Abstract
The transcription factor peroxisome proliferator-activated receptor α (PPARα) is an important regulator of hepatic lipid metabolism. While PPARα is known to activate transcription of numerous genes, no comprehensive picture of PPARα binding to endogenous genes has yet been reported. To fill this gap, we performed Chromatin immunoprecipitation (ChIP)-chip in combination with transcriptional profiling on HepG2 human hepatoma cells treated with the PPARα agonist GW7647. We found that GW7647 increased PPARα binding to 4220 binding regions. GW7647-induced binding regions showed a bias around the transcription start site and most contained a predicted PPAR binding motif. Several genes known to be regulated by PPARα, such as ACOX1, SULT2A1, ACADL, CD36, IGFBP1 and G0S2, showed GW7647-induced PPARα binding to their promoter. A GW7647-induced PPARα-binding region was also assigned to SREBP-targets HMGCS1, HMGCR, FDFT1, SC4MOL, and LPIN1, expression of which was induced by GW7647, suggesting cross-talk between PPARα and SREBP signaling. Our data furthermore demonstrate interaction between PPARα and STAT transcription factors in PPARα-mediated transcriptional repression, and suggest interaction between PPARα and TBP, and PPARα and C/EBPα in PPARα-mediated transcriptional activation. Overall, our analysis leads to important new insights into the mechanisms and impact of transcriptional regulation by PPARα in human liver and highlight the importance of cross-talk with other transcription factors.
INTRODUCTION
Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors (TFs) that play an important role in the regulation of numerous biological processes, including lipid metabolism, adipocyte differentiation, cell proliferation and inflammation (1). Three different highly conserved PPAR isotypes have been identified: PPARα (NR1C1), PPARβ/σ (NR1C2) and PPARγ (NR1C3). The PPARα isotype has been shown to govern expression of numerous genes involved in fatty acid oxidation, ketogenesis, gluconeogenesis, cholesterol catabolism and lipoprotein metabolism (2,3). Additionally, PPARα has anti-inflammatory effects by suppressing pro-inflammatory genes (4,5). Consistent with its prominent function in lipid metabolism, PPARα is activated by various fatty acids and fatty acid derivates, as well as by synthetic agonists such as fenofibrate, WY14,643 and GW7647 (6). Analogous to several other nuclear receptors, PPARs form heterodimers with retinoid X receptors (RXRs) (7), which occurs independently of ligand or DNA binding (8). PPARs bind to DNA by recognizing specific cis-acting PPAR responsive elements (PPREs) present in the regulatory regions of PPAR target genes. The consensus PPRE consists of a direct repeat of the hexameric sequence AGGTCA separated by one less conserved spacer nucleotide. PPARα was shown to bind to the 5′ motif of the PPRE, whereas RXR binds to the 3′ motif (9). Full activation of gene transcription by the DNA-bound PPAR–RXR complex is ultimately dependent on the formation of a larger transcription initiation protein complex via recruitment of a number of co-activator proteins and RNA polymerase II (10). Additionally, PPARα has been shown to down-regulate gene expression by interfering with the activity of other TFs (11,12).
Recent studies using chromatin immunoprecipitation (ChIP) in combination with genomic tilling microarrays or sequencing (ChIP-chip or ChIP-Seq) have provided important new information on the requirements for DNA-binding by PPARs and other nuclear receptors. One of the most interesting findings was that genes that are activated by PPARγ show an enrichment of modules consisting of a PPRE-like motif together with a C/EBP binding element (13,14). It was found that knocking-down of either PPARγ or C/EBP reduced expression of several PPAR target genes, which was further reduced when these genes where knocked down simultaneously (13). A similar enrichment of a TF module was shown for the estrogen receptor (ER), which clustered together with forkhead, Oct1 and C/EBP motifs (15), as well as for the androgen receptor (AR) and the glucocorticoid receptor (GR) (16,17). These ChIP-chip studies thus reveal a complex interplay between nuclear receptors and other TFs. This cross-talk might be an important mechanism in gene regulation by nuclear receptors and may be responsible for transcriptional regulation of specific sets of genes.
While several studies have mapped the genomic binding regions of PPARγ, there are no reports available that describe the use of ChIP-chip or ChIP-Seq to investigate genomic binding of PPARα. Accordingly, in the present study we generated genome-wide maps of PPARα binding regions in the HepG2 human hepatoma cell line using ChIP combined with human promoter tiling arrays. We used HepG2 cells because they represent the most widely used cellular model for human liver cells, despite the modest effects of PPARα activation on gene expression. Our analysis was targeted towards the discovery of promoter sites showing increased PPARα binding in response to PPARα activation by ligand. To investigate the relation between promoter occupancy by PPARα and regulation of gene expression, results of ChIP-chip analysis were coupled to gene expression data.
MATERIALS AND METHODS
Cell culture
HepG2 cells were grown in phenol red-free Dulbecco’s modified medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 0.1 mg/ml streptomycin and 100 U/ml penicillin. Cells were split the day before experiments. Cells were kept at 37°C and 5% CO2. The following day cells were treated with either 100 nM of the PPARα agonist GW7647 or control vehicle (DMSO). Cells used for ChIP-chip analysis were harvested after 2 h of GW7647 treatment. Cells used for gene expression analysis were harvested after 6 h of GW7647 treatment.
ChIP
ChIP was performed as described previously (18). Briefly, protein–DNA complexes were cross-linked in 1% formaldehyde for 10 min at room temperature. Cross-linking was stopped by addition of 1 M glycine to a final concentration of 125 mM for 5 min. Cells were washed twice with PBS, scraped and collected by centrifugation. The cell pellet was dissolved in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–HCl, pH 8.0). Extracts were sonicated using the Bioruptor (Diagenode) at high power until DNA fragments of ∼500–1000 bp were formed. Sonicated chromatin was diluted in five volumes of dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris–HCl, pH 8.0). The immuno-complexes were precipitated using antibodies against PPARα, STAT1, STAT3 or STAT6 or normal serum IgG (sc-9000, sc-592, sc-482, sc-981, sc-2027, Santa Cruz Biotechnologies). Precipitated complexes were reverse cross-linked and proteins digested with proteinase K (Fermentas) overnight at 65°C. DNA fragments were purified using the Affymetrix GeneChIP Clean-up module. Purified DNA was used for qPCR and ChIP-chip analysis. ChIP experiments were run in quadruplicate.
ChIP-chip
To obtain sufficient DNA for hybridization, purified ChIP DNA was amplified and reamplified with the WGA (re)amplification kit (Sigma-Aldrich). Amplified DNA was fragmented with DNAse I to ∼50 bp fragments. The fragments were labeled with biotin according to Affymetrix instructions. Biotinylated DNA fragments were hybridized to Human Promoter 1.0R Arrays (Affymetrix). Arrays were washed and scanned according to instructions from manufacturer. Arrays were run in quadruplicate for both untreated and GW7647 treated cells.
