Abstract
Small nucleolar RNAs (snoRNAs) and small Cajal body-specific RNAs (scaRNAs) are non-coding RNAs whose main function in eukaryotes is to guide the modification of nucleotides in ribosomal and spliceosomal small nuclear RNAs, respectively. Full-length sequences of Arabidopsis snoRNAs and scaRNAs have been obtained from cDNA libraries of capped and uncapped small RNAs using RNA from isolated nucleoli from Arabidopsis cell cultures. We have identified 31 novel snoRNA genes (9 box C/D and 22 box H/ACA) and 15 new variants of previously described snoRNAs. Three related capped snoRNAs with a distinct gene organization and structure were identified as orthologues of animal U13snoRNAs. In addition, eight of the novel genes had no complementarity to rRNAs or snRNAs and are therefore putative orphan snoRNAs potentially reflecting wider functions for these RNAs. The nucleolar localization of a number of the snoRNAs and the localization to nuclear bodies of two putative scaRNAs was confirmed by in situ hybridization. The majority of the novel snoRNA genes were found in new gene clusters or as part of previously described clusters. These results expand the repertoire of Arabidopsis snoRNAs to 188 snoRNA genes with 294 gene variants.
INTRODUCTION
In eukaryotes and achaebacteria, small nucleolar RNAs (snoRNAs) form an abundant group of non-coding RNAs (ncRNAs) which act as guide RNAs to determine the sites of 2′-O-ribose methylation and pseudouridylation of ribosomal RNA (rRNA), spliceosomal small nuclear RNAs (snRNAs) and tRNAs (1–3 for reviews). There are two major structurally different families of snoRNAs: box C/D snoRNAs responsible for 2′-O-ribose methylation and box H/ACA snoRNAs responsible for pseudouridylation (1–3). Box C/D snoRNAs contain conserved sequences: box C (RUGAUGA) and D (CUGA) near their 5′ and 3′ ends, respectively, and one or two regions of complementarity to their cognate RNAs. Box H/ACA snoRNAs can be folded into stem-loop structures in the 5′ and 3′ halves of the RNA which are followed by the conserved internal box H (ANANNA) and the 3′-terminal box ACA (ACANNN). One or both of the stem-loops contain an internal loop sequence with two regions of complementarity to their target RNA flanking a uridine residue which is modified to pseudouridine (1–3). Each class of mature snoRNA is associated with four different core proteins required for stability and function of the snoRNP, although other proteins are required in snoRNP assembly (3,4). The core proteins fibrillarin (box C/D) and NAP57/Cbf5p (box H/ACA) are thought to confer methylase and pseudouridylase activities, respectively. A related class of RNAs are the small Cajal-body-specific RNAs (scaRNAs) which target modification of spliceosomal snRNAs. ScaRNAs contain conserved sequences and structures of box C/D and H/ACA snoRNAs but can have complex combinations of box C/D and H/ACA sequences and are retained in Cajal bodies (CBs) by virtue of CAB box sequences (5–8).
Processing of pre-ribosomal RNAs (pre-rRNAs) into 18S, 5.8S and 25/28S rRNAs involves a series of cleavage reactions and exonucleolytic trimming (9). A small number of snoRNPs are required for pre-rRNA cleavage reactions in humans and yeast: U3, U8, U14, U17, U22, MRP, snR10 and snR30 but the majority of snoRNPs determine the sites of nucleotide modifications of rRNA. Most of the ∼200 2′-O-ribose methylation and pseudouridylation sites in higher eukaryotes are in the active site of the ribosome and are thought to influence the efficiency of the ribosome and protein translation (10–12). The introduction of modifications in non-natural positions impaired cell growth in yeast through less efficient processing and increased turnover of pre-rRNAs, and reduced ribosome activity (13,14).
In the majority of higher eukaryotes studied to date ‘orphan’ snoRNAs have been described which are expressed but do not have complementarity to rRNA or snRNAs (15–26). Many of these orphan snoRNAs have potential mRNA targets suggesting other functions besides modifying rRNAs and snRNAs. Of particular interest are tandem arrays of orphan snoRNAs in a maternally imprinted region (IC-SNURF-SNRPN) of the human genome, many of which are conserved in other mammals. Some of these snoRNAs have brain-specific expression and loss of paternal expression of genes in this region is associated with developmental and behavioural problems (15,16). One of the genes, HBII-52, has complementarity to a brain-specific serotonin 2C receptor (5HT2CR) and may affect an RNA editing event in the mRNA (16,27). In addition, HBII-52 influences alternative splicing of the serotonin 2C receptor by blocking a splicing silencer sequence to promote inclusion of an alternative exon (28). Computational analysis of putative mRNA targets of human orphan snoRNAs shows a significant preference to exon sequences and association with alternatively spliced genes suggesting a role in modulating alternative splicing of many mRNAs (25). More recently, two examples of snoRNAs being processed to microRNAs (miRNAs) have been described (29,30). A human box H/ACA snoRNA is processed by Dicer to generate small RNAs which were associated with Argonaute proteins and caused reduced expression of gene targets (29). Similarly, snoRNAs in the ancient eukaryote, Giardia lamblia, were processed to miRNAs capable of translational repression of target mRNAs (30). Finally, mining of small RNA libraries from different organisms including Arabidopsis has identified numerous snoRNA-derived small RNAs which are associated with components of RNA silencing pathways (31). Thus, orphan snoRNAs can modulate mRNA expression by directly affecting alternative splicing or being processed to miRNAs or sRNAs with the potential to base-pair with specific target mRNAs.
Identification of snoRNAs in plants has concentrated on the model species Arabidopsis and rice. Computational prediction of plant box C/D snoRNAs showed a high degree of sequence diversity in primary gene sequence and in the frequency of gene variants, and a number of snoRNAs specific to plants were identified (17,18,22,32). The gene variants usually show some sequence diversity outside of the conserved box C and D sequences and the regions of complementarity to rRNA and snRNAs (18,33). The occurrence of gene variants reflects either major chromosomal duplication or rearrangements following hybridization or polyploidization (17,18,32,34). In contrast, very few box H/ACA genes have been identified by prediction (18) and our current knowledge of Arabidopsis box H/ACA snoRNAs is based largely on the molecular cloning of small ncRNAs from Arabidopsis seedlings (21). This RNomics approach identified 39 box H/ACA genes as well as novel box C/D snoRNAs, and the first plant scaRNAs (21). Recently, mining of Arabidopsis small RNA sequences identified 31 new snoRNA genes including scaRNAs (35).
One of the main features of plant snoRNA genes is that the majority are organized into polycistronic gene clusters (17,18,22,32,34,36). Plant polycistronic clusters are transcribed as a polycistronic precursor snoRNA (pre-snoRNA) which is then processed to release mature snoRNAs. Processing is thought to involve endonucleolytic activity followed by exonucleolytic trimming to produce the mature snoRNA (32,36). The detection of polycistronic precursor snoRNAs in CBs and the nucleolus by in situ hybridization suggests that processing occurs in both locations and/or that pre-snoRNAs traffic to the nucleolus via CBs (37).
