Abstract
Rna15 is a core subunit of cleavage factor IA (CFIA), an essential transcriptional 3′-end processing factor from Saccharomyces cerevisiae. CFIA is required for polyA site selection/cleavage targeting RNA sequences that surround polyadenylation sites in the 3′-UTR of RNA polymerase-II transcripts. RNA recognition by CFIA is mediated by an RNA recognition motif (RRM) contained in the Rna15 subunit of the complex. We show here that Rna15 has a strong and unexpected preference for GU containing RNAs and reveal the molecular basis for a base selectivity mechanism that accommodates G or U but discriminates against C and A bases. This mode of base selectivity is rather different to that observed in other RRM-RNA structures and is structurally conserved in CstF64, the mammalian counterpart of Rna15. Our observations provide evidence for a highly conserved mechanism of base recognition amongst the 3′-end processing complexes that interact with the U-rich or U/G-rich elements at 3′-end cleavage/polyadenylation sites.
INTRODUCTION
The 3′-end processing of pre-mRNAs is an essential step in the maturation of the transcripts of eukaryotic genes. In this orchestrated process, an initial site-specific cleavage in the 3′-UTR of the pre-mRNA is followed by the addition and subsequent trimming of a homopolymeric polyadenylate tail. Cleavage of the transcript stimulates transcriptional termination, whilst addition of the poly(A) is important for regulating mRNA stability, nuclear export and translation, reviewed (1–3).
The 3′-end processing events in Saccharomyces cerevisiae are coordinated by the interaction of two multiprotein processing complexes with a series of conserved RNA sequence elements located in the proximity of the transcript cleavage site (1,4). Cleavage factor IA (CFIA), comprises four core subunits Rna14, Rna15, Pcf11 and Clp1 and is required for both site selection and transcript cleavage (5). A larger complex, cleavage and polyadenylation factor (CPF) contains a core of eight subunits Cft1, Cft2, Ysh1, Pta1 Mpe1, Pfs2, Fip1 and Yth1 and is required in both the cleavage and polyadenylation reactions (6). Additionally, holo-CPF, a complex containing a further six subunits Ref2, Pti1, Swd2, Glc7, Ssu72 and Syc1 has also been identified (7). Details of the composition of these protein complexes are presented in Supplementary Figure 1. The RNA sequences that are targeted by these complexes are the cleavage/poly(A) site itself, comprising a pyrimidine base followed by a run of adenines (PyAn) (8), the efficiency element (EE) a series of UA repeats located at a variable number of nucleotides 5′ to the cleavage site (9) and a positioning element (PE) an A-rich sequence located ∼20 nucleotides 5′ to the cleavage site (10,11). The polyadenylation site is also frequently surrounded by upstream and downstream U-rich regions that effect the efficiency of cleavage and polyadenylation (12). In mammals, the 3′-UTRs of mRNAs contain similar, but not identical conserved recognition elements including the GU- and U-rich elements located 10–30 nucleotides downstream of the polyadenylation site (13–15).
In order to understand how 3′-end processing complexes recognize RNA sequence elements and direct transcript cleavage/polyadenylation, it is necessary to identify and examine the protein–RNA interactions made by the subunits of processing complexes. Here, we focus on RNA recognition by the Rna15 subunit of CFIA. RNA binding is mediated by an RNA recognition motif (RRM) contained within the N-terminal 100 residues of the protein. We analyse the sequence preference of this domain and provide a detailed model for the interaction using a combination of X-ray crystallography, NMR and fluorescence spectroscopy. Our data demonstrate that the RRM of Rna15 displays a strong preference for GU-rich RNA, mediated by a binding pocket that is entirely conserved in both yeast and mammalian Rna15 orthologues. Moreover, we reveal a nucleobase discrimination mechanism that utilizes base pair like interactions between base edges and the protein main-chain. Based on our observations, we proffer that the G/U binding pocket constitutes a common RNA recognition mechanism utilized by processing factors to target the U- and G/U-rich sequence elements that surround cleavage/polyadenylation sites.
MATERIALS AND METHODS
Protein constructs and Ribo-oligonucleotides
DNA sequences coding for Rna15 residues M1-S94, P16-S103 and P16-N111 were isolated by PCR amplification from the S. cerevisiae genome. Fragments were cloned into Escherichia coli expression vectors. Details of cloning, mutagenesis, expression and purification are in Supplementary Methods and Supplementary Figure 2. Ribo-oligonucleotides were purchased 2′ protected, from Dharmacon and de-protected following the manufacturers instructions. RNAs were reconstituted in small volumes of Rnase free buffer [25 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5 mM Tris(2-Carboxyethyl) phosphine hydrochloride (TCEP)] and stored at –20°C. RNAs that had incorporated a 5′-Tetrachloro-fluorescein (TET) fluorophore were stored in a dark box at –20°C.
Crystallization and structure determination
Complexes of Rna15-RRM and RNA ∼600 µM were prepared in 150 mM NaCl, 25 mM Tris–HCl pH 8.0, 0.5 mM TCEP and crystallized by sitting drop vapour diffusion. The structure of the Rna15(16-103-ht)-GUUGU complex was determined using single wavelength anomalous dispersion (SAD) using data collected on a crystal of seleno-methionine substituted protein. The Rna15(16-111)-GUGUU and free Rna15(16-111) structures were solved using molecular replacement. Details of the crystallization conditions and structure determination are described in Supplementary Methods.
Fluorescence spectroscopy
Titrations were carried out using an ISS photon counting spectrofluorimeter using 5′-TET labelled hexa-ribo-oligonucleotides. Typically, fluorescence was measured from an RNA maintained at a concentration of 125 nM (λex, 515 nm; λem, 545 nm) upon addition of increasing amounts (0–100 µM) of w.t. Rna15(16-111) or substitution mutants. Details of data fitting are described in Supplementary Methods.
NMR spectroscopy
NMR spectra for sequence assignment were recorded at 27°C on ∼0.5–1.0 mM samples of Rna15(1-94)-ht, Rna15(16-103)-ht and Rna15(16-111) in a 90% H2O/10% D2O mixture containing 10 mM Tris-HCl pH 7.4, 50 mM NaCl, 0.5 mM TCEP. Titration experiments were performed by addition of increasing amounts of RNAs to 0.025 mM protein samples. The recording and processing of NMR experiments, assignment of spectra, fitting of the relaxation data, and quantification of chemical shift perturbation (CSP) were carried out using standard methods, described in Supplementary Methods.