Quantitative real-time PCR
Quantitative real-time PCR was performed on enriched regions of LPIN1, HMGCR and AGPAT9. Equal amounts of amplified ChIP material was used and measurements were performed on the iCycler (BioRad) using platinum Taq polymerase (Invitrogen) and the double stranded DNA dye SYBR green. The following primers where used: LPIN1: sense primer 5′-ATTGGGGGTGTTGTGGTATG-3′ and anti-sense primer 5′-ATAACAAATGCTGGCAAACG-3′. AGPAT9: sense primer 5′-CATCTAATACACAAACCAAGG-3′ and anti-sense primer 5′-AAGCCAAACAAAGACTATTCG-3′. HMGCR: sense primer 5′-ACGCTGATTTGGGTCTATGG-3′ and anti-sense primer 5′-GTGTAAATGGCTCCGGTCAC-3′. qPCRs were performed in duplicate and on all ChIP samples used for ChIP-Chip. Other primer sequences are available upon request.
Data analysis ChIP-chip
Affymetrix microarray CEL files were acquired and normalized using the Model-based analysis of tiling-arrays (MAT) algorithm (19). MAT was also used to identify PPARα binding regions induced by GW7647 treatment. Tiling-array probe intensities from ChIP performed on HepG2 cells treated for 2 h with GW7647 were compared with probe intensities from vehicle-treated HepG2 cells. The analysis was performed with a MAT score of 2.4 or higher, which was based on MAT scores found for promoters of several known PPARα target genes. The human NCBIv36 (hg18) was used as a mapping file. In our analysis we used MaxGap of 400 bp and a bandwidth of 400 bp.
Transcriptomics
Treatment of HepG2 cells with GW7647 for gene expression analysis by transcriptomics was performed in triplicate. Total RNA was extracted from HepG2 cells with TRIzol reagent (Invitrogen) and subsequently purified using the SV Total RNA Isolation System (Promega). RNA quality was measured on an Agilent 2100 bioanalyzer (Agilent Technologies) using 6000 Nano Chips according to manufacturer’s instructions. RNA was judged as suitable for array hybridization only when samples showed intact bands corresponding to the 18S and 28S rRNA subunits, displayed no chromosomal peaks or RNA degradation products and had a RNA integrity number above 8.0. Five micrograms of RNA were used for one cycle cRNA synthesis (Affymetrix). Hybridization, washing and scanning of Affymetrix human genome 133 2.0 plus arrays was carried out according to standard Affymetrix protocols. Scans of the Affymetrix arrays were processed using packages from the R/Bioconductor project. Arrays were normalized with quantile normalization and expression levels of probe sets were calculated using the robust multichip average method. Differentially expressed probe sets were identified using Limma and genes were considered to be significantly changed when raw q-value <0.05 and fold-change >1.2. Genes regulated with a annotated ChIP-chip peak in its promotor region are listed in Supplementary Table S1.
Characterization of PPARα binding regions
For annotation of the ChIP peaks to adjacent genes, we used the Genomatix tool RegionMiner. The genes with the shortest distance to the ChIP enriched binding site were selected. For identification of new TF modules the software tool FrameWorker was used (20). GW7647-induced PPARα binding regions that were assigned to the top 25 differentially regulated genes (both up- and down-regulated) were analyzed. The minimal occurrence of a module in the binding regions studied was set at 20% and a maximal distance bandwidth variation between the motives was set at 75 bp. Another Genomatix tool, ModelInspector, was used to scan all the PPARα binding regions linked to GW7647-regulated genes. As a control, we scanned approximately all human promoter regions present in the Genomatix promoter database with sizes between 1 and 1.5 kb, which corresponds with the average size of the binding sites identified by ChIP-chip. We used a two-proportion z-test to analyze significant enrichment of modules. Samples with p-values values below 0.05 were considered significantly different.
Motif analysis
To investigate the presence of de novo DNA motifs, the top 25 PPARα binding regions linked to up- or down-regulated genes were loaded into the MEME tool (http://meme.sdsc.edu/meme4_1/cgi-bin/meme.cgi). The length for the output motif was varied between 12 and 20 bp. Enriched matrixes of output motifs were compared to existing motif matrixes available in both the TRANSFAC and the JASPAR database with the use of the STAMP tool (http://www.benoslab.pitt.edu/stamp/index.php).
Data release
The ChIP-chip and expression array data have been submitted to GEO and the data will be released upon publication.
RESULTS
Mapping of PPARα binding regions to adjacent transcripts
PPARα represents a ligand-induced TF that in the absence of ligand does not seem to act as transcriptional repressor, unlike several other nuclear receptors (21). For the analysis presented here we assumed that induction of PPARα target gene expression by PPARα ligand is associated with increased binding of PPARα to the DNA, which is supported by ample experimental data (22–27). Therefore, to identify the complete repertoire of hepatic PPARα target genes, we treated HepG2 cells with the synthetic ligand GW7647 for 2 h and performed ChIP-chip analysis using an antibody against PPARα, with vehicle-treated HepG2 cells serving as control. After amplification and fluorescent labeling, the immunoprecipitated chromatin templates were hybridized to Affymetrix human promoter tiling arrays. These arrays cover promoter regions of 7.5 kb upstream and 2.5 kb downstream of the transcription start site (TSS). Statistical analysis of four replicate experiments provided evidence for increased binding of PPARα to 4220 binding regions in response to treatment with GW7647.
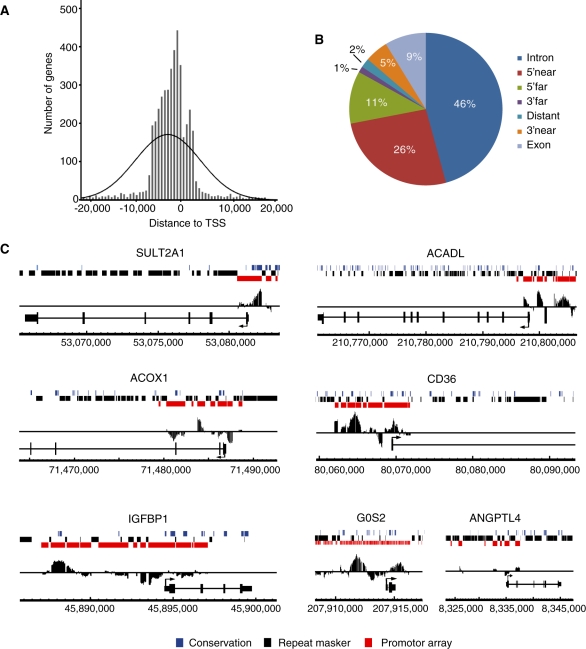
We used the Genomatix tool RegionMiner to map the genes closest to the 4220 GW7647-induced PPARα binding regions in four genomic directions and selected the gene with the shortest distance to the binding region. The 4220 PPARα binding regions were linked to 3670 unique genes, of which 2875 were present on the expression arrays used for gene expression analysis. Analogous to other nuclear receptors, PPARα binding regions showed a bias towards the TSS (Figure 1A) (15,16,28,29). To further analyze the location of PPARα binding regions, the online tool PinkThing (http://pinkthing.cmbi.ru.nl/cgi-bin/index50.pl) was used, which categorizes binding regions based on distance relative to the TSS (Figure 1B). We found that 46% of the binding regions were located within introns, which matches very well with a similar analysis done for PPARγ (14). Since we used promoter arrays, binding regions located to introns are in most cases present in the first intron. It is possible that some of these intronic binding regions actually surround the TSS of an alternative splice variant of a gene. Most other binding regions were identified upstream of the TSS with 26% within 5 kb and 11% between 5 and 25 kb. The more distal binding regions are not covered by promoter tiling arrays and therefore binding regions were not expected for the categories 3′ far (5–25 kb) and distant (>25 kb). Hence, the 1 and 2% of binding regions located to the 3′ far and distant categories, respectively, likely reflect misclassification by the PinkThing tool.