RNomics approaches for analysing small RNA constituents have been successful in a number of species and provide a means of validating computational identification of small RNAs (19,21). We have previously used isolated nucleoli from Arabidopsis cell cultures to show the presence of exon junction complex proteins and mRNAs, and aberrant mRNAs in the nucleolus (38,39). Due to the nucleolar localization of snoRNAs and the processing patterns of plant pre-snoRNAs, we have used RNA from purified nucleoli to construct cDNA libraries to identify capped and uncapped snoRNAs and expressed orphan snoRNAs. The majority of the cDNAs isolated were full-length snoRNAs and allowed the identification of 31 novel box C/D and box H/ACA snoRNA genes including a U13 orthologue and eight putative orphan snoRNAs.
MATERIALS AND METHODS
cDNA libraries for small RNAs from the Arabidopsis nucleoli
Nucleoli were isolated from Arabidopsis Col0 cell cultures as described previously (38). Total RNA was extracted from isolated nucleoli using an RNeasy kit (Qiagen). The RNA was then 3′ polyadenylated using the poly(A) kit (Invitrogen) according to the manufacturer’s instructions. In brief, 1 μg RNA was incubated in a buffer containing 50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2, 10 mM MnCl2, 1 mM EDTA, 1 mM DTT, 1 mM ATP and 5 units Escherichia coli poly A polymerase (Invitrogen) for 8 min at 37°C. Three different cDNA libraries were generated using 250 ng polyadenylated RNA using the GeneRacer system (Invitrogen) following the manufacturer’s instructions. (i) RNA was dephosphorylated by calf intestinal alkaline phosphatase and then 5′ decapped by tobacco acid pyrophosphatase prior to ligation to RNA oligonucleotide adaptor (5′CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA3′) by E. coli RNA ligase to specifically enrich for capped RNAs. (ii) RNA was 5′ decapped prior to oligo adaptor ligation as above to clone both capped and uncapped RNAs. (iii) RNA was directly ligated to the RNA oligonucleotide adaptor to enrich for uncapped RNAs. First-strand cDNA was generated using an oligo(dT)-adaptor primer [5′GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)18–24] and Superscript III RT. The second strand cDNA was generated using primers specific to 3′ and 5′ oligonucleotide adaptor sequences and Platinum Taq polymerase High Fidelity (Invitrogen). cDNA fragments smaller than 400 bp were eluted from agarose gels, inserted into the pCR4 TA-cloning vector (Invitrogen) and sequenced.
Northern analysis and RT–PCR for snoRNA clusters
Total RNA was extracted from cultured cells and Arabidopsis rosette leaves using TRI-reagent (Invitrogen) according to the manufacturer’s protocol. Twenty micrograms RNA was fractionated on 8% polyacrylamide gels containing 8 M urea, transferred to nylon membrane and probed by DIG-uridine-labelled riboprobes. The membrane was washed and visualized by chemi-luminescence (Roche). For RT–PCR, first-strand cDNA was produced using 5 μg of whole-cell RNA and oligo-dT20 primer and Superscript III RT. One-twentieth of the mixture was taken for generation of second-strand cDNA by cycling reactions using Platinum Taq polymerase High Fidelity (Invitrogen) and the primers specific to up- and downstream snoRNA clusters. The reaction mixture was fractionated on 1.2% agarose gel and analysed.
Determination of putative modification sites
Putative modification sites were determined for novel box C/D snoRNAs by searching for complementarity of sequences upstream of the D or D′ boxes to Arabidopsis rRNA and snRNA sequences using BLAST (40). For box H/ACA genes, the sequences were folded using MFOLD (41) and putative pseudouridylation pockets compared against rRNA and snRNA sequences. Orthologues of snoRNA genes were identified by BLAST, displayed and aligned using Clustal on Jalview (42) and conserved sequences identified.
RNA probe preparation for in situ hybridization
Templates for in vitro transcription were generated by PCR using T3-5′ and T7-3′ primers to add promoter sequences for T3 and T7 polymerases, respectively, to the sequences of interest. Primer sequences were: T3-5′ adapter—GAATTAACCCTCACTAAAGGGAGGACACTGACATGGACTGAAGGAGTA and T7-3′ adapter—TGTAATACGACTCACTATAGGGCGCTACGTAACGGCATGACAGTG. Probes were prepared by in vitro transcription (43). PCR was performed with the following cycles: 94°C for 3 min; then 30 cycles of 94°C for 45 s, 63°C for 45 s and 72°C for 1.5 min; and a final extension of 72°C for 6 min. In vitro transcription, using 1 in 10 dilution of the PCR product, was performed for 2 h at 37°C in the presence of digoxigenin-UTP nucleotides (0.35 mM) (43). Hydrolysis was performed immediately in 100 mM carbonate buffer, pH 10.2, at 60°C for 30 min and the products were precipitated in 2.5 M ammonium acetate and three volumes of 100% ethanol for 1 h at 48°C. The product was pelleted by centrifugation at 4000 rpm for 30 min, and the pellets were resuspended in 30 µl of 100 mM Tris, 10 mM EDTA buffer. Probe label incorporation was checked by dot-blotting (43).
Specimen preparation and in situ hybridisation
Four-day-old Arabidopsis Col0 seedlings were fixed in 4% formaldehyde solution, freshly prepared from paraformaldehyde (Sigma-Aldrich, Gillingham, UK, Gillingham, UK) in TBS (TBS: 10 mM Tris, 140 mM NaCl, pH adjusted to 7.4 with HCl) containing 0.1% Igepal CA-630 (Sigma-Aldrich, Gillingham, UK) for 1 h at room temperature. Penetration of fixative throughout the root was ensured by vacuum infiltration, prior to the incubation. Seedlings were washed several times in TBS to remove fixative, then the root tips were laid across wells of multi-well slides pre-treated with aminopropyltri-ethoxysilane (APTES—Sigma-Aldrich, Gillingham, UK), and the rest of the seedling was excised with a razor blade. The root tips were allowed to dry for several hours or overnight before being treated with a cell-wall degrading enzyme mixture consisting of 1% driselase (Sigma-Aldrich, Gillingham, UK), 0.5% cellulase (Onozuka R10, Yakult, Japan) and 0.025%, pectolyase Y23 (Duchefa Biochemie, Haarlem Netherlands) in TBS for 15 min at room temperature. Then the roots were washed several times with TBS alone.
