Abstract
Polo-like kinases (Plk1-4) are emerging as an important class of proteins involved in many aspects of cell cycle regulation and response to DNA damage. Here, we report the cloning of a fifth member of the polo-like kinase family named Plk5. DNA and protein sequence analyses show that Plk5 shares more similarities with Plk2 and Plk3 than with Plk1 and Plk4. Consistent with this observation, we show that mouse Plk5 is a DNA damage inducible gene. Mouse Plk5 protein localizes predominantly to the nucleolus, and deletion of a putative nucleolus localization signal (NoLS) within its N-terminal moiety disrupts its nucleolar localization. Ectopic expression of Plk5 leads to cell cycle arrest in G1, decreased DNA synthesis, and to apoptosis, a characteristic it shares with Plk3. Interestingly, in contrast to mouse Plk5 gene, the sequence of human Plk5 contains a stop codon that produces a truncated protein lacking part of the kinase domain.
INTRODUCTION
Polo-like kinases (Plk’s) are a family of conserved serine/threonine kinases that are highly related in their catalytic domains. To date, there are four known Plks (Plk1, Plk2, Plk3 and Plk4). Plks 1-3 possess two conserved ‘polo box’ motifs while Plk4 (Sak), a divergent member of the Plk family, possesses only one of the two bipartite polo-box motifs. The two polo boxes of Plk1, which comprise the polo box domain (PBD), have been reported to coordinate protein–protein interactions and subcellular localization (1–7). Similar to Plk1, the PBDs of Plk2 and Plk3 preferentially bind phosphoserine and phosphothreonine motifs (8–15). The Plks have been proposed to regulate entry into mitosis, cell cycle progression, cytokinesis and the cellular response to DNA damage deduced by their localization to mitotic structures and phosphorylation of specific substrates via their PBD and catalytic domains, respectively (5,6,16–20).
The best characterized member of the Plk family is Plk1, a key positive regulator of mitosis, meiosis and cytokinesis (18,21). Plk1 is overexpressed in various types of cancers and its elevated expression is associated with poor prognosis. Thus, Plk1 is a validated target for anti-cancer therapy (22–26). In recent years, a range of Plk1 molecular targets has been identified that reveal the signaling pathways by which Plk1 can regulate mitotic entry, DNA damage checkpoint responses, spindle formation and mitotic exit. Plk2 (serum-inducible kinase, Snk) and Plk3 (FGF-inducible kinase, Fnk or Prk) are early-response genes in quiescent mouse fibroblasts that are induced when cells are stimulated by serum (27–30). Plk2 is also reported to be elevated in cancer cells and is a transcriptional target of p53. Plk2 interacts with Chk1 and Chk2 kinases and participates in S phase arrest (31). Plk2 and Plk4 appear to be involved in centriole duplication (32–37).
Plk3 is considered to be a tumor suppressor and regulator of cellular response to DNA damage and angiogenesis (29,30,38,39). Some studies report that Plk3 expression remains relatively constant during normal cell cycle progression (29), while others argue that Plk3 is mainly activated during S phase entry (40) or in mitosis (41). While Plk1 expression is downregulated upon DNA damage (42), Plk3 expression is induced and accompanied by an activating phosphorylation that is ATM dependent (28–30). Activated Plk3 mediates Chk2 phosphorylation by ATM and subsequent checkpoint activation (43), and it participates in the onset of mitosis via inhibitory phosphorylation of Cdc25c (44).
Although Plk1 and Plk3 share some common substrates in vitro and both can rescue the Saccharomyces cerevisiae Cdc5 temperature sensitive mutant (45,46), they differ greatly in their effect when transfected into mammalian cells. Transfection and overexpression of Plk1 in mammalian cells results in cell transformation (47). Overexpression of Plk3 results in cell death (48). Plk3 has been proposed to regulate apoptosis, in part, through the p53 pathway (28), and Plk3 knockout mice suggest that Plk3 may be a tumor suppressor (39). Aging mice that lack Plk3 are reported to have accelerated tumor development, larger tumor size, and more pronounced angiogenesis than their wild-type litter mates (39).
Consistent with their proposed diverse roles, the Plks are differentially expressed, exhibit distinct subcellular localization patterns, and are subject to different types of post-translational modification (18,49,50). The mRNA and protein levels of Plk1 and Plk2 appear to be coordinately expressed during the cell cycle. In contrast, there is little reported concordance regarding the expression of Plk3 mRNA and protein levels.
Here, we report the cloning of a fifth member of the polo-like kinase family designated Plk5. Based on its nucleotide sequence there are two apparent distinct domains within the Plk5 protein. The amino-terminal portion has features characteristic of the catalytic domain of a serine/threonine kinase and shows strong homology to other polo family kinases. The carboxy-terminal portion presumably is the regulatory domain, which contains the PBD and shares extensive homology with the carboxy-terminal domains of the other Plk proteins. Based on sequence similarities both at the DNA and protein levels, Plk5 shares greater similarity to Plk2 and Plk3 than to Plk1 and Plk4. Consistent with this observation, we show that mouse Plk5 gene is activated following DNA damage. The Plk5 promoter region contains several p53 binding motifs but p53 does not appear to regulate Plk5 expression. Interestingly, human Plk5 but not mouse Plk5 contains a stop codon at position 807 of the nucleotide sequence (exon 6) that leads to a truncated protein. A second open reading frame (ORF) starts just after this stop codon and extends through the end of the human Plk5 gene. Unlike other Plk proteins, Plk5 contains five SQ/TQ motifs, three of which are within the PBD, indicating that ATM, ATR or DNA-PK may be involved in its phosphorylation and activation. Furthermore, Plk5 mRNA levels become elevated following introduction of DNA damage or microtubule disruption, suggesting that Plk5 is a stress induced protein that is involved in preserving cell integrity and that responds to a broad range of insults. Mouse Plk5 localizes predominantly in the nucleolus and deletion of a putative nucleolar localization signal (NoLS) within its N-terminal moiety disrupts this subcellular localization. Overexpression of Plk5 leads to G1 cell cycle arrest, decreased DNA synthesis, and apoptosis, a characteristic it shares with Plk3.
MATERIALS AND METHODS
Cloning of Plk5 cDNA, cell culture, transfection and treatment
Plk5 cDNA (6330514A18Rik) clone MGC: 106082, which was isolated from retina of mouse strain C57Bl\6 was purchased from Open Biosystems (Huntsville, AL, USA). The Plk5 ORF (1797 bp) was PCR amplified and cloned in frame with GFP protein in pEGFP-C1 vector (Invitrogen, CA, USA). For cell culture, all cells were cultured in Dulbecco's; modified Eagle's; medium (DMEM) supplemented with 10% heat inactivated fetal bovine serum (FBS; Eurobio, Les Ulis, France), 2 mM glutamine, penicillin (100 U ml−1) and streptomycin (100 µg ml−1) in a humidified atmosphere containing 5% CO2 at 37°C. Transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's; recommendations. For introduction of DNA damage, cells were either treated with etoposide (40 µM) for 12 h or hydroxyurea (HU) (0.5 µM) for 18 h. Nocodazole was added to media at 100 ng/ml for 18 h. Pifithrin-α (Sigma-Aldrich Inc.) was added simultaneously with etoposide at a concentration of 10 µM. Serum starvation was achieved by growing cells in DMEM supplemented with 0.1% FBS.
