patients with localized prostate cancer undergoing radical prostatectomy is unique and important for several reasons. First, it is the largest published series in which methylation was found to be independently associated with an increased risk of biochemical recurrence even when all of the currently accepted clinico-pathological variables were incorporated into a multivariable analysis.13,18 In addition, our cohort included a heterogeneous group of patients. Finally, our approach had a 99% success rate on paraffin specimens, suggesting this could be done on routinely collected tissues.
CONCLUSIONS
Methylation of CDH13 by itself or in combination with ASC is related to recurrence in patients undergoing radical prostatectomy. Given the high sensitivity, high negative predictive value, and low negative likelihood ratio, tumors without methylation of ASC and CDH13 were associated with a significantly reduced risk of recurrence after radical prostatectomy. While promising, these results should be validated on a separate cohort of patients with prostate cancer treated with radical prostatectomies at another institution. Second, methylation of other, novel genes may aid in identifying patients at risk of recurrence. Finally, improved prognostic information needs to be linked to changes in care. This will require advances in adjuvant therapies, so that better risk stratification tools, such as DNA methylation, do not simply increase anxiety without increasing survival.
Acknowledgments
The Johns Hopkins Prostate Spore CA P50-58236 (JJA, MAC, AWP, AMD, and JGH), an ASCO Young Investigator Award (JJA), the Patrick C. Walsh Prostate Cancer Research Fund (JJA), a FAMRI Young Clinical Scientist Award (JJA), and EDRN-NIH/NCI 2U01-CA86323 (EBH, LAM, and AWP) supported this work. Special thanks go to Mr. and Mrs. Bernard Schwartz for their support of prostate cancer research, to Helen Fedor for her help in obtaining the specimens from the Brady Urological Institute Prostate Specimen Tissue Procurement and Repository Facility, and also to Mandy Burns and Susanne McGlothlin for their help in preparing this manuscript.
Footnotes
Disclosures: JGH is a paid consultant to OncoMethylome Sciences.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
BIBLIOGRAPHY
- 1.American Cancer Society. Cancer Facts and Figures. 2007. [Google Scholar]
- 2.Brawer MK, Chetner MP, Beatie J, Buchner DM, Vessella RL, Lange PH. Screening for prostatic carcinoma with prostate specific antigen. J Urol. 1992;147:841–845. doi: 10.1016/s0022-5347(17)37401-3. [DOI] [PubMed] [Google Scholar]
- 3.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. Jama. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 4.Roberts WW, Bergstralh EJ, Blute ML, Slezak JM, Carducci M, Han M, Epstein JI, Eisenberger MA, Walsh PC, Partin AW. Contemporary identification of patients at high risk of early prostate cancer recurrence after radical retropubic prostatectomy. Urology. 2001;57:1033–1037. doi: 10.1016/s0090-4295(01)00978-5. [DOI] [PubMed] [Google Scholar]
- 5.Kattan MW, Eastham JA, Stapleton AM, Wheeler TM, Scardino PT. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 6.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–4296. [PubMed] [Google Scholar]
- 7.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 8.Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, Gasser TC, Koivisto P, Lack EE, Kononen J, et al. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–6115. [PubMed] [Google Scholar]
- 9.Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lou W, Krill D, Dhir R, Becich MJ, Dong JT, Frierson HF, Jr, Isaacs WB, Isaacs JT, Gao AC. Methylation of the CD44 metastasis suppressor gene in human prostate cancer. Cancer Res. 1999;59:2329–2331. [PubMed] [Google Scholar]