Figure 1.
Mapping of PPARα binding regions enriched upon GW7647 treatment. (A) Positional distribution of all identified PPARα binding regions relative to TSSs of the nearest gene. (B) Identification of the genomic location of PPARα binding regions using PinkThing. The following classification criteria were used: distant (>25 kb), 5′ far (25–5 kb), 5′ near (5–0 kb), intron (intronic), exon (exonic), 3′ near (0–5 kb) and 3′ far (5–15 kb). (C) Enrichment of promoter regions in PPARα target genes. Enriched ChIP-chip signals were visualized using Affymetrix integrated genome browser. Coverage of promoter tiling array is indicated in red, repetitive sequences in black, and conserved sequence in blue. PPARα target genes SULT2A1, ACOX1, IGFBP1, ACADL, CD36 and G0S2 all show positive enrichment within promoter regions. No enrichment is observed in the promoter of ANGPTL4 as the known PPRE is present within the (non-covered) intron 3.
Several genes that are established targets of PPARα, such as SULT2A1, ACOX1, ACADL, CD36, IGFBP1 and G0S2 (18,27,30–34), showed GW7647-induced PPARα binding regions in the promoter (Figure 1C). However, many other known PPARα target genes did not show any ligand-induced binding regions in their promoter. For these genes, PPARα is likely bound to a binding site not covered by the promoter array or, less likely, their PPRE may be located in repetitive sequences, which are excluded from Affymetrix human promoter tiling arrays using RepeatMasker. For example, the PPARα target gene ANGPTL4 carries a ligand-induced PPARα binding site in the third intron (24), which is out of the range of 2.5 kb downstream of the TSS covered by the array. As a result, no significant peak is present within the ANGPTL4 promoter (Figure 1C). Finally, it cannot be excluded that for some PPAR targets the degree of promoter occupancy by PPARα is not influenced by PPARα agonist and therefore no signals are detected, since our analysis concentrated on GW7647-induced PPARα binding regions.
Overlap between ChIP-chip and expression array
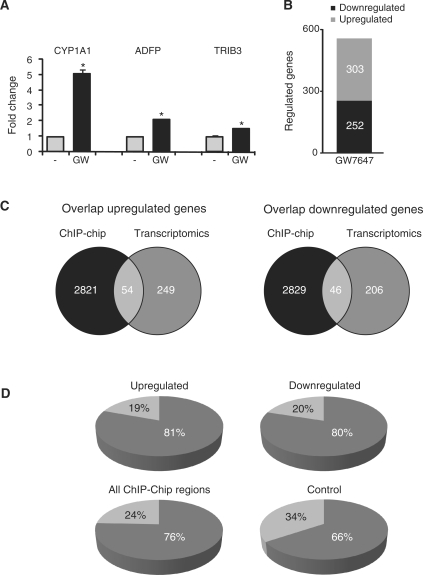
To investigate the relation between genes assigned to a GW7647-induced PPARα binding region and expression of that particular gene, we performed expression microarray analysis on HepG2 cells treated with GW7647 for 6 h. Confirming activation of PPARα, established PPARα targets CYP1A1, ADFP and TRIB3 were significantly induced by GW7647 (Figure 2A). Genes were considered significantly regulated if the mean fold change exceeded 1.2 and q-value <0.05. The low cut-off for fold change was used because the magnitude of induction of PPARα target genes in HepG2 cells is limited. Using these criteria 555 genes were differentially regulated after 6 h GW7647 treatment (Figure 2B). Slightly more genes were up-regulated than down-regulated. Differentially regulated genes were compared with the genes assigned to the GW7647-induced PPARα binding regions. It was found that 54 genes up-regulated by 6 h GW7647 treatment were linked to at least one PPARα binding region, representing 17.8% of all up-regulated genes (Figure 2C and Supplementary Data). In comparison, 16.2% of all genes on the expression array were linked to a PPARα binding region, which indicates that genes up-regulated by GW7647 showed minimal enrichment for PPARα binding. Surprisingly, a PPARα binding region was also linked to 46 genes down-regulated by 6 h treatment with GW7647, representing 18.3% of all down-regulated genes (Figure 2C, Supplementary Data). Expression of the far majority of genes assigned to a GW7647-induced PPARα binding region was not altered by GW7647 treatment.
Figure 2.
Overlap between GW7647-induced PPARα binding and GW7647-induced changes in expression. (A) Significant induction of PPARα targets by GW7647 treatment. (B) Number of genes significantly altered upon GW7647 treatment as determined by microarray analysis using criteria: fold change >1.2 and q-value <0.05. (C) Overlap between genes assigned to GW7647-induced PPARα binding regions and genes altered after treatment with GW7647 as determined by transcriptomics. (D) Percentage of GW7647-induced PPARα binding regions linked to either up- or down-regulated genes that contain at least one V$PERO site, as determined using Genomatix. Similar analysis was done for all GW7647-induced PPARα binding regions as well as a control set of promoter regions in the Genomatix promoter database with similar size range as the binding regions identified by ChIP-chip (1000–1500 bp).
With the use of the Genomatix tool RegionMiner, the PPARα binding regions linked to differentially expressed genes were scanned for the presence of a V$PERO site, which represents a PPRE matrix created by Genomatix (20). At least one V$PERO site was present in 81 and 80% of the PPARα binding regions linked, respectively, to the significantly up- or down-regulated genes (Figure 2D). Similarly, 76% of all PPARα binding regions identified by ChIP-chip contained a V$PERO site. In contrast, only 66% of control promoter regions contained a V$PERO site, for which we selected promoter regions in the Genomatix promoter database with similar size range as the binding regions identified by ChIP-chip (1000–1500 bp). These data suggest modest enrichment of PPARα binding regions for PPREs predicted with the V$PERO matrix.
