Abstract
Although Duchenne muscular dystrophy is primarily classified as a neuromuscular disease, cardiac complications play an important role in the course of this X-linked inherited disorder. The pathobiochemical steps causing a progressive decline in the dystrophic heart are not well understood. We therefore carried out a fluorescence difference in-gel electrophoretic analysis of 9-month-old dystrophin-deficient versus age-matched normal heart, using the established MDX mouse model of muscular dystrophy-related cardiomyopathy. Out of 2,509 detectable protein spots, 79 2D-spots showed a drastic differential expression pattern, with the concentration of 3 proteins being increased, including nucleoside diphosphate kinase and lamin-A/C, and of 26 protein species being decreased, including ATP synthase, fatty acid binding-protein, isocitrate dehydrogenase, NADH dehydrogenase, porin, peroxiredoxin, adenylate kinase, tropomyosin, actin, and myosin light chains. Hence, the lack of cardiac dystrophin appears to trigger a generally perturbed protein expression pattern in the MDX heart, affecting especially energy metabolism and contractile proteins.
1. Introduction
The identification of the gene responsible for the primary defect in X-linked muscular dystrophy [1] led to the discovery of its protein product, the large membrane cytoskeletal element dystrophin [2]. The full-length Dp427 isoform of dystrophin is almost completely missing from the dystrophic surface membrane [3], which triggers the characteristic dystrophic phenotype of progressive skeletal muscle weakness and fibre wasting [4]. Mutations in the dystrophin gene cause the most prevalent and lethal gender-specific genetic disease of childhood, Duchenne muscular dystrophy (DMD), and its more benign and less frequent counterpart Becker's muscular dystrophy, as well as X-linked dilated cardiomyopathy [5]. Dystrophin forms a supramolecular complex with various surface glycoproteins [6] that are involved in cellular signaling, receptor clustering and stabilization of the cellular periphery [7]. Tissue-specific variations in dystrophin isoforms delegate the composition and subcellular localization of the dystrophin-glycoprotein complex, and hence the function of the specific membrane assembly may vary between the brain, heart, and skeletal muscles [8]. In muscular dystrophy, deficiency in dystrophin causes a significant reduction in the surface-associated glycoprotein complex, which severely impairs the integrity of the sarcolemma [9] and causes a broad spectrum of downstream alterations such as enhanced proteolytic destruction of muscle proteins and abnormal Ca2+-handling [10]. This classifies DMD primarily as a genetic disorder of skeletal muscle, but pathological alterations are also present in cardiac muscle and diaphragm, as well as the peripheral and central nervous system [11].
The majority of DMD patients develop clinical cardiac symptoms during the second decade of life [12], including arrhythmias, cardiomyopathy, and regional wall abnormalities, leading to fatal cardiac complications in about 20% of cases [13]. On the cellular level, the gradual replacement of cardiac fibres by connective and fatty tissue is a hallmark of the dystrophic heart [14]. While dystrophin-deficient skeletal muscles undergo cycles of fibre degeneration and regeneration, dystrophic heart fibres exhibit only a limited regenerative capacity causing a progressive decline in the cardiomyocyte population of the dystrophic heart [15]. Since cardiomyopathy is a frequent occurrence in inherited muscular dystrophies [16], a number of novel therapeutic approaches are tested to specifically address the cardiac symptoms [17, 18], especially membrane-stabilizing agents such as tri-block poloxamers [19]. In this study, we used the X-linked muscular dystrophy (MDX) mouse, a well-established animal model of DMD [20] that is missing dystrophin isoform Dp427 [21]. Although the dystrophic phenotype of young MDX hearts does not represent a perfect replica of DMD-related cardiomyopathic complications [22], in aged MDX hearts the pathological changes are of considerable clinical relevance [23–25] and cardiac MDX fibres are frequently used to evaluate new treatment strategies to counteract cardiomyopathic complications [26–29].
The absence of cardiac dystrophin is clearly associated with necrotic changes, extensive infiltration of inflammatory cells, increase in adipose tissue, interstitial fibrosis, tachycardia, and impaired contractile properties in the MDX heart [22, 30, 31]. In analogy to DMD hearts, the expression of dystrophin-associated glycoproteins is also greatly reduced in dystrophin-deficient MDX heart cells [32]. Secondary abnormalities in cardiac fibres from the MDX mouse include the drastic reduction in key luminal Ca2+-binding proteins [33] and abnormal stress-induced Ca2+-influx into the cytosol [34]. Interestingly, both physical exercise and aging seem to accelerate the dystrophic process in cardiac MDX tissue [35, 36], whereby a recent study by Spurney et al. [37] revealed that heart dysfunction was most prominent at 9 months of age. This makes this age group of MDX hearts a suitable DMD model for investigating global changes in the cardiac MDX protein complement and was therefore chosen in this study.
The development of mass spectrometry-based proteomics for the swift identification of proteins has decisively enhanced the analytical capability of comparative biomedical studies [38]. In addition, the introduction of fluorescent dyes has drastically increased the total number of identifiable two-dimensional spots, whereby fluorescence difference in-gel electrophoresis (DIGE) can be considered one of the most advanced biomedical tools for studying two different sets of soluble protein complements [39]. DIGE analysis generates highly reproducible findings due to greatly reduced gel-to-gel variations [40]. We have therefore used this technique here for the proteomic profiling of the dystrophic MDX heart. The usefulness of proteomics for determining the biomarker signature of animal disease models of muscular disorders has recently been reviewed [41]. An important aspect of genetic mouse models is that they show, due to their inbred status, genetically much less interindividual differences than human patients. Therefore, considerably fewer experimental repeats are capable of producing meaningful proteomic data. With respect to X-linked muscular dystrophy, proteomic studies have mostly focused on MDX skeletal muscles and have identified numerous biomarker candidates, such as adenylate kinase [42], calsequestrin [43], regucalcin [44], and the small stress protein cvHsp [45]. In general, dystrophic skeletal muscles show an extremely perturbed protein expression pattern. Although the initial pathobiochemical consequence of a primary genetic abnormality in dystrophin is a severe reduction in dystrophin-associated proteins, this seems to result in a variety of downstream alterations in various classes of muscle proteins involved in fibre contraction, chaperone function, ion homeostasis, cytoskeleton formation and metabolism [41].
In contrast, relatively little is known about global alterations in the dystrophic heart. Building on the recent findings of a combined metabolomic and proteomic investigation into dystrophin deficiency in cardiac muscle [46], this report describes the detailed DIGE analysis of the expression levels of fluorescently-tagged dystrophic versus normal cardiac proteins using both pH 4–7 and pH 6–11 range gels. Following the densitometric determination of the expression pattern of 2,509 cardiac protein spots, mass spectrometry identified 26 proteins being decreased, including various myosin light chains, tropomyosin, actin, adenylate kinase, creatine kinase, vimentin, ATP synthase, fatty acid binding-protein, isocitrate dehydrogenase, NADH dehydrogenase, myozenin, porin, and peroxiredoxin, and 3 proteins being increased, including lamin-A/C and nucleoside diphosphate kinase. These findings suggest that the loss in dystrophin causes abnormalities in cardiac metabolism, the cellular stress response, the cytoskeleton, and the contractile machinery. In the future, these results will be useful for complementing studies into the molecular mechanisms of muscular dystrophy and for evaluating the effects of novel drugs, genetic modifications, or cell-based therapies on disease progression.
2. Materials and Methods
2.1. Materials
For gel electrophoretic analyses, analytical grade chemicals and materials were purchased from Amersham Biosciences/GE Healthcare, Little Chalfont, Buckinghamshire, UK (CyDye DIGE fluor minimal dyes Cy3 and Cy5; 24 cm pH 4–7 and 18 cm pH 6–11 immobilized pH gradient (IPG) strips; IPG buffer; iodoacetamide), Biorad Laboratories, Hemel-Hempstead, Hertfordshire, UK (Laemmli-type buffer system; protein molecular mass markers), National Diagnostics, Atlanta, GA, USA (ultrapure Protogel acrylamide stock solution), Cosmo Bio Company, Tokyo, Japan (2D silver stain II kit), and Perbio Science, Northumberland, UK (Coomassie Brilliant Blue G-250). Protease inhibitors and chemiluminescence substrate were obtained from Roche Diagnostics (Mannheim, Germany). Sequencing grade-modified trypsin was purchased from Promega (Madison, WI, USA). Nitrocellulose transfer stacks were from Invitrogen (Carlsbad, CA, USA). Primary antibodies were obtained from Abcam Ltd., Cambridge, UK (ab28172 to prohibitin; ab36329 to isocitrate dehydrogenase; ab14734 to porin isoform VDAC1; ab54824 to adenylate kinase isoform AK1; ab5432 to ATP synthase, ab16915 to cardiac fatty acid binding protein), Sigma Chemical Company, Dorset, UK (mAb NOQ7.5.4D to slow/cardiac myosin heavy chain; pAb to laminin), Visionbiosystems Novocastra, Newcastle upon Tyne, UK (mAb NCL-DYS1 against the Dp427 isoform of dystrophin), Thermo Fisher Scientific Inc., Rockford, IL, USA (mAb Rd301 to desmin) and Santa Cruz Biotechnology, Santa Cruz, CA, USA (sc27992 to succinate dehydrogenase). All secondary antibodies were purchased from Chemicon International (Temecula, CA, USA). For confocal microscopy, Superfrost Plus positively-charged microscope slides were purchased from Menzel Glaesser (Braunschweig, Germany). Optimum cutting temperature (OCT) compound was from Sakura Finetek Europe B.V (Zoeterwoude, Netherlands) and p-phenylenediamine (PPD)-glycerol was purchased from Citifluor Ltd. (London, UK). MitoTracker Red CMXRos dye and Alexa Fluor-conjugated secondary antibodies were from Invitrogen Molecular Probes (Bio Sciences Ltd., Dun Laoghaire, Ireland). The DNA binding dye diamidino-phenylindole (DAPI) and all other chemicals used were of analytical grade and purchased from Sigma Chemical Company, Dorset, UK.
2.2. Dystrophic MDX Animal Model
Hearts from male 9-month old C57 control mice and age-matched dystrophic mdx mice were obtained through the Bioresource Unit of NUI Maynooth and the Animal House of the University of Bielefeld. Animals were kept under standard conditions and all procedures were performed in accordance with German and Irish guidelines on the use of animals for scientific experiments. Mice were killed by cervical dislocation and their hearts removed and snap-frozen [32]. As previously demonstrated by immunoblotting, the MDX mouse cohort used in this study lacks the 427 kDa isoform of cardiac dystrophin and shows a drastic reduction in dystrophin-associated glycoproteins, such as dystroglycans and sarcoglycans [32, 33].
2.3. Preparation of Total Heart Muscle
For the proteomic profiling of crude cardiac extracts, three normal and three age-matched dystrophic hearts were separately prepared by washing with distilled water to remove excess blood. Muscle tissue was pulverized by grinding in liquid nitrogen using a mortar and pestle. Ground hearts were solubilized in lysis buffer with the ratio 100 mg wet weight to 1 mL lysis buffer consisting of 9.5 M urea, 2% CHAPS, 0.8% IPG buffer pH 3–10, 1% (w/v) DTT and protease inhibitors [45]. Samples for DIGE analysis were placed in buffer containing only urea and CHAPS. Samples were gently rocked for 30 minutes and protein concentration was determined using the Bradford assay system [47]. For immunoblot analysis samples were placed into homogenization buffer (0.5 M Hepes pH 7.4, 200 mM EGTA, 10% (w/v) sucrose, 3 mM MgCl2, and 0.1% (w/v) NaN3). The buffer was supplemented with a protease inhibitor cocktail to prevent protein degradation [45]. Extracts were shaken at 4°C on a Thermomixer from Eppendorf (Hamburg, Germany) for 4 hours and then centrifuged at 4°C for 20 minutes at 20,000× g in an Eppendorf 5417R bench centrifuge.