The digoxigenin-labelled RNA probes were diluted (1 in 20) into a hybridisation mix containing 50% formamide (Sigma-Aldrich, Gillingham, UK), 10% dextran sulphate (Sigma-Aldrich, Gillingham, UK), 1 mg/ml tRNA (Sigma-Aldrich, Gillingham, UK), 1× Denhardts solution (Sigma-Aldrich, Gillingham, UK), 0.33 M NaCl, 0.01 M Tris–HCl, 0.01 M NaPO4 and 5 mM EDTA (pH 6.8), then denatured at 80°C for 2 min, before being cooled on ice. Twenty microlitres of probe was applied to each well and slides incubated in a humid chamber at 37°C overnight. Slides were washed sequentially in 2× SSC (SSC: 20× stock solution consists of 3 M sodium chloride and 300 mM trisodium citrate, adjusted to pH 7.0 with HCl) at room temperature for 10 min, 2× SSC/50% formamide at 45°C for 15 min, 1× SSC/50% formamide at 45°C for 15 min and 2× SSC at room temperature for 5 min. A blocking solution of 3% Bovine Serum Albumin (Sigma-Aldrich, Gillingham, UK) in TBS was applied for 15 min, followed by a primary antibody solution containing monoclonal mouse anti-digoxin (Sigma-Aldrich, Gillingham, UK) diluted 1 in 5000 in blocking solution. Slides were incubated at room temperature for 90 min, washed 3 × 5 min in TBS and a second antibody solution containing goat anti-mouse IgG Alexa Fluor 488 (Invitrogen) diluted 1 in 200 in blocking solution was applied and incubated for 90 min at room temperature. Slides were washed in TBS, 3 × 10 min and counterstained with DAPI (4′,6-diamidino-2-phenylindole: Sigma-Aldrich, Gillingham, UK) (1 µg/ml TBS) for 5 min. Finally slides were washed for 5 min in TBS and mounted in Vectashield (Vector Laboratories Ltd, Peterborough PE2 6XS, UK).
Microscopy
Slides were viewed using a 60× objective (NA 1.4, oil immersion) on a Nikon Eclipse 600 epifluorescence microscope equipped with a Hamamatsu Orca ER cooled CCD digital camera, a motorized xy stage and a z-focus drive. Raw data stacks were deconvolved using AutoDeBlur and Autovisualise software version 9.3 (Autoquant, MediaCybernetics, Marlow, Buckinghamshire, UK). The deconvolved data stacks were then analysed with ImageJ (a public domain program by W. Rasband available from http://rsb.info.nih.gov/ij/). Final figures were prepared using Adobe Photoshop (Adobe Systems Inc., Mountain View, CA).
RESULTS
Isolation of full-length snoRNAs from total RNA of isolated nucleoli
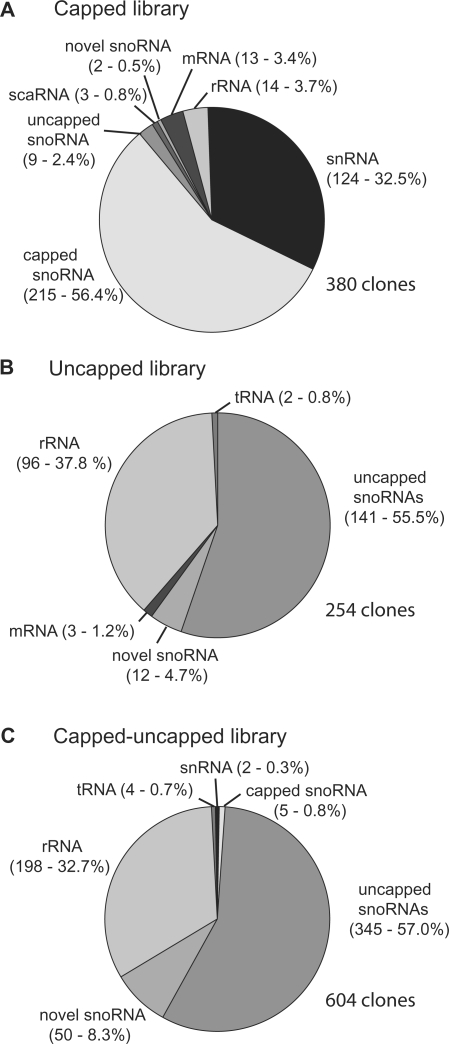
The mode of expression of different small RNAs determines whether transcripts are likely to be capped or uncapped. For example, plant snRNAs, and U3 and MRP snoRNAs are transcribed from their own promoters by RNA polymerase II or III and are capped (44). Most plant snoRNAs are transcribed as polycistronic pre-snoRNAs which are processed to generate mature box C/D and H/ACA snoRNAs and are expected to be uncapped (34,36). Similarly, dicistronic tsnoRNAs (tRNA-snoRNA) are processed to generate uncapped variants of snoR43 (45,46). To identify capped and uncapped snoRNAs and other small RNA transcripts from nucleoli, three different small RNA cDNA libraries were constructed from total RNA isolated from purified nucleoli. The first library was enriched for capped RNAs by treating the total nucleolar RNA with phosphatase to remove 5′ phosphates before treating with decapping enzyme prior to first strand synthesis with reverse transcriptase and cDNA production. The second library was enriched for uncapped RNAs by carrying out cDNA synthesis directly on the total RNA. The third library was designed to contain a mixture of both capped and uncapped RNAs by treating the total nucleolar RNA with decapping enzyme before proceeding to cDNA synthesis. The three libraries are called ‘capped’, ‘uncapped’ and ‘capped–uncapped’ and 380, 254 and 604 clones were fully sequenced from them, respectively (total of 1238 clones sequenced) (Figure 1).
Figure 1.
Classes of small RNAs isolated in cDNA libraries from nucleolar RNA. The number of clones and relative abundance (expressed as a percentage) are given in brackets for the different classes of small RNAs isolated in the capped (A), uncapped (B) and mixed capped–uncapped (C) libraries.
The main RNA constituents of the capped library were small nuclear RNAs (U1, U2, U4 and U5snRNA variants) and capped snoRNAs (Figure 1A). The capped snoRNA population contained a single U3snoRNA variant and multiple copies of three other snoRNAs (snoR105, snoR108 and snoR146). The latter three snoRNAs made up more than half of the clones from the capped library suggesting that these snoRNAs are capped. In addition, 63.3% of the clones were snoR146, 33.3% were snoR108 and only 3.3% were snoR105 suggesting that the abundance of these snoRNAs varies greatly with snoR146 being the most abundant in Arabidopsis cell cultures. The low recovery of U3snoRNAs may reflect cloning bias due to the RNA size fractionation. The capped library also contained two clones of snoR102 and one of snoR109. SnoR102 is a scaRNA and snoR109 is an orphan snoRNA with no known target RNA, identified by Marker et al. (21). Only a small number of uncapped snoRNAs (including some novel snoRNAs) and rRNA fragments were isolated confirming the enrichment of this library for capped RNAs (Figure 1A).
The composition of the uncapped RNA library was distinctly different from that of the capped library (Figure 1B). The two major fractions were uncapped snoRNA variants and rRNA fragments. The rRNA fragments were from many different regions of the rRNA suggesting that they are breakdown products of rRNA degradation or artefacts of cloning. No capped small RNAs were isolated in the uncapped library. One clone each of variants of two orphan snoRNAs, snoR28-1c (C/D) and snoR110-1 (H/ACA) were obtained. To be able to compare the capped with the uncapped library, we also generated a mixed library by decapping before cDNA synthesis. This capped–uncapped library also had uncapped snoRNAs and rRNA fragments as the two major fractions, but contained two clones of capped snRNAs and five of capped snoRNAs including MRP (Figure 1C). Thus, although the capped snoRNAs are the major constituent of the capped library, their abundance on the basis of clone representation in the capped-uncapped library is probably of a similar order to the majority of uncapped snoRNAs. Taking all three libraries together, 134 snoRNA variants of 90 different snoRNA genes were cloned in this study. The majority of these variants are expressed from polycistronic gene clusters and thereby form a major fraction of the uncapped and capped–uncapped libraries. The vast majority of the snoRNA clones of previously identified snoRNAs were consistent with the sizes of computationally predicted gene sequences such that most sequences generated here represent full-length snoRNA sequences.
