Abstract
Background
Nutrigenetics studies the role of genetic variation on interactions between diet and health aimed at providing more personalized dietary advice. However, replication has been very low. Our aim was to study the interaction between a functional APOA2 polymorphism, food intake and body mass index (BMI) in independent populations to replicate findings and to increase their evidence level.
Methods
Cross-sectional, follow-up (20 years) and case-control analyses were undertaken in three independent populations. We analyzed gene-diet interactions between the APOA2 -265T>C polymorphism and saturated fat (SATFAT) intake on BMI and obesity in 3,462 subjects from three American populations: Framingham (1,454 Whites), GOLDN (1,078 Whites) and Boston-Puerto Rican studies (930 Hispanics of Caribbean origin).
Results
Prevalence of CC subjects ranged from 11-15%. We identified statistically significant interactions between the APOA2 -265T>C and SATFAT on BMI in all three populations. Thus, the magnitude of the difference in BMI between the CC and TT+TC subjects differed by SATFAT. A mean increase of 6.2% BMI (ranging from 4.3%-7.9%; P<0.05), was observed between genotypes with high (>=22g/d), but not with low SATFAT intake in all studies. Likewise, the CC genotype was significantly associated with higher obesity prevalence in all populations only in the high-SATFAT stratum. Meta-analysis estimations of obesity for CC compared with TT+TC subjects were: OR=1.84, 95%CI:1.38-2.47; P<0.0001 in the high-SATFAT stratum, but no association was detected in the low-SATFAT stratum (OR=0.81, 95%CI:0.59-1.11;P=0.181).
Conclusions
For the first time, a gene-diet interaction influencing BMI and obesity has been strongly and consistently replicated in three independent populations.
Introduction
Genomics is revolutionizing biomedical research and providing great expectations on disease prevention and treatment (1). The classical candidate gene approach and the newer genome-wide association (GWA) studies (2) have identified genetic variants that predispose to common diseases. If the current trends continue, most common disease-predisposing polymorphisms will be soon identified. Thus, a major remaining research challenge will be to characterize gene-environment interactions as these are essential for translating genomics into clinical medicine and public health (3). Diet is one of the most important environmental factors interacting with the genome to modulate disease risk (4) and better understanding of these interactions has the potential to support disease prevention via modification of dietary recommendations. However, progress in this area has been slow due to the low evidence level achieved so far. Although studies reported enticing gene-diet interactions, their level of replication has been extremely low (4-7). Thus, a vast proportion of gene-diet interactions have not been replicated; only a very small number has been replicated twice; and, to the best of our knowledge, none has been replicated in three or more independent populations. This problem has plagued classical genotype-phenotype association studies, and currently, consistency is thought a crucial causal criterion of credibility of GWAS (8). Therefore, the NCI-NHGRI Working Group on Replication in Genotype-Phenotype Associations (9) supports replication as the most reliable approach to increase evidence level and subsequent clinical applications. Consistent with these recommendations, our major aim was to conduct a replication study in nutrigenetics. For this purpose, we focused on our recently reported association between the functional −265T>C SNP (10,11) in the apolipoprotein A-II (APOA2) gene promoter, food intake and obesity risk in non-Hispanic-White-Americans (12). APOA2, the second major HDL apolipoprotein, is an enigmatic protein in search of a function (13). Although animal models have found that over-expression of APOA2 results in hypertriglyceridemia, obesity and insulin resistance (14,15), its role in humans remains controversial (10,11,16,17). Therefore, our goals were: 1) to analyze the association between the APOA2 −265T>C SNP and obesity-related variables in the Framingham Study, focusing on gene-diet interactions with fat intake; and 2) to study the replication of these gene-diet interactions in other American populations.
Patients and Methods
We studied 3,462 subjects from three independent populations. All participants provided written informed consent.
The Framingham Study
We included 1,454 unrelated non-Hispanic Whites (716 men and 738 women), aged 26-80 years who participated in the fifth-examination visit of the Framingham Offspring Study (FOS) (18) and had complete data for the genetic, clinical, dietary and anthropometric variables analyzed. These subjects were obtained from a standard previously plated set of unrelated FOS DNAs in which only one individual from each pedigree was randomly selected. The Institutional Review Boards (IRB) for Human Research at Boston University and Tufts University/New England Medical Center approved the protocol. Alcohol, tobacco smoking, diabetes status and physical activity were defined previously (19-21). For longitudinal analysis, we included 1,087 unrelated subjects (540 men and 547 women) who attended each of the first five exams: exam 1 (1971-1975), exam 2 (1979–1983), exam 3 (1984-1987), exam 4 (1987-1991) and exam 5 (1991-1995). Anthropometric and demographic variables were measured at each cycle.
The Genetics of Lipid Lowering Drugs and Diet Network (GOLDN) Study
1,200 adult individuals of European ancestry were recruited from two National Heart, Lung and Blood Institute (NHLBI) Family Heart Study (FHS) field centers (Minneapolis, MN and Salt Lake City, UT) as previously reported (12). We included 1,078 subjects (514 men and 564 women) for whom data for all examined variables were complete. The protocol was approved by the IRB at the Universities of Alabama, Minnesota, Utah, and Tufts.
The Boston-Puerto Rican CPHHD Study
Comprising about 1,200 free living Puerto Rican (Hispanics of Caribbean origin) subjects, aged 45-75 years, in the greater Boston area (22), this is one of the NIH-funded Centers on Population Health and Health Disparities (CPHHD). We analyzed 930 (263 men and 667 women) subjects with complete data. The protocol was approved by the IRB at Tufts University.
In these populations subjects included did not differ from those excluded due to incomplete data with regard to the variables analyzed.
Anthropometric, physical activity and biochemical determinations
Anthropometric variables including height, weight and waist were measured in all cohorts by standard techniques (12,17,20,22). Body mass index (BMI) was calculated as weight (kg)/height2(m). Obesity was defined as BMI>=30kg/m2. Physical activity in Framingham was assessed with the physical activity index (PAI) calculated at exam 4 from the number of hours spent each day sleeping and performing sedentary, slight, moderate, or heavy physical activities, weighted according to the estimated oxygen consumption required (21). In GOLDN, a non-validated questionnaire containing questions on the number of hours/day dedicated to different activities was used (22). In the Boston-Puerto Rican study, a physical activity score based on the Paffenbarger questionnaire of the Harvard Alumni Activity Survey (23) was estimated. Fasting glucose, triglycerides, total cholesterol and HDL-C were measured by standard methods (12,19,20,22). For FOS samples, plasma APOA1 and APOA2 concentrations were determined by turbidimetric immunoassays (Wako Chemical, Richmnond, VA, USA).
