Abstract
Nonsyndromic cleft lip with or without cleft palate (NSCLP) is a common birth anomaly that requires prolonged multidisciplinary rehabilitation. Although variation in several genes has been identified as contributing to NSCLP, most of the genetic susceptibility loci have yet to be defined. To identify additional contributory genes, a high-throughput genomic scan was performed using the Illumina Linkage IVb Panel platform. We genotyped 6008 SNPs in nine non-Hispanic white NSCLP multiplex families and a single large African-American NSCLP multiplex family. Fourteen chromosomal regions were identified with LOD>1.5, including six regions not previously reported. Analysis of the data from the African-American and non-Hispanic white families revealed two likely chromosomal regions: 8q21.3–24.12 and 22q12.2–12.3 with LOD scores of 2.98 and 2.66, respectively. On the basis of biological function, syndecan 2 (SDC2) and growth differentiation factor 6 (GDF6) in 8q21.3–24.12 and myosin heavy-chain 9, non-muscle (MYH9) in 22q12.2–12.3 were selected as candidate genes. Association analyses from these genes yielded marginally significant P-values for SNPs in SDC2 and GDF6 (0.01≤P<0.05). Evidence for an altered transmission was found for four MYH9 SNPs (P<0.01). SNP rs1002246 exhibited altered transmission by all analytic methods. However, analysis of two SNP MYH9 haplotypes did not identify a single high-risk haplotype. Our results confirm a previous report that 8q21.3–24.12 may harbor a clefting gene and identify 22q12.2–12.3 as a new candidate region that contains MYH9. Most importantly, we confirm the previous report of an association with MYH9.
Keywords: cleft lip and palate, genome scan, SDC2, GDF6, MYH9
Introduction
Nonsyndromic cleft lip with or without cleft palate (NSCLP) is a common congenital anomaly with a prevalence of 1/500–1/1400 live births and varies with ethnicity.1, 2, 3 Non-Hispanic white and Hispanic populations have a higher prevalence than African-American populations but a lower prevalence than Asian populations.1, 3, 4 NSCLP is considered to be a multifactorial disorder, that is, caused by both genes and environmental exposures, neither of which has been fully defined. Anywhere from 2 to 14 genes have been proposed to contribute to the NSCLP phenotype and association and linkage studies have identified some of these putative genes.5, 6, 7
Genome scans have proven to be a useful tool for identifying NSCLP candidate chromosomal regions. Seven NSCLP genome scans have been reported involving populations from East Asia, India, Europe, Syria and Turkey.8, 9, 10, 11, 12, 13, 14 In addition, a meta-analysis was performed on data generated from 13 NSCLP studies, including 5 of the aforementioned studies and 8 additional unpublished data sets.15 The individual genomic scans have identified 3–20 candidate regions (Supplementary Table 1), and 1p31–36, 2q32–37, 6q21–27 and 16q21–24 were common to at least four of the eight studies listed in Supplementary Table 1.8, 9, 10, 11, 12, 13, 14, 15
To confirm previously reported chromosomal regions and to identify new chromosomal regions likely to contain NSCLP genes, we subjected 10 multiplex NSCLP families to genomic screening utilizing the Illumina Linkage IVb 6K SNP Linkage Panel (Illumina Inc., San Diego, CA, USA). Of these, nine families are from our well-characterized non-Hispanic white data set and one family was African American. We identified one new region from each ethnic group with an LOD >2.5; one from chromosome 8q21.3–24.12 (2.98; African American) and one from chromosome 22q12.2–q12.3 (2.66; non-Hispanic white). Two candidate genes from the chromosome 8 region and one from the chromosome 22 region were further evaluated.
Inspection of the region(s) with the highest LOD score from each ethnic group identified three genes of interest: growth differentiation factor 6 (GDF6) and syndecan 2 (SDC2) in 8q21.3–24.12 and myosin heavy chain 9 (MYH9) in 22q12.2–12.3.16 SDC2 and GDF6 on chromosome 8 were considered to be good candidate genes because of their known biological function. SDC2 is involved in cell signaling, migration and cell–matrix interactions and interacts with members of the transforming growth factor-β family.17, 18, 19 GDF6 is a member of the bone morphogenetic protein group that forms part of the transforming growth factor-β superfamily and is expressed in the craniofacial region during embryogenesis20, 21, 22, 23, 24 MYH9 on chromosome 22 is of particular interest because it is highly expressed in the palatal shelves before fusion.25 Moreover, in a recent study of NSCLP in a group of Italians, an association was identified with an SNP in MYH9.26 In this study, we identified SNPs spanning the candidate genes and evaluated them for evidence and/or association.
Materials and methods
Data set
The data set consists of 123 families with a history of NSCLP. Of these, there are 81 extended multiplex families (65 non-Hispanic white, 15 Hispanic and 1 African American) and 42 multiplex trios/duos (23 non-Hispanic white and 19 Hispanic) with a positive history of NSCLP. In addition, there are 316 simplex trios/duos (226 non-Hispanic white, 82 Hispanic and 8 African American) with no family history of NSCLP. These families, methods of ascertainment and inclusion criteria have been reported earlier.27 Blood or saliva samples were collected after obtaining informed consent. DNA was extracted from blood using Roche DNA Isolation Kit for Mammalian Blood (Roche, Basel, Switzerland) or from saliva using Oragene Purifier (DNA Genotek Inc., Ottawa, ON, Canada) following the manufacturer's protocol.