2.4. Difference in-Gel Electrophoretic Analysis
Potential differences in the expression pattern of the soluble proteome from normal and MDX cardiac muscle were analysed using the fluorescence difference in-gel electrophoretic DIGE technique [39], as recommended by Karp and Lilley [48]. The fluorescent dyes Cy3 and Cy5 were reconstituted as a stock solution of 1 mM in fresh dimethylformamide, then diluted to a 0.2 mM solution prior to use. Labeling was performed with 200 pmols of Cy3 fluor dye per 25 μg protein. An internal pooled standard of every sample was prepared with the Cy5 fluor. Labelled samples were then vortexed and incubated for 30 minutes on ice in the dark. The reaction was quenched with 10 mM lysine on ice for 10 minutes [49]. For isoelectric focusing, pH 6–11 IPG strips were rehydrated for 12 hours in rehydration buffer (8 M Urea, 0.5% CHAPS, 0.2% DTT, and 0.2% ampholytes) with the addition of 1.2% (v/v) DeStreak rehydration solution (Amersham Biosciences/GE Healthcare, Little Chalfont, Buckinghamshire, UK). Sample and equal volumes of reducing lysis buffer (9.5 M Urea, 2% CHAPS, 2% DTT, and 1.6% ampholytes) were added by anodic cup loading. For pH 4–7 focusing, IPG strips were in-gel rehydrated with sample and equal volumes of reducing lysis buffer made up in rehydration buffer. The strips were run on an Amersham IPGphor IEF system with the following running conditions: 3500 V for 21.3 hours, a 3500 to 8000 V gradient over 10 minutes in, and 8000 V for 2 hours. For first dimension separation in the pH 6–11 range the following protocol was employed: 80 V for 4 hours, 100 V for 2 hours, 500 V for 1.5 hours, 1000 V for 1 hour, 2000 V for 1 hour, 4000 V for 1 hour, 6000 V for 2 hours, and finally 8000 V for 2.5 hours. Following isoelectric focusing, the IPG strips were equilibrated for 20 minutes firstly in buffer containing 6 M urea, 30% (w/v) glycerol, 2% (w/v) SDS, 100 mM Tris-HCl, pH 8.8 with the addition of 100 mM DTT and subsequently for 20 minutes in buffer supplemented with 0.25 M iodoacetamide [49]. Strips were briefly washed in sodium dodecyl sulfate-containing running buffer (125 mM Tris, 0.96 M glycine, 0.1% (w/v) SDS) and placed on top of 12.5% (w/v) resolving gels. To seal the strip gels, they were overlaid with the above-described running buffer containing 1% (w/v) agarose. The separation of the cardiac proteins was carried out in the second dimension by standard SDS polyacrylamide gel electrophoresis using the DodecaCell system from Bio-Rad Laboratories (Hemel Hempstead, Herts., UK). Slab gels contained 50 μg protein per gel. Electrophoresis was carried out overnight for approximately 18 hours in the dark at 1.5 V per 21 cm-gel until the Bromophenol Blue tracking dye just ran off the gel.
2.5. Protein Visualization and Data Analysis
Fluorescent CyDye-labelled cardiac proteins were visualized using a Typhoon Trio variable mode imager form Amersham Biosciences/GE Healthcare (Little Chalfont, Bucks., UK). For image acquisition, Cy5- and Cy3-labelled proteins were scanned at a wavelength of λ = 650 nm and λ = 550 nm, respectively. Photomultiplier tube (PMT) values were optimised so that the volume of the most abundant spot was between 80,000 and 99,000 when scanned at a resolution of 100 μm. This guaranteed that no spot would be saturated on the gel, therefore interfering with accurate analysis. The Cy3 images were then analysed using Progenesis SameSpots software version 3.2.3 from NonLinear Dynamics (Newcastle upon Tyne, UK) and normalised against their corresponding Cy5 image. Gels were aligned to the reference image. Following detection of spots, the gels were placed into groups (MDX versus normal) and analysed to determine significant differences in 2D spot abundance. An ANOVA score of 0.5 was required for spots to be included in the subsequent analysis. Then principal component analysis (PCA) was verified with changes displaying power of <0.8 being removed from the analysis. All remaining changed spots that met the significance criteria were visually checked on the aligned gels to ensure feasibility and were subsequently identified by LC-MS/MS analysis.
2.6. Mass Spectrometric Identification of Cardiac Proteins
Preparative gels containing 400 μg of protein (1:1 MDX to normal) were stained with the dye Ruthenium II Bathophenanthroline Disulfonate Chelate (RuBPs) [50]. RuBPs-stained gels were scanned at a wavelength of λ = 550 nm using the criteria outlined above. Prior to identification via LC-MS/MS technology, 2D protein spots of interest were excised using the automated Ettan spot picker system from Amersham Biosciences (Little Chalfont, Bucks., UK). Spots were digested as outlined by Shevchenko et al. [51]. Briefly, gel pieces were destained before the addition of 400 ng trypsin per spot, incubated for 30 minutes at 4°C before being digested overnight at 37°C. 100 μL of extraction buffer (1:2 v/v of 5% formic acid/acetonitrile) was added to the peptides and incubated for 15 minutes at 37°C. All supernantant was then transferred into fresh tubes and extracts were dried down completely in a vacuum centrifuge. Peptides were reconstituted in 12 μL of 0.1% formic acid, vortexed, sonicated, and the mixture was centrifuged for 20 minutes in cellulose spin filter tubes to remove any gel particles. The remaining solution was placed into LC-MS vials. Samples were analysed on an Agilent 6340 Ion Trap LC mass spectrometer using electrospray ionization (Agilent Technologies, Santa Clara, CA, USA) on a 10-minute gradient of 5–100% acetonitrile/0.1% formic acid with a post run of 1 minute [52]. Separation of peptides was performed with a nanoflow Agilent 1200 series system, equipped with a Zorbax 300 SB C18 μm, 4 mm 40 nl pre-column. Mobile phases used were A: 0.1% formic acid, B: 90% acetonitrile and 0.1% formic acid. Samples (8 μL) were loaded onto the enrichment column with capillary flow rate set at 4 μL/min with a mix of A : B at a ratio of 19 : 1. Elution was carried out with the nano pump flow rate set at 0.6 μl/minute. Database searches were carried out using Mascot MS/MS Ion search. Criterion for each search was set at (i) species Mus musculus, (ii) two missed cleavages by trypsin, (iii) variable modification: oxidation of methioine, (iv) fixed modification: carboxymethylation of cysteines, and (v) mass tolerance of precursor ions ±2Da and product ions ±1Da.
2.7. Immunoblot Analysis
In order to verify changes in the expression of select cardiac proteins, 1-D gel electrophoresis, Coomassie staining and immunoblotting was performed as previously described in detail [32]. Gels were transferred to an “iblot” transfer unit from Invitrogen (Carlsbad, CA, USA) for semi-dry blotting. Electrophoretic transfer was carried out for 6 min. Blocking of nitrocellulose sheets was achieved with a milk protein solution (5% (w/v) fat-free milk powder in 0.9% (w/v) NaCl, 50 mM sodium phosphate, pH 7.4) for 1 hour. Incubation with sufficiently diluted primary antibody was carried out overnight with gentle agitation. Nitrocellulose sheets were washed and then incubated for 1 hour with secondary peroxidase-conjugated antibodies, diluted in blocking solution. Immuno-decorated bands were visualized by the enhanced chemiluminescence method using BM Chemiluminescence Blotting substrate from Roche Diagnostics (Mannheim, Germany). Densitometric scanning of immunoblots was performed on a Molecular Dynamics 300 S computing densitometer (Sunnyvale, CA, USA) with ImageJ (NIH, USA) and GraphPad Prism (San Diego, CA, USA) software.
2.8. Confocal Microscopy
For the localization of nuclei, mitochondria and select marker proteins, confocal microscopy was used as previously described in detail [45]. Normal and dystrophic MDX hearts were mounted on cyrocassettes at −20°C and transverse cryosections of 10 μm thickness were cut on a Shandon Cryotome (Life Sciences International, Cheshire, UK). Unfixed tissue sections were transferred to Superfrost Plus positively-charged microscopy slides. For labeling of mitochondria, freshly cut tissue sections were incubated with a 1 : 100 diluted solution of MitoTracker Red Chloromethyl-X-Rosamine (CMXRos) dye for 30 minute, briefly washed in phosphate-buffered saline and then immediately examined by microscopy [53]. CMXRos is a well-established lipophilic cationic fluorescent dye that is highly specific for mitochondria [54] and is routinely used to measure mitochondrial function and morphology in cell culture and in whole tissues [55]. For immunofluorescence microscopy, cryosections were briefly fixed in ice-cold acetone for 5 minutes, followed by submersion in blocking solution (0.2% (w/v) bovine serum albumin, 0.2% (v/v) Triton X-100, and 2.5% (v/v) goat serum in phosphate-buffered saline) for 30 minute, and then incubated with primary antibodies diluted in 0.2% (w/v) bovine serum albumin and 0.2% (v/v) Triton X-100 in phosphate-buffered saline for 4 hours. Tissue sections were washed twice for 30 minute in above solution omitting antibodies and then labelled with secondary antibodies for 1 hour, followed by the above described washing procedure. In order to determine the number of nuclei in normal versus MDX hearts, cardiac tissue sections were labelled with 1 μg/ml diamidino-phenylindole (DAPI) for 30 minute. One drop of p-phenylenediamine (PPD)-glycerol was applied to the immuno-decorated tissue sections and then coverslips carefully placed over the sections, avoiding the introduction of air bubbles. Fluorescent labelling patterns were visualised with an Olympus IX81 microscope (Olympus Life and Material Science Europe, Hamburg, Germany).
3. Results
3.1. Comparative Proteomic Profiling of MDX Versus Normal Heart Muscle
In contrast to non-fluorescent protein dye methodology, comparative studies using fluorescent CyDyes have an enhanced dynamic range of protein coverage [39]. DIGE analysis represents a highly accurate quantitative technique that enables multiple protein samples to be separated on the same two-dimensional gel. This reduces the introduction of potential artifacts due to gel-to-gel variations, making comparative DIGE approaches one of the most powerful analytical tools for conducting comparative biomedical investigations [40]. In order to determine global changes in the heart due to deficiency in the dystrophin isoform Dp427, we have carried out a DIGE analysis of crude tissue extracts from 9-month old normal versus age-matched dystrophic MDX mice. Due to the high cost of breeding and maintaining mice to old age, as well as the considerable cost of fluorescent tagging of large protein populations, the number of biological repeats and analytical DIGE gels was kept to a minimum. In order to satisfy the statistical requirements for the generation of proper proteomic data sets and perform an optimized DIGE analysis of normal versus affected proteomes, we have followed the recommendations by Urfer et al. [56] and Karp and Lilley [48], respectively. In the first dimensional separation step via isoelectric focusing, both a pH 4–7 and a pH 6–11 range were employed to cover as many cardiac proteins with differing charges as possible. Figure 1 illustrates the neutral and the more acidic range of protein species and Figure 2 shows the separation of neutral and more basic heart proteins. Shown are representative two-dimensional gels of Cy3-labelled normal muscle (Figure 1(a); Figure 2(a), Cy3-labelled dystrophic muscle (Figure 1(c); Figure 2(c) and corresponding Cy5-labelled pooled standards (Figures 1(b),1(d); Figures 2(b), 2(d), which were analysed with the help of a Typhoon Trio variable imager and Progenesis 2D analysis software. Overall, 2509 distinct protein species were recognized on the lower and higher pH-range gels. Recently Raddatz et al. [57] have catalogued the soluble murine heart protein complement. The protein spot pattern presented here agrees in large parts with their comprehensive 2D proteomic map of cardiac tissue. Out of 2048 detectable protein spots on the pH 4–7 gels and 487 detectable protein spots on pH 6–11 gels, 79 protein spots showed a drastic differential expression pattern, with 3 proteins being increased and 26 distinct protein species being decreased. Electrospray ionization MS/MS analysis was carried out to unequivocally identify the cardiac proteins with a changed abundance in dystrophic heart fibres.