Nucleolar isolation
Nucleoli were prepared from NIH3T3 cell nuclei using a method based on that first described by Busch and coworkers in 1963 (51). Aliquots (250 μl) containing ∼1 × 108 nuclei were washed three times with PBS, resuspended in 5 ml buffer A [10 mM HEPES–KOH (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT], and dounce homogenized 10 times using a tight pestle. Dounced nuclei were centrifuged at 228g for 5 min at 4°C. The nuclear pellet was resuspended in 3 ml 0.25 M sucrose, 10 mM MgCl2, and layered over 3 ml 0.35 M sucrose, 0.5 mM MgCl2, and centrifuged at 1430g for 5 min at 4°C. The clean, pelleted nuclei were resuspended in 3 ml 0.35 M sucrose, 0.5 mM MgCl2, and sonicated for 6 × 10 s using a microtip probe and a Misonix XL 2020 sonicator at power setting 5. The sonicate was checked using phase contrast microscopy, ensuring that there were no intact cells and that the nucleoli were readily observed as dense, refractile bodies. The sonicated sample was then layered over 3 ml 0.88 M sucrose, 0.5 mM MgCl2 and centrifuged at 2800g for 10 min at 4°C. The pellet contained the nucleoli, while the supernatant consisted of the nucleoplasmic fraction. The nucleoli were then washed by resuspension in 500 μl of 0.35 M sucrose, 0.5 mM MgCl2, followed by centrifugation at 2000g for 2 min at 4°C.
Electrophoresis and immunoblotting
For 1D SDS/PAGE, purified nucleoli were dissolved in 1 × TGN lysis buffer containing 50 mM Tris (pH 7.5), 50 mM ß-glycerophosphate, 150 mM NaCl, 10% glycerol, 1% Tween 20, 1 mM NaF, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 2 µg/ml pepstatin A, 5 µg/ml leupeptin, 10 µg/ml aprotinin and 1 mM DTT, and heated at 70°C for 10 min. Nucleolar proteins were then separated on 10% SDS Tris–acetate gel and subsequently transferred onto nitrocellulose membrane. The membranes were then incubated with one of the following antibodies: mouse anti-GFP (Boehringer Mannheim, Inc., Indianapolis, IN, USA), mouse anti-NPM [B23 (FC-8791) sc-32256, Santa Cruz, CA, USA] or a Plk5 ployclonal antibody raised against two peptides (KEVPCLEGPIHLVAQ and SFSGVPAQLVLSGEG) common to both mouse and human Plk5. Bound antibody was probed using anti-mouse HRP conjugate (1:1000 dilution) (Pierce Chemical Co.) in PBS containing 5% milk powder and 0.05% Tween 20, and detected via chemiluminescence with ECL (Amersham Pharmacia Biotech).
Annexin V staining for flow cytometry
Apoptosis of pEGF and pEGFP-Plk5 transfected cells was measured using the Annexin-V-Cy5.5 (Invitrogen) and propidium iodide (PI). Briefly, harvested cells were washed in PBS and resuspended in Annexin-V binding buffer (10 mM HEPES, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). The 2 × 105 of cells were incubated with 5 μl Annexin V-Cy5.5 and PI (1 mg/ml) for 10 min in the dark at room temperature and analyzed by flow cytometry.
Cell staining and fluorescence microscopy
Cells were cultured on coverslips in DMEM, 10% FBS. Cells were washed with PBS, fixed with 4% paraformaldehyde and permeabilized for 10 min at room temperature with 0.2% Triton X-100 in PBS. After washing, coverslips were blocked with 10% horse serum, 1% bovine serum albumin, 0.02% NaN3 in 1% PBS. Cells were incubated with antibody against NPM [B23 (FC-8791) sc-32256, Santa Cruz] or the Plk5 antibody for 1 h at 37°C. Coverslips were washed extensively with PBS and further incubated with 13.2 nM of the appropriate secondary antibody conjugated to Alexafluor568 phalloidin (Invitrogen, Eugene, OR, USA) in PBS for 30 min at room temperature, washed three times with PBS and mounted with Gelmount (Fisher Scientific, Pittsburgh, PA, USA). Cells were visualized with the use of a LSM 510 laser scanning confocal microscope (Zeiss, Oberkochen, Germany).
For microscopy analysis of apoptotic morphology, fixed cells were stained with 1 µg/ml of DAPI (Invitrogen), mounted on slides and examined under fluorescence microscope. At least 200 cells per sample were counted in three independent experiments.
Determination of Plk5 mRNA levels by quantitative PCR
RNA was prepared according to instructions using Tri Reagent® (Molecular Research Center Inc., Cincinnati, OH, USA). Two micrograms of total RNA was reverse transcribed using the TaqMan Reverse Transcription Reagent kit (Applied Biosystems, Foster City, CA, USA). An aliquot of the cDNA was amplified for 40 cycles on an ABI Prism 7900HT Sequence Detection System with gene-specific primers designed using the Primer Express software (Applied Biosystems). SYBR Green dye was used for signal detection. All analyses were carried out in triplicate, and nontemplate controls and dissociation curves were used to ensure specific template amplification. Serial dilutions of a control cDNA were used to determine standard curves, and curves with R2 > 0.97 used to determine the mRNA levels in individual samples. The expression level of Plk5 was calculated as a ratio of the mRNA level relative to the mRNA level for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in the same cDNA. A list of primers used for qPCR is included in the supplementary data section (Supplementary Figure S4).
Cell cycle, BrdU incorporation and GFP content analyses
HEK293 cells were cultivated to ∼50% confluency and transfected with either pEGFP or pEGFP-Plk5 constructs. After 24 h cells were trypsinized, washed twice in PBS, and fixed overnight in cold 0.25% paraformaldehyde in PBS. Next day, cells were resuspended in 70% ethanol and stored at 4°C until analyzed. Fixed cells were then washed with PBS and stained with solution of 10 μg/ml propidium iodide (Molecular Probes) and 40 μg/ml RNase A (Sigma) in PBS for 30 min at 37°C. Cells were analyzed on BD LSR II flow cytometer system (Becton Dickinson, Franklin Lakes, NJ, USA).
For BrdU incorporation analysis, 30 min prior to harvest 1 mg/ml of BrdU was added to culture medium. Following fixation procedure described for cell cycle analysis (2% paraformaldehyde was used), cells were washed with PBS. DNA denaturation was performed using 2 M HCl with 0.5% Triton X-100 at 37°C for 1 h. Following 10 min neutralization in 0.1M NaBO4 cells were incubated with BrdU-specific antibody (Calbiochem, EMD, Darmstadt, Germany) in 1% BSA in PBS overnight at 4°C on shaking platform. Cells were washed once with PBS and incubated 1 h at room temperature with secondary antibody conjugated with Alexa Fluor 680 (Invitrogen). Thirty minutes prior to flow cytometric analysis, PI (10 mg/ml) and RNAse (40 µg/ml) were added and cells were incubated in dark at 37°C.
The fraction of GFP positive cells was determined by flow cytometry using similar fixation protocol similar to that for cell cycle analysis. At least 104 cells per sample in three independent experiments were analyzed.