Genome scan
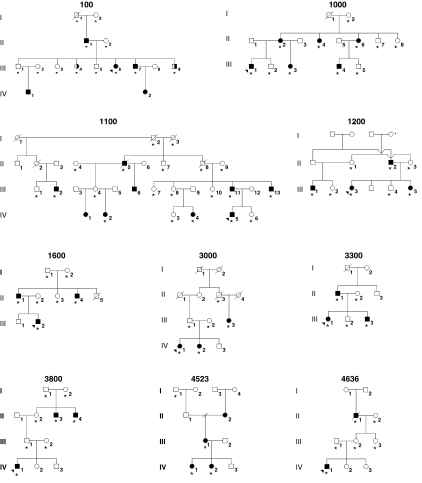
Ten multiplex families were subjected to a genome scan using the 6 K Illumina Linkage IVb mapping panel. These families were selected from our data set on the basis of the number of affected individuals and availability of DNA. Nine families were non-Hispanic white and one (F1100) was African American (Figure 1). Under our dominant model, four of the non-Hispanic white families, 100, 1000, 1200 and 3000, can yield maximum LOD scores greater than 1 (1.6, 1.5, 1.1 and 1.2, respectively). A maximum LOD score of 2.98 is possible for the African-American family. Among the non-Hispanic white families, there were 30 affected individuals of 67 individuals. In family 100 (F100), there were two siblings (III-4 and III-9) who were reported to have a notched gum in childhood, a finding that could indicate a microform cleft lip. However, these diagnoses could not be clinically confirmed. Therefore, the linkage analysis was performed twice: first, assuming these individuals to be affected and then coding them as unknown with regard to the cleft phenotype. There were 27 genotyped individuals in the African-American family, 7 of whom were affected.
Figure 1.
Families included in genome scan. Family 1100 is African American; all the remaining families are non-Hispanic white. *DNA included in the genome scan; arrow denotes proband. Filled symbols denote affected individuals; half-filled symbols denote notched gum; empty symbols denote unaffected individual.
The Illumina Linkage IVb mapping panel consisting of six 008 SNPs was used on a BeadStation system (Illumina Inc.). Allele detection and genotype calling were performed using the BeadStudio software (Illumina Inc.).
Candidate genes
Thirty-seven SNPs spanning the three candidate genes identified in the genome scan (GDF6 – 6; SDC2 – 21; and MYH9 – 10) were subsequently typed in the entire NSCLP data set with the exclusion of the African-American families; selection criteria for these SNPs were discussed earlier.27 GDF6 and SDC2 SNPs were chosen with minor allele frequency >0.3, preferentially selecting coding and TagSNPs to obtain maximum coverage of these genes. Six of the MYH9 SNPs were chosen on the basis of a previous publication;26 the remaining four SNPs in the MYH9 gene were selected using the above criteria. TaqMan Assays (Applied Biosystems; Foster City, CA, USA) were used for genotyping; alleles were detected using the ABI 7900HT Sequence Detection System. All genotyping results were imported into Progeny Lab (South Bend, Indiana, USA), and PedCheck was run on all SNPs to identify Mendelian inconsistencies.28
Analysis
Results of genotyping from the genome scan were subjected to parametric and non-parametric linkage analyses using MERLIN.29 Linkage parameters were as previously calculated, that is, dominant model with penetrances of 0.24 in females and 0.32 in males and a phenocopy rate of 0.001.30 An r2 of 0.19 was the cutoff value for clustering SNPs to remove the effect of linkage disequilibrium. Family 1100 was analyzed separately because of its different race/ethnicity. Analysis of the candidate gene SNPs was restricted to our Hispanic and non-Hispanic white families, as the number of families of other ethnicities was small. For analyses of the candidate gene SNPs, probands/families were stratified by ethnicity alone or by ethnicity and family history. In addition, the Hispanic and non-Hispanic white families were analyzed together for the MYH9 SNPs, as the allele frequencies and LD patterns did not differ between the two ethnicities. Allele frequencies and the Hardy–Weinberg equilibrium (HWE) were calculated using SAS (v9.1). Pairwise linkage disequilibrium values (D′ and r2) were calculated using GOLD.31 To extract the maximum amount of information from the data set, multiple approaches for assessing linkage and/or association were used. Parametric and non-parametric linkage analyses were performed as discussed above. The pedigree disequilibrium test (PDT), Geno-PDT (G-PDT) and association in the presence of linkage (APL) test were used to evaluate evidence for association.32, 33, 34 This panel of analytic tools was chosen because each has different strengths and assumptions. PDT is an extension of the transmission disequilibrium test and allows for the incorporation of extended pedigrees in the analysis of allelic association. The G-PDT examines the association between marker genotypes and disease, whereas APL allows for missing parental genotypes. Each program may be more powerful for certain genetic models and for different pedigree structures. In addition, APL can be used to examine multi-marker haplotypes; it was used to look for the overtransmission of two-marker haplotypes within genes; all possible pairwise combinations of SNPs were included.
Results
Genome scan
Ten families were subjected to the Illumina Linkage IVb SNP genome-wide panel. Nine non-Hispanic white families generated 11 chromosomal regions with an LOD >1.5 with parametric and/or non-parametric analysis (Table 1). The highest LOD score for either methodology was two SNPs in the chromosome 22q12.2–q12.3 region (rs762883 and rs9862). Three of the families, 100, 1000 and 3000, had maximum LOD score >1.0 in the region (1.05, 1.11 and 1.12, respectively), although not for the same set of SNPs (data not shown). Three chromosomal regions with an LOD >1.5 were identified in the large African-American family (Table 2). The highest LOD score (2.98) was found in an SNP in chromosome 8q21.3–24.12. This is the maximum possible LOD score for this family. There was no overlap in the regions identified in these two groups. The multipoint graphs for chromosomes with LOD scores >1.5 are shown in Supplementary Figures 2 and 3.
Table 1. Chromosomal regions with LOD >1.5 in nine non-Hispanic white families.
| Maximum LOD score | |||||||
|---|---|---|---|---|---|---|---|
| Parametric (hLOD) | Non-parametric (LOD) | ||||||
| Chromosome | Regiona | dbSNP | cM | Affb | Unkc | Affb | Unkc |
| 3 | 3p23 | rs342758 | 58.08 | 0.35 | 0.85 | 0.92 | 1.64 |
| 3 | 3p21.1 | rs11235 | 71.53 | 0.87 | 1.58 | 0.49 | 1.03 |
| 3 | 3p13–14.1a | rs736333 | 94.05 | 0.24 | 0.67 | 0.65 | 1.53 |
| 5 | 5p13.3a | rs1966983 | 51.74 | 0.94 | 1.64 | 0.72 | 1.19 |
| 5 | 5q35.2 | rs1875189 | 192.51 | 0.57 | 0.53 | 1.59 | 1.04 |
| 6 | 6q27 | rs727811 | 177.17 | 0.85 | 1.66 | 0.28 | 0.45 |
| 7 | 7q36.2–36.3a | rs7795368 | 182.8 | 1.7 | 2.59 | 0.57 | 1.16 |
| 9 | 9q22.2–22.32a | rs1048510 | 97.53 | 1.78 | 1.27 | 0.96 | 0.82 |
| 9 | 9q33.2–33.3 | rs1984001 | 132.41 | 1.46 | 0.95 | 1.87 | 1.73 |
| 22 | 22q11.23–12.1 | rs763281 | 21.41 | 1.75 | 1.22 | 1.46 | 0.99 |
| 22 | 22q12.2–12.3a | rs762883 | 36.94 | 2.66 | 2.12 | 2.49 | 2.17 |
Novel region identified.