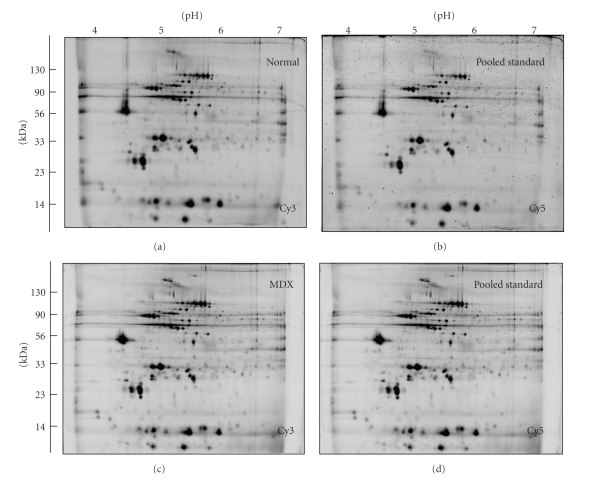
Figure 1.
2D gel electrophoretic analysis of the dystrophic heart using the pH 4–7 range. Shown are Cy3-labelled gels of the soluble fraction from normal (a) and dystrophic MDX (c) cardiac muscle, as well as Cy5-labelled gels containing pooled standards ((b), (d)). Representative fluorescent DIGE gels with electrophoretically separated proteins are shown for the pH 4–7 range. The pH-values of the first dimension gel system and molecular mass standards (in kDa) of the second dimension are indicated on the top and on the left of the panels, respectively.
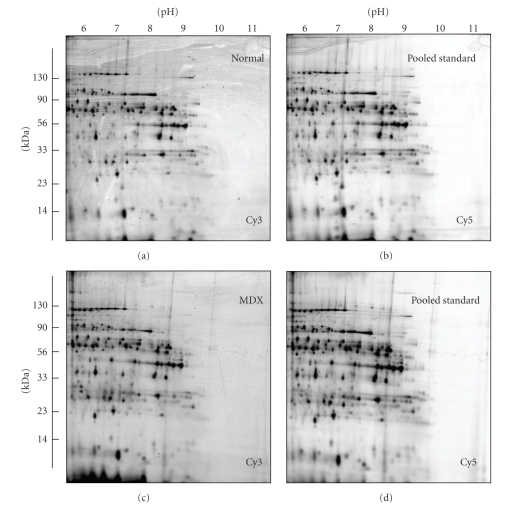
Figure 2.
2D gel electrophoretic analysis of the dystrophic heart using the pH 6–11 range. Shown are Cy3-labelled gels of the soluble fraction from normal (a) and dystrophic MDX (c) cardiac muscle, as well as Cy5-labelled gels containing pooled standards ((b), (d)). Representative fluorescent DIGE gels with electrophoretically separated proteins are shown for the pH 6–11 range. The pH-values of the first dimension gel system and molecular mass standards (in kDa) of the second dimension are indicated on the top and on the left of the panels, respectively.
3.2. DIGE Analysis of the Dystrophin-Deficient Heart
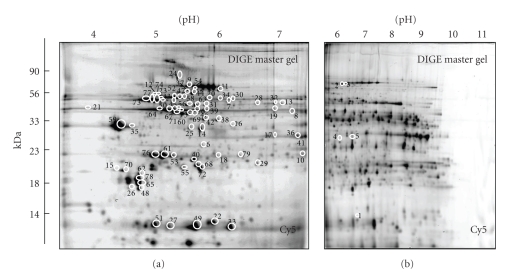
A list of the cardiac protein species with a drastically altered expression level in dystrophic MDX tissue is shown in Table 1. The information on the 79 DIGE-identified protein spots combine data from both pH 4–7 and pH 6–11 gels and contain the spot number, protein name, protein ID, number of matched peptides, percentage sequence coverage, the relative molecular mass, pI-value, Mascot score and fold-change of individual proteins affected by deficiency in dystrophin. Since the MS-based analysis identified a large number of matched peptide sequences, these data are not shown. The majority of identified cardiac proteins belonged to the main classes of metabolic or contractile proteins. Components of the actomyosin apparatus, enzymatic and regulatory elements of mitochondria, glycolytic enzymes, metabolic transporters, cellular stress proteins and cytoskeletal components were clearly identified by MS analysis. Cardiac proteins with a dystrophy-related change in abundance ranged in molecular mass from apparent 14 kDa to 90 kDa and covered a pI-range from approximately 5 to 9. DIGE Cy5 master gels of both the pH 4–7 and the pH 6–11 range are shown in Figure 3(a), 3(b), so that it is possible to correlate MS-identified protein species, listed in Table 1, with distinct two-dimensional spots of altered density in the dystrophic heart. An increased expression level was shown for 3 cardiac proteins, that is, nucleoside diphosphate kinase B (spot 1), the nuclear lamina matrix protein lamin A/C (spot 2), and a component of the electron-transferring flavoprotein dehydrogenase (spot 3). All other identified protein species exhibited a decreased abundance in dystrophin-deficient myocardial tissue. These cardiac proteins are marked and numbered 4 to 79 in the Cy5-labelled master gels of Figure 3. The protein species with the highest decrease in concentration was identified as the anti-oxidant enzyme peroxiredoxin-6 (spot 79). In addition, mitochondrial ATP synthase (spots 50, 55, 69, 72, 73, 74, 77), isocitrate dehydrogenase (spot 38, 42), NADH dehydrogenase (spot 40), pyruvate dehydrogenase (spots 11, 14, 15, 20, 25, 34), isovaleryl-CoA dehydrogenae (spots 8, 19), dienoyl-CoA isomerase (spot 17), oxoglutarate dehydrogenase (spot 7), prohibitin (spot 6), electron-transferring flavoprotein (spots 5, 17), myozenin-2 (spot 4), voltage-dependent anion-selective channel protein VDAC1 (spot 41), the slow/cardiac isoform of myosin light chain MLC2 (spots 26, 48, 62, 65, 70, 78), myosin light chain MLC3 (spots 53, 61, 76), cardiac alpha-actin (spots 21, 39, 45, 46, 56, 60, 64, 66, 67, 71, 75), alpha-1 tropomyosin (spots 35, 59), adenylate kinase AK1 (spot 68), creatine kinase (spot 16), cytochrome c oxidase (spots 22, 51), cytochrome b-c complex (spots 10, 30, 57, 63), desmin (spot 43, 47, 52, 54, 58), vimentin (spots 12, 23), cardiac fatty acid binding-protein FABP-3 (spots 27, 33, 49), heat shock protein Hsp27 (spot 18), heat shock protein Hsp60 (spots 31, 37, 44), enolase (spots 13, 28, 32), DJ-1 protein (spot 29), valsolin-containing protein VCP (spot 24), and albumin (spot 9) were identified. The results for 2 decreased protein spots have not been included in Table 1, since only 1 matched peptide per cardiac protein could be determined by MS analysis. It is international standard that proteomic studies only list proteins with sequence coverage of less than 10% when at least 2 separate matched peptide sequences are available for an independent identification. The peptide sequences LGANSLLDLVVFGR (2% sequence coverage) and LTFDSSFSPNTGK (4% sequence coverage) identified the nonlisted cardiac proteins as succinate dehydrogenase (73.6 kDa; pI 7.1) and porin isoform VDAC1 (32.5 kDa; pI 8.6), respectively.
Table 1.
List of DIGE-identified proteins with a changed abundance in 9-month old dystrophic MDX heart muscle.