Novel snoRNAs
From the analysis of nucleolar RNAs we have identified 31 new Arabidopsis snoRNA genes (38 variants) and 15 new variants of known snoRNA genes (Table 1). Of the 31 new genes, 19 were obtained by direct cloning from nucleolar RNA of which 16 represented novel snoRNAs and a further three represented full-length clones of previously identified small RNA fragments with no known sequence motifs [(21) Supplementary Table S1)]. By analysing the genomic sequences flanking all of these genes and determining their gene cluster organization, we were also able to predict the full-length sequences of 12 other new genes of which four were novel snoRNA genes and eight corresponded to partial sequences of small RNAs to which no function was assigned (21) (Table 1). We also isolated full-length clones of 22 snoRNA genes previously identified as partial sequences (21) for which only partial sequences were available, and by BLAST analyses identified 14 previously unidentified variants of these genes. For example, snoR145 was cloned here and corresponded to the unknown sequence, Ath-122 (21). In the flanking regions of snoR145, other genes—snoR68, snoR159 and two copies of snoR135 were identified (Figure 2A). SnoR159 and snoR135 also corresponded to short RNA sequences with no known motifs, Ath-319 and Ath-118 (21), and upstream of snoR68 another putative box H/ACA gene (snoR157) was identified (Figure 2A). Similarly, two snoR88 gene variants were found upstream of two previously identified gene clusters containing snoR19, snoR20 and snoR38Y (18) (Figure 2B). In addition, snoR64 was found in the first cluster and between snoR19-1 and snoR64, another putative box H/ACA gene was predicted (snoR136). Thus, the cloning of novel snoRNAs and examination of flanking sequences has defined novel gene clusters or extended previously described gene clusters (18) (Figure 2A and B; Supplementary Figures S1 and S2). Although the majority of the new genes were in gene clusters, snoR151 and three variants of snoR155 were found in introns of ribosomal protein genes (Figure 2C; Supplementary Figure S3), and four of the novel genes appeared to be single genes. Recently, 31 new Arabidopsis snoRNA genes (44 gene variants) were identified from assembly of high throughput short sequence reads (35). Comparison of these genes with the 31 novel genes (38 variants) and 15 new variants of known snoRNAs obtained here from direct cloning and predictions from gene organization, only 12 genes (15 variants) were common to both studies (Table 1). In addition, we identified a second variant of snoR137 (35). We have adopted the snoRNA gene numbers of Chen and Wu (35) and used the next consecutive numbers for our novel genes. From previous studies (17,18,21,32) along with this study and that of Chen and Wu (35), to date, Arabidopsis contains 188 different snoRNA/scaRNA genes with 294 gene variants.
Table 1.
Novel snoRNAs and snoRNA gene variants
| SnoRNAa | Size (nt) | RNA class | Modification site | Rep. cloneb | Gene copy | Chrc | Yeast/humand | Organization | Orthologues—representative ESTs | |
|---|---|---|---|---|---|---|---|---|---|---|
| Novel box C/D snoRNAs | ||||||||||
| AtsnoR146 (U13) | 127 | capped C/D | Unknown | At304 | 1 | 1 | – | Single gene | ESTs: monkey flower | |
| AtsnoR147 | 83 | C/D | D 25S Cm1471 13bp | At502 | 1 | 1 | Not modified | Cluster: snoR163/snoR147/snoR95 | ESTs: tobacco, chicory, oat | |
| AtsnoR148 | 84 | C/D | D 25S Cm2673 18bp | At649 | 1 | 5 | Not modified | Single gene | Genomic: Poplar ESTs: tomato, Nicotiana benthamiana, Asparagus, etc. | |
| AtsnoR149 | 80 | C/D | Unknown | At870 | 1 | 2 | – | Cluster: snoR158a/snoR39BYa/snoR21a/snoR149 | ESTs: poplar, safflower, barley, cocoa | |
| AtsnoR116 | 84 | C/D | D’ 18S Um123 11bp | At938 | 1 | 1 | Mgh18S-121; Z17B | Cluster: snoR117/snoR116/snoR85a/snoR85b | ESTs: apple, Jerusalem artichoke, dandelion | |
| AtsnoR117 | 72 | C/D | D’ 25S Um2103 12bp | At359 | 1 | 1 | Not modified | Cluster: snoR117/snoR116/snoR85a/snoR85b | ESTs: lettuce, grape, cotton,etc. | |
| AtsnoR118 | 98 | C/D | D′ 25S Cm2187 12bp | Ath-19b— predicted | 1 | 3 | snR76(Y); HB-II 180A,B,C | Cluster: snoR161/snoR66/snoR118 | Genomic: poplar; ESTs: lettuce, Jerusalem artichoke | |
| AtsnoR120 | 72 | C/D | D 25S Um 44 12bp | At838; Ath-475 | 1 | 5 | Not modified | Cluster: snoR133/snoR120 | None found | |
| AtsnoR133 | 101 | C/D | Unknown | Predicted | 1 | 5 | – | Cluster: snoR133/snoR120 | ESTs: Columbine, rice, maize | |
| Novel box H/ACA snoRNAs | ||||||||||
| AtsnoR150 | 150 | H/ACA | P1:25S 2174 | At213 | 1 | 1 | Not modified | Cluster: snoR150/snoR138 | Genomic: poplar; ESTs: Poplar, Melon, Papaya, etc. | |
| AtsnoR151 | 151 | H/ACA | P1: 25S 973 | At549 | 1 | 4 | snR43(Y); ACA9 | Intronic snoRNA: in intron 2 of At4g17380.1 (RPL15B) | None found | |
| AtsnoR152a AtsnoR152b | 163 | H/ACA | P2: 18S 1531 | At675 | 2 | 2 | Target sequence not conserved | Cluster: snoR152a/snoR152b | Genomic: poplar; EST: poplar, cotton, orange, etc. | |
| AtsnoR153 | 155 | H/ACA | Unknown | At740 | 1 | 5 | – | Cluster: snoR153/snoR12-2/AtU24-2 | ESTs: cocoa, poplar, orange | |
| AtsnoR154 | 156 | H/ACA | P1: 18S 1479 | At696 | 1 | 4 | ACA54 | Cluster: AtU19-2/snoR154 | ESTs: orange, papaya | |