- 11.Maruyama R, Toyooka S, Toyooka KO, Virmani AK, Zochbauer-Muller S, Farinas AJ, Minna JD, McConnell J, Frenkel EP, Gazdar AF. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2002;8:514–519. [PubMed] [Google Scholar]
- 12.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 13.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 14.Wissmann C, Wild PJ, Kaiser S, Roepcke S, Stoehr R, Woenckhaus M, Kristiansen G, Hsieh JC, Hofstaedter F, Hartmann A, et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J Pathol. 2003;201:204–212. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- 15.Toyota M, Sasaki Y, Satoh A, Ogi K, Kikuchi T, Suzuki H, Mita H, Tanaka N, Itoh F, Issa JP, et al. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci U S A. 2003;100:7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway KE, McConnell BB, Bowring CE, Donald CD, Warren ST, Vertino PM. TMS1, a novel proapoptotic caspase recruitment domain protein, is a target of methylation-induced gene silencing in human breast cancers. Cancer Res. 2000;60:6236–6242. [PubMed] [Google Scholar]
- 17.Jarrard DF, Kinoshita H, Shi Y, Sandefur C, Hoff D, Meisner LF, Chang C, Herman JG, Isaacs WB, Nassif N. Methylation of the androgen receptor promoter CpG island is associated with loss of androgen receptor expression in prostate cancer cells. Cancer Res. 1998;58:5310–5314. [PubMed] [Google Scholar]
- 18.Rosenbaum E, Hoque MO, Cohen Y, Zahurak M, Eisenberger MA, Epstein JI, Partin AW, Sidransky D. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res. 2005;11:8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. [DOI] [PubMed] [Google Scholar]
- 19.Catalona WJ, Smith DS. 5-year tumor recurrence rates after anatomical radical retropubic prostatectomy for prostate cancer. J Urol. 1994;152:1837–1842. doi: 10.1016/s0022-5347(17)32397-2. [DOI] [PubMed] [Google Scholar]
- 20.Guo M, Ren J, House MG, Qi Y, Brock MV, Herman JG. Accumulation of promoter methylation suggests epigenetic progression in squamous cell carcinoma of the esophagus. Clin Cancer Res. 2006;12:4515–4522. doi: 10.1158/1078-0432.CCR-05-2858. [DOI] [PubMed] [Google Scholar]
- 21.Glaessgen A, Hamberg H, Pihl CG, Sundelin B, Nilsson B, Egevad L. Interobserver reproducibility of modified Gleason score in radical prostatectomy specimens. Virchows Arch. 2004;445:17–21. doi: 10.1007/s00428-004-1034-0. [DOI] [PubMed] [Google Scholar]
- 22.Epstein JI, Chan DW, Sokoll LJ, Walsh PC, Cox JL, Rittenhouse H, Wolfert R, Carter HB. Nonpalpable stage T1c prostate cancer: prediction of insignificant disease using free/total prostate specific antigen levels and needle biopsy findings. J Urol. 1998;160:2407–2411. [PubMed] [Google Scholar]
- 23.Takeichi M. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 1991;251:1451–1455. doi: 10.1126/science.2006419. [DOI] [PubMed] [Google Scholar]
- 24.Lee SW. H-cadherin, a novel cadherin with growth inhibitory functions and diminished expression in human breast cancer. Nat Med. 1996;2:776–782. doi: 10.1038/nm0796-776. [DOI] [PubMed] [Google Scholar]
- 25.Toyooka S, Toyooka KO, Harada K, Miyajima K, Makarla P, Sathyanarayana UG, Yin J, Sato F, Shivapurkar N, Meltzer SJ, et al. Aberrant methylation of the CDH13 (H-cadherin) promoter region in colorectal cancers and adenomas. Cancer Res. 2002;62:3382–3386. [PubMed] [Google Scholar]
- 26.Wang XD, Wang BE, Soriano R, Zha J, Zhang Z, Modrusan Z, Cunha GR, Gao WQ. Expression profiling of the mouse prostate after castration and hormone replacement: implication of H-cadherin in prostate tumorigenesis. Differentiation. 2007;75:219–234. doi: 10.1111/j.1432-0436.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 27.Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T, Sagara J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274:33835–33838. doi: 10.1074/jbc.274.48.33835. [DOI] [PubMed] [Google Scholar]