Aff, Analysis of F100 individuals with notched gums classified as affected.
Unk, Analysis of F100 individuals with notched gums classified as unknown.
Table 2. Chromosomal regions with LOD>1.5 in the African-American family.
| Maximum LOD score | |||||
|---|---|---|---|---|---|
| Chromosome | Regiona | dbSNP | cM | Parametric (hLOD) | Non-parametric (LOD) |
| 2 | 2p22a | rs1446297 | 62.97 | 2.27 | 1.07 |
| 3 | 3p26 | rs11130180 | 15.16 | 2 | 0.71 |
| 8 | 8q21.3–24.12 | rs536161 | 97.66 | 2.98 | 1.47 |
Indicates novel region identified.
Candidate genes
All 37 SNPs from the three candidate genes, MYH9, GDF6 and SDC2, were in HWE. The allele frequencies of most SNPs in GDF6 and SDC2 were significantly different between the non-Hispanic whites and Hispanics, even after Bonferroni correction (P=0.001) (Table 3A). Therefore, the data were stratified by ethnicity for the analysis of these genes. In contrast, the differences in the MYH9 SNPs did not meet this criterion (Table 3B). For this reason, MYH9 SNPs were analyzed with the two ethnicities combined.
Table 3A. Allele frequencies of chromosome 8 dbSNPs.
| Gene | dbSNPa | Base pair | Location | MAFb | HCFc,d |
|---|---|---|---|---|---|
| SDC2 | rs2452993 | 97 574 572 | Upstream | 0.42 | 0.59*** |
| SDC2 | rs2437772 | 97 582 153 | Intron 1 | 0.31 | 0.27 |
| SDC2 | rs2437780 | 97 588 852 | Intron 1 | 0.30 | 0.26 |
| SDC2 | rs2575738 | 97 599 578 | Intron 1 | 0.29 | 0.20* |
| SDC2 | rs7003874 | 97 607 128 | Intron 1 | 0.46 | 0.35* |
| SDC2 | rs874643 | 97 611 319 | Intron 1 | 0.19 | 0.40*** |
| SDC2 | rs2440681 | 97 613 757 | Intron 1 | 0.35 | 0.22*** |
| SDC2 | rs2582831 | 97 620 954 | Intron 1 | 0.28 | 0.44*** |
| SDC2 | rs2582839 | 97 627 443 | Intron 1 | 0.29 | 0.46*** |
| SDC2 | rs1444573 | 97 633 837 | Intron 1 | 0.37 | 0.58*** |
| SDC2 | rs1984456 | 97 636 282 | Intron 1 | 0.35 | 0.48*** |
| SDC2 | rs2582846 | 97 642 858 | Intron 1 | 0.20 | 0.43*** |
| SDC2 | rs12156240 | 97 646 383 | Intron 1 | 0.49 | 0.69*** |
| SDC2 | rs2575702 | 97 654 007 | Intron 1 | 0.34 | 0.46** |
| SDC2 | rs2651472 | 97 665 849 | Intron 1 | 0.49 | 0.58* |
| SDC2 | rs2028086 | 97 673 354 | Intron 1 | 0.24 | 0.14** |
| SDC2 | rs1836910 | 97 680 704 | Intron 2 | 0.26 | 0.17* |
| SDC2 | rs1042381 | 97 683 837 | Exon 3 | 0.18 | 0.14 |
| SDC2 | rs2651454 | 97 690 146 | Intron 4 | 0.23 | 0.38*** |
| SDC2 | rs2651458 | 97 694 626 | Downstream | 0.23 | 0.38*** |
| SDC2 | rs714046 | 97 700 749 | Downstream | 0.19 | 0.2 |
| GDF6 | rs2247792 | 97 219 221 | Upstream | 0.41 | 0.21*** |
| GDF6 | rs2440199 | 97 225 587 | Exon 1 | 0.41 | 0.63*** |
| GDF6 | rs2255182 | 97 234 080 | Intron 1 | 0.49 | 0.28*** |
| GDF6 | rs2514527 | 97 238 503 | Intron 1 | 0.38 | 0.64*** |
| GDF6 | rs2514531 | 97 243 999 | Downstream | 0.45 | 0.26*** |
| GDF6 | rs714552 | 97 252 618 | Downstream | 0.40 | 0.61*** |
Bold dbSNPs are also genotyped in genome scan.
Minor allele frequency of non-Hispanic white data set.
Corresponding frequency in Hispanic of non-Hispanic white minor allele.
P-value less than *0.05, **0.001 and ***0.0001.
Table 3B. Allele frequencies of chromosome 22 dbSNPs.
| Gene | dbSNP | Base pair | Location | MAFa | HCFb,c |
|---|---|---|---|---|---|
| MYH9 | rs7078 | 35 007 860 | Downstream | 0.30 | 0.25 |
| MYH9 | rs4821478 | 35 020 059 | Intron 28 | 0.35 | 0.35 |
| MYH9 | rs2009930 | 35 029 252 | Intron 19 | 0.35 | 0.37 |
| MYH9 | rs2239783 | 35 035 074 | Intron 15 | 0.26 | 0.27 |
| MYH9 | rs3752462 | 35 040 129 | Intron 13 | 0.36 | 0.37 |
| MYH9 | rs1002246 | 35 044 605 | Intron 10 | 0.34 | 0.33 |
| MYH9 | rs2071731 | 35 048 804 | Intron 5 | 0.37 | 0.34 |
| MYH9 | rs713659 | 35 058 638 | Intron 3 | 0.42 | 0.37 |
| MYH9 | rs713839 | 35 063 884 | Intron 3 | 0.38 | 0.35 |
| MYH9 | rs739097 | 35 076 025 | Intron 1 | 0.44 | 0.36 |
Minor allele frequency of non-Hispanic white data set.