Dietary intake
Diet was measured by validated questionnaires in each specific population (24-26): The Willett (24) semiquantitative food frequency questionnaire (FFQ), administered at exam 5, in FOS; the Diet History Questionnaire (DHQ), in GOLDN (12, 25); and a specially designed and validated FFQ (26) in the Boston-Puerto Rican Study. Nutrient data were derived from: the Harvard University food composition database, the USDA database and the Minnesota Nutrient System, respectively.
Genetic analyses
DNA was isolated from blood (Qiagen, Hilden, Germany). We performed the APOA2 −265T>C genotyping (rs5082) using a Taqman assay with allele-specific probes on the ABIPrism 7900HT Sequence Detection System (Applied Biosystems) (12). Quality control measures were applied. Genotype frequencies were consistent with Hardy-Weinberg equilibrium in all populations.
Statistical analyses
Chisquare tests were used to test percentages. Normality of continuous variables was examined. Triglycerides were log-transformed and alcohol and PUFA were square-root transformed. We applied ANOVA and Student′s t-test to compare crude means. Considering the results obtained in our previous GOLDN study (12), in which similar effects were found for TT and TC subjects, recessive effects for the APOA2−265T>C polymorphism were considered in this analysis after having checked the validity of this model in the other populations. We also tested the statistical homogeneity by sex, and men and women were analyzed together. To study gene-diet interactions in determining BMI, we used multivariate linear regression models including main effects and interaction terms. We fitted separate models for each population including the same variables for the interaction terms and for the multivariate adjustments. Saturated fat (SATFAT) intake was considered as continuous as well as categorical (low or high). 22g/d was established as the cut-off point to classify the low or the high SATFAT intake based on the Framingham results. In addition to the unadjusted models, we adjusted analyses for sex, age, tobacco smoking, alcohol consumption, diabetes, lipid medication and total energy intake (basic models). Additional adjustments of basic models for physical activity were considered for each population. In GOLDN, additional adjustments for family relationships were undertaken as previously described (12). In the Boston-Puerto Rican study further adjustment for admixture using the first component variable derived from the analysis of 100 ancestry informative markers (27) was undertaken To study the specificity of the effect, we sequentially adjusted for other nutrients (total fat, carbohydrates and proteins) as indicated.
When the APOA2-SATFAT interaction was considered as continuous, it was depicted by computing the predicted values for each individual from the adjusted regression model and plotting these values against SATFAT intake by the APOA2 genotype. Regression coefficients were estimated in stratified analyses by genotype. When SATFAT was considered as categorical (below or above 22g/d), stratified analyses were conducted. To increase the consistency, we have also undertaken internal replication analysis on the same population. GOLDN was stratified by study center and the Boston-Puerto Rican study, by diabetes status. In FOS we also analyzed the APOA2-SATFAT interaction on BMI across 20-year follow-up in a general lineal multivariate model for repeated measures with interaction terms. Five direct measures of BMI were considered (at exams 1,2,3,4 and 5) as dependent variables. The APOA2 polymorphism, SATFAT (as dichotomous) and age at baseline were covariates. Main effects and interaction terms were tested.
In all populations, logistic regression models including main effects and interaction terms were fitted to test the APOA2-SATFAT interaction in determining the odds ratio (OR) of obesity. Study-specific OR and 95% confidence interval (CI) were estimated for each stratum of SATFAT. Multivariate adjustments were done as indicated.
We also performed a meta-analysis of study-specific estimates of OR for the two strata of SATFAT intake. Heterogeneity was tested by the Cochran Q–statistic and quantified by I². We pooled study-specific estimates according to the inverse-variance fixed effect. Statistical analyses were conducted with SAS software (v.9.1; SAS Institute, Cary, NC), SPSS software (v.15.0; SPSS Inc, Chicago, IL) and MIX software v.1.7 for meta-analysis. Standard regression diagnostic procedures were used to ensure the appropriateness of the fitted models. All reported probability tests were 2-sided. Differences were considered significant at P <0.05. Considering the magnitude of the effect, the allele frequency and the standard type I error (5%), our study has a power>=80% to detect statistically significant interactions in each population (28).
Results
We studied 3,462 subjects from three independent American cohorts (FOS, GOLDN and Boston-Puerto Rican studies). Table 1 shows demographic, anthropometric, clinical, biochemical, dietary and lifestyle characteristics of participants according to the APOA2 −265T>C SNP for each population. Prevalence of CC subjects did not differ between FOS (15%) and GOLDN (15%). A statistically significant lower prevalence (10.5%) was found in the Boston-Puerto Rican study. Demographic characteristics and physical activity did not differ between CC and T-allele carriers in either of the three populations (Table 1). Among White populations, prevalence of obesity was higher in GOLDN. Likewise, mean fat intake, mainly SATFAT was higher in GOLDN than in FOS. No significant association of the APOA2 −265T>C SNP with HDL-C was found in either of the three populations. APOA2 and APOA1 concentrations were only determined in FOS. In FOS, plasma APOA2 concentrations (Supplemental Table 1) were significantly lower in CC subjects, whereas no effects were observed for APOA1, supporting the functionality and specificity of this SNP.
Table 1.