RESULTS
Cloning of Plk5 and sequence comparison to other members of the Plk family
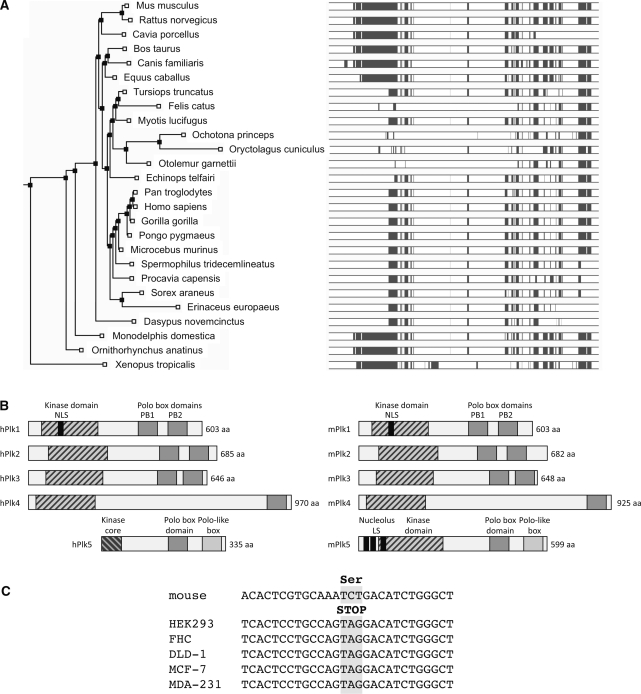
A database search [ENSEMBL (www.ensembl.org)] for proteins with a PBD was initiated after a western blot for Plk3 using commercially available antibodies unexpectedly detected a band of approximately correct size in cell extracts of mouse embryo fibroblasts derived from Plk3 knockout mice (to be described elsewhere). The search identified an unannotated murine gene on Chromosome 10 [RIKEN cDNA 6330514A18 gene (Marker Symbol; Acc: MGI: 3026984)] with an apparent polo-box domain in the C-terminal part of the encoded protein that was designated Plk5. The RIKEN cDNA 6330514A18 was first identified as part of a retinal transcriptome screen and described as a sequence weakly similar to polo-like kinase 2 from Xenopus laevis (52,53). The Plk5 polo box has 37% amino acid similarity to the duplicated polo boxes of Plk3 (Supplementary Figure S1). Although Plk5 is conserved among vertebrate species, the degree of peptide sequence homology between mouse and human (57%) is considerably lower than interspecific mouse to human homologies of other Plks (95%, 96%, 79% and 78% for Plk1, Plk2, Plk3 and Plk4, respectively). The Plk5 gene sequence from different organisms seems to cluster depending on phylogenetic distance (Figure 1A). The most striking difference between human, chimpanzee and orangutan Plk5 on the one hand, and mouse, chicken or Xenopus on the other is that the former contain a nonsense codon in the 5′ third of the gene in exon 6 resulting in a truncation in the N-terminal part of the protein (Figure 1B). Translation of the immediately adjacent ORF encodes a peptide that contains the PBD but lacks two thirds of the kinase domain (218 amino acids) including the ATP binding site. To validate the truncating stop codon in hPlk5, the region containing this nonsense codon was amplified by RT–PCR and the DNA sequence from multiple cell lines of tumor and nontumor origin was determined (Figure 1C). The murine sequence was determined independently and used as a reference. The Plk5 DNA sequence from all five human cell lines contained the TAG stop codon whereas the mouse sequence did not (Figure 1C).
Figure 1.
Structural organization of mouse and human Plk5. (A) A rooted tree was constructed using PHYLIP (70) showing one model of evolutionary descent. (B) Shared domains between mPlk5, hPlk5 and the other Plks. NoLS indicates the nucleolus localization signal. (C) Plk5 DNA sequence from different human cell lines of cancer and noncancer origin showing the presence of the nonsense codon in exon 6. The cell lines include: HEK-293 (Human embryonic kidney cell line), FHC (normal human fetal colon cell line), DLD-1 (human colon carcinoma), MCF-7 (breast cancer cell line) and MDA-231 (breast cancer cell line). The mouse Plk5 sequence is also shown for comparison.
The mPlk5 protein sequence differs from that of the other Plks in that it contains three putative NoLS localization sequences and five SQ/TQ motifs (Figure 1B). Unlike the promoter region of the other Plks, the promoter region of both human and mouse Plk5 sequences contains p53 consensus response elements and putative CpG methylation islands.
Another difference between Plk5 and the other polo-like kinases is that Plk5 is not induced following serum stimulation. NIH 3T3 cells were rendered quiescent by serum deprivation and 48 h later, cells were stimulated to enter S phase and proliferate by addition of serum. The levels of mRNA for Plk3 and Plk5 were compared. As expected, the levels of Plk3 mRNA increased up to 10-fold, consistent with previous findings (27). The level of Plk5 mRNA, however, remained unchanged indicating that Plk5 is not serum inducible (Supplementary Figure S2).
Both mouse and human Plk5 proteins are expressed in vivo
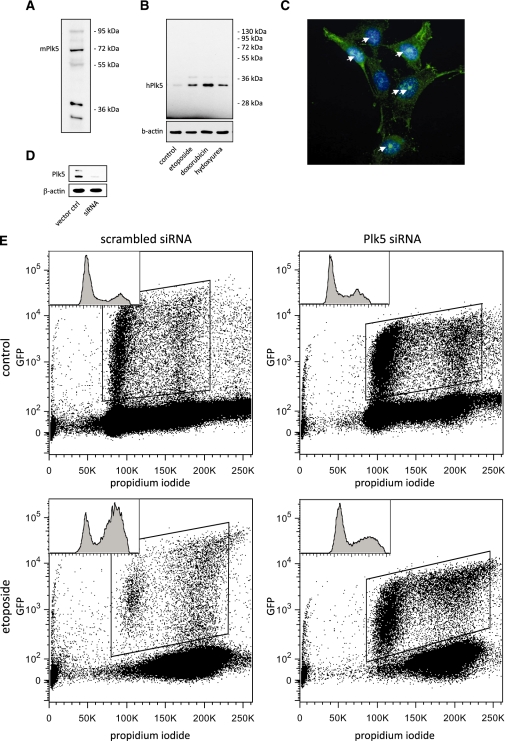
To test whether human and mouse Plk5 proteins were expressed in vivo, a polyclonal antibody was raised against two small peptides conserved in both human and mouse Plk5. In vivo expression of both proteins was tested by western blot, immunofluorescence and immunoprecipitation followed by mass spectrometry identification. Both mouse and human Plk5 proteins were detected by western blot at the expected sizes (Figure 2A and B). The mouse Plk5 protein was detected around the predicted size of 72 kDa and the identity of the protein was confirmed by immunoprecipitation followed by mass spectrometry (Supplementary Figure S3). The mouse Plk5 was also detected by immunofluorescence which shows that mPlk5 is localized in discrete foci in vivo (Figure 2C). The human Plk5 splice variant was also detected by western blot as a doublet at about 34 kDa, roughly equal to the computed molecular weight. The identity of the human Plk5 protein was confirmed by shRNA-mediated depletion, followed by western blot, showing that the detected band corresponds to the human Plk5 protein (Figure 2D). To test whether the human Plk5 protein is functional in vivo, we used a shRNA to deplete Plk5 and analyzed its effect on cell cycle distribution in the presence and absence of DNA damage. While, the cell cycle profile remained mostly intact in the absence of DNA damage, cells depleted of Plk5 show a defect in the G2/M checkpoint following treatment with the DNA damaging agent, etoposide (Figure 2E). This result indicates that the truncated human Plk5 is not only expressed in vivo, but functional as well.