Corresponding frequency in Hispanic of non-Hispanic white minor allele.
P-value less than *0.05, **0.001 and ***0.0001.
Two-point parametric and non-parametric analyses in the combined data set found evidence for linkage to a single SNP, rs1002246, in MYH9 (HLOD=1.58 and P=0.0006, respectively) (Table 4). There was no evidence for linkage by multipoint analysis (data not shown). However, all three methods of association analysis identified evidence for altered transmission of this same SNP, rs1002246 (Table 5). In addition, PDT detected evidence for an altered transmission of three additional SNPs. When the data were stratified by family history, a marginally significant association was still present for the SNP, rs1002246, in the simplex data set (P=0.03). Inspection of all two-SNP haplotypes found that most haplotypes constructed with rs1002246 and containing the ‘A' allele were overtransmitted, but generally did not reach significance, even when stratified by ethnicity and/or family history (data not shown). The minor allele frequency (ie, frequency of the ‘A' allele) was 0.34 in the non-Hispanic whites. Haplotypes with the ‘G' rs1002246 allele were either undertransmitted or demonstrated the expected transmission.
Table 4. Parametric and non-parametric linkage analysis for MYH9.
| Parametric | Non-parametric | |||||
|---|---|---|---|---|---|---|
| dbSNP | cM | Base pair | hLOD | α | LOD | P-valuea |
| rs7078 | 35.008 | 35 007 860 | 0.018 | 0.083 | 0.01 | 0.4 |
| rs4821478 | 35.02 | 35 020 059 | 0.033 | 0.109 | 0.02 | 0.4 |
| rs2009930 | 35.029 | 35 029 252 | 0 | 0 | −0.07 | 0.7 |
| rs2239783 | 35.035 | 35 035 074 | 0.174 | 0.213 | 0 | 0.5 |
| rs3752462 | 35.04 | 35 040 129 | 0.108 | 0.205 | 0.12 | 0.2 |
| rs1002246 | 35.045 | 35 044 605 | 1.578 | 0.694 | 2.26 | 0.0006 |
| rs2071731 | 35.049 | 35 048 804 | 0 | 0 | 0 | 0.5 |
| rs713659 | 35.059 | 35 058 638 | 0 | 0 | 0 | 0.5 |
| rs713839 | 35.064 | 35 063 884 | 0 | 0.005 | 0 | 0.5 |
| rs739097 | 35.076 | 35 076 025 | 0 | 0 | −0.01 | 0.6 |
P-value <0.05 bolded.
Table 5. Results for MYH9 from current and previously reported study.
| Martinelli et al | Current dataa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Italian sample | Entire sample | Multiplex families | Simplex families | ||||||||
| dbSNP | Location | TDTb | PDT | G-PDT | APL | PDT | G-PDT | APL | PDT | G-PDT | APL |
| rs5995288 | Intron 1 | 0.82 | — | — | — | — | — | — | — | — | — |
| rs739097 | Intron 1 | 0.5 | 0.17 | 0.395 | 0.142 | 0.617 | 0.862 | 0.948 | 0.114 | 0.295 | 0.067 |
| rs713839 | Intron 3 | — | 0.067 | 0.191 | 0.11 | 0.452 | 0.558 | 0.596 | 0.043 | 0.151 | 0.094 |
| rs713659 | Intron 3 | — | 0.211 | 0.397 | 0.098 | 0.756 | 0.894 | 0.545 | 0.091 | 0.17 | 0.014 |
| rs2071731 | Intron 5 | 0.88 | 0.12 | 0.186 | 0.165 | 0.607 | 0.663 | 0.456 | 0.065 | 0.144 | 0.026 |
| rs1002246 | Intron 10 | 1 | 0.02 | 0.043 | 0.009 | 0.056 | 0.149 | 0.127 | 0.185 | 0.262 | 0.033 |
| rs3752462 | Intron 13 | 0.02 | 0.035 | 0.117 | 0.602 | 0.114 | 0.216 | 0.662 | 0.162 | 0.382 | 0.353 |
| rs2239783 | Intron 15 | — | 0.681 | 0.834 | 0.369 | 0.478 | 0.726 | 0.143 | 0.86 | 0.61 | 0.857 |
| rs2009930 | Intron 19 | 0.4 | 0.039 | 0.131 | 0.177 | 0.195 | 0.386 | 0.718 | 0.087 | 0.257 | 0.065 |
| rs4821478 | Intron 28 | — | 0.023 | 0.077 | 0.381 | 0.053 | 0.163 | 0.899 | 0.223 | 0.465 | 0.328 |
| rs2269529 | Exon 34 | 0.69 | — | — | — | — | — | — | — | — | — |
| rs7078 | Downstream | 0.34 | 0.836 | 0.832 | 0.345 | 0.564 | 0.712 | 0.137 | 0.766 | 0.879 | 0.770 |
—, not genotyped.
P-values have not been corrected for multiple testing.
P-value <0.05 bolded.
There was no evidence for linkage to SNPs in SDC2. Marginal significance for altered transmission was detected in both the non-Hispanic white and Hispanic samples (Table 6A and B and Supplementary Tables 2 and 3). In the simplex non-Hispanic white sample, rs1042381 was marginally significant (Table 6A). In the Hispanic sample, significant association was detected in the entire data set for rs198456, in the multiplex families for rs2437772 and rs2437780, and for the simplex families for rs2582831 and rs2582846 (Table 6B). Two SDC2 haplotypes demonstrated excess transmission in the non-Hispanic white group and three in the Hispanic group (Table 7).