| Spot No. | Protein name | Protein ID | Peptides matched | Coverage (%) | Molecular mass (kDa) | Isoelectric point (pI) | Mascot score | Fold change |
|---|---|---|---|---|---|---|---|---|
| 1 | Nucleoside diphosphate kinase B | NDKB_MOUSE | 4 | 34 | 17.5 | 6.9 | 69 | 4.4 |
| 2 | Lamin-A/C | LMNA_MOUSE | 4 | 6 | 74.5 | 6.5 | 111 | 1.9 |
| 3 | Electron transfer flavoprotein-ubiquinone oxidoreductase, mitochondrial | ETFD_MOUSE | 8 | 15 | 68.9 | 7.3 | 45 | 1.8 |
| 4 | Myozenin-2 | MYOZ2_MOUSE | 4 | 20 | 29.8 | 8.5 | 145 | 0.67 |
| 5 | Electron transfer flavoprotein subunit alpha, mitochondrial | ETFA_MOUSE | 2 | 12 | 35.3 | 8.6 | 89 | 0.67 |
| 6 | Prohibitin | PHB_MOUSE | 8 | 41 | 29.9 | 5.6 | 290 | 0.67 |
| 7 | Dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | ODO2_MOUSE | 3 | 6 | 49.3 | 9.1 | 86 | 0.63 |
| 8 | Isovaleryl-CoA dehydrogenase, mitochondrial | IVD_MOUSE | 7 | 23 | 46.7 | 8.5 | 89 | 0.59 |
| 9 | Serum albumin | ALBU_MOUSE | 3 | 5 | 70.7 | 5.8 | 82 | 0.59 |
| 10 | Cytochrome b-c1 complex subunit Rieske, mitochondrial | UCRI_MOUSE | 4 | 13 | 29.6 | 8.9 | 84 | 0.56 |
| 11 | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | ODP2_MOUSE | 10 | 20 | 68.5 | 8.8 | 117 | 0.53 |
| 12 | Vimentin | VIME_MOUSE | 11 | 20 | 53.7 | 5.1 | 118 | 0.53 |
| 13 | Beta-enolase | ENOB_MOUSE | 8 | 19 | 47.3 | 6.7 | 111 | 0.50 |
| 14 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | ODPB_MOUSE | 8 | 31 | 39.3 | 6.4 | 279 | 0.50 |
| 15 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | ODPB_MOUSE | 3 | 19 | 39.3 | 6.4 | 134 | 0.50 |
| 16 | Creatine kinase M-type | KCRM_MOUSE | 2 | 4 | 43.3 | 6.6 | 47 | 0.50 |
| 17 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | ECH1_MOUSE | 4 | 24 | 36.4 | 7.6 | 201 | 0.50 |
| 18 | Heat shock protein beta-1 Hsp27 | HSPB1_MOUSE | 2 | 8 | 23.1 | 6.1 | 50 | 0.50 |
| 19 | Isovaleryl-CoA dehydrogenase, mitochondrial | IVD_MOUSE | 7 | 17 | 46.7 | 8.5 | 98 | 0.50 |
| 20 | Pyruvate dehydrogenase protein X component, mitochondrial | ODPX_MOUSE | 5 | 11 | 54.3 | 7.6 | 63 | 0.48 |
| 21 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 6 | 19 | 42.3 | 5.2 | 104 | 0.48 |
| 22 | Cytochrome c oxidase subunit 5B, mitochondrial | COX5A_MOUSE | 2 | 14 | 14.1 | 8.7 | 52 | 0.48 |
| 23 | Vimentin | VIME_MOUSE | 13 | 27 | 53.7 | 5.1 | 122 | 0.48 |
| 24 | Transitional endoplasmic reticulum ATPase (valosin-containing protein VCP) | TERA_MOUSE | 5 | 10 | 89.9 | 5.1 | 171 | 0.46 |
| 25 | Pyruvate dehydrogenase E1 component subunit beta, mitochondrial | ODPB_MOUSE | 4 | 14 | 39.3 | 6.4 | 100 | 0.44 |
| 26 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | MLRV_MOUSE | 9 | 52 | 18.9 | 4.9 | 209 | 0.44 |
| 27 | Fatty acid-binding protein FABP-3, heart | FABPH_MOUSE | 5 | 37 | 14.8 | 6.1 | 149 | 0.44 |
| 28 | Enolase 1, alpha | ENOA_MOUSE | 2 | 3 | 47.5 | 6.4 | 43 | 0.44 |
| 29 | DJ-1 protein | PARK7_MOUSE | 4 | 26 | 20.2 | 6.3 | 90 | 0.44 |
| 30 | Cytochrome b-c1 complex | QCR1_MOUSE | 6 | 17 | 53.4 | 5.8 | 83 | 0.44 |
| 31 | 60 kDa heat shock protein, Hsp60, mitochondrial | CH60_MOUSE | 2 | 6 | 61.1 | 5.9 | 63 | 0.42 |
| 32 | Beta-enolase | ENOB_MOUSE | 3 | 7 | 47.3 | 6.7 | 92 | 0.42 |
| 33 | Fatty acid binding protein, FABP-3, heart | FABPH_MOUSE | 7 | 52 | 14.8 | 6.1 | 162 | 0.42 |
| 34 | Pyruvate dehydrogenase protein X component, mitochondrial | ODPX_MOUSE | 2 | 4 | 54.3 | 7.6 | 66 | 0.42 |
| 35 | Tropomyosin alpha-1 chain | TPM1_MOUSE | 15 | 43 | 32.7 | 4.7 | 302 | 0.40 |
| 36 | Electron transfer flavoprotein subunit alpha, mitochondrial | ETFA_MOUSE | 8 | 33 | 35.3 | 8.6 | 235 | 0.40 |
| 37 | 60 kDa heat shock protein, Hsp60, mitochondrial | CH60_MOUSE | 12 | 32 | 61.1 | 5.9 | 346 | 0.40 |
| 38 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | IDH3A_MOUSE | 9 | 29 | 40.1 | 6.3 | 164 | 0.40 |
| 39 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 12 | 31 | 42.3 | 5.2 | 317 | 0.39 |
| 40 | NADH dehydrogenase [ubiquinone] flavoprotein 2, mitochondrial | NDUV2_MOUSE | 6 | 29 | 27.6 | 7.0 | 370 | 0.39 |
| 41 | Voltage-dependent anion-selective channel protein VDAC1 | VDAC1_MOUSE | 10 | 29 | 32.5 | 8.6 | 292 | 0.39 |
| 42 | Isocitrate dehydrogenase [NAD] subunit alpha, mitochondrial | IDH3A_MOUSE | 13 | 39 | 40.1 | 6.3 | 376 | 0.39 |
| 43 | Desmin | DESM_MOUSE | 30 | 50 | 53.5 | 5.2 | 1184 | 0.39 |
| 44 | 60 kDa heat shock protein, Hsp60, mitochondrial | CH60_MOUSE | 25 | 38 | 61.1 | 5.9 | 842 | 0.37 |
| 45 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 10 | 26 | 42.3 | 5.2 | 276 | 0.36 |
| 46 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 4 | 14 | 42.3 | 5.2 | 88 | 0.36 |
| 47 | Desmin | DESM_MOUSE | 15 | 37 | 53.5 | 5.2 | 349 | 0.36 |
| 48 | Myosin regulatory light chain 2, atrial isoform | MLRA_MOUSE | 9 | 57 | 19.6 | 4.8 | 299 | 0.36 |
| 49 | Fatty acid-binding protein, FABP-3, heart | FABPH_MOUSE | 12 | 61 | 14.8 | 6.1 | 425 | 0.35 |
| 50 | ATP synthase subunit beta, mitochondrial | ATPB_MOUSE | 18 | 40 | 56.3 | 5.2 | 1093 | 0.35 |
| 51 | Cytochrome c oxidase subunit 5A | COX5A_MOUSE | 7 | 28 | 16.3 | 6.1 | 286 | 0.35 |
| 52 | Desmin | DESM_MOUSE | 26 | 62 | 53.5 | 5.2 | 1657 | 0.35 |
| 53 | Myosin light chain 3 | MYL3_MOUSE | 6 | 41 | 22.5 | 5.0 | 90 | 0.35 |
| 54 | Desmin | DESM_MOUSE | 12 | 30 | 53.5 | 5.2 | 431 | 0.33 |
| 55 | ATP synthase subunit d, mitochondrial | ATP5H_MOUSE | 3 | 21 | 18.8 | 5.5 | 48 | 0.33 |
| 56 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 16 | 40 | 42.3 | 5.2 | 598 | 0.32 |
| 57 | Cytochrome b-c1 complex | QCR1_MOUSE | 12 | 27 | 53.4 | 5.8 | 330 | 0.32 |
| 58 | Desmin | DESM_MOUSE | 36 | 68 | 53.5 | 5.2 | 1289 | 0.32 |
| 59 | Tropomyosin alpha-1 chain | TPM1_MOUSE | 21 | 46 | 32.7 | 4.7 | 605 | 0.32 |
| 60 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 18 | 59 | 42.3 | 5.2 | 450 | 0.31 |
| 61 | Myosin light chain 3 | MYL3_MOUSE | 11 | 63 | 22.5 | 5.0 | 231 | 0.30 |
| 62 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | MLRV_MOUSE | 13 | 73 | 18.9 | 4.9 | 118 | 0.29 |
| 63 | Cytochrome b-c1 complex | QCR1_MOUSE | 12 | 31 | 53.4 | 5.8 | 342 | 0.29 |
| 64 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 7 | 36 | 42.3 | 5.2 | 160 | 0.29 |
| 65 | Myosin regulatory light chain 2, ventricular/cardiac muscle isoform | MLRV_MOUSE | 7 | 46 | 18.9 | 4.9 | 354 | 0.29 |
| 66 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 4 | 24 | 42.3 | 5.2 | 91 | 0.28 |
| 67 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 17 | 62 | 42.3 | 5.2 | 294 | 0.27 |
| 68 | Adenylate kinase isoenzyme 1 | KAD1_MOUSE | 5 | 31 | 21.6 | 5.7 | 174 | 0.26 |
| 69 | ATP synthase subunit beta, mitochondrial | ATPB_MOUSE | 4 | 21 | 56.3 | 5.2 | 92 | 0.26 |
| 70 | Myosin regulatory light chain 2 | MLRA_MOUSE | 6 | 47 | 19.6 | 4.8 | 168 | 0.25 |
| 71 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 18 | 61 | 42.3 | 5.2 | 302 | 0.25 |
| 72 | ATP synthase subunit d, mitochondrial | ATP5H_MOUSE | 5 | 36 | 18.8 | 5.5 | 69 | 0.25 |
| 73 | ATP synthase subunit beta, mitochondrial | ATPB_MOUSE | 18 | 51 | 56.3 | 5.2 | 640 | 0.25 |
| 74 | ATP synthase subunit beta, mitochondrial | ATPB_MOUSE | 25 | 71 | 56.3 | 5.2 | 1102 | 0.25 |
| 75 | Actin, alpha cardiac muscle 1 | ACTC_MOUSE | 6 | 16 | 42.3 | 5.2 | 119 | 0.25 |
| 76 | Myosin light chain 3 | MYL3_MOUSE | 6 | 30 | 22.5 | 5.0 | 99 | 0.25 |
| 77 | ATP synthase subunit beta, mitochondrial | ATPB_MOUSE | 24 | 70 | 56.3 | 5.2 | 871 | 0.24 |
| 78 | Myosin light chain 2, ventricular/cardiac | MLRV_MOUSE | 11 | 77 | 18.9 | 4.9 | 206 | 0.24 |
| 79 | Peroxiredoxin-6 | PRDX6_MOUSE | 3 | 19 | 24.9 | 5.7 | 102 | 0.19 |
Figure 3.
DIGE analysis of normal versus dystrophic MDX cardiac muscle. Shown are Cy5-labelled master gels of the soluble fraction from mouse heart muscle, covering both the pH 4–7 (a) and pH 6–11 (b) range. Protein spots with a drastically different expression level are marked by circles and are numbered 1 to 79. See Table 1 for a detailed listing of cardiac muscle proteins with a changed abundance in dystrophin-deficient tissue. The pH-values of the first dimension gel system and molecular mass standards (in kDa) of the second dimension are indicated on the top and on the left of the panels, respectively.
3.3. Decreased Proteins in Dystrophic Heart Muscle
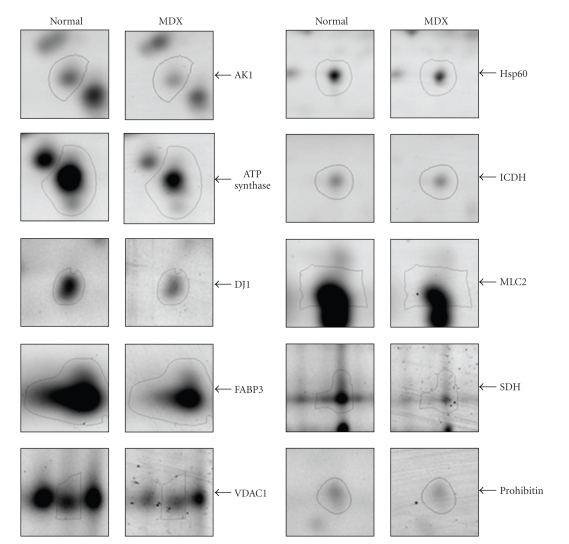
To illustrate representative examples of key cardiac proteins with a drastically decreased expression in dystrophin-deficient fibres, enlarged images of DIGE-identified protein spots are shown in Figure 4. The direct comparison of fluorescently labeled 2-D protein spots, which have been unequivocally identified by mass spectrometric analysis (Table 1), shows the cytosolic AK1 isoform of adenylate kinase, the mitochondrial proteins ATPase synthase, porin isoform VDAC1, isocitrate dehydrogenase, succinate dehydrogenase, and prohibitin, as well as DJ1-protein, the stress protein Hsp60 and myosin light chain isoform MLC2. A large proportion of the cardiac proteins identified in this study by DIGE analysis belong to the contractile apparatus, the cellular stress response and the cytoskeletal network, as well as mitochondrial metabolism including oxidative phosphorylation, the citric acid cycle and fatty acid transportation.
Figure 4.