RESULTS
Rna15 preferentially binds GU-rich RNAs
Previously, it has been proposed that, in vitro, in the absence of other protein factors Rna15 binds RNA sequences only weakly and/or non-specifically (16,17). This observation, combined with the lack of a strong Rna15 consensus-binding site in S. cerevisiae 3′-UTRs suggests it is unlikely that the domain recognizes a long specific RNA sequence within the mRNA 3′-end. However, one possibility is that Rna15 displays weak sequence preference for the multiple low complexity sequences flanking the cleavage/polyadenylation sites. To test this notion, we examined the interaction of the core RRM domain, Rna15(1-94)-ht, with two dissimilar pentameric ribo-oligonucleotides UAUUU and UGGCG using NMR spectroscopy, Figure 1A. These data show that the same residues mediate the interaction with both sequences and that the complexes are in a moderately fast regime of exchange. However, because of the low complexity of these RNA sequences it is likely that multiple binding frames are present where small chemical shift differences between resonances of proteins bound in different frames are averaged by the fast regime of exchange. Accordingly, the titrations likely represent the interaction of Rna15(1-94)-ht with a base-averaged sequence rather than a unique RNA–protein interaction. Indeed, considering the low complexity of target sequences in the 3′-UTR multiple binding frames may be important to enhance the RNA binding affinity of Rna15 in vivo. Nevertheless, regardless of the dynamics, quite unexpectedly, the induced chemical shift perturbations are significantly larger upon interaction with UGGCG than they are with the UAUUU sequence, indicating that Rna15(1-94)-ht binds to the G-rich sequence with a higher affinity than the AU sequence.
Figure 1.
The Rna15 RRM-RNA interaction. (A) Superimposition of three 15N-1H correlation NMR experiments recorded during titrations of Rna15(1-94)-ht with either the UGGCG or UAUUU ribo-oligonucleotides. The spectrum free Rna15(1-94)-ht (magenta), together with 1: 3 molar ratios of Rna15(1-94)-ht:UAUUU (blue) and Rna15(1-94)-ht:UGGCG (gold) are shown. Arrows connect the free and bound positions of two representative resonances. The resonances of the same residues are perturbed by interaction with the two RNAs, but larger shifts occur when the UGCGG sequence is added. (B) RNA binding affinity for different sequences measured by changes in fluorescence intensity of 5′-Tetrachloro-fluorescein labelled RNAs. Titration curves for UGUUGU, UUUUUU, UGUUUG, UCUUCU, UAUUAU, AGAAGA and ACAACA are shown. The fraction of bound RNA (ΔF/Fmax) is plotted against total Rna15(16–111) concentration. Equilibrium association constants determined by fitting a hyperbolic binding isotherm to the titration data are shown below.
To more precisely characterize the RNA binding specificity, we examined the RNA binding activity of the Rna15 RRM using fluorescence spectroscopy employing 5′-end dye-labelled short ribo-oligonucleotides. Fluorescence titrations and derived equilibrium association constants for GU rich, U rich together with other C and A containing sequences are shown in Figure 1B. These data show that Rna15 has a strong preference for GU- and U-rich sequences over C- or A-rich sequences. The equilibrium association constant for UGUUGU is in the order of 2.1 × 105 M−1. Similarly, the affinities for the UUUUUU and UGUUUG ribo-oligonucleotides are 1.1 × 105 M−1 and 1.0 × 105 M−1, respectively. However, where U and G bases were replaced individually by C or A bases (UCUUCU, UAUUAU, AGAAGA), the association constant was reduced by 4- to 8-fold. Furthermore, in a titration that employed a ribo-oligonucleotide containing only C and A bases (ACAACA), complex formation was barely detectable. The affinity of Rna15(16-111)-UGUUGU interaction was also measured using isothermal titration calorimetry (ITC) (Supplementary Figure 3). The association constant (KA = 1.7 × 105 M−1) is wholly consistent with that determined by fluorescence and so based on these titration experiments we conclude that the Rna15 RRM has a strong GU preference and binds to these sequences with affinities in the order of 105 M−1.
Structure of the Rna15 RRM
In an effort to understand the molecular basis of Rna15 sequence specificity, we have determined three independent crystal structures. The first comprises a C-terminally his-tagged Rna15 RRM domain, Rna15(P16-S103)-ht, bound to the penta-ribonucleotide sequence GUUGU. The other two structures comprise Rna15(16-111), an RRM domain that contains only an additional N-terminal glycine, in both the free form and bound to the GUUGU penta-ribonucleotide. The Rna15(P16-S103)-ht-GUUGU structure was determined from a single wavelength anomalous diffraction experiment recorded on the selenium absorbance edge. The other two structures were solved by molecular replacement using the Rna15(P16-S103)-ht as a search model. Details of the structure determination and refinement are presented in Table 1.
Table 1.
Statistics of data collection, phasing and refinement
| Se peak (Complex 1) Rna15(16-103-ht)- GUUGU (SAD) | Native (Complex 1) Rna15(16-103-ht)- GUUGU | Native (Complex 2) Rna15(16-111)- GUUGU | Free protein Rna15(16-111) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P4 | P4 | P21212 | P212121 |
| Cell dimensions a, b, c (Å) | 56.9, 56.9, 28.9 | 56.7, 56.7, 29.2 | 37.0, 75.0, 31.5 | 27.0 36.8, 92.8 |
| Wavelength (Å) | 0.9708 | 1.54 | 1.54 | 1.54 |
| Unique reflections | 4509 | 12488 | 5957 | 9150 |
| Resolution range (Å) | 20–2.8 | 20–1.6 | 25–2.05 | 20–1.8 |
| Rmerge (%)a,b | 5.7 (10.7) | 4.6 (41.9) | 11.4 (35.8) | 5.9 (31.5) |
| I/σ(I)b | 35.3 (21.8) | 52.1 (3.1) | 14.8 (3.3) | 31.6 (5.1) |
| Completeness (%)b | 99.2 (95.2) | 99.3 (92.5) | 99.3 (94.5) | 99.7 (96.6) |
| Redundancyb | 10.5 (9.3) | 13.0 (6.0) | 6.3 (4.6) | 7.4 (5.9) |
| Phasing | ||||
| No. of sites | 2 | / | / | / |
| Mean FOM solve (12–2.9Å) | 0.4 | |||
| No. of copies in AU | 1 | 1 | 1 | 1 |
| Mean FOM (resolve) (12–2.9Å) | 0.72 | |||
| Refinement | ||||
| Resolution (Å) | 20–1.6 | 25–2.05 | 20–1.8 | |
| Rfactor/Rfree (%)c | 20.9/24.0 | 20.6/25.8 | 18.3/22.6 | |
| No residues/atoms | ||||
| Rna15 | 94/741 | 87/666 | 87/676 | |
| RNA | 2/40 | 1/24 | / | |
| Phosphate | / | / | 1/5 | |
| Water | 123 | 44 | 147 | |
| Average B-factor (Å2) | 23.5 | 24.7 | 21.0 | |
| Rna15 | 21.1 | 23.0 | 17.2 | |
| RNA | 22.4 | 54.1 | / | |
| Phosphate | / | / | 30.7 | |
| Water | 38.4 | 34.7 | 38.2 | |
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.007 | 0.008 | 0.010 | |
| Bond angles (°) | 1.068 | 1.107 | 1.160 | |
| Ramachandran plot | ||||
| Preferred regions (%) | 100 | 100 | 98.8 | |
| Allowed regions (%) | 0 | 0 | 1.2 |
aRmerge=ΣhΣi|Ih,i – <Ih>|/ΣhΣiIh,i, where Ih,i is the intensity of the individual reflections and <Ih> is the mean intensity over symmetrically equivalent reflections.
bValues in parentheses are for the highest resolution shell.
cR=Σ||Fobs| – | Fcalc|| / Σh|Fobs|, where Fobs and Fcalc are observed and calculated structure factures, respectively. Rfree and Rfactor were calculated from the test reflection and working sets, respectively. Rfree was calculated with 5% of the data.