Table 6. Association results for SDC2 and GDF6 on chromosome 8.
| All NHWa,b | Simplex NHW | Multiplex NHW | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | dbSNP | PDT | G-PDT | APL | PDT | G-PDT | APL | PDT | G-PDT | APL |
| (A) Non-Hispanic white populationc | ||||||||||
| SDC2 | rs1042381 | 0.796 | 0.822 | 0.632 | 0.046 | 0.029 | 0.289 | 0.123 | 0.075 | 0.349 |
| GDF6 | rs2247792 | 0.804 | 0.223 | 0.149 | 0.804 | 0.223 | 0.147 | 0.631 | 0.013 | 0.244 |
| GDF6 | rs2440199 | 0.955 | 0.348 | 0.597 | 0.955 | 0.348 | 0.587 | 0.449 | 0.05 | 0.887 |
| GDF6 | rs2255182 | 0.582 | 0.036 | 0.764 | 0.582 | 0.036 | 0.763 | 0.679 | 0.019 | 0.678 |
| Alla,b | Simplex | Multiplex | ||||||||
| Gene | dbSNP | PDT | G-PDT | APL | PDT | G-PDT | APL | PDT | G-PDT | APL |
| (B) Hispanic populationc | ||||||||||
| SDC2 | rs2437772 | 0.522 | 0.168 | 0.819 | 0.336 | 0.487 | 0.716 | 1 | 0.035 | 0.915 |
| SDC2 | rs2437780 | 0.796 | 0.139 | 0.95 | 0.578 | 0.839 | 0.975 | 0.858 | 0.04 | 0.959 |
| SDC2 | rs2582831 | 0.655 | 0.89 | 0.147 | 1 | 0.793 | 0.04 | 0.564 | 0.668 | 0.878 |
| SDC2 | rs1984456 | 0.753 | 0.939 | 0.037 | 0.706 | 0.837 | 0.154 | 0.9 | 0.974 | 0.086 |
| SDC2 | rs2582846 | 0.274 | 0.594 | 0.058 | 0.132 | 0.281 | 0.036 | 0.715 | 0.804 | 0.548 |
P-value <0.05 bolded.
P-values have not been corrected for multiple testing.
Results for SNPs with at least one test statistic P<0.05.
Table 7. Overtransmitted haplotypes for SDC2 and GDF6.
| Gene | dbSNP1 | dbSNP2 | Ethnicity | P-value | Haplotype |
|---|---|---|---|---|---|
| SDC2 | rs2452993 | rs7003874 | NHW | 0.01 | 1–2 |
| SDC2 | rs2452993 | rs12156240 | NHW | 0.003 | 2–1 |
| SDC2 | rs2575738 | rs2651472 | H | 0.04 | 2–1 |
| SDC2 | rs2575738 | rs1836910 | H | 0.04 | 1–2 |
| SDC2 | rs2582846 | rs12156240 | H | 0.016 | 2–1 |
| GDF6 | rs2440199 | rs2514527 | H | 0.008 | 2–1 |
NHW, non-Hispanic white; H, Hispanic.
No evidence for linkage to SNPs in GDF6 was detected. Again, altered transmission was only marginally significant for two SNPs: rs2255182 in the complete non-Hispanic white sample and both the multiplex and simplex subgroups (P=0.04, P=0.02 and P=0.04, respectively) and rs2247792 in the non-Hispanic white multiplex families (P=0.03) (Table 6A). Overtransmission of one haplotype involving rs24401199 and rs2514527 was observed in the Hispanic sample (P=0.008) (Table 7).
Discussion
This study was undertaken to identify additional genes that contribute to the NSCLP phenotype. One African-American and nine non-Hispanic white families were analyzed by a 6 K genome scan. Fourteen regions were identified with LOD scores >1.5 and four regions had LOD scores >2 (2p22, 7q36.2–36.3, 8q21.3–24.12 and 22q12.1-q12.3) (Tables 1 and 2). Of these, only the 8q21.3–24.12 region had previously been identified by genomic scanning (Supplementary Table 1).11, 15 The region from each ethnicity that resulted in the highest LOD score was subjected to candidate gene analysis.
Eleven regions on six chromosomes with LOD scores >1.5 were identified in the non-Hispanic white families (Table 2). The highest LOD score was found in chromosomal region 22q12.2–12.3 and the total linkage area spans over 20 cM. This region contains the MYH9 gene, which is expressed in the developing palate and was found to be associated with NSCLP in an Italian population.25, 26 To further evaluate the role of MYH9 in NSCLP, 10 intragenic and flanking SNPs, which included six of the SNPs genotyped in the Italian families, were interrogated in our NSCLP sample.
Linkage was found between a single SNP, rs1002246, in the MYH9 gene and our entire data set (Table 5), consistent with the prior identification of this region in our genome scan. PDT, G-PDT and APL analyses, all identified this SNP in our entire NSCLP sample. rs1002246 differs from the SNP found in the Italian NSCLP sample (Table 5). rs1002246 is in intron 10 of the MYH9 gene but this is not in a region associated with intron–exon splicing. Although intronic SNPs do not typically alter protein structure, an association with intronic variants has been reported for a number of complex diseases.27, 35, 36, 37, 38, 39, 40 rs1002246 is located in a region of reduced LD that can complicate identifying at-risk haplotypes (Supplementary Table 3). In fact, we were unable to identify a high-risk haplotype. Interestingly, rs3752462, the SNP identified by the Italian group, was only marginally significant in both data sets (Table 5). Our finding of linkage by parametric and non-parametric analyses and association strongly suggests that MYH9 may play a causal role in NSCLP. Nevertheless, we cannot conclusively exclude the possibility that another gene in the 22q12.2–12.3 chromosomal region is contributing to the positive linkage and association. We are continuing to evaluate this region.
This is the first NSCLP genomic scan to include an African-American family. NSCLP is relatively rare in African Americans with a birth prevalence of 1/2500, when compared with 1/1400 in non-Hispanic whites and Hispanics in Texas.3 Although earlier genome scans have been performed on non-Hispanic white, Indian, Chinese and Syrian populations, it is a rarity to find a large African-American family with segregating NSCLP8, 9, 10, 11, 12, 13, 14 Using this family, three chromosomal regions, 2p22, 3p26 and 8q21.3–24.12, were identified with LOD scores >1.5, of which only 2p22 is new (Table 2).
The most significant linkage (LOD=2.98) was to 8q21.3–24.12 region, the same region identified by the meta-analysis, providing further support that this region contains an NSCLP gene.15 SDC2 and GDF6 were found in this region and, because of their known biological function, they were considered to be good candidate genes. Nevertheless, linkage was not detected between either of these genes and NSCLP in our data set. Marginal evidence for association (0.01<P<0.05) was detected for SNPs in both of these genes in the non-Hispanic white and Hispanic samples. In SDC2, SNPs rs2437772 and rs2437780 were both significant in the multiplex non-Hispanic white data set using the G-PDT; however, neither PDT nor APL detected a significant association. Therefore, these results are marginal at best. Haplotype analysis revealed altered transmission for two SDC2 haplotypes (P<0.01). However, given the large number of haplotypes tested, this does not provide strong evidence for a role of SDC2 in NSCLP.
