Abstract
Summary
Illumina BeadArray platform (Illumina Inc.) is playing an increasing role in cancer research. MBCB, an R package designed for use on Illumina Bead-Array data, allows for microarray data to be pre-processed through various model-based statistical methods. These model-based background-correction methods have proven to be a significant improvement over the traditional methods provided by Illumina in their BeadStudio software. MBCB accepts the summarized bead-type data; the data can then be normalized and background-corrected in a statistically-efficient manner. When compared to the popular Robust multi-array (RMA) background correction approach and the default, Illumina-provided background-correction method, MBCB has shown to lead to more precise determination of gene expression and better biological interpretation of Illumina BeadArray data. The software developed will facilitate molecular biomedical - especially cancer - research.
Availability
This package will soon be available from Bioconductor. Instructions for use are included with the package.
Keywords: Background correction, Microarray, BeadArray
Introduction
Illumina have produced a novel microarray platform – BeadArray - for use in multiple environments: Gene expression studies, CGH, and SNP, among others. Illumina expression BeadArrays often generate high quality data with relatively low cost and less RNA sample input. These features make BeadArray an increasingly popular microarray platform.
One distinguishing feature of the BeadArray platform is that each array contains thousands of non-specific negative control bead types. These negative control beads offer great potential for controlling background noise.
The background-correction method in BeadStudio software provided by Illumina Inc. did not use negative control beads efficiently. It takes the mean of all negative control beads and subtracts that value from all of the other beads. Unfortunately, this method tends to result in a large number of negative expression values which are typically discarded. In certain cases, more than half of the beads on the chip have been negative when using this method. Some studies (Barnes et al., 2005) have suggested that the pre-processing methods offered by Illumina will actually cause such massive data loss that the raw values should be used instead. However, using only the raw values has also shown to be problematic as significant data attenuation is observed when expression ratios are calculated between two expression- level data sets (Ding et al., 2008).
Ding et al., (2008) suggested an alternative model-based background- correction method to address this problem. Xie et al., (2009) proposed three different statistical methods to estimate the parameters in the model. We developed MBCB – an R package – to take advantage of these new methods. By using R, the package is inherently cross-platform, easily distributable and can be easily integrated into existing R tools.
Description
Input
The user provides MBCB with the summarized bead-type data. This summarization can be obtained through BeadStudio. Essentially, the file summarizes the raw, bead-level data and provides the average intensity and variance of each bead type.
Background-correction
The primary contribution of this package is the ability to background- correct the given data in a statistically-efficient manner. The algorithms used no longer cause massive data attenuation; instead, they lead to more accurate measurement of gene expression levels.
The user can select from a list of background-correction methods:
Maximum likelihood estimation
One of the more accurate methods built. Assuming Gaussian distribution for the noise term, MLE iteratively updates parameter estimates by making use of the non-specific beads on the microarray.
Gamma maximum likelihood estimation
This method is similar to Maximum Likelihood Estimation except that the noise term is assumed to have a Gamma distribution. It is preferred when the distribution of the non-specific negative control data is not symmetric.
Bayesian method
Is possibly more extensible than the others (because it allows for extra prior information), but has not consistently outperformed the other methods, despite being, by far, the most computationally-intensive.
Non-Parametric
Avoids the use of assumptions about the parameters of the model. This is the fastest and one of the most accurate methods.
Robust multi-array average
This method (modeled after the methods found in the Affymetrix package) can be used if the users could not provide negative control information.
Normalization
For convenience’ sake, the package also provides the opportunity to normalize the data, if so desired, using either Quantile- Quantile normalization (from the affy package) or global (median) normalization. Obviously, normalization is not mandatory for this package. Users can also apply their own preferred normalization approach after using MBCB for background-correction.
Output
The files created by this package include the background-corrected data (one file per background-correction method used). In addition, the user is given a file detailing the parameter estimations for each correction method.