Figure 2.
Human and mouse Plk5 proteins are both expressed in cultured cells. Western blot showing that mouse Plk5 (A) and human Plk5 (B) proteins are detected in lysates from NIH3T3 and HEK-293 cells, respectively, using a Plk5 polyclonal antibody. (C) Immunofluorescence detection of endogenous mPlk5 in discrete foci (arrows) in NIH3T3 cells. (D) A shRNA that targets hPlk5 mRNA was able to deplete Plk5 protein from HEK-293 cells. (E) HEK-293 cells transiently transfected with vector expressing both shRNA against Plk5 and GFP were analyzed by flow cytometry. The cell cycle distribution of GFP-positive cells is shown in left top corner of each panel. Depletion of hPlk5 from HEK-293cells leads to loss of G2/M checkpoint in response to treatment with the DNA damaging agent, etoposide (lower panel, right) compared to nondepleted cells (lower panel, left).
Ectopic expression of Plk5 induces a G1 arrest and subsequent apoptotic cell death
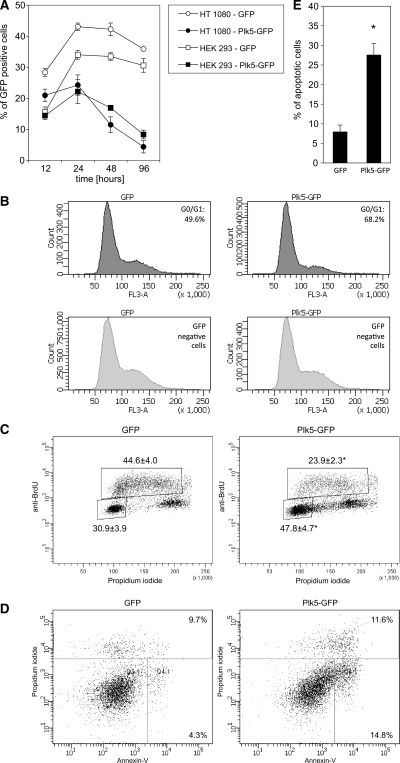
Overexpression of Plk1 induces mitotic abnormalities (47), and has the capacity to transform NIH 3T3 cells (47). Overexpression of Plk3 inhibits proliferation, and induces cell death (48). We have now assessed the effects of Plk5 overexpression on cellular proliferation and cell cycle progression. Transfection of mouse Plk5 cDNA into HEK293 or NIH3T3 cells produced no colonies in either cell line that stably overexpress the protein, suggesting that overexpression of Plk5 is either generally cytostatic or cytotoxic. Growth curves of cells following transfection with either a pEGFP-mPlk5 construct or a pEGFP vector expressing GFP protein alone showed that ectopic expression of mPlk5 in both cell lines inhibited cellular proliferation while expression of GFP alone had no profound effect (Figure 3A). To determine whether cells transfected with Plk5 arrest randomly throughout the cell cycle or accumulate preferentially at one or more cell cycle phases, HEK293 cells were transfected with pEGFP-mPlk5 or pEGFP alone and subjected to flow cytometry to determine cell cycle distribution. As shown in Figure 3B, the cells accumulated in the G1 phase of the cell cycle within 24 h after transfection with mPlk5. Nontransfected cells or those transfected with GFP alone exhibited a normal cell cycle profile, but by 48 h, very few cells transfected with GFP-mPlk5 remained on the culture plate (Figure 3A) indicating that cells overexpressing mPlk5 undergo cell death. Similarly, cells transfected with pEGFP-mPlk5 accumulated in G1 (Figure 3B) and failed to enter S phase as confirmed by reduced staining with BrdU (Figure 3C). The cell cycle distribution of cells transfected with pEGFP alone was unaltered and similar to that of untransfected cells with normal levels of BrdU incorporation (Figure 3B and C).
Figure 3.
Ectopic expression of murine Plk5 induces G1 arrest and cell death by apoptosis. (A) Growth curves of cells transfected by either a pEGFP plasmid control or a pEGFP-mPlk5 construct. (B) Cell cycle profile of cells transfected with either pEGFP or pEGFP-mPlk5 showing G1 arrest in the Plk5 transfected cells. (C) BrdU staining of cells transfected by either pEGFP or pEGFP-mPlk5 confirms the G1 arrest in the Plk5 transfected cells (upper panel, right) compared to GFP transfected cells (upper panel, left). No major change is seen in GFP negative cells (lower panel). (D) Annexin staining of the transfected cells showing increased apoptosis in pEGFP-mPlk5 transfected cells and (E) Quantitave analysis of nuclei with apoptotic morphology as a result of mPlk5 expression confirming the Annexin V staining. Asterisk denotes statistical difference P < 0.05.
Since cells that overexpress Plk5 appear to undergo rapid cell death, we asked whether cell death was due to apoptosis. To this end, cells were transiently transfected with the pEGFP-mPlk5 or the pEGFP control plasmid. Forty-eight hours after transfection, cells were stained for Annexin V, an early marker of apoptosis, and analyzed by flow cytometry. Transfection with pEGFP alone resulted in background levels of Annexin V staining in EGFP-positive cells. Transfection of cells with pEGFP-mPlk5, however, resulted in a dramatic increase in staining with Annexin V (Figure 3D). Consistent with the Annexin V staining, there was a significant increase in the number of cells transfected with pEGFP-mPlk5, but not with pEGFP, that had condensed and/or fragmented nuclei, an independent marker of apoptosis (Figure 3E). These data support the observation that ectopic expression of mPlk5 inhibits cellular proliferation by arresting cells in G1 and inducing apoptosis.
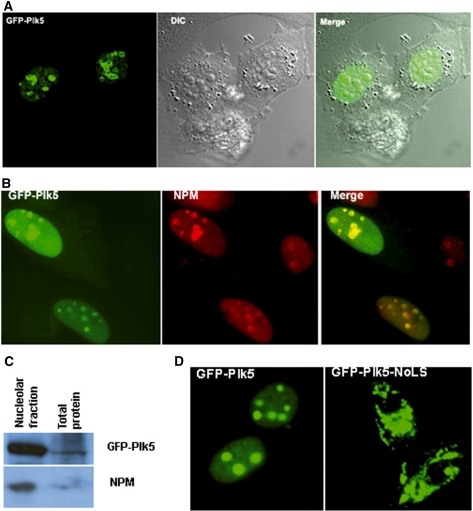
Plk5 localizes predominantly to the nucleolus and its nucleolar localization is not fully required for induction of cell death
Subcellular localization of endogenous Plk5 was determined by immunohistochemistry using a polyclonal Plk5 antibody (Figure 2C) or by ectopically expressing Plk5 following transfection of cells with GFP-Plk5. Plk5 localized predominantly within discrete foci in the nucleus (Figure 4A), and localization to the nucleolus was supported by co-localization with nucleophosmin (NPM), a nucleolar marker (Figure 4B). Nucleolar localization was further confirmed by fractionating isolated nuclei for nucleolar enrichment, and probing for either GFP-Plk5 or NPM. As shown in Figure 4C, Plk5 and NPM were both greatly enriched in the nucleolar fraction. Sequence analysis of the mPlk5 protein revealed three separate nucleolar localization signal (NoLS) motifs in the N-terminal domain of mPlk5 (missing in hPlk5). Deletion analysis of each of the NoLS motifs individually revealed that the absence of only one NoLS led to loss of nucleolar localization (Figure 4D). Nucleolar localization of Plk5 is not fully required for inducing cell death by apoptosis. The nucleolar mutant, although much less toxic to the cells than wild type, is still able to induce more apoptosis compared to GFP control as indicated by the percentage of Annexin V positive cells (Supplementary Figure S5).