Five genes out of 54 genes that were linked to a GW7647-induced PPARα binding region (Figure 3A and data not shown) and were up-regulated by GW7647 (Figure 3B) are direct target genes of the SREBP TFs: HMGCS1, HMGCR, FDFT1, SC4MOL and LPIN1, while AGPAT9 is a candidate target gene based on its role in triacylglycerol synthesis. Enrichment by GW7647 treatment of PPARα binding regions linked to LPIN1, AGPAT9 and HMGCR was confirmed by normal ChIP and qPCR (Figure 3C). As expression of several other established SREBP target genes was also up-regulated by GW7647 (Figure 3D) (35), these data suggest possible cross-talk between PPARα and SREBP signaling.
Figure 3.
Cross-talk between PPARα- and SREBP-dependent gene-regulation. (A) Enriched ChIP-chip signals for HMGCS1, HMGCR, LPIN1 and AGPAT9 genes were visualized using Affymetrix integrated genome browser. Coverage of promoter tiling array is indicated in red, repetitive sequences in black and conserved sequences in blue. (B) Gene expression changes after 6 h PPARα agonist treatment of five direct SREBP target genes and possible SREBP target gene AGPAT9. A GW7647-induced PPARα binding region was assigned to each of these genes. Significant differences are indicated with an asterisk (Student’s t-test, p < 0.05). (C) Transcriptional up-regulation of selected SREBP1 target genes involved in lipogenesis after 6 h PPARα agonist treatment represented as a heat map. (D) Enriched DNA binding of PPARα to promoter regions of LPIN1, AGPAT9 and HMGCR after 2 h GW7647 treatment, verified by ChIP-qPCR using primers designed within the binding region found by ChIP-Chip.
Motifs and module searches
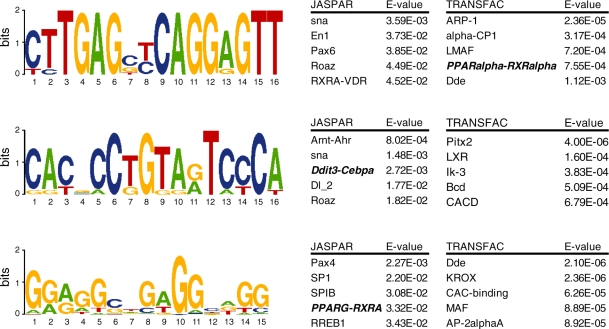
To search for specific DNA motifs in GW7647-induced PPARα binding regions we performed a de novo motif search with MEME (http://meme.sdsc.edu/meme4_1/cgi-bin/meme.cgi). For this analysis we used the binding regions of the 25 most significantly up-regulated genes that were assigned to a GW7647-induced PPARα binding site (Figure 4). Identified motifs were compared to the two major motif databases TRANSFAC and JASPAR to search for similarities to existing TF binding motifs using STAMP (http://www.benoslab.pitt.edu/stamp/index.php). One of the identified motifs matched the PPARα–RXRα motif in the TRANSFAC database. While the classical AGGTCA motif of a PPRE was present, a clear DR1-type tandem repeat was not found. This is in line with the earlier findings indicating that the PPRE 5′ motif is less conserved than the 3′ motif (14,36), suggesting that the site we found is most likely a 3′ motif. A second motif had a similarity hit with the C/EBPα motif in the JASPAR database, indicating that C/EBPα motif is enriched in PPARα binding regions. Interestingly, the C/EBPα motif was recently shown to be enriched in PPARγ binding regions and important for regulation of PPARγ target genes (13).
Figure 4.
De novo motif analysis. GW7647-induced PPARα binding regions were screened for specific DNA motifs via de novo motif search using MEME. The binding regions of the 25 most significantly up-regulated genes assigned to GW7647-induced PPARα binding regions were analyzed. Significantly enriched motifs were compared with motif databases TRANSFAC as well as JASPAR with the use of STAMP. Similarity scores with known TF binding regions are expressed by E-values. One motif identified showed similarity to a PPARα motif within the TRANSFAC database, another motif identified showed similarity to the C/EBPα motif in the JASPAR database.
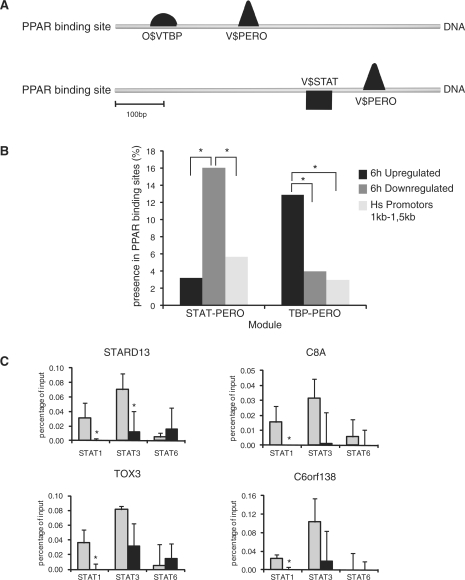
TFs often work in conjunction with other TFs to regulate DNA transcription. Accordingly, we examined whether other TF binding motifs are enriched together with the PPAR binding motif within the set of GW7647-induced PPARα binding regions linked to up- or down-regulated genes, using the Genomatix tool Frameworker. The PPAR matrix V$PERO was set and screening was performed for binding motifs of other TFs. Within the binding regions of the 25 most significantly up-regulated genes linked to a GW7647-induced PPARα binding region, we identified a highly significant (p = 6.94 × 10−8) module composed of the TATA binding protein (TBP, O$TBP) in combination with the PPAR binding motif V$PERO (Figure 5A). Within the binding regions of the 25 most significantly down-regulated genes linked to a GW7647-induced PPARα binding region, we identified a highly significant (p = 1.04 × 10−8) module composed of the binding motif for the TF signal transducers and activators of transcription (STAT) family (V$STAT) together with V$PERO (Figure 5A). To further analyze for the presence of these specific modules, we used ModelInspector to scan all the PPARα binding regions linked to GW7647-regulated genes. As a control, we also scanned all human promoter areas present in the Genomatix promoter database with a size between 1 and 1.5 kb. As shown in Figure 5B, the module STAT-PERO is significantly enriched in the binding regions linked to genes that are down-regulated after 6 h GW7647 treatment compared to binding regions linked to genes up-regulated by GW7647 treatment and to all human promoter regions present in the Genomatix promoter database. In fact, the STAT-PERO module was found in eight out of 46 genes that were linked to a GW7647-induced PPARα binding region and were down-regulated by GW7647. Similarly, the TBP-PERO module is significantly enriched in binding regions linked to up-regulated genes compared to binding regions linked to down-regulated genes and all human promoter regions. These results suggest interaction between PPARα and STAT in PPARα-mediated transcriptional repression and interaction between PPARα and TBP in PPARα-mediated transcriptional activation.