G-PDT analysis of SNPs in GDF6 found borderline evidence for association in the non-Hispanic white data set; PDT and APL tests were negative. rs2255182, in intron 1, was significant in the total non-Hispanic white data set as well as when the data were stratified by family history (Table 3A). In addition, rs2247792, 22.5 kb upstream of the GDF6 gene, was associated in the multiplex non-Hispanic white data set. Interestingly, analysis identified a significant haplotype in the Hispanic data set, but not in the non-Hispanic white sample (Table 7). Although association analyses of the SNPs in SDC2 and GDF6 showed some evidence for altered transmission, the level did not correspond to the LOD score detected in this region. This may be the result of linkage without an association. An alternative explanation is that this region was identified in an African-American family, and genes in this region do not play a significant role in the non-Hispanic white and Hispanic populations. NSCLP displays genetic heterogeneity with a variety of genes playing an etiologic role, and it is likely that these genes vary with ethnicity.41 However, because this region was also identified in other ethnicities, additional testing is underway to interrogate this region in the entire data set. Of particular interest is the Frizzled 6 (Fzd6) gene that is located in this region. Fzd6 is a Wnt receptor whose expression is upregulated during osteogenic differentiation of mesenchymal stem cells.42 Fzd6 functions in the noncanonical Wnt pathway to decrease the amount of binding of TCF/LEF and TCF–β-catenin to target DNA.43 We have recently identified an association between several Wnt genes and NSCLP,44 and studies are underway to evaluate Fzd6 for association with NSCLP and interaction with Wnt genes.
Regions 2p22 and 3p26 were also identified in the African-American family; these are new avenues for discovery of NSCLP genes. 2p22 is a novel chromosomal region, whereas 3p26 was previously reported in a genome scan of Chinese multiplex families.9 Further examination of these regions needs to be undertaken to identify novel NSCLP genes.
In addition to 22q12.2–q12.3 (which contains MYH9), 10 regions were identified in the non-Hispanic white families with LOD scores greater than 1.5 but less than 2. Four of these regions, 3p14.1–p13, 5p13.3, 7q36.2–q36.3 and 9q22.22–q22.32, were novel; however, and there are no obvious candidate genes in two of these regions (3p14.1–p13 and 5p13.3). The remaining six regions have been reported earlier in genomic scans.8, 9, 10, 11, 13, 14, 15
The 7q36.2–36.3 region is novel and contains sonic hedgehog homolog (SHH) and engrailed homeobox 2 (EN2), two known NSCLP candidate genes. SHH plays a critical role in the development and patterning of the craniofacial processes that give rise to the developing lip and palate.45 Mutations in the human SHH gene cause holoprosencephaly-3, which can be associated with cleft lip and palate (OMIM: #142945);46 SHH null mice can have cleft palate.47 However, single-strand conformational polymorphism analysis of the SHH gene in NSCLP patients has not identified any disease-causing mutations.48 EN2 plays a role in the central nervous system development and has also been implicated in autism spectrum disorders (OMIM: #608636).49 Previous RFLP testing found no association between the EN2 gene and NSCLP.50 Although no positive findings have been found, these remain candidate genes of interest.
Two regions on chromosome 9, 9q22.2–22.32 and 9q33.2–33.3, were identified in this genomic scan. The 9q22.2–22.32 region is novel and contains both the BarH-like homeobox 1 (Barx1) and patched homolog 1 (PTCH1). Mouse BARX1 is expressed in the first and second branchial arches that form the craniofacial processes.51 The role of Barx1 in NSCLP has not been evaluated. PTCH1, an SHH receptor, in combination with BARX1, plays a role in mid-face structure formation.52 Mutations in PTCH1 cause basal cell nevus syndrome (BCNS; OMIM: #109400) and 5% of BCNS patients have a cleft lip and palate.53 Interestingly, forkhead box E1 (FOXE1), which plays a role in thyroid morphogenesis, is located just outside this region in 9q22.33.54 The FOXE1 region has been identified in the NSCLP genome scans.55 Mutations in FOXE1 cause the Bamforth–Lazarus syndrome of which cleft palate is an associated finding (OMIM: #241850). However, sequencing of the FOXE1 gene in the NSCLP probands identified two missense mutations, both of which were predicted to be benign.56
There are some limitations to the study. NSCLP is clearly heterogeneous, and among the non-Hispanic white families, none have the potential to yield an LOD score >1.6, making it difficult to obtain conclusive evidence for linkage. The one family that can yield a nearly significant LOD score is African American; this racial group has a significantly lower risk of NSCLP than non-Hispanic whites and Hispanics and very likely has a different etiology. In addition, the sample for the association studies is not large enough to detect SNPs with minor effects. Nonetheless, we have identified new regions of interest through linkage analyses as well as candidate genes through our association studies, both of which warrant further evaluation.
Here, we report six new regions (2p22, 3p13–14.1, 5p13.3, 7q36.2–36.3, 9q22.2–22.32 and 22q12.2–12.3) that can be interrogated for NSCLP genes. We identified eight regions that were previously implicated in other genomic scans in a variety of populations. The strongest linkages were found for the 8q21.3–24.12 and 22q12.2–12.3 regions, with the MYH9 (chromosome 22) gene exhibiting the strongest evidence for a causal role. This gene should be further evaluated in other populations for its possible role in NSCLP. The results of this study are important as they provide additional regions to search for new NSCLP genes and confirm the findings of earlier genomic scans. Our findings also demonstrate that large multiplex families with complex disorders can be successfully used in genome mapping strategies.
Acknowledgments
We thank Maria Elena Serna for expert clinical co-ordination and for managing the databases. We also thank Jacqueline Bui and Priscilla Roche for their technical assistance, Yuki Bradford for data analysis at the Center for Human Genetics Research at Vanderbilt University and all of the NSCLP families who kindly participated in this study. This study was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston (HSC-MS-03-090). This work was funded by grants from the National Institutes of Health (R01-DE011931 to JTH, T32-DE015355-05 trainee BTC).