In all three structures, residues P16 to S94 adopt the same β1–α1–β2–β3–α2–β4–β5 topology and the secondary structural elements pack in a canonical RRM fold, consisting of a four-stranded central β-sheet backed by two α-helices, Figure 2. The only structural difference in this core RRM region is a shift of around 5Å of the β2-β3 loop between the Rna15(P16-S103)-ht structure and the two Rna15(16-111) structures. Solution NMR data recorded on the Rna15(1-94)-ht, Rna15(P16-S103)-ht and Rna15(P16-S111) confirm the core topology observed in the crystal structures. Near complete assignment of the backbone resonances of all three constructs and comparison of the chemical shift of resonances from the β2–β3 loop shows there are no conformational differences between the three constructs (Supplementary Figure 4a). However, backbone 15N T1 and T2 relaxation and heteronuclear NOE data recorded on the Rna15(P16-S111) and Rna15(1-94)-ht proteins (Supplementary Figure 4b) show that the β2-β3 loop is rather flexible likely accounting for the 5Å shift we observe in the crystal structures.
Figure 2.
Crystal structures of the RRM from S. cerevisiae Rna15. (A) Rna15(16-111) free, (B) Rna15(16-111)-GUUGU and (C) Rna15(16-103)ht-GUUGU. The top panels show the structures in cartoon representation, secondary structures and chain termini are labelled. RNA is shown in stick representation, guanine in magenta and uracil in yellow. The bottom panels show the molecules in the same orientation but in semi-transparent surface representation. Residues that interact with the RNA are shown as sticks. In the Rna15(16-103)ht-GUUGU structure, the GU dinucleotide bound at Y21 is from a symmetry related molecule.
The RNA binding site
In the two structures of the Rna15 RRM crystallized in the presence of RNA nucleobase-binding sites are present where there is clear electron density for one/two bound RNA bases, see Supplementary Figure 5a. Both structures contain a base-binding site (Site-I) located on the loop connecting β1 and α1, Figure 3. Within Site-I, the side-chains of Y27 and R87 form the walls of a binding pocket and make base-stacking interactions with either the uracil base in the Rna15(P16-S103)-ht structure, Figure 3A or the guanine base in Rna15(16-111) structure, Figure 3B. In the free Rna15(16-11) structure the Y27 and R87 side chains adopt a different orientation, Figure 2A, indicating Site-I forms only upon RNA binding. In the bound structures, the conformation of the R87 side chain at Site-I is maintained by hydrogen bonding to both the main chain carbonyl and side chain hydroxyl of S24. Binding is further stabilized by Watson–Crick-like hydrogen bonding interactions between functional groups on the edges of the U or G base and the backbone of Y27 and I25. In the Rna15(P16-S103)-ht structure the 4 carbonyl of the uracil base is hydrogen bonded to the backbone amide of Y27, whilst the imino proton of N3 is shared with the carbonyl of I25. In the Rna15(16-111) structure the 6 carbonyl of the guanine is hydrogen bonded to the backbone amide of Y27 and the imino proton of N1 is shared with the carbonyl of I25. At first glance, it seems the two complexes utilize different base–protein interactions. However, at the positions that are involved in hydrogen bonding interactions, the aromatic rings of G and U are in fact superimposable, see Supplementary Figure 5b. Moreover, the hydrogen bonding interactions are made by the functional groups that are utilized in the standard Watson–Crick AU and GC basepairs.
Figure 3.
Structural details of the Site-I and Site-II base-binding pockets. Close up views of bases bound in the Site-I binding pocket in (A) the Rna15(16-103)-ht-GUUGU and (B) the Rna15(16-111)-GUUGU structure. Uracil (yellow) and guanine (magenta) bases along with the side-chains of S24, I25, Y27 and R87 that form the Site-I base-binding pocket are shown in stick representation, hydrogen bonds are shown as black dashed lines. The remainder of the protein is shown as a semi-transparent surface representation with the backbone highlighted as a cartoon. (C) Comparison of the configuration and complementarity of base pair edges involved in base-backbone pairing at Site-I. Bound guanine (magenta) and uracil (yellow) together with adenine (cyan) and cytosine (green) bases modelled in Site-I are shown in stick representation. Base imino and amino protons are shown and hydrogen bonds to the protein backbone in the SIPY loop are shown as black dashed lines. (D) The Site-II base-binding pocket in the Rna15(16-103)ht-GUUGU structure. The bound dinucleotide guanine (magenta) and uracil (yellow) together with protein residues Y21 and Y93 are shown in stick representation.
Based on the similarity of the base–protein recognition in these two structures we propose that Site-I binding mediates either wholly or at least in part the G/U specificity of the Rna15–RNA interaction. This idea is further supported by inspection of the functional groups that are present in these positions on all four RNA bases. Figure 3C shows the hydrogen-bonding configuration of the Site-I binding pocket with either a G or U base bound and also where A and C bases have been modelled by structural alignment with the G and U bases bound at Site-I. Inspection of these alignments with respect to the configuration of functional groups on the base edge clearly reveals the basis for nucleobase selectivity in this system. Both the G and U have a carbonyl (position 6 purine; position 4 pyrimidine) and an imino proton (position 1 purine; position 3 pyrimidine) that are hydrogen-bonded to the amide of Y27 and the carbonyl of I25 respectively. In the case of A and C, an exocyclic amino group replaces the carbonyl and there is no imino proton present. This configuration of the A and C bases means neither base can participate in base-backbone hydrogen bonding interaction that are observed in the G and U bound structures. Moreover, the exocyclic amino group could potentially clash with the amide of Y27 making the interaction of either A or C in the Site-I pocket much more unfavourable than with G or U.
In the Rna15(P16-S103)-ht complex structure a GU dinucleotide is present, bound between two copies of the asymmetric unit. This dinucleotide links Site I of one molecule and a second nucleobase-binding site (Site-II) located on β1 of the central sheet, of a neighbouring molecule, Figure 3D. Here, a guanine base is stacked against the aromatic side chain of Y21 that protrudes from the surface of the β-sheet and is part of the conserved RNP-2 RNA binding motif. This stacking interaction is further stabilized by hydrogen bonding between the exocyclic amino group on the guanine N2 and the backbone carbonyl of Y93 and by the presence of fortuitous interactions between the guanine base and residues from the C-terminal his-tag. In contrast to Site-I, where direct hydrogen bonding between the base edges and protein backbone is the major driver of specificity the Site-II-base interaction is predominantly mediated by a less specific base-aromatic side chain stacking interaction, more typical of that observed in canonical RRM-RNA structures. It is possible that the presence of base–protein contacts derived from the extraneous his-tag sequence might enhance the RNA–protein interaction at Site-II. However, the Y21 base-stacking interaction is supported by NMR experiments that show that Y21 resonances are strongly perturbed when RNA is bound to constructs were the his-tag is absent, see Supplementary Figure 5c.