General characteristics of the studied populations depending on the APOA2 −265T>C polymorphism
| Framingham Study | GOLDN Study | Boston-Puerto Rican Study | ||||
|---|---|---|---|---|---|---|
| TT+TC (n=1217) | CC (n=237) | TT+TC (n=913) | CC (n=165) | TT+TC (n=832) | CC (n=98) | |
| Mean SD | Mean SD | Mean SD | Mean SD | Mean SD | Mean SD | |
| Male/Female (n/n) | 606/611 | 110/127 | 439/474 | 75/90 | 231/601 | 32/66 |
| Age (years) | 55.4 (9.3) | 55.7 (9.8) | 48.7 (16.3) | 49.8 (15.4) | 57.7 (7.7) | 57.9 (7.5) |
| Weight (kg) | 78.5 (16.6) | 78.8 (16.9) | 82.2 (18.) | 86.1 (19.2)* | 80.2 (17.4) | 81.5 (19.7) |
| BMI (kg/m2) | 27.6 (4.9) | 27.5 (4.9) | 28 (5.5) | 29.2 (6.2)* | 32.1 (6.8) | 32.1 (7.2) |
| Waist (m) | 1.0 (.1) | 1.0 (.1) | 0.95 (.2) | 0.99 (.2)* | 1.0 (.1) | 1.0 (.2) |
| Cholesterol (mg/dL) | 204.9 (36.1) | 208.3 (36.9) | 190.8 (39.3) | 189.8 (26.7) | 184.1 (43.) | 182.8 (38.3) |
| LDL-C (mg/dL) | 126.9 (30.8) | 129.1 (32.9) | 121.4 (31.5) | 121.4 (30.1) | 107.2 (34.9) | 109.0 (34.4) |
| HDL-C (mg/dL) | 49.4 (14.8) | 50.1 (16.1) | 47.2 (13.1) | 47.3 (13.2) | 44.5 (12.2) | 44.6 (12.4) |
| Triglycerides (mg/dL) | 149.7 (109.9) | 150.5 (97.7) | 138.4 (101.1) | 131.0 (74.5) | 164.5 (123.7) | 157.8 (87.4) |
| Fasting glucose (mg/dL) | 101.5 (29.2) | 101.5 (29.2) | 101.4 (18.7) | 101.4 (15.7) | 122.8 (53.1) | 117.8 (40.2) |
| Energy intake (Kcal/d) | 1837.8 (611.7) | 1940.9 (649.6)* | 2021.7 (827.4) | 2203.9 (973.2)* | 2120.1 (868.) | 2076.9 (853.4) |
| Total fat (g/d) | 60.5 (25.3) | 62.8 (25.6) | 80.6 (39.8) | 89.8 (44.2)* | 74.0 (34.9) | 71.9 (35.1) |
| SATFAT (g/d) | 21.3 (9.5) | 22.1 (9.7) | 27.2 (14.1) | 29.9 (15.9)* | 23.1 (11.9) | 22.3 (12.2) |
| MUFA (g/d) | 23.1 (10.1) | 23.9 (10.1) | 30.3 (15.1) | 34.1 (17.1)* | 26.8 (13.) | 26.0 (12.9) |
| PUFA (g/d) | 12.1 (5.4) | 12.5 (5.7) | 17.1 (8.5) | 19.2 (9.7)* | 17.8 (8.6) | 17.7 (8.6) |
| Proteins (g/d) | 69.4 (25.7) | 72.0 (27.2) | 79.5 (35.2) | 87.0 (40.1)* | 91.6 (40.7) | 89.8 (43.1) |
| Carbohydrates (g/d) | 234.7 (89.5) | 252.1 (96.1)* | 245.6 (101.2) | 259.3 (123.3) | 272.1 (112.8) | 269.1 (109.8) |
| Physical activity+ | 37.1 (7.3) | 36.6 (6.7) | 34.2 (6.2) | 34.7 (6.4) | 31.5 (4.5) | 31.1 (4.7) |
| Current smokers (%) | (17.6) | (17.6) | (7.3) | (8.5) | (23.7) | (24.7) |
| Current drinkers (%) | (70.1) | (71.3) | (49.3) | (58.2)* | (38.5) | (37.1) |
| Lipid medication (%) | (8.6) | (7.6) | (4.9) | (5.1) | (37.3) | (42.9) |
| Diabetes (%) | (8.7) | (10.1) | (7.7) | (9.7) | (40.5) | (46.9) |
| Obesity (%) | (25.3) | (25.7) | (32.1) | (40.6)* | (56.8) | (57.1) |
BMI: Body mass index; LDL-C: Low-density lipoprotein cholesterol; HDL-C: High density lipoprotein cholesterol; SATFAT: Saturated fatty acids; MUFA: Monounsaturated fatty acids; PUFA: Polyunsaturated fatty acids.
: Physical activity was measured by different physical activity scores in the Framingham, GOLDN and the in Boston-Puerto Rican studies as described in Methods
: Statistically significant differences (P<0.05) between carriers of the T allele and CC subjects for the corresponding variable in each population
We next examined in FOS our previously described association between the APOA2 SNP, food intake and body-weight. In FOS we also found that CC subjects had higher energy intake than T-allele carriers (P=0.017) (Table 1). These results were consistent with our previous finding in GOLDN showing that daily energy intake was ∼200 Kcal/d higher in CC subjects than in T-allele carriers (P=0.005). However, the magnitude of the genotype effect was lower in Framingham (∼100 Kcal/d), and differences in total fat intake, SATFAT, and monounsaturated fatty acids (MUFA) did not reach the statistical significance as they did in GOLDN. This difference could be due to the higher prevalence of obesity, total fat and SATFAT intake in the GOLDN population (Table 1). Therefore, we hypothesized that the APOA2 SNP would have a greater influence in determining food intake in obese subjects. Consistent with this notion, we found that obese CC subjects from FOS had statistically higher intakes of energy, total fat, SATFAT, MUFA, protein, carbohydrates and fructose than T-allele carriers (Supplemental Table 2). The greater carbohydrate and fructose intake in obese CC subjects from Framingham compared to those of GOLDN, could reflect a greater intake of fruit and cereals in those Framingham subjects to satisfy their higher appetite, possible due to the CC genotype, at the same time as following a healthy diet. Further adjustment for physical activity did not affect the statistical significance of results.
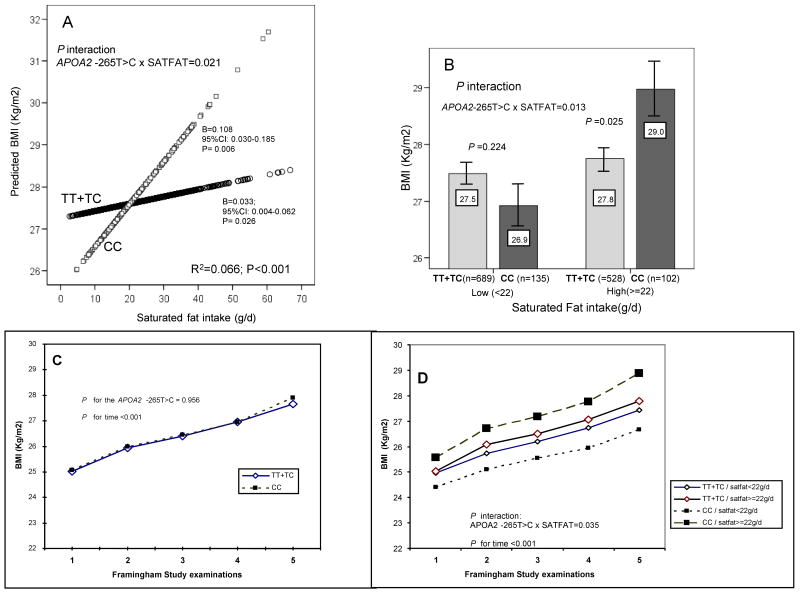
Moreover, in the Framingham population as a whole, the CC genotype was not associated with higher BMI or obesity as previously observed in GOLDN (Table 1). In view of the different dietary fat intakes between these populations, we focused on gene-dietary fat interactions. We found a statistically significant interaction between total fat and the APOA2 SNP (P<0.05). However, on analyzing the different fat types, the interaction was stronger and more significant for SATFAT, indicating a more specific effect of this variable, and then we focused on SATFAT. When we considered SATFAT as continuous in FOS (Figure 1A), CC subjects exhibited a higher association (B=0.108 Kg/m2; P=0.006) than carriers of the T-allele (B=0.033 Kg/m2; P=0.026) between SATFAT intake and BMI (P for interaction=0.021). Thus, the impact of increased SATFAT intake on BMI increase was most noticeable for CC individuals, with the crossing point between the two regression lines at 22g/d of SATFAT, which was approximately the population mean. We next assessed the relationship of the APOA2 SNP with BMI, stratified according to these levels of SATFAT (Figure 1B). We also detected a statistically significant interaction term (P=0.013) between the APOA2 SNP and SATFAT intake as categorical. Among those within the lower SATFAT strata (<22g/d), the APOA2 SNP was not significantly associated with BMI (P=0.224). In contrast, the CC genotype was associated with greater BMI (∼4.3%; P=0.025) in the higher SATFAT strata. Further adjustment of this interaction for physical activity did not alter the statistical significance of results (P<0.05). Furthermore, in FOS we examined whether the APOA2-SATFAT interaction is influenced by plasma APOA2 concentrations. Thus, the basic model was adjusted for APOA2 and we found that the significance of the interaction term remained practically unchanged (P=0.012).