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Conflict of interest
The authors state no conflict of interest.
Supplementary Material
References
- Gorlin RJ, Cohen MM, Hennekam RCM. Syndromes of the Head and Neck. New York: Oxford University Press; 2001. [Google Scholar]
- Wyszynski D. Cleft Lip and Palate: From Origin to Treatment. Oxford: Oxford University Press; 2002. [Google Scholar]
- Hashmi SS, Waller DK, Langlois P, Canfield M, Hecht JT. Prevalence of nonsyndromic oral clefts in Texas: 1995–1999. Am J Med Genet A. 2005;134:368–372. doi: 10.1002/ajmg.a.30618. [DOI] [PubMed] [Google Scholar]
- Canfield MA, Honein MA, Yuskiv N, et al. National estimates and race/ethnic-specific variation of selected birth defects in the United States, 1999–2001. Birth Defects Res A Clin Mol Teratol. 2006;76:747–756. doi: 10.1002/bdra.20294. [DOI] [PubMed] [Google Scholar]
- Schliekelman P, Slatkin M. Multiplex relative risk and estimation of the number of loci underlying an inherited disease. Am J Hum Genet. 2002;71:1369–1385. doi: 10.1086/344779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Moreno LM. Progress toward discerning the genetics of cleft lip. Curr Opin Pediatr. 2005;17:731–739. doi: 10.1097/01.mop.0000185138.65820.7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: an update. Int J Pediatr Otorhinolaryngol. 2007;71:1509–1519. doi: 10.1016/j.ijporl.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Field LL, Ray AK, Cooper ME, Goldstein T, Shaw DF, Marazita ML. Genome scan for loci involved in nonsyndromic cleft lip with or without cleft palate in families from West Bengal, India. Am J Med Genet A. 2004;130:265–271. doi: 10.1002/ajmg.a.30252. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Field LL, Cooper ME, et al. Genome scan for loci involved in cleft lip with or without cleft palate in Chinese multiplex families. Am J Hum Genet. 2002;71:349–364. doi: 10.1086/341944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Field LL, Tuncbilek G, Cooper ME, Goldstein T, Gursu KG. Genome-scan for loci involved in cleft lip with or without cleft palate in consanguineous families from Turkey. Am J Med Genet A. 2004;126:111–122. doi: 10.1002/ajmg.a.20564. [DOI] [PubMed] [Google Scholar]
- Prescott NJ, Lees MM, Winter RM, Malcolm S. Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Hum Genet. 2000;106:345–350. doi: 10.1007/s004390051048. [DOI] [PubMed] [Google Scholar]
- Radhakrishna U, Ratnamala U, Gaines M, et al. Genomewide scan for nonsyndromic cleft lip and palate in multigenerational Indian families reveals significant evidence of linkage at 13q33.1–34. Am J Hum Genet. 2006;79:580–585. doi: 10.1086/507487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley BM, Schultz RE, Cooper ME, et al. A genome-wide linkage scan for cleft lip and cleft palate identifies a novel locus on 8p11–23. Am J Med Genet A. 2007;143:846–852. doi: 10.1002/ajmg.a.31673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski DF, Albacha-Hejazi H, Aldirani M, et al. A genome-wide scan for loci predisposing to non-syndromic cleft lip with or without cleft palate in two large Syrian families. Am J Med Genet. 2003;123A:140–147. doi: 10.1002/ajmg.a.20283. [DOI] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75:161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Klass C, Woods A. Syndecan-2 regulates transforming growth factor-beta signaling. J Biol Chem. 2004;279:15715–15718. doi: 10.1074/jbc.C300430200. [DOI] [PubMed] [Google Scholar]
- Essner JJ, Chen E, Ekker SC. Syndecan-2. Int J Biochem Cell Biol. 2006;38:152–156. doi: 10.1016/j.biocel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Oh ES, Couchman JR. Syndecans-2 and -4; close cousins, but not identical twins. Mol Cells. 2004;17:181–187. [PubMed] [Google Scholar]
- Settle SH, Jr, Rountree RB, Sinha A, Thacker A, Higgins K, Kingsley DM. Multiple joint and skeletal patterning defects caused by single and double mutations in the mouse Gdf6 and Gdf5 genes. Dev Biol. 2003;254:116–130. doi: 10.1016/s0012-1606(02)00022-2. [DOI] [PubMed] [Google Scholar]
- Sena K, Morotome Y, Baba O, Terashima T, Takano Y, Ishikawa I. Gene expression of growth differentiation factors in the developing periodontium of rat molars. J Dent Res. 2003;82:166–171. doi: 10.1177/154405910308200304. [DOI] [PubMed] [Google Scholar]
- Portnoy ME, McDermott KJ, Antonellis A, et al. Detection of potential GDF6 regulatory elements by multispecies sequence comparisons and identification of a skeletal joint enhancer. Genomics. 2005;86:295–305. doi: 10.1016/j.ygeno.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Hanel ML, Hensey C. Eye and neural defects associated with loss of GDF6. BMC Dev Biol. 2006;6:43. doi: 10.1186/1471-213X-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy ME, McDermott KJ, Antonellis A, et al. Detection of potential GDF6 regulatory elements by multispecies sequence comparisons and identification of a skeletal joint enhancer. Genomics. 2005;86:295–305. doi: 10.1016/j.ygeno.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Marigo V, Nigro A, Pecci A, et al. Correlation between the clinical phenotype of MYH9-related disease and tissue distribution of class II nonmuscle myosin heavy chains. Genomics. 2004;83:1125–1133. doi: 10.1016/j.ygeno.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Martinelli M, Di Stazio M, Scapoli L, et al. Cleft lip with or without cleft palate: implication of the heavy chain of non-muscle myosin IIA. J Med Genet. 2007;44:387–392. doi: 10.1136/jmg.2006.047837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Lidral AC, Stal S, et al. CRISPLD2: A novel NSCLP candidate gene. Hum Mol Genet. 2007;16:2241–2248. doi: 10.1093/hmg/ddm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell JR, Weeks DE. The VITESSE algorithm for rapid exact multilocus linkage analysis via genotype set-recoding and fuzzy inheritance. Nat Genet. 