Decreased expression of key proteins in the dystrophin-deficient MDX heart. Shown are expanded views of 2-D gels of normal versus dystrophic MDX heart tissue. Shown are the AK1 isoform of adenylate kinase, ATP synthase, DJ1-protein, fatty acid binding protein FABP3, porin isoform VDAC1, stress protein Hsp60, isocitrate dehydrogenase (ICDH), myosin light chain MLC2, succinate dehydrogenase (SDH) and prohibitin. The position of 2-D spots representing distinct mitochondrial proteins is indicated by arrowheads on the right.
3.4. Immunoblot Analysis of Dystrophic Heart Muscle
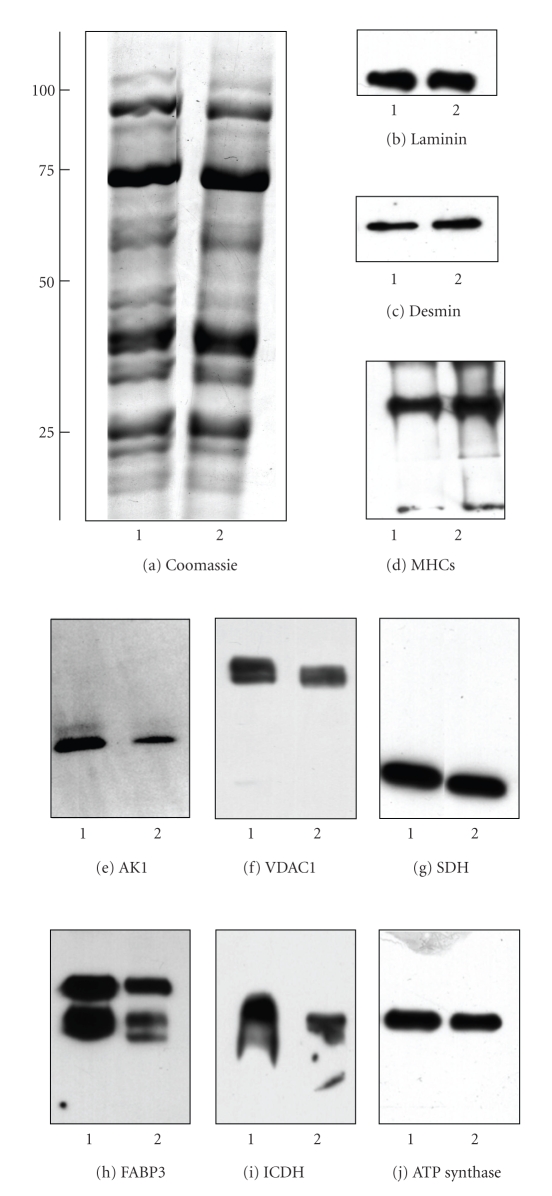
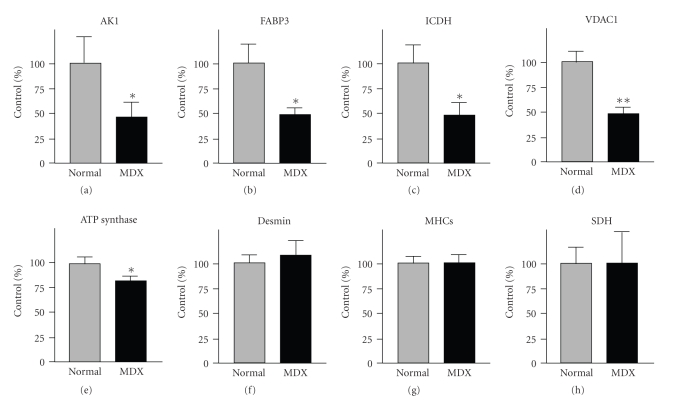
Previous studies have clearly shown that the deficiency in dystrophin triggers a drastically reduced concentration of the dystrophin-associated α/β-dystroglycan complex in the MDX heart [32]. Immunoblotting has established distinct alterations down-stream of the cardiac dystrophin-glycoprotein complex, such as the reduction in the luminal Ca2+-binding proteins calsequestrin and sarcalumenin [33]. In analogy, we employed Western blotting here to verify the results of our DIGE analysis by immuno-decoration analysis of a select group of cardiac proteins. In this respect, it is important to stress that the mass spectrometry-based proteomic analysis of individual 2D spots identifies changes in distinct subspecies of proteins and not alterations in the total population of a protein isoform. The recent large-scale cataloguing of the heart proteome has shown that cardiac tissues contain thousands of different protein species, whereby many proteins are represented by a large number of distinct 2D-spots in analytical gels [57–59]. It is one of the great advantages of modern proteomics that this analytical approach can differentiate between differently charged or sized subspecies of individual proteins. Since 1D immunoblotting or immunofluorescence microscopy often do not recognize all forms of a specific protein due to a lack of antibody specificity, proteomic findings on an individual 2D protein spot sometimes do not correlate fully with the findings from biochemical or cell biological analyses of total protein cohorts. These apparent differences in the fate of individual subspecies of a protein versus the total protein population probably reflect the complexity of disease-related changes in the cardiac proteome. Figure 5(a) shows a Coomassie-stained gel of crude preparations from normal and dystrophic hearts and illustrates that no major differences exist in the overall protein expression levels between the two different phenotypes. The immunoblot analysis of laminin, desmin, the slow/cardiac myosin heavy chain and succinate dehydrogenase revealed no major differences in the concentration of these abundant cardiac proteins in normal versus MDX specimens (Figures 5(b)–5(d) and 5(g)). In contrast, immunoblotting of the enzyme adenylate kinase AK1, the fatty acid transporter FABP3, isocitrate dehydrogenase, ATP synthase and porin isoform VDAC1 showed a reduced abundance in MDX preparations (Figures 5(e), 5(f), 5(h)–5(j)). Figure 6 summarizes the statistical evaluation of the immunoblot survey in a graphical presentation. The reduced expression of adenylate kinase isoform AK1, fatty acid binding protein FABP3, isocitrate dehydrogenase ICDH, porin isoform VDAC1, and ATP synthase was found to be statistically significant, while the concentration of desmin, myosin heavy chain and succinate dehydrogenase was shown not to be significantly different between normal and dystrophic preparations. Thus the overall isoform population of succinate dehydrogenase and desmin does not appear to be reduced in dystrophinopathy, although the above-described DIGE analysis showed a lower level of distinct subspecies of these cardiac elements. In general, the immunoblotting results support the main findings from our DIGE analysis of normal versus dystrophic heart tissue and demonstrate independently that the deficiency in dystrophin has a profound effect on the expression pattern of the cardiac protein complement.
Figure 5.
Immunoblotting survey of cardiac proteins in dystrophic tissue. Shown is a Coomassie-stained gel (a) and corresponding immunoblots with expanded views of immuno-decorated bands ((b)–(j)). Immunoblotting was performed with antibodies to laminin (b), desmin (c), slow/cardiac myosin heavy chain ((d); MHCs), adenylate kinase isoform AK1 (e), porin isoform VDAC1 (f), succinate dehydrogenase ((g); SDH), fatty acid binding protein FABP3 (h), isocitrate dehydrogenase ((i); ICDH), and ATP synthase (j). See Figure 6 for the statistical evaluation of this immunoblotting survey of cardiac proteins. Molecular mass standards (in kDa) are indicated on the left of panel (a). Lanes 1 and 2 represent normal and dystrophic muscle extracts from control and MDX mice, respectively.
Figure 6.
Graphical presentation of the immunoblot analysis of cardiac marker proteins in dystrophic tissue. Shown is the graphical presentation of the statistical evaluation of immuno-decoration using antibodies to adenylate kinase isoform AK1 (a), fatty acid binding protein FABP3 (b), isocitrate dehydrogenase ICDH (c), porin isoform VDAC1 (d), ATP synthase (e), desmin (f), slow/cardiac myosin heavy chain MHCs (G) and succinate dehydrogenase SDH (H). The comparative blotting was statistically evaluated using an unpaired Student's t-test (n = 5; *P < .05; **P < .01). The concentration of desmin, myosin heavy chain and succinate dehydrogenase was found not to be significantly different between normal and dystrophic preparations. Lanes 1 and 2 represent normal and dystrophic muscle extracts from control and MDX mice, respectively.
3.5. Immunofluorescence Analysis of Dystrophic Heart Muscle
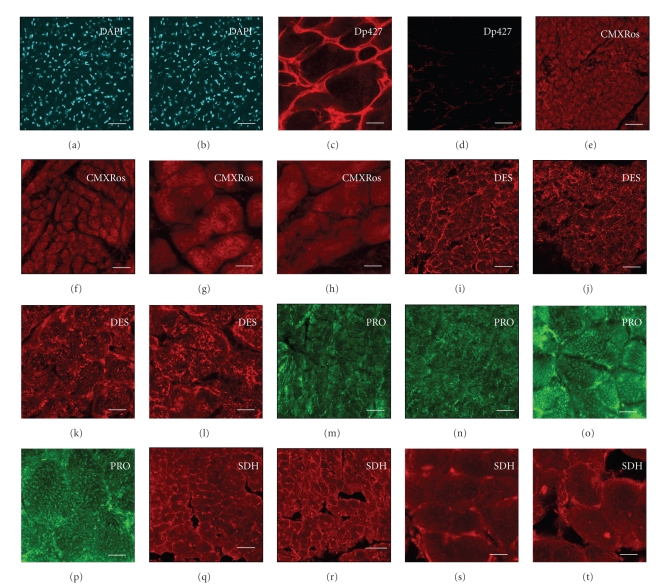
Since the DIGE analysis of the normal versus the dystrophic MDX heart has revealed changes in a variety of mitochondrial components, it was important to determine whether this alteration was due to a reduced number of mitochondria in the dystrophic heart or internal abundance changes in the mitochondrial protein complement. Figure 7 shows representative results from an immunofluorescence survey of mitochondrial content, nuclei and cardiac marker proteins in dystrophic tissue. Cryosections of MDX tissue showed a complete absence of the dystrophin isoform Dp427 (Figures 7(c), 7(d)), confirming the mutant status of the dystrophic hearts used in this study. DAPI staining revealed 229 ± 6 and 270 ±18 nuclei per examined tissue section in normal versus dystrophic tissue sections, respectively (Figures 7(a), 7(b)). The slight increase of nuclei in MDX preparations was not statistically significant (n = 4). Thus the proteomic finding of a dystrophy-dependent increase in lamin-A/C is therefore not directly related to a drastic change in the number of nuclei per cardiac tissue unit. Fluorescent labeling of mitochondria with the MitoTracker dye CMXRos showed a characteristic internal staining pattern in cardiac fibres (Figures 7(e)–7(h)). Fluorescent intensity values of 1470 ± 36 and 1384 ± 40 for normal versus MDX preparations were found not to be significantly different (n = 4). Hence, lower levels of mitochondrial enzymes do not seem to be a consequence of a drastic decrease in mitochondrial density in the dystrophic MDX heart. In addition, confocal microscopy was employed for the localization of cardiac marker proteins. The immunofluorescence labeling of desmin (Figures 7(i)–7(l)), prohibitin (Figures 7(m)–7(p)) and succinate dehydrogenase (Figures 7(q)–7(t)) was shown to be comparable between control and pathological samples. The microscopical localization of desmin and succinate dehydrogenase revealed unexpectedly not a decrease of these proteins in dystrophic tissue sections, as found by DIGE analysis. Hence, although distinct subspecies of certain cardiac proteins are affected in X-linked muscular dystrophy as determined by mass spectrometry-based proteomics, the overall isoform complement of these elements is not drastically altered. This indicates that no major differences exist in the expression levels of desmin-containing intermediate filament structures and the population of cardiac mitochondria in normal versus dystrophin-deficient cells.
Figure 7.