Figure 5.
Enrichment of TF modules in PPARα binding regions. (A) The binding regions of the 25 most significantly up-regulated genes assigned to GW7647-induced PPARα binding regions were analyzed for TF modules using the Genomatix tool Frameworker. Two modules were identified: TBP-PERO and STAT-PERO in the binding regions linked to up- or down-regulated genes, respectively. (B) The modules TBP-PERO and STAT-PERO were scanned for the relative presence in all GW7647-induced PPARα binding regions located near transcriptional regulated as well as all human promoter regions present in the Genomatix database. A two-proportion z-test was used to analyze significant enrichment of modules. p-values values below 0.05 were considered significantly different. (C) Loss of DNA binding by STAT3 and STAT1 upon PPARα activation. HepG2 cells were treated with GW7647 for 2 h and ChIP performed using antibodies against STAT3, STAT1 and STAT6 with vehicle-treated HepG2 cells serving as control. Precipitated chromatin was subsequently amplified using primers around the predicted STAT-PERO site found in four genes that were linked to a GW7647-induced PPARα binding region and were down-regulated by GW7647. Significant differences are indicated with an asterisk (Student's; t-test, p<0.05).
Members of the STAT family bind to similar DNA sequences. Expression profiling indicated that expression of STAT3 was highest in HepG2 cells, followed by STAT1 and STAT6 (data not shown). Accordingly, we focused on those three proteins to experimentally validate the suggested interaction between PPARα and STATs on the promoter of the aforementioned eight genes. HepG2 cells were treated with GW7647 for 2 h and ChIP performed using antibodies against STAT3, STAT1 and STAT6 with vehicle-treated HepG2 cells serving as control. Precipitated chromatin was subsequently amplified using primers around the predicted STAT-PERO site. Consistent with interaction between PPARα and STATs, for seven out of eight genes STAT1 was released in response to GW7647 treatment (Figure 5C and Supplementary Figure S1). A similar picture emerged for STAT3. In contrast, STAT6 showed minor binding which generally was not altered by GW7647. These data show that PPARα activation and DNA binding causes the release of STAT3 and STAT1 from gene promoters concurrent with down-regulation of gene expression.
Biological clustering
Finally, within up-regulated genes that were assigned to GW7647-induced PPARα binding regions we analyzed for functional biological clusters using DAVID (http://niaid.abcc.ncifcrf.gov/home.jsp). We found significant over-representation of genes in the biological cluster of sterol and lipid biosynthetic process (Table 1), in line with binding of PPARα to the putative promoter of several SREBP targets. Within down-regulated genes that were assigned to GW7647-induced PPARα binding regions we found significant over-representation of genes in the biological cluster of humoral and innate immune response, which is consistent with the known suppressive effect of PPARα on inflammation (Table 1).
Table 1.
Enrichment of biological processes in GW7647-regulated genes assigned to a PPARα binding region
| Biological process | p-value |
|---|---|
| Up-regulated genes | |
| Sterol biosynthetic process | 1.1E-4 |
| Apoptosis | 1.9E-2 |
| Developmental process | 2.2E-2 |
| Lipid biosynthetic process | 4.0E-2 |
| Down regulated genes | |
| γ-Hexachlorocyclohexane | 1.8E-3 |
| Humoral immune response | 1.2E-2 |
Up-regulated genes assigned to GW7647-induced PPARα binding regions were analyzed for functional biological clusters using DAVID.
DISCUSSION
It is well established that PPARα is a major transcriptional regulator of fatty acid metabolism in liver. Numerous genes involved in fatty acid oxidation and other metabolic processes have been identified as direct target genes of PPARα in mouse, characterized by the presence of a functional PPRE (2,33). In the past few years, genomic binding regions of several nuclear receptors including PPARγ have been mapped (13,14). However, no reports are available that were aimed at mapping the binding regions for PPARα. In our study we used ChIP-chip to investigate ligand-induced PPARα binding to genomic regions within HepG2 human hepatoma cells.
A total number of 4220 ligand-induced PPARα binding regions were identified, which were assigned to 3670 unique genes. Although this number may appear exceptionally high, the threshold for inclusion was set based on inspection of a number of known PPARα binding regions. Our assignment of PPARα binding regions relative to the nearest gene showed a distance-to-TSS distribution centered around the TSS. Categorizing these distances revealed that the majority of the binding regions are located within an intron or in the 5′ proximal region. These findings are in line with various studies showing that TF binding regions are normally distributed around the TSS (29,37–39). The preponderance of binding regions around the TSS may also reflect DNA looping allowing nuclear receptors bound to distal sites to contact the basal transcription machinery. Previous genome-wide analysis of ERα and PPARγ binding sites indicate that a large fraction of the binding regions are located distal from the TSS (13,15). Since we used promoter tiling arrays, our analysis was unable to detect binding to distant binding regions. Some of these distal elements have been shown to indeed function as functional PPREs (24,40–42). However, many distant binding regions are likely misclassified and are actually located in proximal promoters of new transcripts (43,44). Furthermore, many regions identified by ChIP-based methods do not show any effect on the expression of the gene closest to that region. These issues raise questions about the functional relevance of many of these binding regions (45).
We compared our ChIP-chip data with mRNA expression data collected by expression microarray and found that ∼18% of the genes induced by ligand were linked to a GW7647-induced PPARα binding region. The other 82% may not represent direct target genes, may bind PPARα equally in absence and presence of ligand, may bind PPARα via a site located outside the range covered by the array, or the actual PPARα binding region was linked to a different gene.
Importantly, the overwhelming majority of PPARα binding regions were linked to genes that were not significantly altered upon GW7647 treatment, even after more prolonged treatment (data not shown). It is well recognized that the majority of binding regions found by ChIP-chip or ChIP-Seq experiments do not have any effect on the genes they were linked to (45). This could be partially due to miss-annotation, which is inevitable when assigning to the nearest gene. However, since the number of GW7647-induced binding regions far exceeds the number of GW7647-regulated genes many regions must bind PPARα without any impact on gene regulation. These findings are consistent with the ‘scanning model’ proposed for PPARs, which states that PPARs scan the genome and transiently bind to PPRE-like sequences without inducing any transcription. According to this model, PPARs only start transcription upon binding to a bona fide PPRE (10). What distinguishes a bona fide PPRE from a PPRE-like sequence is not well understood. Recent ChIP-chip studies on several TFs revealed that clusters of different TF binding elements are enriched in proximity of the binding region for the TF under study (13,15,16,46–48). We searched for these types of clusters in PPARα binding regions that were linked to differentially regulated genes and found several modules of TF binding elements in combination with a PPRE. In PPARα binding regions linked to up-regulated genes we found enrichment for a module composed of the binding sequence for TBP together with that of PPARα. Since TBP is an important component of the basal transcriptional machinery, this finding suggests binding of PPARα adjacent to the core promoter. A second interesting module we found was a combination of a PPARα binding sequence and a STAT binding sequence, which was strongly enriched in PPARα binding regions linked to genes that were down-regulated by PPARα activation. STAT TFs function downstream in the signaling pathway of a large number of cytokines, growth factors and hormones (49). Follow-up analysis by normal ChIP showed that binding of STAT3 and also STAT1 to these genes was reduced upon GW7647 treatment. Our data suggest that loss of STAT binding is dependent on binding of PPARα adjacent to the STAT binding site. Inhibition of STAT-dependent transcriptional activity and DNA binding has been previously documented for PPARγ and PPARβ/σ (50–54), and may also partially account for the anti-inflammatory action of PPARα activation (17). Overall, down-regulation of gene expression by PPARα activation in HepG2 cells may be partially mediated by interfering with binding of STAT3 and STAT1 to the DNA.