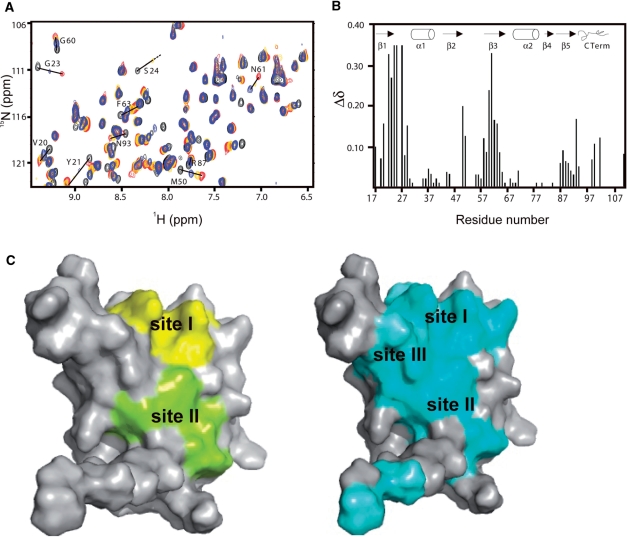
In order to test our structural models the extent of the RNA–protein interface in the Rna15(16-111)-UGUUGU complex was analysed using heteronuclear NMR. Figure 4A shows the 15N-1H HSQC spectrum of Rna15(16-111) recorded at increasing stoichiometric ratios of the UGUUGU ribo-oligonucleotide. Quantification of these CSP data, Figure 4B, reveals the most strongly perturbed residues are V20-G23, M50 and S58-F63 located on strands β1, β2 and β3 of the central sheet, together with S24, I25, Y27 and Q29 in Site-I and on the β1-α1 loop. A much smaller degree of the CSP is also observed for the resonances surrounding R87. In the left panel of Figure 4C, residues that are located within 4Å of the nucleobase binding sites in the crystal structures are displayed on a surface representation of the molecule. In contrast, in the right panel residues that show significant CSP upon complex formation are highlighted. The comparison of these data usefully reveals the extent of the RNA–protein interface. Both the CSP data and crystal structures show that the interface surface extends from the Site-I observed in both complex structures over strand β1 to encompass Site-II in the Rna15(P16-S103)-ht structure. However, additionally the CSP data extends the RNA-binding surface to include much of the β3 strand where resonances of residues Y60-F63 show very strong shifts. These residues are located within a canonical RRM RNP1 motif so in all likelihood the aromatic rings of Y61 and F63 constitute a further nucleobase binding-site (Site-III) interspersing Site-I and Site-II.
Figure 4.
NMR analysis of the RNA–protein interface. (A) Superimposition of the central section of 15N-1H HSQC NMR spectrum of Rna15(16-111), free (black) and after addition of 0.5 (blue), 1.0 (gold) and 2.0 (red) equivalents of a UGUUGU ribo-oligonucleotide. Residues that show significant chemical shift perturbation are labelled. (B) Weighted chemical shift changes (Δδ) of Rna15(16-111) upon binding to UGUUGU. The weighted chemical shift change is plotted against the Rna15 sequence. Secondary structure elements are indicated, above. Resonances assigned in the free protein spectrum that could not be assigned in the bound protein spectrum, were given a weighted chemical shift change value of 0.35 as default. (C) Surface representations of Rna15(16-111). Residues <4Å from the nucleotides at Site-I and II in the X-ray structure are coloured in yellow and green on the left, whilst residues showing significant chemical shift perturbation upon RNA binding are displayed in cyan on the right. Site-I, II and III are indicated.
Mutational analysis of Site-I and Site-II base-binding sites
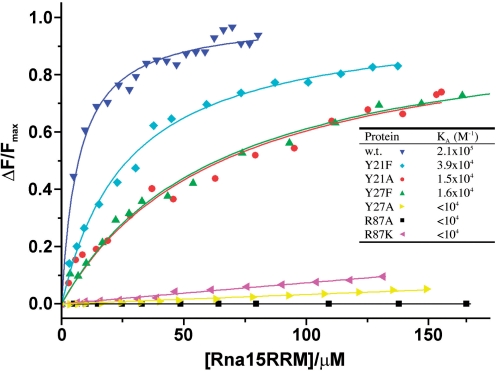
In order test our structural observations and to investigate the contribution that residues in the Site-I and Site-II base-binding pockets make to the affinity and specificity of Rna15-RNA interaction, substitution mutants were prepared. At Site-I, Y27 was replaced by either A or F and R87 by either A or K. At Site-II, Y21 was substituted by A or F. The effects of these mutations on the RNA binding activity of Rna15(16-111) were then examined using fluorescence spectroscopy employing the Tet-UGUUGU ribo-oligonucleotide. Binding isotherms constructed from fluorescence intensity measurements are shown in Figure 5. It is apparent that where substitutions have been made at residues that form the walls of the Site-I base-binding pocket, RNA binding is heavily diminished or even abolished. For instance, Y27A and even the conservative R87K replacement reduce the binding affinity to an immeasurable level in the fluorescence assay. Although, abolition of RNA binding by the Y27A substitution might be anticipated on the basis of the crystal structures, it is somewhat surprising that the lysine mutation also has such strong effect. Presumably, although both K and R maintain a positive charge the absence of delocalization and planarity of lysine side-chain means it is unable to stack against the bound base in the same favourable way as the arginine side-chain does. The conservative substitution Y27F also reduces binding but not to the same degree as the R87 substitutions. Notably, a weak hydrogen bond between the Y27 phenolic hydroxyl and the O4’ of the ribose is observed in the Rna15(P16-S103)-ht complex, so it is possible in Y27F removal of this interaction is responsible for the diminished RNA-binding affinity. At Site-II the Y21F substitution reduces the RNA binding 5-fold and Y21A around 10-fold. However, relative to the larger effects we observe with the Site-I mutations, in Site-II the substitution of a phenolic by an aromatic ring produces only a modest reduction in binding supporting the idea that aromatic ring-base stacking rather than hydrogen bonding is important for the Site-II-nucleobase interaction.
Figure 5.
Examination of the binding affinity of U/G-pocket mutants by fluorescence spectroscopy. The panel shows the titration curves [fraction of bound RNA; ΔF/Fmax versus total Rna15(16-111) concentration] derived from the change in fluorescence intensity for Rna15(16-111) (dark blue) and substitution mutants Y21F, Y21A, Y27F, Y27A, R87A and R87K upon binding to the Tet-UGUUGU ribo-oligonucleotide. The Binding constants in the inset table were determined from these plots by fitting of a hyperbolic binding isotherm to the data.