Immunofluorescence survey of mitochondrial content, nuclei and cardiac marker proteins in dystrophic tissue. Confocal microscopy was used for the localization of nuclei, mitochondria and cellular markers in normal ((a), (c), (e), (g), (i), (k), (m), (o), (q), (s)) versus dystrophic MDX ((b), (d), (f), (h), (j), (l), (n), (p), (r), (t)) heart cryosections. Shown is labeling of nuclei with the DNA binding dye DAPI ((a), (b)), visualization of mitochondria with the red-fluorescent MitoTracker dye CMXRos ((e)-(h)), and antibody labeling of full-length dystrophin isoform Dp427 ((c), (d)), desmin DES ((i)-(l)), prohibitin PRO ((m)-(p)) and succinate dehydrogenase SDH ((q)-(t)). The number of nuclei and the labeling of mitochondria with the MitoTracker dye CMXRos were found not to be significantly different between normal and dystrophic preparations. The bars in panels (a) to (t) equal 30 μm.
4. Discussion
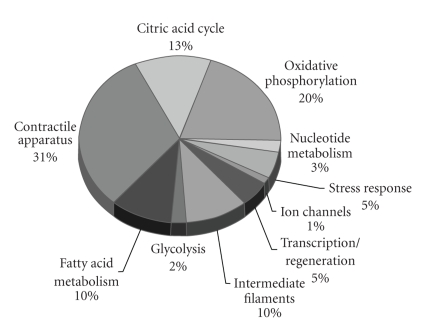
In order to enhance our knowledge of the molecular pathogenesis of cardiomyopathy linked to X-linked muscular dystrophy, a detailed proteomic profiling of the dystrophic heart was carried out. The biochemical establishment of a comprehensive disease-specific biomarker signature for X-linked muscular dystrophy is crucial for (i) the development of superior diagnostic methods, (ii) the identification of new therapeutic targets to address cellular alterations down-stream of the primary abnormality in dystrophin, and (iii) the evaluation of new treatment approaches to counter-act the various pathological aspects of dystrophinopathy [41]. The proteomic profiling of MDX skeletal muscle has recently established altered expression levels of soluble proteins involved in nucleotide metabolism, Ca2+-handling and the cellular stress response [42–45]. The mass spectrometric analysis of experimental exon skipping therapy revealed a partial reversal of dystrophic changes in these new signature molecules [60], illustrating the potential of proteomic technology to decisively enhance the capabilities of biomedical research into the molecular mechanisms that underlie neuromuscular disorders. In contrast to skeletal muscle, very little is known about the global pathobiochemical mechanisms that lead to cardiac complications in the dystrophin-deficient organism. Here, we have successfully applied the fluorescent DIGE method to the large-scale analysis of the dystrophic heart. MS-based proteomics clearly revealed a drastic decrease in key metabolic, regulatory and contractile proteins, as well as components of the cytoskeleton and the cellular stress response. The pie chart of Figure 8 summarizes the apparent functions of DIGE-identified proteins with a differential expression in normal versus dystrophin-deficient heart muscle.
Figure 8.
Overview of biological functions of DIGE-identified proteins with an altered expression in dystrophic heart muscle. The pie chart summarizes the apparent functions of DIGE-identified proteins with a differential expression in normal versus dystrophin-deficient MDX heart muscle. A large proportion of these affected proteins belong to cardiac mitochondria including metabolic elements of oxidate phosphorylation, the citric acid cycle and fatty acid transportation, as well as proteins associated with the contractile apparatus, the cellular stress response, and the cytoskeletal network.
The DIGE analysis presented here showed a moderate increase in lamin-A/C, which might improve nuclear stability, chromatin structure and gene expression in the dystrophic heart. Since comparative DAPI staining indicates comparable numbers of nuclei in normal versus dystrophic fibres, tissue scarring is probably not responsible for an increase in the number of nuclei per tissue unit. Therefore, the elevated lamin concentration does not appear to be associated with increased numbers of nuclei. Cardiac lamins make up the matrix that is located close to the inner nuclear membrane. Interestingly their deficiency causes familial dilated cardiomyopathy [61]. The only drastic increase in a cardiac protein was the 4-fold change in the expression of nucleoside diphosphate kinase B. Nucleoside diphosphate kinases are key regulatory enzymes involved in intracellular di- and tri-phosphonucleoside homeostasis. Their cellular functions are critical for signaling, proliferation, differentiation and bioenergetics [62], which suggests that the up-regulation of one of their isoforms could be considered a counter measure of the dystrophin-deficient heart to stabilize essential cellular functions in severely stressed muscle cells. In contrast to the 4-fold increase in nucleoside diphosphate kinase, our proteomic screening demonstrated a massive decrease in a variety of cardiac proteins in the MDX heart. The dystrophinopathy-associated loss in numerous key proteins, which belong to different cellular pathways and functions in the heart, agrees with the severity of the cardiomyopathic complications observed in aged MDX mice and DMD patients [13–15].
Key proteins involved in cardiac contraction, cytoskeletal integrity, nucleotide metabolism, cellular stress response, mitochondrial metabolism, and fatty acid transportation were shown to be affected by a deficiency in cardiac dystrophin. Interestingly, Braun et al. [63] could show an altered integration of mitochondria and ATPases in slow-twitching MDX muscles. The coupling between mitochondrial creatine kinase and the adenine nucleotide translocase system appears to be weakened in dystrophin-deficient fibres. This seems to result in a rearrangement of intracellular energy transfer in dystrophic muscle [63]. The 6-fold decrease in peroxiredoxin-6 may have drastic consequences for the capability of the dystrophic heart to counter-act the detrimental effects of stressors. Peroxiredoxin-6 is a crucial enzyme that protects cells from oxidative stress [64]. In analogy to the reduced concentration of this antioxidant enzyme in cardiac MDX tissue, the decrease in the small heat shock protein Hsp27, as well as Hsp60 and DJ-1 protein, also agrees with an impaired cellular stress response in muscular dystrophy-associated cardiomyopathy. Small heat shock proteins provide crucial chaperone function in striated muscles and are upregulated in the heart following exercise training [65]. An impaired stress response could result in the detrimental aggregation of misfolded proteins causing cellular dysfunction in the dystrophic heart. The dystrophinopathy-associated decrease in valsolin-containing protein VCP might also impair cardiac function. VCP belongs to the AAA-ATPase superfamily and is involved in various cellular functions such as membrane fusion and cell-cycle control, as well as protein degradation through the ubiquitin-proteasome pathway [66].
The DIGE analysis of the MDX heart presented here has clearly shown a severely altered concentration of numerous key players involved in cardiac contraction and cytoskeletal organization. In contrast to comparable levels of myosin heavy chain, other critical contractile proteins are affected by the lack of dystrophin. The biomolecules that form the thick and thin filaments of the basic contractile units exist in a complex arrangement of myosin heavy chains, myosin light chains, and actins. Muscle myosin consists of a hexameric arrangement that is formed by two heavy chains and two pairs of myosin light chains. Changes in myosin light chains have been implicated in severe cardiac impairments, such as familial hypertrophic cardiomyopathy [67]. Hence, the reduction in the cardiac isoform of myosin light chain MLC2, myosin light chain MLC3, and cardiac alpha-actin might have a severe impact on the contractile apparatus in the dystrophic heart. Contractile weakness observed in dystrophinopathy could be directly related to the altered concentration of myosin light chains and actin. Cardiac muscle cells are activated by Ca2+-binding to TnC-subunit of troponin and is regulated by the interactions of tropomyosin and troponin in the thin filament. Thus, the observed reduction of alpha-1 tropomyosin in the MDX heart might also be of pathophysiological significance. Since the molecular coupling between the myosin head structure and actin filaments, in the presence of ATP, represents the major step that underlies the sliding of thin filaments past thick filaments, abnormal concentrations of regulatory proteins may result in impaired patterns of sarcomeric shortening.
In addition, a close connection exists between the contractile apparatus, the membrane-associated actin network and the cytoskeleton. Intermediate filaments are of crucial importance for the mechanical integrity and elasticity of contractile fibres. Although proteomics showed that the expression levels of distinct subspecies of desmin were reduced in dystrophic preparations, confocal microscopy and immunoblotting revealed that the overall population of desmin isoforms is comparable between normal and MDX heart. However, the deficiency in cardiac dystrophin appears to trigger a concomitant reduction in the density of myozenin-2 and vimentin. Myozenin-2 belongs to the calsarcin family of proteins, a new type of sarcomeric calcineurin-binding protein [68]. Vimentin is a crucial intermediary filament protein and plays a key role in cytoskeletal network formation. The abnormal expression levels of these proteins might therefore cause disturbed interfilament interactions and impaired cystoskeletal arrays, thereby causing cellular instability in the dystrophic heart.
One of the most important findings of this mass spectrometry-based proteomic study is the drastically reduced concentration of numerous mitochondrial proteins and metabolic transporters. Since mitochondria represent the primary site for energy generation via oxidative phosphorylation [69], a reduction in elements belonging to the oxidative phosphorylation complexes, the citric acid cycle and ion homeostasis will have a profound influence on the bioenergetic status of the dystrophic heart. Cardiac mitochondria are involved in intermediary metabolism, cell cycle progression, calcium signaling and the regulation of apoptosis, as well as the production of heme and iron-sulfur clusters. Proteomic investigations suggest that approximately 1,500 different mitochondrial proteins exist [70]. Previous studies have shown that altered expression levels within the mitochondrial proteome influence many aspects of normal development, diseases and the aging process. With respect to the heart, mitochondrial dysfunction appears to be associated with numerous pathologies such as congestive heart failure, ischaemia reperfusion injury and cardiomyopathy [71]. Therefore, the reduced expression of many mitochondrial proteins associated with the matrix, outer membrane and inner membrane system suggests an impaired metabolism in the dystrophic heart. Reduced expression levels within the mitochondrial proteome appear to affect the functioning of the mitochondrial ATP synthase, isocitrate dehydrogenase, NADH dehydrogenase, oxoglutarate dehydrogenase, isovaleryl-CoA dehydrogenase, cytochrome c oxidase and the cytochrome b-c complex, all critical elements involved in mitochondrial function in the heart [69]. This agrees with a previous study by Zhang et al. [72], who reported a decreased citrate synthase activity in the MDX heart. Although a certain degree of cellular scarring and the infiltration of fatty and connective tissue may reduce the number of contractile fibres in the dystrophic heart, as previously shown by histochemical analysis [72], this does not appear to drastically alter the density of muscle-associated mitochondria. The confocal microscopy analysis shown in this report suggests no major changes in the number of mitochondria between normal and dystrophic heart muscle tissue, as judged by labeling with the established chloromethyl-X-rosamine MitoTracker dye which is concentrated inside mitochondria by their negative membrane potential [53–55].