1995;11:402–408. doi: 10.1038/ng1295-402. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin – rapid analysis of dense genetic maps using sparse gene flow trees.[see comment] Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Blanton SH, Bertin T, Patel S, Stal S, Mulliken JB, Hecht JT. Nonsyndromic cleft lip and palate: four chromosomal regions of interest. Am J Med Genet A. 2004;125:28–37. doi: 10.1002/ajmg.a.20423. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO. GOLD – graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Gilbert JR, Pericak-Vance MA, Hauser ER. Genotype-based association test for general pedigrees: the genotype-PDT. Genet Epidemiol. 2003;25:203–213. doi: 10.1002/gepi.10258. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: the pedigree disequilibrium test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RH, Hauser ER, Martin ER. The APL test: extension to general nuclear families and haplotypes and examination of its robustness. Hum Hered. 2006;61:189–199. doi: 10.1159/000094774. [DOI] [PubMed] [Google Scholar]
- Blanton SH, Cortez A, Stal S, Mulliken JB, Finnell RH, Hecht JT. Variation in IRF6 contributes to nonsyndromic cleft lip and palate. Am J Med Genet A. 2005;137:259–262. doi: 10.1002/ajmg.a.30887. [DOI] [PubMed] [Google Scholar]
- Li M, Atmaca-Sonmez P, Othman M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- McWhinney SR, Boru G, Binkley PK, et al. Intronic single nucleotide polymorphisms in the RET protooncogene are associated with a subset of apparently sporadic pheochromocytoma and may modulate age of onset. J Clin Endocrinol Metab. 2003;88:4911–4916. doi: 10.1210/jc.2003-030245. [DOI] [PubMed] [Google Scholar]
- Scapoli L, Palmieri A, Martinelli M, et al. Strong evidence of linkage disequilibrium between polymorphisms at the IRF6 locus and nonsyndromic cleft lip with or without cleft palate, in an Italian population. Am J Hum Genet. 2005;76:180–183. doi: 10.1086/427344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351:769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- Reich DE, Goldstein DB. Detecting association in a case–control study while correcting for population stratification.[see comment] Genet Epidemiol. 2001;20:4–16. doi: 10.1002/1098-2272(200101)20:1<4::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Boland GM, Perkins G, Hall DJ, Tuan RS. Wnt 3a promotes proliferation and suppresses osteogenic differentiation of adult human mesenchymal stem cells. J Cell Biochem. 2004;93:1210–1230. doi: 10.1002/jcb.20284. [DOI] [PubMed] [Google Scholar]
- Wang Y, Guo N, Nathans J. The role of Frizzled3 and Frizzled6 in neural tube closure and in the planar polarity of inner-ear sensory hair cells. J Neurosci. 2006;26:2147–2156. doi: 10.1523/JNEUROSCI.4698-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiquet BT, Blanton SH, Burt A, et al. Variation in WNT genes is associated with non-syndromic cleft lip with or without cleft palate. Hum Mol Genet. 2008;17:2212–2218. doi: 10.1093/hmg/ddn121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Helms JA. The role of sonic hedgehog in normal and abnormal craniofacial morphogenesis. Development. 1999;126:4873–4884. doi: 10.1242/dev.126.21.4873. [DOI] [PubMed] [Google Scholar]
- Verlinsky Y, Rechitsky S, Verlinsky O, et al. Preimplantation diagnosis for sonic hedgehog mutation causing familial holoprosencephaly. N Engl J Med. 2003;348:1449–1454. doi: 10.1056/NEJMoa022652. [DOI] [PubMed] [Google Scholar]
- Rice R, Spencer-Dene B, Connor EC, et al. Disruption of Fgf10/Fgfr2b-coordinated epithelial–mesenchymal interactions causes cleft palate. J Clin Invest. 2004;113:1692–1700. doi: 10.1172/JCI20384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orioli IM, Vieira AR, Castilla EE, Ming JE, Muenke M. Mutational analysis of the Sonic Hedgehog gene in 220 newborns with oral clefts in a South American (ECLAMC) population. Am J Med Genet. 2002;108:12–15. doi: 10.1002/ajmg.10204. [DOI] [PubMed] [Google Scholar]
- Benayed R, Gharani N, Rossman I, et al. Support for the homeobox transcription factor gene ENGRAILED 2 as an autism spectrum disorder susceptibility locus. Am J Hum Genet. 2005;77:851–868. doi: 10.1086/497705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevix-Trench G, Jones K, Green AC, Duffy DL, Martin NG. Cleft lip with or without cleft palate: associations with transforming growth factor alpha and retinoic acid receptor loci. Am J Hum Genet. 1992;51:1377–1385. [PMC free article] [PubMed] [Google Scholar]
- Tissier-Seta JP, Mucchielli ML, Mark M, Mattei MG, Goridis C, Brunet JF. Barx1, a new mouse homeodomain transcription factor expressed in cranio-facial ectomesenchyme and the stomach. Mech Dev. 1995;51:3–15. doi: 10.1016/0925-4773(94)00343-l. [DOI] [PubMed] [Google Scholar]
- Cox TC. Taking it to the max: the genetic and developmental mechanisms coordinating midfacial morphogenesis and dysmorphology. Clin Genet. 2004;65:163–176. doi: 10.1111/j.0009-9163.2004.00225.x. [DOI] [PubMed] [Google Scholar]
- Hahn H, Wicking C, Zaphiropoulous PG, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85:841–851. doi: 10.1016/s0092-8674(00)81268-4. [DOI] [PubMed] [Google Scholar]
- Trueba SS, Auge J, Mattei G, et al. PAX8, TITF1, and FOXE1 gene expression patterns during human development: new insights into human thyroid development and thyroid dysgenesis-associated malformations. J Clin Endocrinol Metab. 2005;90:455–462. doi: 10.1210/jc.2004-1358. [DOI] [PubMed] [Google Scholar]
- Machida J, Moreno LM, Mansilla MA, Bullard SB, et al. The role of FOXE1 in the etiology of cleft lip 2005. 57th Annual Meeting of the American Society of Human GeneticsSan Diego, CA, 470p.
- Vieira AR, Avila JR, Daack-Hirsch S, et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1:e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.