Figure 4.
mPlk5 localizes to the nucleolus. (A) GFP-mPlk5 localizes to discrete foci. (B) GFP-mPlk5 co-localization with endogenous NPM in the nucleolus. (C) Enrichment for mPlk5 in the nucleolar fraction. (D) Loss of nucleolar localization in the NoLS mutant.
Plk5 expression is induced in response to multiple stressors
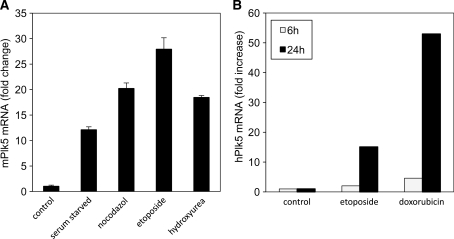
Plk5 has more characteristics in common with Plk2 and Plk3 than with Plk1 and Plk4. We therefore asked whether expression of Plk5 like Plk2 and Plk3, is inducible by DNA damage. Murine NIH3T3 cells were treated with DNA damaging agents and the levels of Plk5 mRNA transcript were assessed by qPCR. The levels of Plk5 mRNA increased markedly following DNA damage by treatment with etoposide, a topoisomerase II inhibitor that induces DNA double strand breaks (Figure 5A and B). Nocodazole, which interferes with microtubule polymerization and activates the spindle checkpoint, also induced Plk5 expression, as did inhibition of replication by HU. Interestingly, serum starvation also elevated levels of Plk5 mRNA (Figure 5A), but this increase was reversed upon addition of serum (data not shown). This finding distinguishes Plk5 from Plk3 which is serum inducible. The broad range of reagents that can induce Plk5 expression indicates that Plk5 is a stress inducible gene with potential involvement in a variety of processes designed to protect cells from a wide range of insults.
Figure 5.
Plk5 expression is induced following different stress stimuli. (A) Murine NIH 3T3 cells were either left untreated (control) or treated for 18 h with the DNA damaging agents etoposide (Etop) or HU, the spindle disassembly agent, nocodazole (Noc) or were serum starved (SS). (B) Human HEK293 cells were either left untreated (control) or treated with DNA damaging agent etoposide or doxorubicin for 6 or 24 h. The Plk5 mRNA level was measured by qPCR in both cases.
Induction of Plk5 in response to DNA damage is not dependent on p53
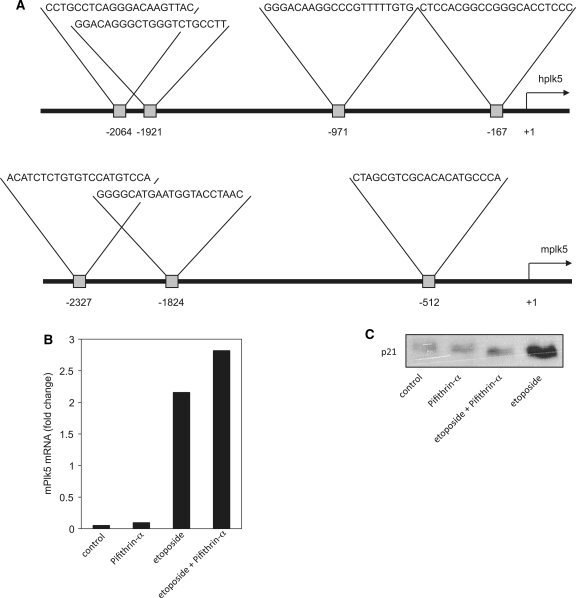
Unlike other Plk genes, mouse and human Plk5 contains p53 response elements (Figure 6A). To determine whether p53 plays a functional role in Plk5 transcriptional activation, NIH 3T3 cells were treated with pifithrin-α, a chemical inhibitor of p53-dependent transcription, and tested for Plk5 response to DNA damage induced by etoposide. The loss of p53 activity was confirmed by the lack of p21 expression following etoposide treatment (Figure 6B). NIH3T3 cells were left untreated, treated with pifithrin-α, etoposide or etoposide plus pifithrin-α and the mRNA level measured by qPCR. As shown in Figure 6C, the levels of Plk5 mRNA did not drop in the etoposide treated cells in the presence of pifithrin-α but rather showed a slight but reproducible increase. These results indicate that Plk5 induction in response to DNA damage is not p53-dependent.
Figure 6.
Plk5 transcriptional activation in response to DNA damage is not p53-dependent. (A) p53 consensus binding sites in Plk5 promoter, (B) mPlk5 mRNA expression levels measured by qPCR following etoposide and pifithrin treatment of NIH 3T3 cells for 18 h. (C) Expression of p21 protein in lysates from cells either nontreated (1), treated with pifithrin (2), etoposide alone (3) or pifithrin and etoposide (4) was analyzed by western blot.
DISCUSSION
The Plk family comprises a group of serine/threonine kinases (54) that can also bind motifs containing a phosphoserine or phosphothreoine residue (8–15). In the case of Plk3, a priming phosphorylation of Chk2 facilitates subsequent phosphorylation by ATM to activate the Chk2 kinase (38). As a family, the Plks contribute to a broad spectrum of cellular roles, many of which are presumably coordinated into the functions of a single polo kinase in flies and lower eukaryotes (5,6,16–20). Drosophila and Caenorhabditis elegans have a single Polo kinase, whereas mammals have four reported Plks (designated Plk1 through Plk4) that encompass roles as diverse as participation in centrosome dynamics, the intra-S-phase checkpoint, spindle formation, cytokinesis, G2/M checkpoints of the cell cycle and the DNA damage response (16–20). Homozygous disruption of Plk1 or Plk4 in mice is fetal lethal (55,36), whereas mice lacking Plk2 and Plk3 are viable (56,39).
When cell extracts from mice in which the promoter and first six exons had been removed by targeted homologous recombination produced a band of predicted size for Plk3 in western blots using a commercial Plk3 antibody, we searched available databases for other sequences that potentially might cross-react. We identified an unannotated sequence, which has been designated 6330514A18RIK (52,53). The protein named Plk5 contains two polo boxes and has additional similarity to the known polo-like kinases. The cDNA sequences for mouse and human Plk5 were cloned by RT–PCR and recombinant protein was produced in Escherichia coli. Analysis of protein and DNA sequences suggested that Plk5 is more similar to Plk2 and Plk3 than to the Plk1 and Plk4 kinases. Functionally, however, mouse Plk5 appears to have unique characteristics. The known Plks are reported to promote spindle formation at G2/M, to activate the anaphase promoting complex (APC) as cells transit through mitosis, to participate in the process of cytokinesis, and to participate in DNA damage checkpoint signaling cascade in G2/M (16–20). The Plk1 kinase has numerous mitotic functions including the regulation of cyclin B sub-cellular localization (21) and recruitment of γ-tubulin to centrosomes (57). The Plk4 kinase, also designated Sak, is required for the destruction of cyclin B by the APC complex and for exit from mitosis in the postgastrula embryo (36). The Plk2 and Plk3 genes are expressed as immediate-early transcripts and their function has been mainly associated with mitosis and DNA damage response (28–31).