The C-terminal region
Crystal structures and solution NMR data demonstrate that residues 16–94 of Rna15 are organized in a canonical RRM fold. However, in the regions both N- and C-terminal of the RRM significant structural flexibility is apparent. The amide resonances from residues 1–15, N-terminal to the core domain, are either not visible or extremely broad in 15N-1H correlation NMR spectra. Our interpretation is that these residues are disordered and undergoing chemical/conformational exchange and notably, constructs containing these first fifteen residues have also proved refractory to crystallization. The residues C-terminal to serine 94 also display conformational flexibility as in each of the three crystal structures this region adopts different secondary structure. In the free Rna15(16-111) structure, residues 96–101 comprise a short turn of α-helix that packs near perpendicularly against strands β1–β3 of the RRM β-sheet, Figure 2A. In the Rna15(16-111) nucleotide-bound structure, the same region (95-102) is in an extended β conformation (Figure 2B) forming a β-sheet with a 2-fold related molecule. In the Rna15(P16-S103)-ht complex, residues S95–S103 are in a loop followed by six residues from the his-tag (LEHHHH) that form a short helix running parallel to β1 and β2 of the central sheet, Figure 2C.
These observations demonstrate that in Rna15, the sequence immediately C-terminal to the RRM has the capacity to adopt multiple conformations. By comparison, in the solution structure of the RRM of CstF64, the human orthologue of Rna15 and a component of cleavage stimulatory factor (CstF), these C-terminal residues form a short α-helix that packs perpendicularly against the central β-sheet. Further, it has been proposed that in CstF64, RNA-binding induces displacement and a concomitant unfolding of this C-terminal helix from the central β-sheet (18,19). The strong functional similarity between CstF64 and Rna15 prompted us to use NMR to investigate the nature of the conformational flexibility in the C-terminal region of Rna15 and answer two key questions. Whether the C-terminal region of Rna15 interacts with residues at the RNA-binding site(s) on the surface of the central sheet and does the C-terminal region adopt a single stable conformation in solution, as in CstF64.
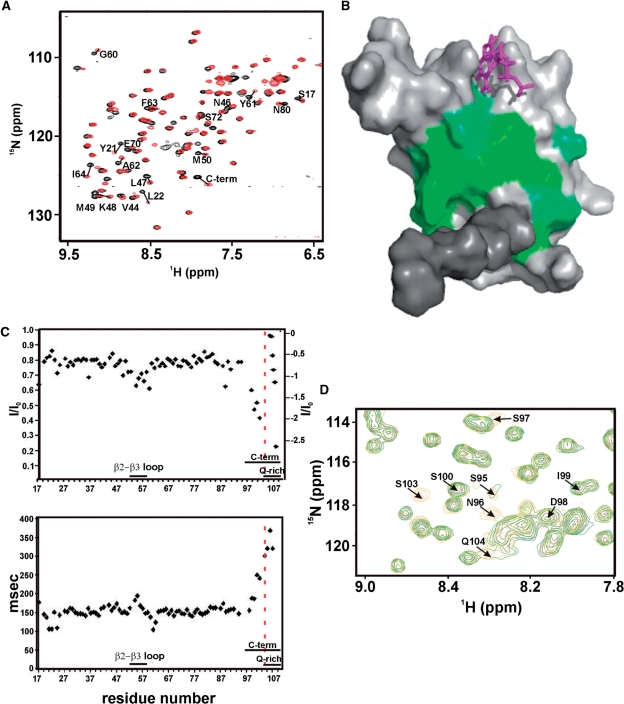
A comparison of the 15N-1H correlation spectra of Rna15(1-94)-ht and Rna15(16-111) reveals that there are substantial chemical shift differences, attributable to the presence of the C-terminal region, Figure 6A. The residues most perturbed by the presence of the C-terminal region map to a large area of the central β-sheet, Figure 6B, and include Y21, L22, Y61 and F63 in the RNP-1 and RNP-2, RNA binding motifs. These data support the notion that the C-terminal region of Rna15 interacts with the RRM sheet similar to what is observed in CstF64. Examination of the 15N-1H-correlation spectrum of Rna15(16-111) also reveals that the amide resonances of several residues in the C-terminal region are absent (S95–S97 and S103), presumably because of chemical or conformational exchange. However, the D98-V102 resonances are present and only slightly broadened relative to those of the core domain. Moreover, the T1, T2 and heteronuclear NOE relaxation data for the backbone amides of D98-V102 report nanosecond time scale motions comparable with, or only slightly faster than those for residues in the flexible β2–β3 loop, Figure 6C. In contrast, the resonances for five, further C-terminal unassigned glutamines (residues 104–109) display relaxation behaviour characteristic of a highly mobile structure. These data indicate substantial flexibility but are inconsistent with the entire C-terminal region rapidly tumbling in solution. More likely, these observations are a result of transient structuring of at least some parts of the C-terminal region together with the interaction of these residues with the core RRM domain. NOESY spectra were examined to determine if, for these residues, patterns of NOEs characteristic of secondary structure elements were present. However, only intra-residue and very weak n+1 backbone-side chain NOE correlations are visible. Further the backbone chemical shifts of residues D98-V102 are close to random coil values, confirming that residues D98-V102 do not form a secondary structure element (data not shown). As the structure of the CstF64 domain was determined at pH 6.0 (18) and our solution studies were carried out at pH 7.4 we sought to establish if the Rna15 C-terminal sequence might form an α-helix at lower pH. 15N-1H HSQC spectra were recorded at pH 7.4, 6.8 and 6.0 together with a single HNCACB spectrum, recorded at pH 6.0. Overall, these HSQC spectra are very similar with the exception that the amide resonances of residues S95, N96, N97 and V103 are present at pH 6.0, (Figure 6D). However, they are in the random coil region and have a linewidth pH dependency observed for exposed amide resonances in exchange with the bulk water. It is therefore very unlikely that a conformational change such as the formation of α-helical structure in the C-terminal region occurs as a result of lowering the pH. Subsequent TALOS (20) and Random Coil Index (RCI) (21) analysis of the backbone chemical shifts also confirms that residues 95–103 are random coil, likely to be flexible and are not engaged in the formation of either α-helical or β-strand secondary structures.
Figure 6.
Conformation of the C-terminal region. (A) Superimposition of the 15N-1H HSQC spectra of RNA15(16-111) (black) and RNA15(1-94)-ht (red). The core domain, residues 16-94, that have a Δδ > 0.06 between the RNA15(16-111) and RNA15(1-94)-ht constructs are labelled. (B) Residues that display significant chemical shift changes (Δδ > 0.06) are plotted on a surface representation of the domain, green. The C-terminal region is shown in dark grey and a G nucleotide bound at Site-I is shown in magenta. (C) Heteronuclear NOEs (top) and 15N T2 (bottom) of the Rna15(16-111) backbone resonances plotted against the domain sequence. Unassigned resonances from Q104-Q109 are inset on the top panel and are ordered by decreasing value of the heteronuclear NOEs, reported on the scale on the right. Fast motions are visible in the β2-β3 loop and in residues 98–103. (D) Superimposition of the 15N-1H HSQC spectra of RNA15(16-111) at pH 7.4 (black), 6.8 (green) and 6.0 (orange) (blow-up). Resonances of residues C-terminal to 94 are labelled. G101 and V102 amide correlations are outside the region shown.