Figure 1. Interaction between the APOA2 -265T>C polymorphism and SATFAT intake on BMI in the Framingham Study.
Panel A, predicted values of BMI at exam 5 by the APOA2 -265T>C polymorphism (n=1217 carriers of the T allele and n=237 CC subjects) depending on the SATFAT consumed (as continuous) in men and women are depicted. Predicted values were calculated from the regression models containing SATFAT intake, the APOA2 polymorphism, their interaction term, and the potential confounders [sex, age (as continuous), tobacco smoking (as categorical), alcohol consumption (as categorical), diabetes status (as categorical), cholesterol medication (as categorical) and total energy intake (as continuous)]. To estimate these predicted values, BMI was first adjusted for covariates and the residuals used as dependent variable in the model containing the interaction variables. Predicted values for this model were obtained for each individual. Round and squared symbols represent estimated values for T-allele carriers and CC subjects, respectively. The P value for the interaction term was obtained in the multivariate interaction model. R2 in Figure refers to all the variables in the model (r2: 0.016; P=0.01 for the interaction variables). In the stratified analysis by genotype, multivariate adjusted regression coefficients (B), 95%CI and the corresponding P values were estimated after adjustment for the described covariates. Further adjustment for other macronutrients including carbohydrates (as continuous), proteins (as continuous) and total fat (as continuous) did not change the statistical significance of the interaction term or of regression coefficients. Panel B represents means of BMI at exam 5 in both men and women depending on the APOA2 -265T>C polymorphism according to the strata of SATFAT intake (below and above 22g/d). Estimated means were adjusted for sex, age (as continuous), tobacco smoking (as categorical), alcohol consumption (as categorical), diabetes status (as categorical), cholesterol medication (as categorical) and total energy intake (as continuous). P value for the interaction term between SATFAT intake (as dichotomous) and the APOA2 polymorphism were obtained in the multivariate interaction model. In the stratified analysis, P values were estimated after multivariate adjustment for the covariates indicated above. Further adjustment for the other macronutrients including carbohydrates (as continuous), proteins (as continuous) and total fat (as continuous) did not change the statistical significance of the interaction term or of the P values in the stratified analyses. Bars indicate standard error (SE) of means. Panels C and D represent the longitudinal analysis of BMI depending on the APOA2 −265T>C polymorphism across 20 years of follow-up in the Framingham Study. We included 1087 subjects who attended each of the first five exams: exam 1 (1971-1975), exam 2 (1979– 1983), exam 3 (1984-1987), exam 4 (1987-991) and exam 5 (1991-1995). Models for repeated measures were fitted and the corresponding P values are shown. The model in panel C did not include the interaction term between the polymorphism and SATFAT and was adjusted for sex and age. Model in panel D additionally included the interaction term with SATFAT (as dichotomous, two strata as in exam 5). Further adjustment for total energy intake did not change the statistical significance of results.
To verify the internal replication of this gene-diet interaction, we analyzed BMI data from 1,087 subjects who attended Framingham examinations 1 through 5 (20 years of follow-up). When the interaction with SATFAT was not considered, no differences in BMI were observed depending on the APOA2 SNP at any exam (Figure 1C). However, if two strata of SATFAT intake were considered (assuming a similar fat intake strata across the exams), a statistically significant interaction between the APOA2 SNP and SATFAT (P=0.035) on BMI across the 20-year follow-up was noted (Figure 1D). Thus, consistent with the results observed for exam 5, CC subjects present a higher BMI than the other genotypes throughout the 20-year follow-up period, only when they have a high saturated fat consumption.
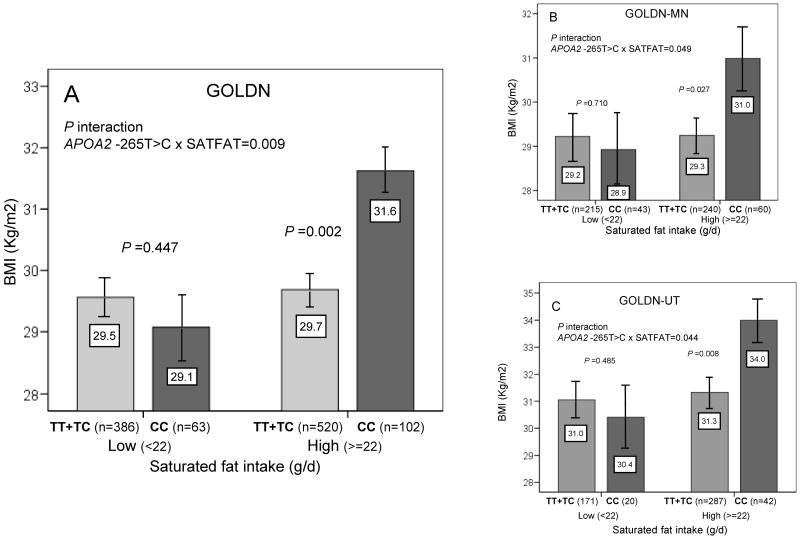
Due to the relevance and novelty of this gene-diet interaction, we examined its replication in GOLDN. Considering the two categories of SATFAT (below and above 22 g/d) that we considered in Framingham, we consistently found the same statistically significant interaction whether considering the unadjusted model or the multivariate basic model (P=0.009) (Figure 2A). Further adjustment of this basic model for family relationships (P=0.040) or for physical activity (P=0.007) did not alter the statistical significance of results. Given that this GOLDN population consumes a higher SATFAT-diet, the APOA2 SNP was generally associated with higher BMI. However, this observation was not present in GOLDN subjects with a low SATFAT intake (P=0.447). In contrast, the CC genotype was strongly associated with greater BMI in subjects with a high SATFAT intake (∼6.4%; P=0.002).