Graphical user interface
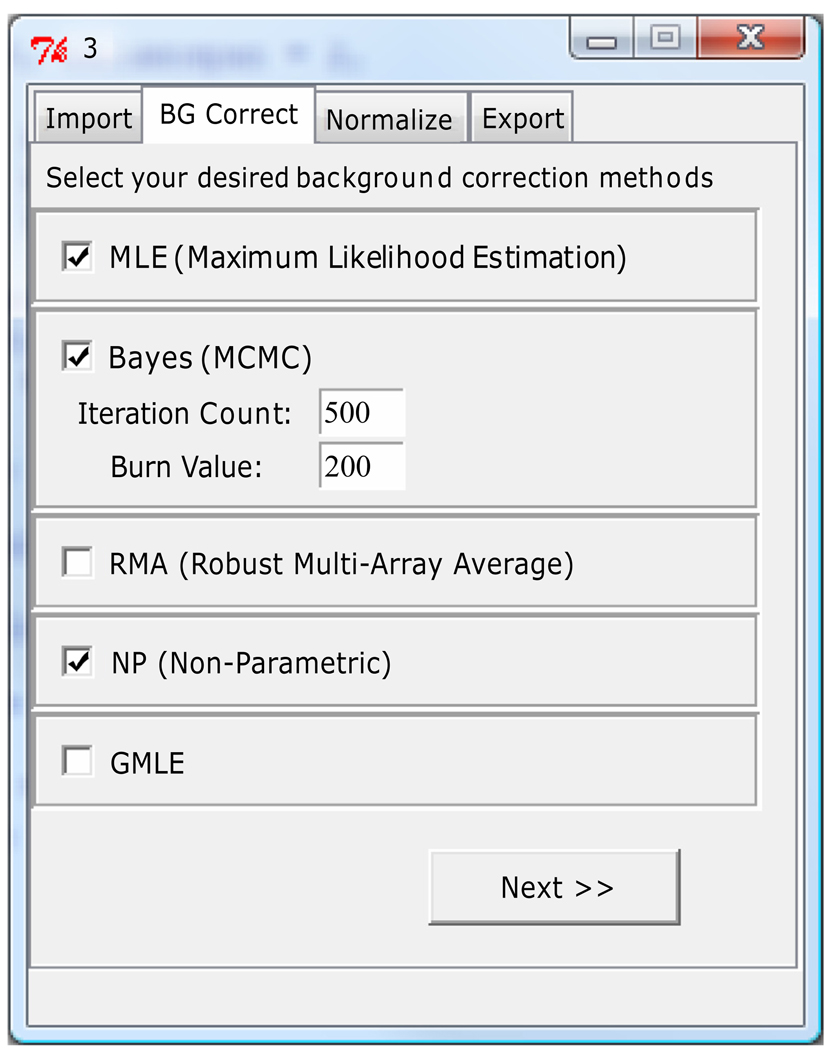
To ensure ease-of-use among non-technical audiences, we provide a graphical interface through which users can accomplish all that they could with the command-level functions. A screenshot is shown in Figure 1. The user can browse to the data files, select one or multiple model-based background-correction methods from a list, then select normalization method(s). The user can then browse to the location at which they’d like to save the output.
Figure 1.
A screenshot of the background-correction options in MBCB.
Discussion
BeadArray technology has a great potential for molecular profiling especially for cancer research; however, because of the analysis issues surrounding the background-correction of the data, the platform has experienced significant data loss/attenuation. These new model-based methods have proven to be more accurate and efficient (Ding et al., 2008; Xie et al., 2009) and the user-friendly R package will simplify the use of this data processing approach.
Because the package is written exclusively in R, it can be used on any Windows or UNIX-based operating system. Many previously-written tools surrounding this research have also been written in R. Due to the open-source nature of most such packages, the tools can easily be combined or modified to meet specific needs.
Acknowledgements
This work was support by NIH UL1 RR024982, NNJ05HD36G, and 1R21DA027592.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Barnes M, Freudenberg J, Thompson S, Aronow B, Pavlidis P. Experimental comparison and cross-validation of the Affymetrix and Illumina gene expression analysis platforms. Nucleic Acids Res. 2005;33:5914–5923. doi: 10.1093/nar/gki890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding LH, Xie Y, Park S, Xiao G, Story MD. Enhanced identification and biological validation of differential gene expression via Illumina whole-genome expression arrays through the use of the model-based background correction methodology. Nucleic Acids Res. 2008;36:e58. doi: 10.1093/nar/gkn234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y, Wang X, Story M. Statistical methods of background correction for Illumina BeadArray data. Bioinformatics. 2009;25:751–757. doi: 10.1093/bioinformatics/btp040. [DOI] [PMC free article] [PubMed] [Google Scholar]