| AtsnoR155a AtsnoR155b AtsnoR155c | 95 | H/ACA | P1: 25S 397 P2: 18S 56 | At801 | 3 | 1 | PI: target sequence not conserved; P2: not modified | Intronic H/ACA cluster in intron 2 of At1g04480.1 (RPL23A): snoR155a/snoR155b/snoR155c | Genomic: poplar EST: orange, tobacco, apple, etc. | |
| AtsnoR156 | 156 | H/ACA | P1:25S 1465 | At1140 | 1 | 5 | Not modified (Y); C in human | Single gene | ESTs: globe artichoke, barnadesia, oil palm | |
| AtsnoR157 | 122 | H/ACA | Unknown | Predicted | 1 | 2 | – | Cluster: snoR157/snoR68/snoR159/snoR145/snoR135a/snoR135b | None found | |
| AtsnoR158a AtsnoR158b | 226 | H/ACA | Unknown | Ath-143— predicted | 2 | 2 | – | Clusters: snoR158a/snoR39BYa/snoR21a/snoR149; snoR158b/snoR39BY/snoR21b | ESTs: cow pea, red clover, asparagus | |
| AtsnoR159 | 225 | H/ACA | Unknown | Ath-319—predicted | 1 | 2 | – | Cluster: snoR157/son R68/snoR159/snoR145/snoR135a/snoR135b | ESTs: melon, American sweet flag, wild radish | |
| AtsnoR160 | 139 | H/ACA | P2: 18S 1306 | Ath-362—predicted | 1 | 1 | Not modified | Cluster: snoR160/snoR16-2/AtU43-2 | ESTs: orange, apple, wild radish | |
| AtsnoR161 | 155 | H/ACA | P2: 18S 1188 | At395; Ath-382 | 1 | 3 | snR36 (Y); ACA36; ACA36B | Cluster: snoR161/snoR66/snoR118 | ESTs: red sage, cowpea, runner bean | |
| AtsnoR162 | 146 | H/ACA | P1: 25S 654 | Ath-408—predicted | 1 | 1 | Not modified | Intron 6/exon 7 of AT1G30970 (Suppressor of FRIGIDA4) | Genomic: poplar; ESTs: eucalyptus, poplar, etc. | |
| AtsnoR163 | 194 | H/ACA | Unknown | Ath-424 | 1 | 1 | – | Cluster: snoR163/snoR147/snoR95 | ESTs: cowpea, orange, cassava | |
| AtsnoR164 | 142 | H/ACA | Unknown | Ath-601—predicted | 1 | 2 | – | Single gene in 3′UTR of At2g37250 Adenosine kinase | Genomic: poplar; ESTs: globe artichoke, Russian dandelion | |
| AtsnoR134-1 AtsnoR134-2 AtsnoR134-3 | 144 | H/ACA | P1: 18S 1104; P2: 18S 1192 | At511 | 3 | 1;1;4 | P1: not modified; P2: snR35(Y); ACA13 | Clusters: snoR134-1/AtU36-1/AtU38-1/snoR6-1/snoR97-1; snoR134-2/AtU36a-2/AtU38-2/snoR6-2/snoR97-2; snoR134-3/AtU36a-3/AtU38-3/snoR6-3 | Genomic: poplar; ESTs: poplar, tomato, morning glory, etc. | |
| AtsnoR139 | 152 | H/ACA | P1: 18S 762 | At654 | 1 | 5 | snR80 (Y); ACA28 | Cluster: snoR82/snoR139 | Genomic: poplar; ESTs: Aristolochia fimbriata | |
| AtsnoR138 | 116 | H/ACA | P1: 25S 2445 | Predicted | 1 | 1 | ACA3 | Cluster: snoR150/snoR138 | ESTs: apple, orange, papaya, Columbine | |
| AtsnoR137-1 AtsnoR137-2 | 158 | H/ACA | P1: 18S 1208 | At1253 | 2 | 3; 4 | Not modified | Single genes | Genomic: poplar; ESTs: Russian dandelion, papaya, Tomato, etc. | |
| AtsnoR136 | 128 | H/ACA | P2: 25S 2833 | Predicted | 1 | 3 | E3 | Cluster:snoR88-2/snoR19-1/snoR136/snoR64/snoR20-1/snoRY38Y-1 | ESTs: soya, tomato | |
| AtsnoR135a AtsnoR135b | 142 | H/ACA | P1: 25S 536 P2: 25S 2181 | Ath118a/b—predicted | 2 | 2 | P1: modified by unknown human snoRNA P2: snR30 (Y); unknown human snoRNA | Cluster: snoR157/snoR68/snoR159/snoR145/snoR135a/snoR135b | Genomic: poplar; ESTs: lettuce, watermelon | |
| AtsnoR145 | 145 | H/ACA | Unknown | At559; Ath-122 | 1 | 2 | – | Cluster: snoR157/snoR68/snoR159/snoR145/snoR135a/snoR135b | ESTs: cowpea | |
| Novel variants of known snoRNAs | ||||||||||
| Organization | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| AtsnoR79a AtsnoR79b AtsnoR79c | 199; 194; 195 | H/ACA | See ref. (21) | AtsnoR79c = At394 | 3 | 1 | Intronic cluster—Intron of At1g31860 (Histidine synthesis bifunctional protein) | ||
| AtsnoR80-1 AtsnoR80-2 AtsnoR80-3 | 141; 139; 139 | H/ACA | See ref. (21) | AtsnoR80-1 = At162 AtsnoR80-2 = At3103 | 3 | 3, 4 | Clusters: snoR37-1/snoR22-1/snoR23-1/snoR80-1; snoR37-2/snoR22-1/snoR23-1/snoR80-2; snoR37-1/snoR22-3a/snoR22-3b/snoR23-3/snoR80-3 | ||
| At snoR85a AtsnoR85b | 139; 140 | H/ACA | See ref. (21) | 2 | 1 | Cluster: snoR117/snoR116/snoR85a/snoR85b | |||
| AtsnoR86-1 AtsnoR86-2 | 139; 143 | H/ACA | See ref. (21) | AtsnoR86-1 = At378 AtsnoR86-2 = At3235 | 2 | 1, 3 | Cluster: snoR14-1/snoR15/snoR86-2; Single gene snoR86-1 | ||
| AtsnoR87-1 AtsnoR87-2a AtsnoR87-2b | 153; 155; 155 | H/ACA | See ref. (21) | AtsnoR87-1 = At589 | 3 | 3 | Intronic—intron 1 of At3g11400 (eIF3G); Intronic cluster—intron 1 of At5g06000 (eIF3G) | ||
| AtsnoR90a AtsnoR90b | 150; 151 | H/ACA | See ref. (21) | 2 | 2 | Intronic cluster: intron 3 of At2g33430 (plastid development protein) | |||
| AtsnoR97-1 AtsnoR97-2 | 134; 134 | H/ACA | See ref. (21) | AtsnoR97-1 = At815 AtsnoR97-2 = At552 | 2 | 3, 4 | Clusters: snoR134-1/U36a-1/U38-1/snoR6-1/snoR97-1; snoR134-2/U36a-2/U38-2/snoR6-2/snoR97-2 | ||
| AtsnoR100a AtsnoR100b AtsnoR100c | 154; 147; 159 | H/ACA | See ref. (21) | 3 | 5 | Intronic cluster—intron 2 of At5g08180 (RPL7Ae/RPL30e/RPS12e) | |||
| AtsnoR110-1 AtsnoR110-2a AtsnoR110-2b | 114; 138; 147 | H/ACA | See ref. (21) | AtsnoR110-1 = At1126 AtsnoR110-2b = At835 | 3 | 1 | Intronic—intron2 of At1g26880 (RPL34a); Intronic cluster—intron2 of At1g69620 (Rpl34B) | ||
asnoRNAs with numbers between 113 and 145 were also identified by Chen and Wu (35).
bRepresentative clone: clone numbers beginning with ‘At’ refer to clones obtained in this study; those beginning in ‘Ath’ refer to Marker et al. (21).
cChromosome location of genes.
dModification sites/snoRNA information from LMBE snoRNABase v3 (49). Not modified, sequence fully or partially conserved in yeast/human but no evidence of modification; Y, yeast snoRNAs.