Figure 2. Interaction between the APOA2 − 265T>C polymorphism and SATFAT intake on BMI in the GOLDN Study.
Means of BMI in the entire GOLDN population (panel A), GOLDN-Minnesota subjects (n=558) (panel B) and GOLDN-Utah subjects (n=520) (panel C) are shown depending on the APOA2 -265T>C polymorphism according to the strata of SATFAT intake (below and above 22g/d). Estimated means were adjusted for sex, age (as continuous), tobacco smoking (as categorical), alcohol consumption (as categorical), diabetes status (as categorical), cholesterol medication (as categorical) and total energy intake (as continuous). P values for the interaction terms between SATFAT intake (as dichotomous) and the APOA2 polymorphism in each population were obtained in the hierarchical multivariate interaction model. In the stratified analysis by SATFAT intake levels, P values for mean comparisons of BMI between APOA2 genotypes were estimated after multivariate adjustment for the covariates indicated above. Further adjustment for other macronutrients including carbohydrates, proteins and total fat (all as continuous) did not change the statistical significance of the interaction term or of the P values in the stratified analyses. Bars indicate standard error (SE) of means.
Further internal replication of this interaction was obtained from separate analyses of the Minnesota- and Utah-based subjects (Figure 2B and 2C).
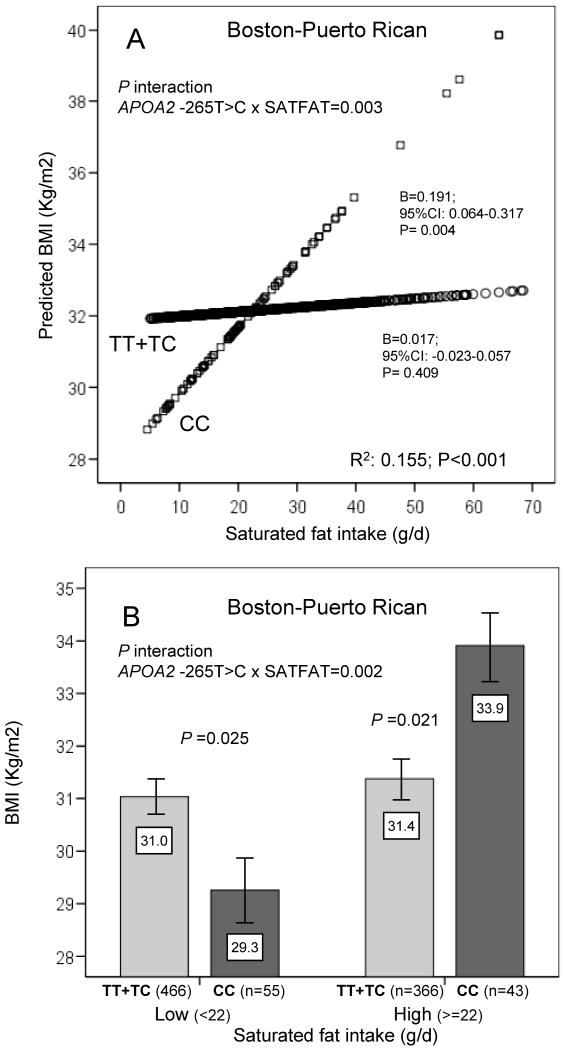
Furthermore, we investigated the replication of this gene-diet interaction in an ethnically different population of US-Hispanics of Caribbean origin living in Boston. We consistently found a statistically significant interaction between the APOA2 SNP and SATFAT on BMI (basic models) whether SATFAT was considered as continuous (P=0.003) or as categorical (P=0.002) (Figure 3 A and B). After additional adjustment for physical activity, the interaction terms remained statistically significant (P=0.004 and P=0.001, respectively). These results were totally in accordance with our previous findings in Whites. Thus, in the Boston-Puerto Rican study, when SATFAT intake was high, CC subjects also had significantly higher BMI than carriers of the T allele (∼7.9%; P=0.021). Moreover, further adjustment of basic models for admixture (27) did not change the statistical significance of the interaction terms (P=0.006 and P=0.003, for continuous and for categorical SATFAT variables, respectively).
Figure 3. Interaction between the APOA2 -265T>C polymorphism and SATFAT intake on BMI in the Boston-Puerto Rican Study.
In Panel A, predicted values of BMI by the APOA2 -265T>C polymorphism (n=832 carriers of the T allele and n=98 CC subjects) depending on the SATFAT consumed (as continuous) in both men and women are depicted. Predicted values were calculated from the regression models containing the SATFAT intake, the APOA2 polymorphism, their interaction term, and the potential confounders [sex, age (as continuous), tobacco smoking (as categorical), alcohol consumption (as categorical), diabetes status (as categorical), cholesterol medication (as categorical) and total energy intake (as continuous). To estimate these predicted values, BMI was first adjusted for covariates and the residuals used as dependent variable in the model containing the interaction variables. The P value for the interaction term was obtained in the multivariate interaction model. Round and squared symbols represent estimated values for T-allele carriers and CC subjects, respectively. R2 in Figure refers to all the variables in the model (r2: 0.012; P=0.008 for the interaction variables). In the stratified analysis by genotype, multivariate adjusted regression coefficients (B), 95%CI and the corresponding P values were estimated after adjustment for the described covariates. Further adjustment for other macronutrients including carbohydrates (as continuous), proteins (as continuous) and total fat (as continuous) did not change the statistical significance of the interaction term or of regression coefficients. Panel B represents means of BMI in both men and women depending on the APOA2 -265T>C polymorphism according to the strata of SATFAT intake (below and above 22g/d). Estimated means were adjusted for sex, age (as continuous), tobacco smoking (as categorical), alcohol consumption (as categorical), diabetes status (as categorical), cholesterol medication (as categorical) and total energy intake (as continuous). P value for the interaction term between SATFAT intake (as dichotomous) and the APOA2 polymorphism were obtained in the multivariate interaction model. In the stratified analysis by SATFAT intake levels, P values for mean comparisons of BMI between APOA2 genotypes were estimated after multivariate adjustment for the covariates indicated above. Further adjustment for the other macronutrients including carbohydrates (as continuous), proteins (as continuous) and total fat (as continuous) did not change the statistical significance of the interaction term or of the P values in the stratified analyses. Bars indicate standard error (SE) of means.
Considering that in the Boston-Puerto Rican population prevalence of diabetes was high (42%), we analyzed if the APOA2-SATFAT interaction was present in both diabetic and non-diabetic subjects. The internal replication of this interaction was also obtained (P for interaction <0.05 in each group, results not shown).