Based on these observations, we conclude that the C-terminal of Rna15 comprises a dynamic ensemble of conformations unlike in CstF64 were a stable α-helical secondary structure element is present. Furthermore, the C-terminal region associates, albeit transiently, with residues on the surface of the RRM β-sheet, likely mediated via the exchange-protected residues D98-V102.
Structural similarity with other RRM–RNA complexes
The prevalence of RRM structures in the PDB means similarity searches using both the DALI search engine (22) and the SSM server (23) identify a large number of structurally similar molecules. However, two of the matches, the RRMs of CstF64 and Fox-1 (feminising locus on X) are of particular interest. CstF64 because of the obvious functional similarity and Fox-1 as quite unexpectedly the Fox-1 RRM displays a remarkable degree of structural conservation at the Site-I base-binding pocket.
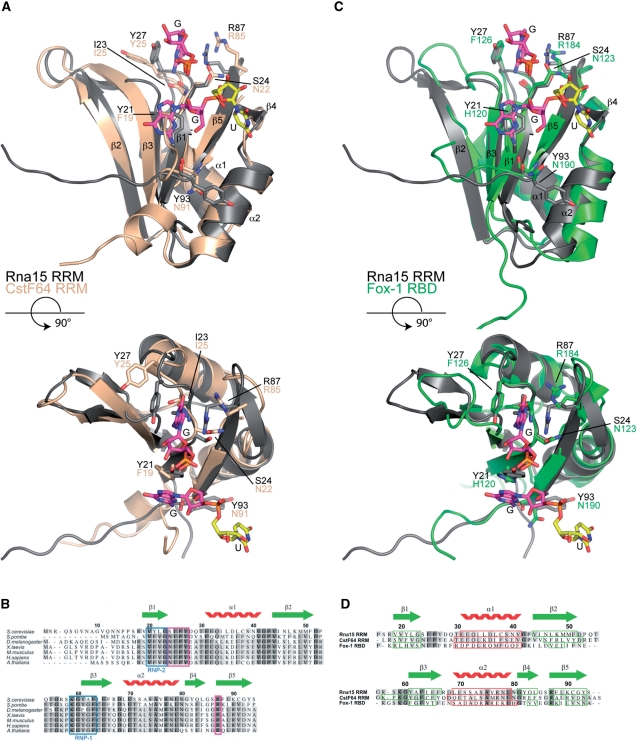
The 3D superposition of the RRMs from CstF64 and Rna15 is depicted in Figure 7A and a primary sequence alignment of Rna15 residues 1–94, with the RRMs of Schizpsaccharomyces pombe Rna15 and the CstF64 of other higher eukaryotes is presented in Figure 7B. The structural alignment reveals the two structures to be highly similar with a primary sequence identity of 44% the r. m. s. d. is 1.25Å over 77 aligned Cα positions. The structural conservation is also apparent at the base-binding sites. At Site-I the conformation of the β1-α1 loop is entirely conserved and residues I25, P26, Y27 and R87 that form the walls and bottom of the Site-I base-binding pocket in Rna15 all have structural counterparts in CstF64 (I23, P24, Y25 and R85). However, in CstF64 the binding pocket is not formed because the Y25 and R85 side-chains are oriented away from each other similar to what is observed in Rna15(16-111) in the absence of RNA. The superposition also aligns Y21 of Rna15 with F19 of CstF64 on the β1 strand supporting the notion that the Site-II RNA binding interaction is also conserved. Examination of the primary sequence alignment of the Rna15 RRM from S. cerevisiae and S. pombe together with those of CstF64 from a diverse range of species including, Drosophila melanogaster, Xenopus laevis, Homo sapiens and Arabiana thaliana, reveals four areas of strong sequence conservation. The first two are the characterized RNP-1 and RNP-2 motifs located on the β3 and β1 strands, respectively. The RNP-1 motif contains the highly conserved residues K59, Y61 and F63 that show strong perturbation of backbone amide chemical shifts upon RNA binding. In the RNP-2 motif, conservation of an aromatic residue Y21 in Rna15 and F19 in CstF64 reinforces the importance of Site-II in both proteins. Alongside these conserved canonical RNA binding sites the β1-α1 loop and R87 that constitute the Site-I base-binding pocket are completely conserved. It is therefore likely that Site-I and the inherent G/U specificity is a feature common to the CFIA and CstF complexes of all eukaryotes.
Figure 7.
Sequence and structure based alignments. (A) Structural superposition of the Rna15(16-111)-GUUGU complex onto the RRM of CstF64 (pdb code:1P1T), (r.m.s.d. Rna15 residues 16-94 is 1.25Å over 77 Cα positions, 44% sequence identity). Rna15(16-111) is shown in grey, CstF64 is coloured wheat. A GU dinucleotide has been modeled at Site-II by superposition of the Rna15(16-111)-GUUGU and Rna15(16-111)ht-GUUGU structures. Bound ribonucleotides and residues that mediate protein–RNA interactions are shown as sticks, secondary structure elements and termini are labelled. (B) Multiple primary sequence alignment of Rna15 RRM, species range from S. cerevisiae to A. thaliana. The RNP-1 and RNP-2 motifs are highlighted in blue, residues that form the Site-I base binding pocket are boxed in magenta and the Rna15 RRM secondary structure elements are shown above. (C) Structural superposition of the Rna15(16-111)-GUUGU complex onto the RRM of Fox-1 (pdb code:2ERR), (r.m.s.d. Rna15 residues 16-94 is 1.55Å over 73 Cα positions, 19.2% sequence identity) Rna15(16-111) and the Fox-1 RRM are shown in grey and green respectively. Bound nucleotides and residues involved in protein–RNA interactions are highlighted as in (A). (D) Structure based sequence alignment of the RRMs from Rna15, CstF64 and Fox-1. Rna15 sequence numbering and secondary structure elements are shown above the aligned sequences. Darker to lighter grey depicts greater to lower sequence conservation, secondary structure elements are boxed in all sequences, β-strands in green and α-helices in red.