Figure 2.
Gene organization of novel snoRNA gene clusters. (A) Novel snoRNA gene cluster: cloning of snoR145 (Ath-122) and examination of flanking sequences identified snoR68 and three snoRNA genes for which partial sequences had been identified previously but were not recognized as snoRNAs (Ath-319, Ath-122, Ath-118a and Ath-118b) (21). (B) Extended snoRNA gene cluster: two clusters containing gene copies of snoR19, snoR20 and snoR38Y (black boxes) were identified previously (18). Further three genes were added to the first cluster: snoR88-1 and snoR64 (21) and snoR136 was predicted. (C) Intronic snoRNA genes and gene cluster: two of the novel H/ACA snoRNAs were found in introns of mRNAs with snoR155 having three variants. White boxes, exons; black boxes, snoRNAs.
The majority of the novel snoRNAs identified here had orthologues in other plant species (identified by BLAST searches of plant ESTs and genomic sequences) providing evidence that the cloned sequences represented bona fide snoRNAs (Table 1). The alignment of orthologous gene sequences and secondary-structures predicted by MFOLD (Supplementary Figure S4) aided the identification of putative modification sites in rRNA and snRNAs for the majority of the novel box C/D and H/ACA snoRNA genes (Table 1; Supplementary Figures S5 and S6, respectively). Similarly, putative modification sites in 25S rRNA and U6snRNA were found for snoR111 and snoR112, respectively (Supplementary Figure S6) which were identified previously as orphan box H/ACA snoRNAs (21). The modification site for snoR112 (U6 position 35) corresponds to the site modified by ACA12 and HBI-100 in human U6 (position 40). However, we were unable to identify complementarity to rRNA or snRNA for the box C/D snoRNAs—snoR149 and snoR133, and the box H/ACA snoRNAs—snoR145, snoR157-snR159, snoR163 or snoR164. These eight snoRNAs therefore represent putative orphan snoRNAs (Table 2).
Table 2.
Orphan snoRNAs in Arabidopsis
| Orphan snoRNA | Type | Expression | Variants | Orthologues to date | Reference |
|---|---|---|---|---|---|
| snoR6 | C/D | No | 3 | No | (18) |
| snoR28 | C/D | Yes | 6 | Yes | (18, present study) |
| snoR106 | C/D | Yes | 2 | No | (21) |
| snoR107 | C/D | Yes | 1 | No | (21) |
| snoR110 | H/ACA | Yes | 3 | No | (18, 21, present study) |
| snoR149 | C/D | Yes | 1 | Yes | Present study |
| snoR133 | C/D | No | 1 | Yes | Present study |
| snoR157 | H/ACA | No | 1 | No | Present study |
| snoR145 | H/ACA | Yes | 1 | Yes | (21, present study) |
| snoR158 | H/ACA | Yes | 1 | Yes | (21, present study) |
| snoR159 | H/ACA | Yes | 1 | Yes | (21, present study) |
| snoR163 | H/ACA | Yes | 1 | Yes | (21, present study) |
| snoR164 | H/ACA | Yes | 1 | Yes | (21, present study) |
Capped box C/D snoRNAs are U13 orthologues
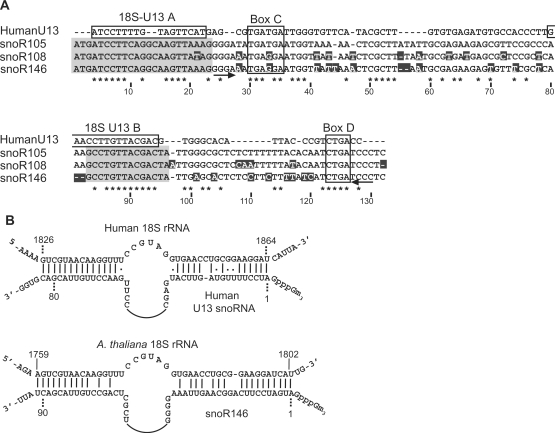
Of particular interest was the demonstration that three snoRNA species were highly abundant in the capped library. SnoR105, snoR108 and snoR146 are related monocistronic box C/D snoRNAs. SnoR105 and snoR108 were identified previously as partial sequences which contained recognized promoter elements of plant snRNA genes [an upstream sequence element (USE) at ∼−90 and a TATA-box at ∼30 bp upstream of the transcription start site—ref. (44)] in the upstream region of their genomic sequences (21). Here, we obtained full-length sequences of snoR105 and snoR108 as well as the related snoR146 (Figure 3). The genomic sequences upstream of all three genes have USE and TATA promoter elements in the RNA polymerase II configuration (44). The presence of snRNA promoter elements as well as their efficient isolation from the capped library strongly suggest that these snoRNAs are capped while the vast majority of Arabidopsis snoRNAs are processed from polycistronic snoRNA precursors and are uncapped. The three genes contain conserved C and D boxes except that the box C sequence is internal in the coding sequence lying ∼30 nt from the 5′ end (Figure 3). Like most box C/D snoRNAs, short inverted repeats are present directly up and downstream of the box C and D sequences, respectively, which may facilitate the formation of a K-turn in the C/D motif to which the core p15.5 kDa protein binds as the first stage in box C/D snoRNP assembly (47). Alignment of the three snoRNA sequences show two regions of high conservation: the first 24 nt which shows only a single nucleotide change and positions 83–96 (Figure 3A). The structural features of these snoRNAs were reminiscent of animal U8 and U13 snoRNAs which contain complementarity to rRNA sequences (48). Alignment with the human U13 sequence (49) clearly showed similarity in the two most highly conserved regions (above) (Figure 3A). The Arabidopsis sequences were complementary to the 3′-end of 18S rRNAs and formed similar putative base-pairing interactions (Figure 3B). A number of other plant orthologues of the U13 snoRNAs have been identified in EST libraries and all have complementarity to the 3′-end of 18S rRNA (results not shown).
Figure 3.
Sequence alignment of U13 snoRNAs. (A) The three plant U13 orthologues are aligned with human U13. Identical sequences are indicated by asterisks; for the three plant genes, sequence differences from snoR105 are highlighted white on black. Box C and D sequences are boxed; the two highly conserved regions in the plant genes are shaded grey; the two regions of complementarity to 18S rRNA in human U13 are boxed and labelled A and B [following human model: ref. (48)] and putative inverted repeats adjacent to boxes C and D are shown by arrows. (B) Putative base-pairing interactions between human U13snoRNA and Arabidopsis U13 (snoR146) with the 3′ regions of their cognate 18S rRNAs.