The mouse Plk5 kinase appears to reside in the nucleolus. When GFP-tagged Plk5 is introduced into cultured cells, it quickly localizes to the nucleolus and subsequently causes the cells to undergo apoptosis. Nucleolar localization is not fully required for Plk5-induced apoptosis since a deletion mutant lacking a functional NoLS also induces cell death although much less efficiently. The endogenous Plk5 can also be visualized within the nucleolus with the Plk5 antibody. The Plk5 protein has three nucleolar localization motifs at its amino end, only one of which appears to be required for nucleolar localization. These data suggest that in mice Plk5 is either sequestered within nucleoli and participates in ribosome biogenesis or other nucleolar function, or that it remains in nucleoli until cells are stressed by DNA damage at which time it is released to participate in the cellular response to such damage. Indeed, recent reports indicate that the nucleolus also functions in the regulation of DNA-damage response, primarily through sequestration of regulators, such as p53, MDM2, CDC14b, pRB, pRB2, E2F4, p107 and p14ARF (58–65). However, we were unable to see any change in Plk5 localization following any form of stress to the cell. Consistent with its putative involvement in DNA damage response, the Plk5 gene contains three p53 regulatory elements in the promoter region. The gene, however, appears to be responsive to DNA damage, but not in a p53-dependent manner. The level of Plk5 mRNA is elevated following treatment with etoposide, a topoisomerase II inhibitor which causes double strand DNA breaks, as well as nocodazol, a mitotic inhibitor. It is also elevated after treatment with HU, which causes replication forks to stall. Treatment of cells with pifithrin, an inhibitor of p53 function, prevents an increase in p21 protein but has little effect on the induction of Plk5 mRNA in cells treated with etoposide or HU. These data suggest that mouse Plk5 is inducible following DNA damage but that despite the presence of p53 response elements within the Plk5 promoter; its induction is not dependent on p53.
This report has emphasized mouse Plk5 since the gene encoding the human protein has a mutation in exon 6 that introduces a stop codon into the Plk5 reading frame, terminating the protein within the putative kinase domain. The ensuing ORF encodes a protein that retains the remainder of the kinase domain and the polo box domain. This short protein is expressed in vivo and its expression is induced in response to stress and its overexpression induces cell death although at lesser extent compared to full-length mouse Plk5. Depletion of the hPlk5 leads to loss of the G2/M checkpoint in response to DNA damage indicating that this truncated form of Plk5 still retains some Plk5 functions.
The mouse and human Plk5 genes are quite similar in sequence, but their function appears to have diverged within recent evolutionary times. The truncating stop codon is found in the great apes and apparently occurred at the time of or just before the divergence of the orangutan from the other primates, including Homo sapiens. Whether retention of the inactivating stop codon was maintained due to selective pressure or absence of selective advantage is unclear, but it represents an apparent example of recent evolutionary divergence. Loss of a functional gene and gene product in humans is not unique to Plk5. In some cases, evolutionary loss of a gene in humans can predispose to disease. Loss of the gene encoding l-gulono-gamma-lactone oxidase that is involved in synthesis to ascorbic acid in most eukaryotes renders primates prone to scurvy in the absence of dietary vitamin C (66). Similarly, primates, including humans, have lost the gene encoding urate oxidase predisposing them to hyperuricemia, gout and renal stones (67). The human threonine dehydrogenase gene also contains an inactivating nonsense mutation (68) which may account for observed differences in metabolism and growth kinetics of mouse and human embryonic stem cells (69). One unusual aspect of human Plk5 is that the inactivating stop codon is immediately followed by a methionine and a sizable ORF that may not only retain some of the intact Plk5 functions, but has the potential for serving a cellular function different than that of Plk5. It is unclear whether loss of Plk5 has a selective advantage in humans and other hominids, but the abbreviated peptides encoded by the hominid Plk5 gene, and their potentially altered function, may have been sufficient to confer the necessary selective advantage for the mutated Plk5 to persist.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R03 ES015307 to E.M.B., R01 ES012695 and R01 ES016625 to P.J.S.) in part; the Center for Environmental Genetics (P30-ES006096). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Sandy Schwemberger and Dr George Babcock from Cincinnati Shrine's; Hospital for their help with Flow cytometry and Drs Anil Jegga and Bruce Aronow from Cincinnati Children’s Hospital for sequence analyses.
REFERENCES
- 1.Lee KS, Grenfell TZ, Yarm FR, Erikson RL. Mutation of the polo-box disrupts localization and mitotic functions of the mammalian polo kinase Plk. Proc. Natl Acad. Sci. USA. 1998;95:9301–9306. doi: 10.1073/pnas.95.16.9301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Logarinho E, Sunkel CE. The Drosophila POLO kinase localises to multiple compartments of the mitotic apparatus and is required for the phosphorylation of MPM2 reactive epitopes. J. Cell Sci. 1998;111(Pt 19):2897–2909. doi: 10.1242/jcs.111.19.2897. [DOI] [PubMed] [Google Scholar]
- 3.Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qian YW, Erikson E, Li C, Maller JL. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol. Cell Biol. 1998;18:4262–4271. doi: 10.1128/mcb.18.7.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moutinho-Santos T, Sampaio P, Amorim I, Costa M, Sunkel CE. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol. Cell. 1999;91:585–596. [PubMed] [Google Scholar]
- 6.Qian YW, Erikson E, Maller JL. Mitotic effects of a constitutively active mutant of the Xenopus polo-like kinase Plx1. Mol. Cell Biol. 1999;19:8625–8632. doi: 10.1128/mcb.19.12.8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van de Weerdt BC, Littler DR, Klompmaker R, Huseinovic A, Fish A, Perrakis A, Medema RH. Polo-box domains confer target specificity to the Polo-like kinase family. Biochim. Biophys. Acta. 2008;1783:1015–1022. doi: 10.1016/j.bbamcr.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Lin CY, Madsen ML, Yarm FR, Jang YJ, Liu X, Erikson RL. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc. Natl Acad. Sci. USA. 2000;97:12589–12594. doi: 10.1073/pnas.220423497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutterlin C, Lin CY, Feng Y, Ferris DK, Erikson RL, Malhotra V. Polo-like kinase is required for the fragmentation of pericentriolar Golgi stacks during mitosis. Proc. Natl Acad. Sci. USA. 2001;98:9128–9132. doi: 10.1073/pnas.161283998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yarm FR. Plk phosphorylation regulates the microtubule-stabilizing protein TCTP. Mol. Cell Biol. 2002;22:6209–6221. doi: 10.1128/MCB.22.17.6209-6221.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elia AE, Cantley LC, Yaffe MB. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science. 2003;299:1228–1231. doi: 10.1126/science.1079079. [DOI] [PubMed] [Google Scholar]