The superimposition of the RRM of Fox-1 on Rna15 residues 16–94 is shown in Figure 7C and a structure-based sequence alignment of Fox-1 with Rna15 and CstF64 is shown in Figure 7D. Although in this case the sequence identity is lower at 19.2%, the r. m. s. d. for the alignment is still only 1.55Å over 73 Cα positions and the topological arrangement of secondary structures is conserved. Along with the sequence and structural conservation at the RNP-1 and RNP-2 motifs, surprisingly we also find Fox-1 contains a structurally conserved Site-I base-binding pocket. In Fox1 F126 and R184 form the walls of a G binding pocket and I124, P125 and F126 form the turn that makes same backbone base edge interactions observed in Rna15 and CstF64. Based on these structure and sequence alignments it is apparent that the Site-I base binding pocket is an important structural feature that is found not only in the Rna15 of yeasts and the CstF64 of higher eukaryotes but also in other unrelated RRMs. In all cases, the conformation of the β1-α1 loop is maintained and the two conserved residues forming the walls of the Site-I binding pocket are located in the same position in each of the structures.
DISCUSSION
G/U recognition at the RNA binding site
Our structural analysis of Rna15–RNA complexes reveals several features of RNA recognition by this 3′-end processing factor. The most striking observation is the presence of the Site-I base-binding pocket located in the β1-α1 loop interspersing the central β-sheet of the RRM. More usually, the RNA binding interfaces of RRMs are located almost entirely on the face of the central β-sheet (24). However, although these canonical RRM binding sites are important in Rna15, the main driver of G/U sequence specificity is the Site-I binding pocket where sequence specificity is attained by direct readout of the base-edges through recognition of the pattern of amide and carbonyl main-chain atoms in the β1-α1 SIPY loop. The same direct readout pocket is also present in the human orthologue CstF64, presumably meaning the same mode of base recognition operates in both yeast CFIA and mammalian CstF processing complexes. More startling, we find that a Site-I binding pocket is present in the RRM of the functionally unrelated splicing regulator Fox-1, a system characterized by very tight binding (KA > 109 M−1) and a high degree of sequence specificity for GCAUG recognition sites. In the structure of a Fox1 RRM–RNA complex (25), the specific RNA–protein interaction is mediated, at least in part, by a network of RNA–protein interactions involving residues in the β1–α1 loop of the RRM. Our comparison of the RRMs from Fox1 and Rna15 revealed that the β1-α1 loop is structurally conserved and that in Fox1, F126 and R184 occupy the same position as Y27 and R87 in Rna15, forming the walls of Site-I binding pocket. Mutational analyses of Rna15 and Fox1 also demonstrate the importance of Site-I residues in RNA binding. F126A substitution in Fox1 results in a large decrease in the RNA binding affinity (>1000-fold). Similarly, although the isolated RRM of Rna15 has a modest affinity for its RNA targets (KA ∼ 1–2 × 105 M−1), mutation of either Y27 or R87 results in a loss of RNA binding activity to immeasurable levels.
The structural conservation of Site-I in Fox1 and Rna15/CstF64 highlights the remarkable versatility of this RNA recognition module. The same α1–β1 loop region of the RRM is utilized to mediate both the high affinity Fox1–RNA interaction and the substantially lower affinity G/U recognition observed in Rna15. In Fox1, synergistic intra-nucleotide hydrogen bonding is combined with Site-I specific base edge–side chain interactions to enhance the specificity and affinity of the interaction. Whereas in Rna15, intra-nucleotide interactions are entirely absent and sequence specificity in this system is mediated only through the main-chain base hydrogen bonding interactions that select for G and U bases. These observations highlight that although canonical RNP-1 and RNP-2 binding sites are extensively utilized in RNA recognition by RRMs it is apparent that in Rna15 and CstF64 exquisite base selectivity is achieved by Site-I, a completely separate part of the molecule.
The C-terminal region and RNA binding
Another feature of the Rna15 RRM is the contribution that protein sequences C-terminal to the P16-S94 RRM core make to RNA binding. In CstF64, a C-terminal α-helix packs onto the central β-sheet. A mechanism for RNA recognition has been proposed where RNA binding displaces the C-terminal helix from the face of the central β-sheet. As a result of helix displacement, the aromatic rings of F19 and F61 are exposed and make base-stacking interactions with uracil bases (18). Our NMR data demonstrates that in Rna15 there is also an interaction between the C-terminal region and the surface of the central sheet. However, these data together with the X-ray structures provide evidence that unlike CstF64 the C-terminal region of Rna15 does not form a stable secondary structure, but instead samples many conformations. Interestingly, despite these conformational differences, presumably attributable to sequence variation, both C-terminal regions of Rna15 and CstF64 RRM appear to perform the same function. In the absence of RNA, both C-terminal elements occlude RNA binding sites by packing onto the surface of the RRM β-sheet. Upon RNA binding, they are displaced from the sheet but in Rna15 the coupled unfolding of a C-terminal structure does not occur. Why there is a necessity for reversible occlusion of the RNA binding interface in these 3′-end processing factors is unclear. Nevertheless, whilst the structure in C-terminal region structure has diverged the mechanism remains conserved from yeast to mammals.
Rna15 targets G/U sequence elements in the 3′-UTR
Typically, S. cerevisiae polyadenylation sites are surrounded by upstream and downstream U-rich sequences (9,26). Similarly, GU-rich downstream-elements are also present at metazoan 3′-ends. These elements constitute target recognition sites for CstF and interact with the RRM of the CstF subunit, CstF64 (13,27,28). However, although there is a clear structural and evolutionary relationship between the RRMs of Rna15 and CstF64, RNA recognition sequences for Rna15 in S. cerevisiae 3′-UTRs are not as well defined. A U-rich sequence has been proposed using SELEX (13) but it has also been shown that in the presence of HrpI, a complex of Rna15 and Rna14 binds to an A-rich positioning element in the GAL7 3′-UTR (16). However, other studies have underlined the importance of both upstream and downstream U-rich sequences in CFIA-CPF-dependent cleavage of polyA sites in S. cerevisiae (12,29). Here, mutation or deletion of surrounding U-rich sequences in the CYC1 and ADH1 3′-UTR drastically reduces CFIA-CPF-mediated cleavage. Moreover, introduction of a U-rich element downstream of the polyA site in the poorly processed GAL7 3′-UTR greatly enhances transcript cleavage by CPF-CFIA. Our crystal structures together with our observations from solution NMR and fluorescence spectroscopy now clearly demonstrate that the RRM of Rna15 contains a specific G/U base-binding pocket. These observations support the existing biological data for the requirement of distal U-rich elements for efficient CPF-CFIA-directed transcript cleavage. Moreover, they raise the possibility that the proposed requirement for U-rich sequences in S. cerevisiae might be extended to include G/U-rich sequences. The existence of the same G/U recognition pocket in the RRM of CstF64 and its complete sequence conservation in the Rna15 and CstF64 proteins that mediate 3′-end recognition in organisms ranging from S. cerevisiae to A. thaliana further supports the notion that the RRMs of Rna15 and CstF64 are functionally and mechanistically equivalent. These yeast–human similarities also extend to the higher order quaternary structure of CstF and CFIA. It has been demonstrated that Rna15 and Rna14 assemble into a Rna142–Rna152 tetramer, mediated through Rna14 homodimerization (17). Similarly, CstF77 is homodimeric (30,31) suggesting that CstF contains two molecules of CstF64, so also contains two RNA binding domains. In all likelihood, the presence of two copies of the RNA binding domain and the ability of Site I to rapidly exchange between adjacent G/U bases is important to enhance the modest affinity observed for a single Rna15 RRM. However, based upon the elongated nature of the CstF77 dimer there is the potential to interact with two non-contiguous G/U elements in a 3′-UTR, perhaps even spanning the polyA cleavage site. We now propose that target recognition sequences for both CstF and CFIA are multiple G/U and/or U-rich sequences that surround polyadenylation sites. This notion is primarily based on our discovery of a structurally conserved G/U selectivity pocket in Rna15 and CstF64 that has pan-eukaryote sequence conservation. This hypothesis is further strengthened by the observation that multiple copies of U- and/or GU-rich sequences are found at polyA sites and that mutation diminishes or abolishes CFIA-dependent transcript cleavage whilst introduction of U-rich elements can enhance cleavage in weakly processed transcripts. The idea that the CstF and CFIA processing complexes utilize the same base recognition and C-terminal displacement mechanism is compelling because it unifies the process of cleavage site selection by yeast and mammalian polyadenylation factors. Further structural similarities between the yeast and mammalian system include the conservation of the Pcf11-Clp1 and Rna14/CstF77 heterodimer and homodimer interfaces (30–32). More structural and biochemical data from both the yeast and mammalian systems will be required to determine just how far this commonality extends.