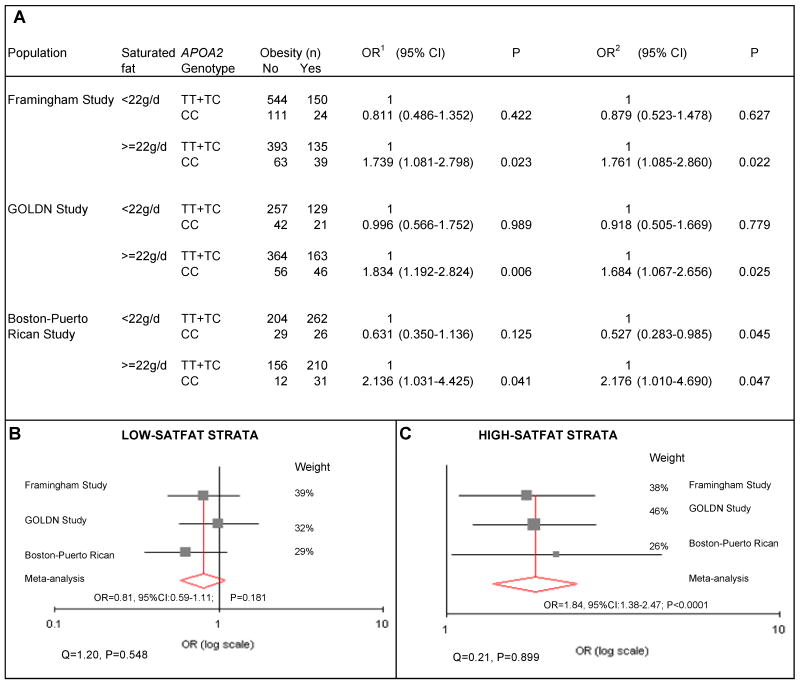
Finally, we examined the APOA2-SATFAT interaction in determining obesity in the three populations independently and pooled in a meta-analysis (Figure 4). We found consistent gene-diet interactions across all three populations. The CC genotype was only associated with a higher obesity in subjects in the high-SATFAT stratum. If SATFAT consumption was low, the CC genotype was not associated with obesity. In the meta-analysis, we observed no significant heterogeneity either for the high-SATFAT (I²=0%, P=0.899) or for the low-SATFAT stratum (I²=0%, P=0.548). The overall association meta-analysis in the high-SATFAT group showed a statistically higher OR of obesity for CC homozygotes of OR: 1.84 (95%CI: 1.38, 2.47; P<0.0001), using the fixed effect model. However, in the low-SATFAT group no increased OR for obesity was found for CC homozygotes in comparison with carriers of the T allele (OR =0.81; 95%CI:0.59-1.11; P=0.181).
Figure 4. Interaction between the APOA2 -265T>C polymorphism and SATFAT intake in determining obesity risk in three independent populations (the Framingham Study, the GOLDN Study and the Boston-Puerto Rican Study).
Separated and pooled analyses depending on the SATFAT intake strata (below and above 22g/d). Panel A represents logistic regression estimation in determining obesity risk in each independent population according to the SATFAT intake strata (below and above 22g/d). Study-specific odds ratios (OR) and 95% confidence interval (CI) were estimated for each strata of SATFAT intake. Two separate multivariate adjustments were performed. Model 1 was adjusted for sex, age (as continuous), tobacco smoking (as categorical), alcohol consumption (as categorical), diabetes status (as categorical), lipid medication (as categorical). Model 2 was additionally adjusted for energy intake (as continuous) and for the other macronutrients including carbohydrates (as continuous), proteins (as continuous) and total fat (as continuous). Further adjustment for physical activity did not change the statistical significance of the results in either of the three populations. Moreover, additional adjustment for family relationships in the GOLDN study or for admixture in the Boston-Puerto Rican study did not change the statistical significance of Models 1 or 2.
Considering that the adjustment for other macronutrients, physical activity or for the other variables did not change the statistical significance of results, the meta-analysis was undertaken with study-specific estimates obtained from model 1. Panel B and Panel C show study-specific estimates of OR and the pooled estimation of obesity risk in CC subjects depending on the two strata of SATFAT intake (low and high respectively) in comparison with carriers of the T allele. Heterogeneity was tested by the χ2-based Q–statistic. We pooled study-specific estimates according to the inverse-variance fixed effect. We obtained statistically significant interaction terms (P<0.05) in the logistic regression models between the SATFAT strata and the APOA2 polymorphism in both the White-Americans and the US-Hispanic of Caribbean origin.
Comment
We have replicated a gene-diet interaction influencing body-weight in three independent US-populations. This is the first time that such consistent replication is found in nutrigenetic studies. This novel and reliable interaction involves the APOA2 −265T>C SNP and SATFAT intake on BMI and obesity. When SATFAT intake is low, the APOA2 −265T>C SNP does not affect BMI. However, when SATFAT intake is high, this SNP is strongly associated with BMI and obesity. Therefore, this APOA2-SATFAT interaction may clarify previous controversial associations reported for this promoter polymorphism (10,12,16,17). The APOA2 SNP can be considered as a thrifty genotype, as depending on the presence of an “obesogenic” (high-SATFAT diet) or “restrictive” (low-SATFAT diet) environment, the phenotypic expression is different. We have selected the cut-off point of 22g/d to define the two SATFAT strata based on the Framingham results and considering that this amount of fat represents 10% of daily energy intake in a standard 2.000 Kcal/d diet. This figure has been largely reported as the threshold between low and high-SATFAT diets (29). Moreover, we have demonstrated a linear dose-effect in the interaction that contributes to its independence from a fixed cut-off level. Another strength of this study is the replication of the interaction, not only in White-Americans, but also in US-Hispanics of Caribbean origin, with a lower C-allele prevalence, which contribute to its external validity and reinforce the notion that CC subjects are especially susceptible to the detrimental effect of high-SATFAT diets on obesity prevalence. Furthermore, the magnitude of the association with obesity was very homogeneous across populations, and higher (OR=1.84 in the meta-analysis) than usually reported in relevant genetic studies (30). Our findings also demonstrated a good internal replication within each population. Of note is the interesting longitudinal observation found in the Framingham Study over 20-year follow-up. One limitation of this analysis is the assumption of a similar classification of subjects into lower or higher SATFAT strata for the whole period. However, in previous work analyzing Framingham diets at exams 3 and 5, we demonstrated stable patterns of consumption over time, specifically for intakes of SATFAT (31).
On the other hand, the association between SATFAT intake and obesity risk is controversial and has been the subject of intense debate (32,33). One explanation may be the different response to SATFAT depending on the individual genotype. Here we demonstrated that the effect of SATFAT on BMI and obesity is highly dependent on the APOA2 −265T>C genotype. Furthermore, although this gene-diet interaction only applies to 10-15% of the population (those with the CC genotype), other genes could have similar interactions, contributing to the diversity and complexity of obesity. Considering that efforts to manage obesity have been soundly defeated, new perspectives to reenergize prevention and treatment are needed. Thus, studying the interaction effects of dietary factors and genes on obesity may define the new challenge of this field. Moreover, recent studies (34) have outlined the fact that Mediterranean, low-carbohydrate or other diets may be effective alternatives to low-fat diets to lose weight. However, these studies did not analyze genetic factors and so did not identify which subjects could respond better to each diet. Such information will be crucial in the new era of obesity research.