Using Genomatix we found that at least one V$PERO site was present in 81% of the PPARα binding regions linked to the significantly up-regulated genes. A similarly high percentage was found for all the GW7647-induced PPARα binding regions resulting from the ChIP-chip analysis. While this result would suggest that the majority of PPAR binding detected by our ChIP-chip analysis conforms to the general paradigm of PPRE-dependent DNA binding, it should be emphasized that V$PERO sites are found at relatively high frequency throughout the genome and may have limited specificity. As ChIP is capable of detecting any type of binding of PPARα to DNA, which includes indirect binding via other TFs, it is difficult to provide a good estimate of the relative importance of PPRE-dependent and -independent binding to DNA. Binding to DNA of TFs in the absence of a consensus motif has been a common observation in recent ChIP-chip and ChIP-seq studies. Several explanation may account for this apparent discrepancy as elaborated by Farnham (45), including binding at a distal site that contains a consensus motif and looping to the site in question through protein–protein interactions; ‘piggyback’ binding that is mediated by protein–protein interactions with a second factor and that does not involve the DNA binding domain of the first factor; or assisted binding to a site that is similar to the consensus site, which is enhanced by protein–protein interaction with another site specific DNA binding factor or with a specifically modified histone.
Within the set of GW7647-induced PPARα binding regions linked to up-regulated genes, no established direct PPARα targets were present. One problem is that HepG2 cells, despite their broad use, poorly reflect gene regulation by PPARα in other cultured cells such as rat FAO hepatoma cells (55) and primary human hepatocytes. Indeed, we found remarkably little overlap in gene regulation by PPARα agonist between HepG2 cells and primary human hepatocytes (van der Meer et al., manuscript in preparation). Unfortunately, ChIP-chip analysis in primary human hepatocytes is practically unfeasible. Clearly, an ideal system to study PPARα-dependent gene regulation in human is lacking.
The TFs SREBP-1 and SREBP-2 are important regulators of hepatic lipid and cholesterol synthesis (56). Previously, it was shown that PPARα is involved in the normal circadian regulation of target genes of SREBPs, including HMGCR (57). Furthermore, synthetic PPARα agonists were found to induce expression of SREBP targets in liver, which was completely abolished in SREBP-1−/− mice and PPARα−/− mice (58,59). The effect of PPARα agonists on SREBP targets was attributed to increased activation of SREBP-1c via enhanced proteolytic cleavage, and was not mediated by changes in SREBP-1 mRNA (58). In the present article, we find induction of expression of several SREBP targets by PPARα activation in human hepatoma HepG2 cells. Furthermore, in these cells PPARα agonist stimulated binding of PPARα to the putative promoter of SREBP targets HMGCS1, HMGCR, FDFT1, SC4MOL and LPIN1 genes. These data suggest important cross-talk between PPARα and SREBP signaling. The exact nature of the cross-talk requires further investigation but one possibility is that PPARα is recruited to promoters of SREBP targets via direct physical interaction with SREBP. It could be envisioned that through this interaction PPARα may promote SREBP activity, perhaps by assisting with recruitment of transcriptional co-activators. In addition to the above-mentioned genes, a PPARα binding region was assigned to the AGPAT9 gene, which can be suspected to be a SREBP target as well. LPIN1 and AGPAT9 encode enzymes that catalyze the second and third step in the triacylglycerol synthesis pathway. So far, several genes involved in either fatty acid or triacylglycerol synthesis have been identified as direct PPARα target genes including Δ5 and Δ6 desaturases and malic enzyme [summarized in refs (2,3)]. Indeed, the involvement of PPARα in lipogenesis appears to be much more extensive that previously understood (60). Although this notion is seemingly at odds with increased hepatic TG observed upon PPARα deletion, regulation of fatty acid metabolism by PPARα is probably more subtle than generally envisioned. According to data presented here, PPARα may also indirectly impact lipid biosynthesis via cross-talk with SREBP.
In aggregate, it can be concluded that ChIP-chip represents a powerful tool to investigate whole genome binding of PPARα. Our data indicate that PPARα agonists trigger the binding of PPARα to a large number of genomic sites and provide novel insights into the mechanisms of PPARα-dependent transcriptional regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research was funded by the Nutrigenomics Consortium, the European Nutrigenomics Organisation and the Graduate School VLAG.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Desvergne B, Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr. Rev. 1999;20:649–688. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- 2.Mandard S, Muller M, Kersten S. Peroxisome proliferator-activated receptor alpha target genes. Cell Mol. Life Sci. 2004;61:393–416. doi: 10.1007/s00018-003-3216-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lefebvre P, Chinetti G, Fruchart JC, Staels B. Sorting out the roles of PPAR alpha in energy metabolism and vascular homeostasis. J. Clin. Invest. 2006;116:571–580. doi: 10.1172/JCI27989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zandbergen F, Plutzky J. PPARalpha in atherosclerosis and inflammation. Biochim. Biophys. Acta. 2007;1771:972–982. doi: 10.1016/j.bbalip.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalik L, Wahli W. PPARs mediate lipid signaling in inflammation and cancer. PPAR Res. 2008;2008:134059. doi: 10.1155/2008/134059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu HE, Lambert MH, Montana VG, Parks DJ, Blanchard SG, Brown PJ, Sternbach DD, Lehmann JM, Wisely GB, Willson TM, et al. Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 7.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feige JN, Gelman L, Tudor C, Engelborghs Y, Wahli W, Desvergne B. Fluorescence imaging reveals the nuclear behavior of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. J. Biol. Chem. 2005;280:17880–17890. doi: 10.1074/jbc.M500786200. [DOI] [PubMed] [Google Scholar]