Our DIGE analysis revealed that individual protein spots representing electron-transferring flavoprotein showed both increased and decreased abundance. Electron transfer flavoprotein ubiquinone oxidoreductase provides a crucial link between electrons derived from fatty acid oxidation and certain amino acids to the main respiratory chain system of mitochondria [73]. The differential effect on different isoforms of this protein is difficult to interpret, but dystrophin deficiency must affect distinct subspecies of this metabolic element with differing post-translational modifications in a dissimilar way. The observed reduction in the rate-limiting enzyme pyruvate dehydrogenase could have severe impact on the utilization of energy carriers and thereby impair the overall bioenergetics of the MDX heart. Isoforms of cardiac prohibitin are present in the inner mitochondrial membrane and are responsible for the efficient assembly of mitochondrial respiratory chain enzymes and perform chaperone activity. Interestingly, a proteomic survey of mitochondrial proteins in cardiomyocytes from chronic stressed rats has shown a drastic increase in prohibitin [74]. Although the overall complement of prohibitin isoforms is not drastically affected in muscular dystrophy, a reduced expression of distinct subspecies of prohibitin in the dystrophic heart might be associated with an impaired cellular stress response. The only affected glycolytic enzyme appears to be enolase, indicating that mitochondrial metabolism is more severely affected than glycolysis in dystrophy-related cardiomyopathy. Interestingly, the abundance of the extracellular and intracellular fatty acid transporters albumin and FABP-3, which are considered limiting factors of oxidative metabolism in striated muscles, is decreased in the dystrophin-deficient heart. Thus, dystrophic heart cells would be starved of essential fuel supplies, which might trigger a decline in contractile strength. A mitochondrial receptor present in the outer mitochondrial membrane was identified as a porin isoform that forms voltage-dependent anion channels. These ion channels belong to a family of pore-forming proteins that provide large membrane-spanning aqueous channels [75]. The decreased concentration in the porin transport protein would indicate a dystrophy-associated decrease in mitochondrial function and abnormal ion homeostasis.
In contrast to a recent combined metabolomic and proteomic study of the MDX heart [46], and in agreement with two comprehensive proteomic studies on MDX skeletal muscle [42, 45], we could show here a decreased abundance of the AK1 isoform of adenylate kinase. The proteomic finding was clearly verified by immunoblot analysis. Nucleotide metabolism, involving adenylate kinase and creatine kinase appears to be disturbed in the dystrophic MDX heart, and this finding might be useful for the future development of a comprehensive biomarker signature of cardiomyopathy associated with muscular dystrophy. Overall, our DIGE-based screening of the soluble proteome from dystrophic MDX hearts has revealed a severely perturbed protein expression pattern due to deficiency in dystrophin. The observed changes in essential proteins involved in cardiac contraction, mitochondrial metabolism, the cellular stress response and nucleotide metabolism might be useful for the future improvement of differential diagnostic procedures, the assessment of cardiac disease progression in X-linked muscular dystrophies, the identification of novel therapeutic targets to treat cardiomopathic complications in Duchenne muscular dystrophy, and the evaluation of novel treatment strategies to counter-act the loss in cardiac dystrophin.
Acknowledgments
Research was supported by a project Grant from Muscular Dystrophy Ireland (MDI-125437), a postgraduate stipend from the Irish Research Council for Science, Engineering and Technology, Fonds der Chemischen Industrie and a Hume scholarship from NUI Maynooth, as well as equipment Grants from the Irish Health Research Board and the Higher Education Authority (HRB-EQ/2003/3, HEA-RERGS-07-NUIM, HRB-EQ/2004/2). The authors thank Dr. Peter Heimann (University of Bielefeld) for his professional help obtaining MDX muscle, Ms. Caroline Batchlor (NUI Maynooth) for assistance with mass spectrometry, and Dr. Ica Dix (NUI Maynooth) for her help with confocal microscopy. The Irish Higher Education Authority and the Deutscher Akademischer Austauschdienst supported laboratory visits of the Maynooth team in Bielefeld.
Abbreviations
- DIGE:
Difference in-gel electrophoresis
- DMD:
Duchenne muscular dystrophy
- Dp:
Dystrophin protein
- MDX:
X-linked muscular dystrophy.
References
- 1.Koening M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50(3):509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman EP, Brown RH, Jr., Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51(6):919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Bonilla E, Samitt CE, Miranda AF, et al. Duchenne muscular dystrophy: deficiency of dystrophin at the muscle cell suface. Cell. 1988;54(4):447–452. doi: 10.1016/0092-8674(88)90065-7. [DOI] [PubMed] [Google Scholar]
- 4.Deconinck N, Dan B. Pathophysiology of Duchenne muscular dystrophy: current hypotheses. Pediatric Neurology. 2007;36(1):1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Dalkilic I, Kunkel LM. Muscular dystrophies: genes to pathogenesis. Current Opinion in Genetics and Development. 2003;13(3):231–238. doi: 10.1016/s0959-437x(03)00048-0. [DOI] [PubMed] [Google Scholar]
- 6.Ohlendieck K. Towards an understanding of the dystrophin-glycoprotein complex: linkage between the extracellular matrix and the membrane cytoskeleton in muscle fibers. European Journal of Cell Biology. 1996;69(1):1–10. [PubMed] [Google Scholar]
- 7.Ervasti JM. Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochimica et Biophysica Acta. 2007;1772(2):108–117. doi: 10.1016/j.bbadis.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nature Genetics. 1993;3(4):283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 9.Cohn RD, Campbell KP. Molecular basis of muscular dystrophies. Muscle & Nerve. 2000;23(10):1456–1471. doi: 10.1002/1097-4598(200010)23:10<1456::aid-mus2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 10.Batchelor CL, Winder SJ. Sparks, signals and shock absorbers: how dystrophin loss causes muscular dystrophy. Trends in Cell Biology. 2006;16(4):198–205. doi: 10.1016/j.tcb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 11.Anderson JL, Head SI, Rae C, Morley JW. Brain function in Duchenne muscular dystrophy. Brain. 2002;125(1):4–13. doi: 10.1093/brain/awf012. [DOI] [PubMed] [Google Scholar]
- 12.Connuck DM, Sleeper LA, Colan SD, et al. Characteristics and outcomes of cardiomyopathy in children with Duchenne or Becker muscular dystrophy: a comparative study from the Pediatric Cardiomyopathy Registry. American Heart Journal. 2008;155(6):998–1005. doi: 10.1016/j.ahj.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finsterer J, Stollberger C. The heart in human dystrophinopathies. Cardiology. 2003;99(1):1–19. doi: 10.1159/000068446. [DOI] [PubMed] [Google Scholar]
- 14.Cox GF, Kunkel LM. Dystrophies and heart disease. Current Opinion in Cardiology. 1997;12(3):329–343. [PubMed] [Google Scholar]
- 15.Muntoni F. Cardiomyopathy in muscular dystrophies. Current Opinion in Neurology. 2003;16(5):577–583. doi: 10.1097/01.wco.0000093100.34793.81. [DOI] [PubMed] [Google Scholar]
- 16.Sultan A, Fayaz M. Prevalence of cardiomyopathy in Duchenne and Becker’s muscular dystrophy. Journal of Ayub Medical College, Abbottabad. 2008;20(2):7–13. [PubMed] [Google Scholar]
- 17.McNally EM. New approaches in the therapy of cardiomyopathy in muscular dystrophy. Annual Review of Medicine. 2007;58(1):75–88. doi: 10.1146/annurev.med.58.011706.144703. [DOI] [PubMed] [Google Scholar]
- 18.Duan D. Challenges and opportunities in dystrophin-deficient cardiomyopathy gene therapy. Human Molecular Genetics. 2006;15(2):R253–R261. doi: 10.1093/hmg/ddl180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Townsend DW, Yasuda S, Metzger J. Cardiomyopathy of Duchenne muscular dystrophy: pathogenesis and prospect of membrane sealants as a new therapeutic approach. Expert Review of Cardiovascular Therapy. 2007;5(1):99–109. doi: 10.1586/14779072.5.1.99. [DOI] [PubMed] [Google Scholar]
- 20.Grounds MD, Radley HG, Lynch GS, Nagaraju K, De Luca A. Towards developing standard operating procedures for pre-clinical testing in the MDX mouse model of Duchenne muscular dystrophy. Neurobiology of Disease. 2008;31(1):1–19. doi: 10.1016/j.nbd.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the MDX mouse: a point mutation. Science. 1989;244(4912):1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 22.Bridges LR. The association of cardiac muscle necrosis and inflammation with the degenerative and persistent myopathy of MDX mice. Journal of the Neurological Sciences. 1986;72(2-3):147–157. doi: 10.1016/0022-510x(86)90003-1. [DOI] [PubMed] [Google Scholar]
- 23.Chu V, Otero JM, Lopez O, et al. Electrocardiographic findings in MDX mice: a cardiac phenotype of Duchenne muscular dystrophy. Muscle and Nerve. 2002;26(4):513–519. doi: 10.1002/mus.10223. [DOI] [PubMed] [Google Scholar]
- 24.Quinlan JG, Hahn HS, Wong BL, Lorenz JN, Wenisch AS, Levin LS. Evolution of the MDX mouse cardiomyopathy: physiological and morphological findings. Neuromuscular Disorders. 2004;14(8-9):491–496. doi: 10.1016/j.nmd.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Khairallah M, Khairallah R, Young ME, Dyck JRB, Petrof BJ, Des Rosiers C. Metabolic and signaling alterations in dystrophin-deficient hearts precede overt cardiomyopathy. Journal of Molecular and Cellular Cardiology. 2007;43(2):119–129. doi: 10.1016/j.yjmcc.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 26.Yue Y, Li Z, Harper SQ, Davisson RL, Chamberlain JS, Duan D. Microdystrophin gene therapy of cardiomyopathy restores dystrophin-glycoprotein complex and improves sarcolemma integrity in the MDX mouse heart. Circulation. 2003;108(13):1626–1632. doi: 10.1161/01.CIR.0000089371.11664.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wehling-Henricks M, Jordan MC, Roos KP, Deng B, Tidball JG. Cardiomyopathy in dystrophin-deficient hearts is prevented by expression of a neuronal nitric oxide synthase transgene in the myocardium. Human Molecular Genetics. 2005;14(14):1921–1933. doi: 10.1093/hmg/ddi197. [DOI] [PubMed] [Google Scholar]
- 28.Wu B, Moulton HM, Iversen PL, et al. Effective rescue of dystrophin improves cardiac function in dystrophin-deficient mice by a modified morpholino oligomer. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):14814–14819. doi: 10.1073/pnas.0805676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buyse GM, Van der Mieren G, Erb M, et al. Long-term blinded placebo-controlled study of SNT-MC17/idebenone in the dystrophin deficient MDX mouse: cardiac protection and improved exercise performance. European Heart Journal. 2009;30(1):116–124. doi: 10.1093/eurheartj/ehn406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sapp JL, Bobet J, Howlett SE. Contractile properties of myocardium are altered in dystrophin-deficient MDX mice. Journal of the Neurological Sciences. 1996;142(1-2):17–24. doi: 10.1016/0022-510x(96)00167-0. [DOI] [PubMed] [Google Scholar]