These results should stimulate more mechanistic research to explain such epidemiological interactions. On this issue, there are some lines of evidence supporting our findings. Thus, genetic linkages between body weight and lipoprotein metabolism in mice are strongly suggested by a QTL for body weight pointing to Apoa2. Moreover, studies in mice have revealed a role of APOA2 gene expression on insulin resistance, obesity and atherosclerosis (10,13,14,35), but with controversial results that have been attributed to dietary interactions (36). In addition, our further in silico analysis of the -265 APOA2 region indicates the possibility of allele-specific binding of the transcription factor CEBPA [CCAAT/enhancer binding protein (C/EBP), alpha], which has been involved in adipogenesis (37). Our results also suggest that APOA2 acts as a satiety signal, as described for APOA4 in other studies (38), given the significant associations between the APOA2 −265T>C SNP and food intake in Framingham and in GOLDN (12).
In conclusion, we have consistently replicated a gene-diet interaction on BMI and obesity in three US-populations, by which APOA2 CC subjects seem more susceptible to increased BMI and obesity when consuming a high-SATFAT diet. Therefore, if no unmeasured confounders exist, and these results are replicated in subsequent trials, personalized nutritional recommendations in terms of specific reductions of SATFAT intake in CC-subjects may be a future nutrigenetics application.
Supplementary Material
Table 1 (Supplemental material). Association between the APOA2 -265T>C polymorphism and plasma APOA2 or APOA1 concentrations in the whole population and stratified by gender and by obesity in the Framingham Study
Table 2 (Supplemental material). Association between the APOA2 -265T>C polymorphism and dietary intake in men and women depending on the obesity status in the Framingham Study
Acknowledgments
Funding/Support: This work was supported by National Heart, Lung, and Blood Institute grants U 01 HL72524 and HL-54776, National Institute on Aging, Grant Number 5P01AG023394-05 and by contracts 53-K06-5-10 and 58-1950-9-001 from the US Department of Agriculture Research Service, the Ministerio de Ciencia e Innovación (PR2008-268), CNIC06, CIBER CB06/03/0035 and RD07/0067/0006 from the ISCIII, Spain.
Role of the Sponsor: No funding organization or sponsor aside from the National Heart, Lung, and Blood Institute played a role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Footnotes
Author Contributions: Drs Ordovas and Corella had full access to all of the data and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Corella, Ordovas
Acquisition of data: Arnett, Tucker,
Analysis and interpretation of data: Corella, Cupples, Demissie, Ordovas, Peloso,
Drafting of the manuscript: Corella, Ordovas
Critical revision of the manuscript for important intellectual content: Coltell, Lai, Parnell
Statistical analysis: Corella, Cupples, Demissie, Peloso,
Obtained funding: Arnett, Corella, Ordovas, Tucker,
Administrative, technical, or material support: Lee
Study supervision: Ordovas, Corella
Financial Disclosures: None reported.
References
- 1.Guttmacher AE, Porteous ME, McInerney JD. Educating health-care professionals about genetics and genomics. Nat Rev Genet. 2007;8:151–7. doi: 10.1038/nrg2007. [DOI] [PubMed] [Google Scholar]
- 2.Pearson TA, Manolio TA. How to interpret a genome-wide association study. JAMA. 2008;299:1335–44. doi: 10.1001/jama.299.11.1335. [DOI] [PubMed] [Google Scholar]
- 3.Corella D, Ordovas JM. Integration of environment and disease into ‘omics’ analysis. Curr Opin Mol Ther. 2005;7:569–76. [PubMed] [Google Scholar]
- 4.Ordovas JM, Corella D. Nutritional genomics. Annu Rev Genomics Hum Genet. 2004;5:71–118. doi: 10.1146/annurev.genom.5.061903.180008. [DOI] [PubMed] [Google Scholar]
- 5.Afman L, Müller M. Nutrigenomics: from molecular nutrition to prevention of disease. J Am Diet Assoc. 2006;106:569–76. doi: 10.1016/j.jada.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 6.Corella D, Ordovas JM. Single nucleotide polymorphisms that influence lipid metabolism: Interaction with Dietary Factors. Annu Rev Nutr. 2005;25:341–90. doi: 10.1146/annurev.nutr.25.050304.092656. [DOI] [PubMed] [Google Scholar]
- 7.Loos RJ, Rankinen T. Gene-diet interactions on body weight changes. J Am Diet Assoc. 2005;105:S29–34. doi: 10.1016/j.jada.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Ioannidis JP, Boffetta P, Little J, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 9.NCI-NHGRI Working Group on Replication in Association Studies. Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 10.van 't Hooft FM, Ruotolo G, Boquist S, de Faire U, Eggertsen G, Hamsten A. Human evidence that the apolipoprotein A-II gene is implicated in visceral fat accumulation and metabolism of triglyceride-rich lipoproteins. Circulation. 2001;104:1223–8. doi: 10.1161/hc3601.095709. [DOI] [PubMed] [Google Scholar]
- 11.Takada D, Emi M, Ezura Y, et al. Interaction between the LDL-receptor gene bearing a novel mutation and a variant in the apolipoprotein A-II promoter: molecular study in 1135-member familial hypercholesterolemia kindred. J Hum Genet. 2002;47:656–664. doi: 10.1007/s100380200101. [DOI] [PubMed] [Google Scholar]
- 12.Corella D, Arnett DK, Tsai MY, et al. The -256T>C polymorphism in the apolipoprotein A-II gene promoter is associated with body mass index and food intake in the genetics of lipid lowering drugs and diet network study. Clin Chem. 2007;53:1144–52. doi: 10.1373/clinchem.2006.084863. [DOI] [PubMed] [Google Scholar]
- 13.Kalopissis AD, Pastier D, Chambaz J. Apolipoprotein A-II: beyond genetic associations with lipid disorders and insulin resistance. Curr Opin Lipidol. 2003;14:165–72. doi: 10.1097/00041433-200304000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Castellani LW, Goto AM, Lusis AJ. Studies with apolipoprotein A-II transgenic mice indicate a role for HDLs in adiposity and insulin resistance. Diabetes. 2001;50:643–51. doi: 10.2337/diabetes.50.3.643. [DOI] [PubMed] [Google Scholar]