- 9.IJpenberg A, Jeannin E, Wahli W, Desvergne B. Polarity and specific sequence requirements of peroxisome proliferator-activated receptor (PPAR)/retinoid X receptor heterodimer binding to DNA. A functional analysis of the malic enzyme gene PPAR response element. J. Biol. Chem. 1997;272:20108–20117. doi: 10.1074/jbc.272.32.20108. [DOI] [PubMed] [Google Scholar]
- 10.Feige JN, Gelman L, Michalik L, Desvergne B, Wahli W. From molecular action to physiological outputs: peroxisome proliferator-activated receptors are nuclear receptors at the crossroads of key cellular functions. Prog. Lipid Res. 2006;45:120–159. doi: 10.1016/j.plipres.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Bougarne N, Paumelle R, Caron S, Hennuyer N, Mansouri R, Gervois P, Staels B, Haegeman G, De Bosscher K. PPARalpha blocks glucocorticoid receptor alpha-mediated transactivation but cooperates with the activated glucocorticoid receptor alpha for transrepression on NF-kappaB. Proc. Natl Acad. Sci. USA. 2009;106:7397–7402. doi: 10.1073/pnas.0806742106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delerive P, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors in inflammation control. J. Endocrinol. 2001;169:453–459. doi: 10.1677/joe.0.1690453. [DOI] [PubMed] [Google Scholar]
- 13.Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, Feng D, Zhuo D, Stoeckert C.J., Jr, Liu XS, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 16.Bolton EC, So AY, Chaivorapol C, Haqq CM, Li H, Yamamoto KR. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 2007;21:2005–2017. doi: 10.1101/gad.1564207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Joe EH, Jou I. PPAR-alpha activators suppress STAT1 inflammatory signaling in lipopolysaccharide-activated rat glia. Neuroreport. 2005;16:829–833. doi: 10.1097/00001756-200505310-00010. [DOI] [PubMed] [Google Scholar]
- 18.Degenhardt T, Matilainen M, Herzig KH, Dunlop TW, Carlberg C. The insulin-like growth factor-binding protein 1 gene is a primary target of peroxisome proliferator-activated receptors. J. Biol. Chem. 2006;281:39607–39619. doi: 10.1074/jbc.M605623200. [DOI] [PubMed] [Google Scholar]
- 19.Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl Acad. Sci. USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 21.Semple RK, Meirhaeghe A, Vidal-Puig AJ, Schwabe JW, Wiggins D, Gibbons GF, Gurnell M, Chatterjee VK, O’Rahilly S. A dominant negative human peroxisome proliferator-activated receptor (PPAR){alpha} is a constitutive transcriptional corepressor and inhibits signaling through all PPAR isoforms. Endocrinology. 2005;146:1871–1882. doi: 10.1210/en.2004-1405. [DOI] [PubMed] [Google Scholar]
- 22.Donelson E, Chen L, Zhang X, Goswami P, Song BJ, Hardwick JP. Genomic structure and regulation of the rat hepatic CYP4F1 gene by peroxisome proliferators. Arch. Biochem. Biophys. 2008;472:1–16. doi: 10.1016/j.abb.2008.01.018. [DOI] [PubMed] [Google Scholar]
- 23.Dongol B, Shah Y, Kim I, Gonzalez FJ, Hunt MC. The acyl-CoA thioesterase I is regulated by PPARalpha and HNF4alpha via a distal response element in the promoter. J. Lipid Res. 2007;48:1781–1791. doi: 10.1194/jlr.M700119-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Mandard S, Zandbergen F, Tan NS, Escher P, Patsouris D, Koenig W, Kleemann R, Bakker A, Veenman F, Wahli W, et al. The direct peroxisome proliferator-activated receptor target fasting-induced adipose factor (FIAF/PGAR/ANGPTL4) is present in blood plasma as a truncated protein that is increased by fenofibrate treatment. J. Biol. Chem. 2004;279:34411–34420. doi: 10.1074/jbc.M403058200. [DOI] [PubMed] [Google Scholar]
- 25.Nagasawa M, Hara T, Kashino A, Akasaka Y, Ide T, Murakami K. Identification of a functional peroxisome proliferator-activated receptor (PPAR) response element (PPRE) in the human apolipoprotein A-IV gene. Biochem Pharmacol. 2009;78:523–530. doi: 10.1016/j.bcp.2009.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Viswakarma N, Yu S, Naik S, Kashireddy P, Matsumoto K, Sarkar J, Surapureddi S, Jia Y, Rao MS, Reddy JK. Transcriptional regulation of Cidea, mitochondrial cell death-inducing DNA fragmentation factor alpha-like effector A, in mouse liver by peroxisome proliferator-activated receptor alpha and gamma. J. Biol. Chem. 2007;282:18613–18624. doi: 10.1074/jbc.M701983200. [DOI] [PubMed] [Google Scholar]
- 27.Zandbergen F, Mandard S, Escher P, Tan NS, Patsouris D, Jatkoe T, Rojas-Caro S, Madore S, Wahli W, Tafuri S, et al. The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem. J. 2005;392:313–324. doi: 10.1042/BJ20050636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong H, Yauk CL, Rowan-Carroll A, You SH, Zoeller RT, Lambert I, Wade MG. Identification of thyroid hormone receptor binding sites and target genes using ChIP-on-chip in developing mouse cerebellum. PLOS one. 2009;4:e4610. doi: 10.1371/journal.pone.0004610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hamza MS, Pott S, Vega VB, Thomsen JS, Kandhadayar GS, Ng PW, Chiu KP, Pettersson S, Wei CL, Ruan Y, et al. De-novo identification of PPARgamma/RXR binding sites and direct targets during adipogenesis. PLOS one. 2009;4:e4907. doi: 10.1371/journal.pone.0004907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang HL, Strom SC, Cai H, Falany CN, Kocarek TA, Runge-Morris M. Regulation of human hepatic hydroxysteroid sulfotransferase gene expression by the peroxisome proliferator-activated receptor alpha transcription factor. Mol. Pharmacol. 2005;67:1257–1267. doi: 10.1124/mol.104.005389. [DOI] [PubMed] [Google Scholar]
- 31.Heinaniemi M, Uski JO, Degenhardt T, Carlberg C. Meta-analysis of primary target genes of peroxisome proliferator-activated receptors. Genome Biol. 2007;8:R147. doi: 10.1186/gb-2007-8-7-r147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Motojima K, Passilly P, Peters JM, Gonzalez FJ, Latruffe N. Expression of putative fatty acid transporter genes are regulated by peroxisome proliferator-activated receptor alpha and gamma activators in a tissue- and inducer-specific manner. J. Biol. Chem. 1998;273:16710–16714. doi: 10.1074/jbc.273.27.16710. [DOI] [PubMed] [Google Scholar]