- 31.Bia BL, Cassidy PJ, Young ME, et al. Decreased myocardial nNOS, increased iNOS and abnormal ECGs in mouse models of duchenne muscular dystrophy. Journal of Molecular and Cellular Cardiology. 1999;31(10):1857–1862. doi: 10.1006/jmcc.1999.1018. [DOI] [PubMed] [Google Scholar]
- 32.Lohan J, Culligan K, Ohlendieck K. Deficiency in cardiac dystrophin affects the abundance of the α-/β-dystroglycan complex. Journal of Biomedicine and Biotechnology. 2005;2005(1):28–36. doi: 10.1155/S111072430440103X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohan J, Ohlendieck K. Drastic reduction in the luminal Ca2+-binding proteins calsequestrin and sarcalumenin in dystrophin-deficient cardiac muscle. Biochimica et Biophysica Acta. 2004;1689(3):252–258. doi: 10.1016/j.bbadis.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 34.Fanchaouy M, Polakova E, Jung C, Ogrodnik J, Shirokova N, Niggli E. Pathways of abnormal stress-induced Ca2+ influx into dystrophic MDX cardiomyocytes. Cell Calcium. 2009;46(2):114–121. doi: 10.1016/j.ceca.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lefaucheur JP, Pastoret C, Sebille A. Phenotype of dystrophinopathy in old MDX mice. Anatomical Record. 1995;242(1):70–76. doi: 10.1002/ar.1092420109. [DOI] [PubMed] [Google Scholar]
- 36.Nakamura A, Yoshida K, Takeda S, Dohi N, Ikeda S-I. Progression of dystrophic features and activation of mitogen-activated protein kinases and calcineurin by physical exercise, in hearts of MDX mice. FEBS Letters. 2002;520(1–3):18–24. doi: 10.1016/s0014-5793(02)02739-4. [DOI] [PubMed] [Google Scholar]
- 37.Spurney CF, Knoblach S, Pistilli EE, Nagaraju K, Martin GR, Hoffman EP. Dystrophin-deficient cardiomyopathy in mouse: expression of Nox4 and Lox are associated with fibrosis and altered functional parameters in the heart. Neuromuscular Disorders. 2008;18(5):371–381. doi: 10.1016/j.nmd.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Hoog CL, Mann M. Proteomics. Annual Review of Genomics and Human. Genetics. 2004;5(1):267–293. doi: 10.1146/annurev.genom.4.070802.110305. [DOI] [PubMed] [Google Scholar]
- 39.Viswanathan S, Unlu M, Minden JS. Two-dimensional difference gel electrophoresis. Nature Protocols. 2006;1(3):1351–1358. doi: 10.1038/nprot.2006.234. [DOI] [PubMed] [Google Scholar]
- 40.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Analytical and Bioanalytical Chemistry. 2005;382(3):669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 41.Doran P, Gannon J, O’Connell K, Ohlendieck K. Proteomic profiling of animal models mimicking skeletal muscle disorders. Proteomics. 2007;1(9):1169–1184. doi: 10.1002/prca.200700042. [DOI] [PubMed] [Google Scholar]
- 42.Ge Y, Molloy MP, Chamberlain JS, Andrews PC. Proteomic analysis of MDX skeletal muscle: great reduction of adenylate kinase 1 expression and enzymatic activity. Proteomics. 2003;3(10):1895–1903. doi: 10.1002/pmic.200300561. [DOI] [PubMed] [Google Scholar]
- 43.Doran P, Dowling P, Lohan J, McDonnell K, Poetsch S, Ohlendieck K. Subproteomics analysis of Ca2+-binding proteins demonstrates decreased calsequestrin expression in dystrophic mouse skeletal muscle. European Journal of Biochemistry. 2004;271(19):3943–3952. doi: 10.1111/j.1432-1033.2004.04332.x. [DOI] [PubMed] [Google Scholar]
- 44.Doran P, Dowling P, Donoghue P, Buffini M, Ohlendieck K. Reduced expression of regucalcin in young and aged MDX diaphragm indicates abnormal cytosolic calcium handling in dystrophin-deficient muscle. Biochimica et Biophysica Acta. 2006;1764(4):773–785. doi: 10.1016/j.bbapap.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Doran P, Martin G, Dowling P, Jockusch H, Ohlendieck K. Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics. 2006;6(16):4610–4621. doi: 10.1002/pmic.200600082. [DOI] [PubMed] [Google Scholar]
- 46.Gulston MK, Rubtsov DV, Atherton HJ, et al. A combined metabolomic and proteomic investigation of the effects of a failure to express dystrophin in the mouse heart. Journal of Proteome Research. 2008;7(5):2069–2077. doi: 10.1021/pr800070p. [DOI] [PubMed] [Google Scholar]
- 47.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Analytical Biochemistry. 1976;72(1-2):248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 48.Karp NA, Lilley KS. Maximising sensitivity for detecting changes in protein expression: experimental design using minimal CyDyes. Proteomics. 2005;5(12):3105–3115. doi: 10.1002/pmic.200500083. [DOI] [PubMed] [Google Scholar]
- 49.Doran P, O’Connell K, Gannon J, Kavanagh M, Ohlendieck K. Opposite pathobiochemical fate of pyruvate kinase and adenylate kinase in aged rat skeletal muscle as revealed by proteomic DIGE analysis. Proteomics. 2008;8(2):364–377. doi: 10.1002/pmic.200700475. [DOI] [PubMed] [Google Scholar]
- 50.Rabilloud T, Strub J-M, Luche S, van Dorsselaer A, Lunardi J. A comparison between Sypro Ruby and ruthenium II tris (bathophenanthroline disulfonate) as fluorescent stains for protein detection in gels. Proteomics. 2001;1(5):699–704. doi: 10.1002/1615-9861(200104)1:5<699::AID-PROT699>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 51.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nature Protocols. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 52.Gannon J, Doran P, Kirwan A, Ohlendieck K. Drastic increase of myosin light chain MLC-2 in senescent skeletal muscle indicates fast-to-slow fibre transition in sarcopenia of old age. European Journal of Cell Biology. 2009;88(11):685–700. doi: 10.1016/j.ejcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 53.Poot M, Zhang Y-Z, Kraemer JA, et al. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. Journal of Histochemistry and Cytochemistry. 1996;44(12):1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- 54.Pendergrass W, Wolf N, Pool M. Efficacy of MitoTracker Green and CMXrosamine to measure changes in mitochondrial membrane potentials in living cells and tissues. Cytometry A. 2004;61(2):162–169. doi: 10.1002/cyto.a.20033. [DOI] [PubMed] [Google Scholar]
- 55.Chazotte B. Labeling mitochondria with fluorescent dyes for imaging. Cold Spring Harbor Protocols. 2009;4(6) doi: 10.1101/pdb.prot4948. [DOI] [PubMed] [Google Scholar]
- 56.Urfer W, Grzegorczyk M, Jung K. Statistics for proteomics: a review of tools for analyzing experimental data. Proteomics. 2006;6(2, supplement):48–55. doi: 10.1002/pmic.200600554. [DOI] [PubMed] [Google Scholar]
- 57.Raddatz K, Albrecht D, Hochgraefe F, Hecker M, Gotthardt M. A proteome map of murine heart and skeletal muscle. Proteomics. 2008;8(9):1885–1897. doi: 10.1002/pmic.200700902. [DOI] [PubMed] [Google Scholar]
- 58.Westbrook JA, Wheeler JX, Wait R, Welson SY, Dunn MJ. The human heart proteome: two-dimensional maps using narrow-range immobilised pH gradients. Electrophoresis. 2006;27(8):1547–1555. doi: 10.1002/elps.200500777. [DOI] [PubMed] [Google Scholar]
- 59.McGregor E, Dunn MJ. Proteomics of the heart: unraveling disease. Circulation Research. 2006;98(3):309–321. doi: 10.1161/01.RES.0000201280.20709.26. [DOI] [PubMed] [Google Scholar]
- 60.Doran P, Wilton SD, Fletcher S, Ohlendieck K. Proteomic profiling of antisense-induced exon skipping reveals reversal of pathobiochemical abnormalities in dystrophic MDX diaphragm. Proteomics. 2009;9(3):671–685. doi: 10.1002/pmic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malhotra R, Mason PK. Lamin A/C deficiency as a cause of familial dilated cardiomyopathy. Current Opinion in Cardiology. 2009;24(3):203–208. doi: 10.1097/HCO.0b013e32832a11c6. [DOI] [PubMed] [Google Scholar]
- 62.Kimura N, Shimada N, Fukuda M, et al. Regulation of cellular functions by nucleoside diphosphate kinases in mammals. Journal of Bioenergetics and Biomembranes. 2000;32(3):309–315. doi: 10.1023/a:1005549315846. [DOI] [PubMed] [Google Scholar]
- 63.Braun U, Paju K, Eimre M, et al. Lack of dystrophin is associated with altered integration of the mitochondria and ATPases in slow-twitch muscle cells of MDX mice. Biochimica et Biophysica Acta. 2001;1505(2-3):258–270. doi: 10.1016/s0005-2728(01)00172-4. [DOI] [PubMed] [Google Scholar]
- 64.Nagy N, Malik G, Fisher AB, Das DK. Targeted disruption of peroxiredoxin 6 gene renders the heart vulnerable to ischemia-reperfusion injury. American Journal of Physiology. 2006;291(6):H2636–H2640. doi: 10.1152/ajpheart.00399.2006. [DOI] [PubMed] [Google Scholar]
- 65.Boluyt MO, Brevick JL, Rogers DS, Randall MJ, Scalia AF, Li ZB. Changes in the rat heart proteome induced by exercise training: increased abundance of heat shock protein hsp20. Proteomics. 2006;6(10):3154–3169. doi: 10.1002/pmic.200401356. [DOI] [PubMed] [Google Scholar]
- 66.Fisher EA, Lapierre LR, Junkins RD, McLeod RS. The AAA-ATPase p97 facilitates degradation of apolipoprotein B by the ubiquitin-proteasome pathway. Journal of Lipid Research. 2008;49(10):2149–2160. doi: 10.1194/jlr.M800108-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Szczesna D. Regulatory light chains of striated muscle myosin. Structure, function and malfunction. Current Drug Targets—Cardiovascular & Haematological Disorders. 2003;3(2):187–197. doi: 10.2174/1568006033481474. [DOI] [PubMed] [Google Scholar]
- 68.Frey N, Richardson JA, Olson EN. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14632–14637. doi: 10.1073/pnas.260501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lemieux H, Hoppel CL. Mitochondria in the human heart. Journal of Bioenergetics and Biomembranes. 2009;41(2):99–106. doi: 10.1007/s10863-009-9211-0. [DOI] [PubMed] [Google Scholar]
- 70.Distler AM, Kerner J, Hoppel CL. Proteomics of mitochondrial inner and outer membranes. Proteomics. 2008;8(19):4066–4082. doi: 10.1002/pmic.200800102. [DOI] [PubMed] [Google Scholar]
- 71.White MY, Edwards AVG, Cordwell SJ, Van Eyk JE. Mitochondria: a mirror into cellular dysfunction in heart disease. Proteomics. 2008;2(6):845–861. doi: 10.1002/prca.200780135. [DOI] [PubMed] [Google Scholar]
- 72.Zhang W, ten Hove M, Schneider JE, et al. Abnormal cardiac morphology, function and energy metabolism in the dystrophic MDX mouse: an MRI and MRS study. Journal of Molecular and Cellular Cardiology. 2008;45(6):754–760. doi: 10.1016/j.yjmcc.2008.09.125. [DOI] [PubMed] [Google Scholar]
- 73.Swanson MA, Usselman RJ, Frerman FE, Eaton GR, Eaton SS. The iron-sulfur cluster of electron transfer flavoprotein-ubiquinone oxidoreductase is the electron acceptor for electron transfer flavoprotein. Biochemistry. 2008;47(34):8894–8901. doi: 10.1021/bi800507p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu XH, Qian LJ, Gong JB, Shen J, Zhang XM, Qian XH. Proteomic analysis of mitochondrial proteins in cardiomyocytes from chronic stressed rat. Proteomics. 2004;4(10):3167–3176. doi: 10.1002/pmic.200300845. [DOI] [PubMed] [Google Scholar]
- 75.De Pinto V, Reina S, Guarino F, Messina A. Structure of the voltage dependent anion channel: state of the art. Journal of Bioenergetics and Biomembranes. 2008;40(3):139–147. doi: 10.1007/s10863-008-9140-3. [DOI] [PubMed] [Google Scholar]