Expression of novel snoRNAs and U13 orthologues
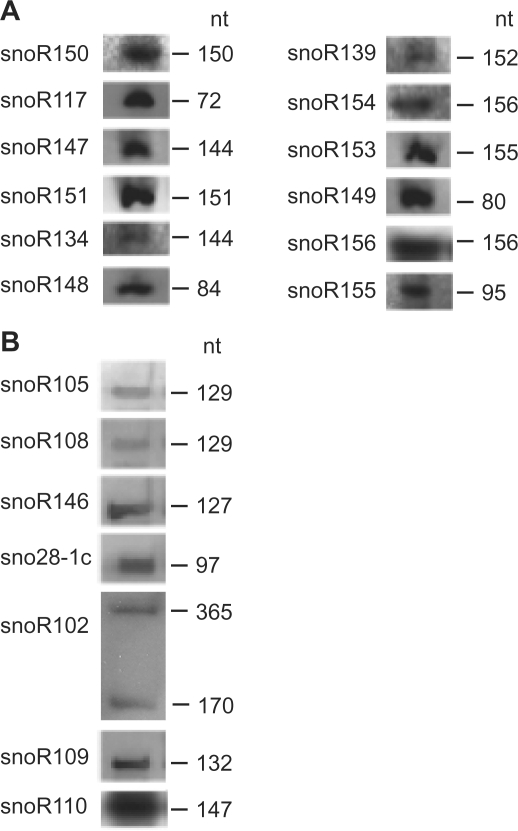
The cloning of the U13 orthologues and novel snoRNAs provides direct evidence of expression. To further demonstrate expression of some of the novel genes, we detected snoRNAs by northern analysis (Figure 4). RNAs of predicted size were obtained with antisense probes to the novel box C/D snoRNAs (snoR117, snoR147–149) and to the novel box H/ACA snoRNAs (snoR134, snoR150–156) (Figure 4A). Expression of the U13 orthologues (snoR105, snoR108 and snoR146), other orphan snoRNAs (snoR28-1c, snoR109, snoR110) and the scaRNA, snoR102, all of which were cloned in the nucleolar libraries, was also confirmed by northern analysis (Figure 4B). SnoR102 showed two bands on the northern analysis of ∼350 and 150–170 bp. The clones of snoR102 obtained from the capped library were 365 bp long. SnoR102 was originally cloned as a 133 bp sequence from the 3′ UTR of a protein-coding gene which gave a product on northern analysis of 185 bp (21) and corresponds to the 3′ half of the clones obtained here and based on the positions of the C/D boxes would have an expected size of ∼156 bp. A BLAST search identified our snoR102 sequence as the antisense of an annotated protein coding gene, At1g68945, of unknown function. It is highly likely that this gene encodes the 365 bp snoR102 transcript and is mis-annotated. The 5′ half of this snoR102 transcript contains some sequences similar to C and D boxes but is not clearly identifiable as a snoRNA. Chen and Wu (35) identified a 166 nt variant of snoR102 (snoR102-2) which is derived from the 3′ UTR of an unknown protein gene, At4g30993. However, there is extensive sequence similarity upstream of the snoR102-2 gene with the 365 bp snoR102 sequence cloned here. Thus, both At1g68945 and At4g30993 contain related sequences with the snoR102 variants in their 3′ halves. Thus, it appears that there are two variants of snoR102 which are each processed from either a longer precursor (cloned here and visible on Northerns) or from the 3′ UTR of the mRNA transcript from At4g30993. SnoR102 is therefore reminiscent of doublet guide RNAs which contain a box C/D snoRNA domain in their 3′ halves and a novel box C/D-like domain in their 5′ halves and where only the capped doublet RNA or the 3′-most box C/D RNA are stable (8). Whether the 5′ half of snoR102 contains a functional domain guiding modification of snRNAs or rRNAs is unknown.
Figure 4.
Northern analysis of novel snoRNAs, U13 orthologues and scaRNAs. (A) Northern analysis of novel snoRNAs and (B) northern analysis of the U13 snoRNAs (snoR105, snoR108 and snoR113), scaRNA (snoR102) and orphan snoRNAs (snoR28-1c, snoR109 and snoR110). Expected sizes of the transcripts are indicated and are consistent with markers (data not shown).
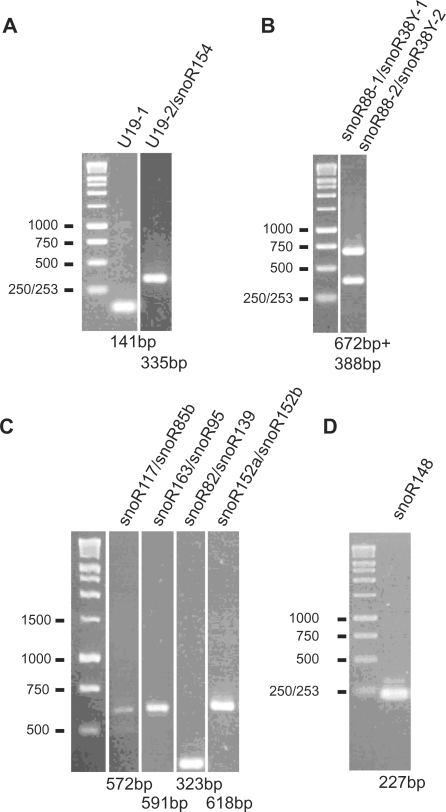
Many of the novel uncapped snoRNAs were part of polycistronic gene clusters (for example—Figure 2A and B). To confirm that the new and updated clusters are indeed transcribed as polycistronic pre-snoRNAs, we performed RT–PCR with primers to genes in various clusters using total RNA from Arabidopsis cell culture cells. Polycistronic precursor snoRNA transcripts of the expected sizes were detected for all clusters tested (Figure 5A–C and Supplementary Figure S7). In addition, four of the novel genes and some variants appeared to be single genes and expression of snoR148 and U19-1 (intronic) was also confirmed by RT–PCR (Figure 5A and D).
Figure 5.
RT–PCR expression analysis of novel and extended polycistronic snoRNA gene clusters. (A–C) RT–PCR of polycistronic snoRNA clusters. Primers were positioned in the genes indicated above the lanes (see also Supplementary Figures S1 and S2) and the expected RT–PCR product sizes are indicated below the lanes and are consistent with size markers. Some primers pairs were able to amplify more than one related precursor RNA (B). (A and D) Transcripts from single genes were amplified using primers to the 5′ and 3′ ends of the coding sequences.