- 15.Castellani LW, Nguyen CN, Charugundla S, et al. Apolipoprotein AII is a regulator of very low density lipoprotein metabolism and insulin resistance. J Biol Chem. 2008;283:11633–44. doi: 10.1074/jbc.M708995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lara-Castro C, Hunter GR, Lovejoy JC, Gower BA, Fernandez JR. Apolipoprotein A-II polymorphism and visceral adiposity in African-American and white women. Obes Res. 2005;13:507–12. doi: 10.1038/oby.2005.53. [DOI] [PubMed] [Google Scholar]
- 17.Xiao J, Zhang F, Wiltshire S, et al. The apolipoprotein AII rs5082 variant is associated with reduced risk of coronary artery disease in an Australian male population. Atherosclerosis. 2008;199:333–9. doi: 10.1016/j.atherosclerosis.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 18.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4:518–25. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 19.Osgood D, Corella D, Demissie S, et al. Genetic variation at the scavenger receptor class B type I gene locus determines plasma lipoprotein concentrations and particle size and interacts with type 2 diabetes: the Framingham Study. J Clin Endocrinol Metab. 2003;88:2869–79. doi: 10.1210/jc.2002-021664. [DOI] [PubMed] [Google Scholar]
- 20.Corella D, Lai CQ, Demissie S, et al. APOA5 gene variation modulates the effects of dietary fat intake on body mass index and obesity risk in the Framingham Heart Study. J Mol Med. 2007;85:119–28. doi: 10.1007/s00109-006-0147-0. [DOI] [PubMed] [Google Scholar]
- 21.Kiely DK, Wolf PA, Cupples LA, Beiser AS, Kannel WB. Physical activity and stroke risk: the Framingham Study. Am J Epidemiol. 1994;140:608–20. doi: 10.1093/oxfordjournals.aje.a117298. [DOI] [PubMed] [Google Scholar]
- 22.Lai CQ, Tucker KL, Parnell LD, et al. PPARGC1A variation associated with DNA damage, diabetes, and cardiovascular diseases: the Boston Puerto Rican Health Study. Diabetes. 2008;57:809–16. doi: 10.2337/db07-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee IM, Paffenbarger RS., Jr Physical activity and stroke incidence: the Harvard Alumni Health Study. Stroke. 1998;29:2049–2054. doi: 10.1161/01.str.29.10.2049. [DOI] [PubMed] [Google Scholar]
- 24.Rimm EB, Giovannucci EL, Stampfer MJ, Colditz GA, Litin LB, Willett WC. Reproducibility and validity of an expanded self-administered semiquantitative food frequency questionnaire among male health professionals. Am J Epidemiol. 1992;135:1114–26. doi: 10.1093/oxfordjournals.aje.a116211. [DOI] [PubMed] [Google Scholar]
- 25.Subar AF, Thompson FE, Kipnis V, et al. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires: the Eating at America's Table Study. Am J Epidemiol. 2001;154:1089–99. doi: 10.1093/aje/154.12.1089. [DOI] [PubMed] [Google Scholar]
- 26.Tucker KL, Bianchi L, Maras J, Bermudez OI. Adaptation of a food frequency questionnaire to assess diets of Puerto Rican and non-Hispanic adults. Am J Epidemiol. 1998;148:507–18. doi: 10.1093/oxfordjournals.aje.a009676. [DOI] [PubMed] [Google Scholar]
- 27.Lai CQ, Tucker KL, Choudhry S, Parnell LD, Mattei J, García-Bailo B, Beckman K, Burchard EG, Ordovás JM. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. Hum Genet. 2009;125:199–209. doi: 10.1007/s00439-008-0612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luan JA, Wong MY, Day NE, Wareham NJ. Sample size determination for studies of gene-environment interaction. Int J Epidemiol. 2001;30:1035–40. doi: 10.1093/ije/30.5.1035. [DOI] [PubMed] [Google Scholar]
- 29.Lichtenstein AH, Kennedy E, Barrier P, et al. Dietary fat consumption and health. Nutr Rev. 1998;56:S3–19. doi: 10.1111/j.1753-4887.1998.tb01728.x. [DOI] [PubMed] [Google Scholar]
- 30.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Millen BE, Quatromoni PA, Pencina M, et al. Unique dietary patterns and chronic disease risk profiles of adult men: the Framingham nutrition studies. J Am Diet Assoc. 2005;105:1723–34. doi: 10.1016/j.jada.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 32.Moussavi N, Gavino V, Receveur O. Could the quality of dietary fat, and not just its quantity, be related to risk of obesity? Obesity (Silver Spring) 2008;16:7–15. doi: 10.1038/oby.2007.14. [DOI] [PubMed] [Google Scholar]
- 33.Gardner CD, Kiazand A, Alhassan S, et al. Comparison of the Atkins, Zone, Ornish, and LEARN diets for change in weight and related risk factors among overweight premenopausal women: the A TO Z Weight Loss Study: a randomized trial. JAMA. 2007;297:969–77. doi: 10.1001/jama.297.9.969. [DOI] [PubMed] [Google Scholar]
- 34.Shai I, Schwarzfuchs D, Henkin Y, et al. Weight loss with a low-carbohydrate, Mediterranean, or low-fat diet. N Engl J Med. 2008;359:229–41. doi: 10.1056/NEJMoa0708681. [DOI] [PubMed] [Google Scholar]
- 35.Blanco-Vaca F, Escolà-Gil JC, Martín-Campos JM, Julve J. Role of apoA-II in lipid metabolism and atherosclerosis: advances in the study of an enigmatic protein. J Lipid Res. 2001;42:1727–39. [PubMed] [Google Scholar]
- 36.Escolà-Gil JC, Marzal-Casacuberta A, Julve-Gil J, et al. Human apolipoprotein A-II is a pro-atherogenic molecule when it is expressed in transgenic mice at a level similar to that in humans: evidence of a potentially relevant species-specific interaction with diet. J Lipid Res. 1998;39:457–62. [PubMed] [Google Scholar]
- 37.Krempler F, Breban D, Oberkofler H, et al. Leptin, peroxisome proliferator-activated receptor-gamma, and CCAAT/enhancer binding protein-alpha mRNA expression in adipose tissue of humans and their relation to cardiovascular risk factors. Arterioscler Thromb Vasc Biol. 2000;20:443–9. doi: 10.1161/01.atv.20.2.443. [DOI] [PubMed] [Google Scholar]
- 38.Tso P, Liu M. Apolipoprotein A-IV, food intake, and obesity. Physiol Behav. 2004;83:631–43. doi: 10.1016/j.physbeh.2004.07.032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1 (Supplemental material). Association between the APOA2 -265T>C polymorphism and plasma APOA2 or APOA1 concentrations in the whole population and stratified by gender and by obesity in the Framingham Study
Table 2 (Supplemental material). Association between the APOA2 -265T>C polymorphism and dietary intake in men and women depending on the obesity status in the Framingham Study