- 33.Rakhshandehroo M, Sanderson LM, Matilainen M, Stienstra R, Carlberg C, de Groot PJ, Muller M, Kersten S. Comprehensive analysis of PPARalpha-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007;2007:26839. doi: 10.1155/2007/26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varanasi U, Chu R, Huang Q, Castellon R, Yeldandi AV, Reddy JK. Identification of a peroxisome proliferator-responsive element upstream of the human peroxisomal fatty acyl coenzyme A oxidase gene. J. Biol. Chem. 1996;271:2147–2155. doi: 10.1074/jbc.271.4.2147. [DOI] [PubMed] [Google Scholar]
- 35.Reed BD, Charos AE, Szekely AM, Weissman SM, Snyder M. Genome-wide occupancy of SREBP1 and its partners NFY and SP1 reveals novel functional roles and combinatorial regulation of distinct classes of genes. PLoS Genet. 2008;4:e1000133. doi: 10.1371/journal.pgen.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Temple KA, Cohen RN, Wondisford SR, Yu C, Deplewski D, Wondisford FE. An intact DNA-binding domain is not required for peroxisome proliferator-activated receptor gamma (PPARgamma) binding and activation on some PPAR response elements. J. Biol. Chem. 2005;280:3529–3540. doi: 10.1074/jbc.M411422200. [DOI] [PubMed] [Google Scholar]
- 37.O’Geen H, Squazzo SL, Iyengar S, Blahnik K, Rinn JL, Chang HY, Green R, Farnham PJ. Genome-wide analysis of KAP1 binding suggests autoregulation of KRAB-ZNFs. PLoS Genet. 2007;3:e89. doi: 10.1371/journal.pgen.0030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeller KI, Zhao X, Lee CW, Chiu KP, Yao F, Yustein JT, Ooi HS, Orlov YL, Shahab A, Yong HC, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl Acad. Sci. USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Helledie T, Grontved L, Jensen SS, Kiilerich P, Rietveld L, Albrektsen T, Boysen MS, Nohr J, Larsen LK, Fleckner J, et al. The gene encoding the Acyl-CoA-binding protein is activated by peroxisome proliferator-activated receptor gamma through an intronic response element functionally conserved between humans and rodents. J. Biol. Chem. 2002;277:26821–26830. doi: 10.1074/jbc.M111295200. [DOI] [PubMed] [Google Scholar]
- 41.Mandard S, Stienstra R, Escher P, Tan NS, Kim I, Gonzalez FJ, Wahli W, Desvergne B, Muller M, Kersten S. Glycogen synthase 2 is a novel target gene of peroxisome proliferator-activated receptors. Cell Mol. Life Sci. 2007;64:1145–1157. doi: 10.1007/s00018-007-7006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napal L, Marrero PF, Haro D. An intronic peroxisome proliferator-activated receptor-binding sequence mediates fatty acid induction of the human carnitine palmitoyltransferase 1A. J. Mol. Biol. 2005;354:751–759. doi: 10.1016/j.jmb.2005.09.097. [DOI] [PubMed] [Google Scholar]
- 43.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 44.Massie CE, Mills IG. Chromatin immunoprecipitation (ChIP) methodology and readouts. Methods Mol. Biol. 2009;505:123–137. doi: 10.1007/978-1-60327-575-0_7. [DOI] [PubMed] [Google Scholar]
- 45.Farnham PJ. Insights from genomic profiling of transcription factors. Nature Rev. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, et al. Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-alpha responsive promoters. Mol. Cell. 2006;21:393–404. doi: 10.1016/j.molcel.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 47.Smeenk L, van Heeringen SJ, Koeppel M, van Driel MA, Bartels SJ, Akkers RC, Denissov S, Stunnenberg HG, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phuc Le P, Friedman JR, Schug J, Brestelli JE, Parker JB, Bochkis IM, Kaestner KH. Glucocorticoid receptor-dependent gene regulatory networks. PLoS Genet. 2005;1:e16. doi: 10.1371/journal.pgen.0010016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decker T, Kovarik P. Transcription factor activity of STAT proteins: structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol. Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kino T, Rice KC, Chrousos GP. The PPARdelta agonist GW501516 suppresses interleukin-6-mediated hepatocyte acute phase reaction via STAT3 inhibition. Eur. J. Clin. Invest. 2007;37:425–433. doi: 10.1111/j.1365-2362.2007.01796.x. [DOI] [PubMed] [Google Scholar]
- 51.Ricote M, Li AC, Willson TM, Kelly CJ, Glass CK. The peroxisome proliferator-activated receptor-gamma is a negative regulator of macrophage activation. Nature. 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 52.Shipley JM, Waxman DJ. Down-regulation of STAT5b transcriptional activity by ligand-activated peroxisome proliferator-activated receptor (PPAR) alpha and PPARgamma. Mol. Pharmacol. 2003;64:355–364. doi: 10.1124/mol.64.2.355. [DOI] [PubMed] [Google Scholar]
- 53.Li M, Pascual G, Glass CK. Peroxisome proliferator-activated receptor gamma-dependent repression of the inducible nitric oxide synthase gene. Mol. Cell Biol. 2000;20:4699–4707. doi: 10.1128/mcb.20.13.4699-4707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chinetti G, Fruchart JC, Staels B. Peroxisome proliferator-activated receptors (PPARs): nuclear receptors at the crossroads between lipid metabolism and inflammation. Inflamm Res. 2000;49:497–505. doi: 10.1007/s000110050622. [DOI] [PubMed] [Google Scholar]
- 55.Vanden Heuvel JP, Kreder D, Belda B, Hannon DB, Nugent CA, Burns KA, Taylor MJ. Comprehensive analysis of gene expression in rat and human hepatoma cells exposed to the peroxisome proliferator WY14,643. Toxicol. Appl. Pharmacol. 2003;188:185–198. doi: 10.1016/s0041-008x(03)00015-2. [DOI] [PubMed] [Google Scholar]
- 56.Horton JD. Sterol regulatory element-binding proteins: transcriptional activators of lipid synthesis. Biochem. Soc. Trans. 2002;30:1091–1095. doi: 10.1042/bst0301091. [DOI] [PubMed] [Google Scholar]
- 57.Patel DD, Knight BL, Wiggins D, Humphreys SM, Gibbons GF. Disturbances in the normal regulation of SREBP-sensitive genes in PPAR alpha-deficient mice. J. Lipid Res. 2001;42:328–337. [PubMed] [Google Scholar]
- 58.Knight BL, Hebbachi A, Hauton D, Brown AM, Wiggins D, Patel DD, Gibbons GF. A role for PPARalpha in the control of SREBP activity and lipid synthesis in the liver. Biochem. J. 2005;389:413–421. doi: 10.1042/BJ20041896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oosterveer MH, Grefhorst A, van Dijk TH, Havinga R, Staels B, Kuipers F, Groen AK, Reijngoud DJ. Fenofibrate simultaneously induces hepatic fatty acid oxidation, synthesis and elongation in mice. J. Biol. Chem. 2009;284:34036–34044. doi: 10.1074/jbc.M109.051052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kersten S, Desvergne B, Wahli W. Roles of PPARs in health and disease. Nature. 2000;405:421–424. doi: 10.1038/35013000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.