- 12.Elia AE, Rellos P, Haire LF, Chao JW, Ivins FJ, Hoepker K, Mohammad D, Cantley LC, Smerdon SJ, Yaffe MB. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 2003;115:83–95. doi: 10.1016/s0092-8674(03)00725-6. [DOI] [PubMed] [Google Scholar]
- 13.Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 2003;162:863–875. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou T, Aumais JP, Liu X, Yu-Lee LY, Erikson RL. A role for Plk1 phosphorylation of NudC in cytokinesis. Dev. Cell. 2003;5:127–138. doi: 10.1016/s1534-5807(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Zhou T, Kuriyama R, Erikson RL. Molecular interactions of Polo-like-kinase 1 with the mitotic kinesin-like protein CHO1/MKLP-1. J. Cell Sci. 2004;117:3233–3246. doi: 10.1242/jcs.01173. [DOI] [PubMed] [Google Scholar]
- 16.Lane HA, Nigg EA. Antibody microinjection reveals an essential role for human polo-like kinase 1 (Plk1) in the functional maturation of mitotic centrosomes. J. Cell Biol. 1996;135:1701–1713. doi: 10.1083/jcb.135.6.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.do Carmo Avides M, Tavares A, Glover DM. Polo kinase and Asp are needed to promote the mitotic organizing activity of centrosomes. Nat. Cell Biol. 2001;3:421–424. doi: 10.1038/35070110. [DOI] [PubMed] [Google Scholar]
- 18.Golsteyn RM, Schultz SJ, Bartek J, Ziemiecki A, Ried T, Nigg EA. Cell cycle analysis and chromosomal localization of human Plk1, a putative homologue of the mitotic kinases Drosophila polo and Saccharomyces cerevisiae Cdc5. J. Cell Sci. 1994;107(Pt 6):1509–1517. doi: 10.1242/jcs.107.6.1509. [DOI] [PubMed] [Google Scholar]
- 19.Lee KS, Yuan YL, Kuriyama R, Erikson RL. Plk is an M-phase-specific protein kinase and interacts with a kinesin-like protein, CHO1/MKLP-1. Mol. Cell Biol. 1995;15:7143–7151. doi: 10.1128/mcb.15.12.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song S, Grenfell TZ, Garfield S, Erikson RL, Lee KS. Essential function of the polo box of Cdc5 in subcellular localization and induction of cytokinetic structures. Mol. Cell Biol. 2000;20:286–298. doi: 10.1128/mcb.20.1.286-298.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 22.Fabbro D, Garcia-Echeverria C. Targeting protein kinases in cancer therapy. Curr. Opin. Drug Discov. Dev. 2002;5:701–712. [PubMed] [Google Scholar]
- 23.Noble ME, Endicott JA, Johnson LN. Protein kinase inhibitors: insights into drug design from structure. Science. 2004;303:1800–1805. doi: 10.1126/science.1095920. [DOI] [PubMed] [Google Scholar]
- 24.Stout TJ, Foster PG, Matthews DJ. High-throughput structural biology in drug discovery: protein kinases. Curr. Pharm. Des. 2004;10:1069–1082. doi: 10.2174/1381612043452695. [DOI] [PubMed] [Google Scholar]
- 25.Lenart P, Petronczki M, Steegmaier M, Di Fiore B, Lipp JJ, Hoffmann M, Rettig WJ, Kraut N, Peters JM. The small-molecule inhibitor BI 2536 reveals novel insights into mitotic roles of polo-like kinase 1. Curr. Biol. 2007;17:304–315. doi: 10.1016/j.cub.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 26.Steegmaier M, Hoffmann M, Baum A, Lenart P, Petronczki M, Krssak M, Gurtler U, Garin-Chesa P, Lieb S, Quant J, et al. BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol. 2007;17:316–322. doi: 10.1016/j.cub.2006.12.037. [DOI] [PubMed] [Google Scholar]
- 27.Donohue PJ, Alberts GF, Guo Y, Winkles JA. Identification by targeted differential display of an immediate early gene encoding a putative serine/threonine kinase. J. Biol. Chem. 1995;270:10351–10357. doi: 10.1074/jbc.270.17.10351. [DOI] [PubMed] [Google Scholar]
- 28.Xie S, Wu H, Wang Q, Cogswell JP, Husain I, Conn C, Stambrook P, Jhanwar-Uniyal M, Dai W. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J. Biol. Chem. 2001;276:43305–43312. doi: 10.1074/jbc.M106050200. [DOI] [PubMed] [Google Scholar]
- 29.Bahassi el M, Conn CW, Myer DL, Hennigan RF, McGowan CH, Sanchez Y, Stambrook PJ. Mammalian Polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways. Oncogene. 2002;21:6633–6640. doi: 10.1038/sj.onc.1205850. [DOI] [PubMed] [Google Scholar]
- 30.Xie S, Wu H, Wang Q, Kunicki J, Thomas RO, Hollingsworth RE, Cogswell J, Dai W. Genotoxic stress-induced activation of Plk3 is partly mediated by Chk2. Cell Cycle. 2002;1:424–429. doi: 10.4161/cc.1.6.271. [DOI] [PubMed] [Google Scholar]
- 31.Matthew EM, Yen TJ, Dicker DT, Dorsey JF, Yang W, Navaraj A, El-Deiry WS. Replication stress, defective S-phase checkpoint and increased death in Plk2-deficient human cancer cells. Cell Cycle. 2007;6:2571–2578. doi: 10.4161/cc.6.20.5079. [DOI] [PubMed] [Google Scholar]
- 32.Simmons DL, Neel BG, Stevens R, Evett G, Erikson RL. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol. Cell Biol. 1992;12:4164–4169. doi: 10.1128/mcb.12.9.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liby K, Wu H, Ouyang B, Wu S, Chen J, Dai W. Identification of the human homologue of the early-growth response gene Snk, encoding a serum-inducible kinase. DNA Seq. 2001;11:527–533. doi: 10.3109/10425170109041337. [DOI] [PubMed] [Google Scholar]
- 34.Ma S, Charron J, Erikson RL. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol. Cell Biol. 2003;23:6936–6943. doi: 10.1128/MCB.23.19.6936-6943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson JW, Chen L, Fode C, Binkert C, Dennis JW. Sak kinase gene structure and transcriptional regulation. Gene. 2000;241:65–73. doi: 10.1016/s0378-1119(99)00467-9. [DOI] [PubMed] [Google Scholar]
- 36.Hudson JW, Kozarova A, Cheung P, Macmillan JC, Swallow CJ, Cross JC, Dennis JW. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr. Biol. 2001;11:441–446. doi: 10.1016/s0960-9822(01)00117-8. [DOI] [PubMed] [Google Scholar]