ACCESSION NUMBER
The coordinates of RNA15(16-103-ht)-GUUGU, RNA15(16-111)-GUUGU and RNA15(16-111) have been deposited in the Protein Data Bank under accession numbers 2X1F, 2X1A and 2X1B.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by MRC, UK. Funding for open access charge: MRC, UK.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the MRC, UK for support and Dave Hollingworth for critical reading of the article and help with isotopic labelling. NMR spectra were recorded at the MRC biomedical NMR facility, Mill Hill, London, UK.
REFERENCES
- 1.Zhao J, Hyman L, Moore C. Formation of mRNA 3′-ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Proudfoot N. New perspectives on connecting messenger RNA 3′-end formation to transcription. Curr. Opin. Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 4.Guo Z, Sherman F. 3′-end-forming signals of yeast mRNA. Trends Biochem. Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, Moore C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol. Cell Biol. 1992;12:3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Preker PJ, Ohnacker M, Minvielle-Sebastia L, Keller W. A multisubunit 3′-end processing factor from yeast containing poly(A) polymerase and homologues of the subunits of mammalian cleavage and polyadenylation specificity factor. EMBO J. 1997;16:4727–4737. doi: 10.1093/emboj/16.15.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nedea E, He X, Kim M, Pootoolal J, Zhong G, Canadien V, Hughes T, Buratowski S, Moore CL, Greenblatt J. Organization and function of APT, a subcomplex of the yeast cleavage and polyadenylation factor involved in the formation of mRNA and small nucleolar RNA 3′-ends. J. Biol. Chem. 2003;278:33000–33010. doi: 10.1074/jbc.M304454200. [DOI] [PubMed] [Google Scholar]
- 8.Heidmann S, Obermaier B, Vogel K, Domdey H. Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol. Cell Biol. 1992;12:4215–4229. doi: 10.1128/mcb.12.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graber JH, Cantor CR, Mohr SC, Smith TF. Genomic detection of new yeast pre-mRNA 3′-end-processing signals. Nucleic Acids Res. 1999;27:888–894. doi: 10.1093/nar/27.3.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russo P, Li WZ, Hampsey DM, Zaret KS, Sherman F. Distinct cis-acting signals enhance 3′ endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991;10:563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo Z, Sherman F. 3′-end-forming signals of yeast mRNA. Mol. Cell Biol. 1995;15:5983–5990. doi: 10.1128/mcb.15.11.5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dichtl B, Keller W. Recognition of polyadenylation sites in yeast pre-mRNAs by cleavage and polyadenylation factor. EMBO J. 2001;20:3197–3209. doi: 10.1093/emboj/20.12.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol. Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil A, Proudfoot NJ. Position-dependent sequence elements downstream of AAUAAA are required for efficient rabbit beta-globin mRNA 3′-end formation. Cell. 1987;49:399–406. doi: 10.1016/0092-8674(87)90292-3. [DOI] [PubMed] [Google Scholar]
- 15.Salisbury J, Hutchison KW, Graber JH. A multispecies comparison of the metazoan 3′-processing downstream elements and the CstF-64 RNA recognition motif. BMC Genomics. 2006;7:55. doi: 10.1186/1471-2164-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross S, Moore CL. Rna15 interaction with the A-rich yeast polyadenylation signal is an essential step in mRNA 3′-end formation. Mol. Cell Biol. 2001;21:8045–8055. doi: 10.1128/MCB.21.23.8045-8055.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noble CG, Walker PA, Calder LJ, Taylor IA. Rna14-Rna15 assembly mediates the RNA-binding capability of Saccharomyces cerevisiae cleavage factor IA. Nucleic Acids Res. 2004;32:3364–3375. doi: 10.1093/nar/gkh664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perez Canadillas JM, Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deka P, Rajan PK, Perez-Canadillas JM, Varani G. Protein and RNA dynamics play key roles in determining the specific recognition of GU-rich polyadenylation regulatory elements by human Cstf-64 protein. J. Mol. Biol. 2005;347:719–733. doi: 10.1016/j.jmb.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 20.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Berjanskii MV, Wishart DS. A simple method to predict protein flexibility using secondary chemical shifts. J. Am. Chem. Soc. 2005;127:14970–14971. doi: 10.1021/ja054842f. [DOI] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Dali/FSSP classification of three-dimensional protein folds. Nucleic Acids Res. 1997;25:231–234. doi: 10.1093/nar/25.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 24.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr. Opin. Struct. Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Helden J, del Olmo M, Perez-Ortin JE. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chou ZF, Chen F, Wilusz J. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 1994;22:2525–2531. doi: 10.1093/nar/22.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beyer K, Dandekar T, Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 29.Minvielle-Sebastia L, Beyer K, Krecic AM, Hector RE, Swanson MS, Keller W. Control of cleavage site selection during mRNA 3′-end formation by a yeast hnRNP. EMBO J. 1998;17:7454–7468. doi: 10.1093/emboj/17.24.7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bai Y, Auperin TC, Chou CY, Chang GG, Manley JL, Tong L. Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors. Mol. Cell. 2007;25:863–875. doi: 10.1016/j.molcel.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 31.Legrand P, Pinaud N, Minvielle-Sebastia L, Fribourg S. The structure of the CstF-77 homodimer provides insights into CstF assembly. Nucleic Acids Res. 2007;35:4515–4522. doi: 10.1093/nar/gkm458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noble CG, Beuth B, Taylor IA. Structure of a nucleotide-bound Clp1-Pcf11 polyadenylation factor. Nucleic Acids Res. 2007;35:87–99. doi: 10.1093/nar/gkl1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.