- 37.Leung GC, Hudson JW, Kozarova A, Davidson A, Dennis JW, Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat. Struct. Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- 38.Bahassi el M, Myer DL, McKenney RJ, Hennigan RF, Stambrook PJ. Priming phosphorylation of Chk2 by polo-like kinase 3 (Plk3) mediates its full activation by ATM and a downstream checkpoint in response to DNA damage. Mutat. Res. 2006;596:166–176. doi: 10.1016/j.mrfmmm.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Bai J, Shen R, Brown SA, Komissarova E, Huang Y, Jiang N, Alberts GF, Costa M, Lu L, et al. Polo-like kinase 3 functions as a tumor suppressor and is a negative regulator of hypoxia-inducible factor-1 alpha under hypoxic conditions. Cancer Res. 2008;68:4077–4085. doi: 10.1158/0008-5472.CAN-07-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmerman WC, Erikson RL. Polo-like kinase 3 is required for entry into S phase. Proc. Natl Acad. Sci. USA. 2007;104:1847–1852. doi: 10.1073/pnas.0610856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chase D, Feng Y, Hanshew B, Winkles JA, Longo DL, Ferris DK. Expression and phosphorylation of fibroblast-growth-factor-inducible kinase (Fnk) during cell-cycle progression. Biochem. J. 1998;333(Pt 3):655–660. doi: 10.1042/bj3330655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smits VA, Klompmaker R, Arnaud L, Rijksen G, Nigg EA, Medema RH. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000;2:672–676. doi: 10.1038/35023629. [DOI] [PubMed] [Google Scholar]
- 43.Bahassi el M, Penner CG, Robbins SB, Tichy E, Feliciano E, Yin M, Liang L, Deng L, Tischfield JA, Stambrook PJ. The breast cancer susceptibility allele CHEK2*1100delC promotes genomic instability in a knock-in mouse model. Mutat. Res. 2007;616:201–209. doi: 10.1016/j.mrfmmm.2006.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Bahassi el M, Hennigan RF, Myer DL, Stambrook PJ. Cdc25C phosphorylation on serine 191 by Plk3 promotes its nuclear translocation. Oncogene. 2004;23:2658–2663. doi: 10.1038/sj.onc.1207425. [DOI] [PubMed] [Google Scholar]
- 45.Lee KS, Erikson RL. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol. Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ouyang B, Li W, Pan H, Meadows J, Hoffmann I, Dai W. The physical association and phosphorylation of Cdc25C protein phosphatase by Prk. Oncogene. 1999;18:6029–6036. doi: 10.1038/sj.onc.1202983. [DOI] [PubMed] [Google Scholar]
- 47.Smith MR, Wilson ML, Hamanaka R, Chase D, Kung H, Longo DL, Ferris DK. Malignant transformation of mammalian cells initiated by constitutive expression of the polo-like kinase. Biochem. Biophys. Res. Commun. 1997;234:397–405. doi: 10.1006/bbrc.1997.6633. [DOI] [PubMed] [Google Scholar]
- 48.Conn CW, Hennigan RF, Dai W, Sanchez Y, Stambrook PJ. Incomplete cytokinesis and induction of apoptosis by overexpression of the mammalian polo-like kinase, Plk3. Cancer Res. 2000;60:6826–6831. [PubMed] [Google Scholar]
- 49.Sumara I, Gimenez-Abian JF, Gerlich D, Hirota T, Kraft C, de la Torre C, Ellenberg J, Peters JM. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 2004;14:1712–1722. doi: 10.1016/j.cub.2004.09.049. [DOI] [PubMed] [Google Scholar]
- 50.Hanisch A, Wehner A, Nigg EA, Sillje HH. Different Plk1 functions show distinct dependencies on Polo-Box domain-mediated targeting. Mol. Biol. Cell. 2006;17:448–459. doi: 10.1091/mbc.E05-08-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Busch H, Muramatsu M, Adams H, Steele WJ, Liau MC, Smetana K. Isolation of nucleoli. Exp Cell Res. 1963;24(Suppl. 9):150–163. [PubMed] [Google Scholar]
- 52.Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Müller glial cells. J. Comp. Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lowery DM, Lim D, Yaffe B. Structure and function of Polo-like kinases. Oncogene. 2005;24:248–259. doi: 10.1038/sj.onc.1208280. [DOI] [PubMed] [Google Scholar]
- 55.Lu LY, Wood JL, Minter-Dykhouse K, Ye L, Saunders TL, Yu X, Chen J. Polo-like kinase 1 is essential for early embryonic development and tumor suppression. Mol. Cell Biol. 2008;28:6870–6876. doi: 10.1128/MCB.00392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma S, Charron J, Erikson RL. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol. Cell Biol. 2003;23:6936–6943. doi: 10.1128/MCB.23.19.6936-6943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haren L, Stearns T, Lüders J. Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE. 2009;4:e5976. doi: 10.1371/journal.pone.0005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wsierska-Gadek J, Horky M. How the nucleolar sequestration of p53 protein or its interplayers contributes to its (re)-activation. Ann. NY Acad. Sci. 2003;1010:266–272. doi: 10.1196/annals.1299.046. [DOI] [PubMed] [Google Scholar]
- 59.Cerutti L, Simanis V. Controlling the end of the cell cycle. Curr. Opin. Genet. Dev. 2000;10:65–69. doi: 10.1016/s0959-437x(99)00044-1. [DOI] [PubMed] [Google Scholar]
- 60.Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S. Regulation of ubiquitin ligase dynamics by the nucleolus. J. Cell Biol. 2005;170:733–744. doi: 10.1083/jcb.200506030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Argentini M, Barboule N, Wasylyk B. The contribution of the RING finger domain of MDM2 to cell cycle progression. Oncogene. 2000;19:3849–3857. doi: 10.1038/sj.onc.1203737. [DOI] [PubMed] [Google Scholar]
- 62.Rogalsky V, Todorov G, Moran D. Translocation of retinoblastoma protein associated with tumor cell growth inhibition. Biochem. Biophys. Res. Commun. 1993;192:1139–1146. doi: 10.1006/bbrc.1993.1535. [DOI] [PubMed] [Google Scholar]
- 63.Zini N, Trimarchi C, Claudio PP, Stiegler P, Marinelli F, Maltarello MC, La Sala D, De Falco G, Russo G, Ammirati G, et al. pRb2/p130 and p107 control cell growth by multiple strategies and in association with different compartments within the nucleus. J. Cell Physiol. 2001;189:34–44. doi: 10.1002/jcp.1135. [DOI] [PubMed] [Google Scholar]
- 64.Green C, Chatterjee R, McGarrigle HH, Ahmed F, Thomas NS. p107 is active in the nucleolus in non-dividing human granulosa lutein cells. J. Mol. Endocrinol. 2000;25:275–286. doi: 10.1677/jme.0.0250275. [DOI] [PubMed] [Google Scholar]
- 65.Rizos H, Darmanian AP, Mann GJ, Kefford RF. Two arginine rich domains in the p14ARF tumour suppressor mediate nucleolar localization. Oncogene. 2000;19:2978–2985. doi: 10.1038/sj.onc.1203629. [DOI] [PubMed] [Google Scholar]
- 66.Nishikimi M, Fukuyama R, Minoshima S, Shimizu N, Yagi K. Cloning and chromosomal mapping of the human nonfunctional gene for L-gulono-gamma-lactone oxidase, the enzyme for L-ascorbic acid biosynthesis missing in man. J. Biol. Chem. 1994;269:13685–13688. [PubMed] [Google Scholar]
- 67.Wu XW, Lee CC, Muzny DM, Caskey CT. Urate oxidase: primary structure and evolutionary implications. Proc. Natl Acad. Sci. USA. 1989;86:9412–9416. doi: 10.1073/pnas.86.23.9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Edgar AJ. The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet. 2002;3:18. doi: 10.1186/1471-2156-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Felsenstein J. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 1997;46:101–111. doi: 10.1093/sysbio/46.1.101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.