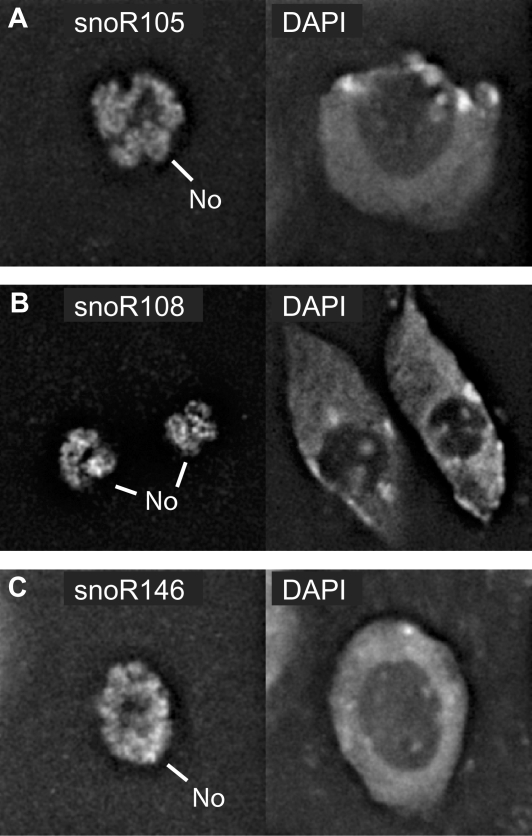
Sub-cellular localization of novel snoRNAs and U13 orthologues
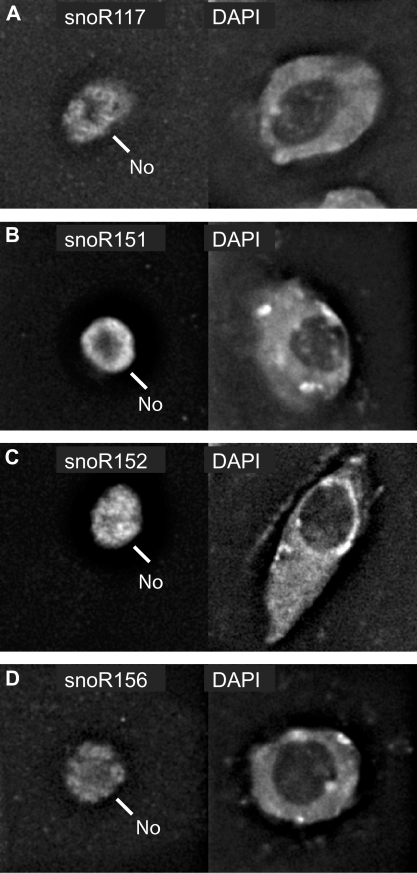
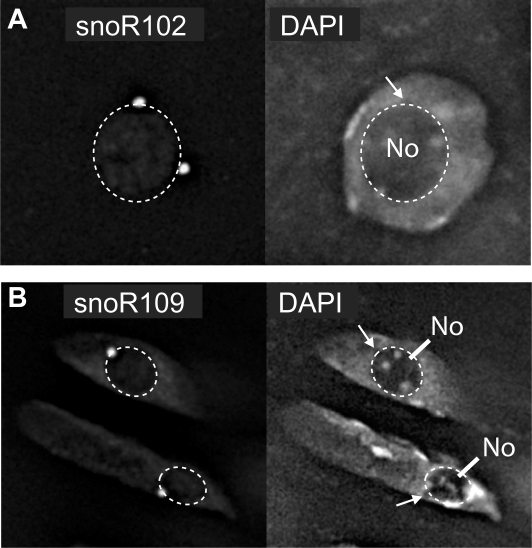
SnoRNAs are expected to be nucleolar. To examine the sub-cellular localization of the new box C/D and H/ACA snoRNAs identified here, antisense RNA probes were generated from some of the cDNA clones and hybridized to Arabidopsis Col-0 cell culture cells. The novel box C/D snoRNA (snoR117) and H/ACA snoRNAs (snoR151, snoR152 and snoR156) localized to the nucleolar region in the nuclei (Figure 6A–D). Similarly, the U13 orthologues also localized to the nucleolus (Figure 7A–C). Finally, to date, few Arabidopsis scaRNAs have been described (21,35) but the subcellular location of plant scaRNAs has not been addressed previously. Therefore, we determined the sub-cellular localization of the scaRNA, snoR102, which was cloned from the capped library. In contrast to the various novel box C/D and H/ACA snoRNAs, snoR102 did not localize to the whole nucleolar area but instead to two intensely labelled foci in and on the periphery of the nucleolus, perfectly consistent with CB labelling (Figure 8A). We also analysed the localization of the orphan snoRNA, snoR109 (21) which had again been cloned in the capped library. snoR109 localized to a single intense spot near the nucleolar periphery (Figure 8B). The labelling of snoR102 and snoR109 were therefore very similar and given that these cells usually show between 1 and 3 CBs, snoR109 may represent a novel scaRNA.
Figure 6.
(A–D) Nucleolar localization of novel snoRNAs. In situ hybridizations with antisense probes of four novel snoRNAs labelled the nucleolus (left panel) compared to nuclear staining with DAPI (right panel). No: nucleolus.
Figure 7.
Nucleolar localization of U13 snoRNAs. In situ hybridizations with antisense probes of the U13 snoRNA variants. (A) snoR105, (B) snoR108 and (C) snoR146 labelled the nucleolus (left panel) compared to nuclear staining with DAPI (right panel). No: nucleolus.
Figure 8.
Subnuclear localization of the scaRNA, snoR102 and orphan snoRNA, snoR109. In situ hybridizations with antisense probes of (A) snoR102 and (B) snoR109 labelled bright foci on the periphery of the nucleolus (left panel) when compared to nuclear staining with DAPI (right panel). Darker regions in the DAPI stained nuclei which coincide with the labelled sites are indicated by white arrows. No: nucleolus.
DISCUSSION
The eukaryotic nucleolus is involved in many aspects of RNA biogenesis and metabolism. The use of isolated plant nucleoli has led to the demonstration that mRNAs are present in nucleolar RNA and aberrantly spliced mRNAs are enriched in nucleoli (39). This is consistent with the detection of exon-junction complex (EJC) proteins in the nucleolus and the dynamic redistribution of a core EJC protein (eIF4A-III) under different growth conditions (38,50). By using isolated nucleoli and sequencing cDNA libraries generated from nucleolar RNA, we have generated full-length snoRNA and scaRNA sequences and identified novel box C/D and H/ACA snoRNA genes including U13 snoRNAs. From this and other studies, a total of 188 different snoRNA/scaRNA genes and 294 snoRNA/scaRNA gene variants are found in Arabidopsis. We provide direct evidence of expression for 40% of these genes/variants by cloning of cDNAs, northern analysis, in situ hybridization or RT–PCR. We have also identified novel orphan snoRNAs raising the possibility of wider functions in other aspects of RNA metabolism or gene regulation.
The previous RNomic study (21) identified many Arabidopsis snoRNA sequences from total seedling RNA and, in particular, significantly added to our knowledge of box H/ACA snoRNAs which are difficult to detect computationally. This study also identified the first plant scaRNAs (snoR101–104) and eight orphan snoRNAs (snoR105–112) as well as many ncRNAs of unknown function. Many of the sequences from Marker et al. (21) were short, partial sequences and in this study we have isolated corresponding full-length clones allowing the mature snoRNA and scaRNA sequences to be defined, and to characterize 11 unknown sequences as snoRNAs (see Supplementary Table S1). Furthermore, by analysing the sequences of the orphan snoRNAs, snoR105–112 (21), snoR111 and snoR112 have putative pseudouridylation sites in 25S rRNA and U6snRNA and could be re-classified as a box H/ACA snoRNA and a new scaRNA, respectively. Similarly, in situ hybridization of snoR102 (scaRNA) and snoR109 strongly suggests that snoR109 is also a new scaRNA.
Three U13 snoRNAs were isolated multiple times from the capped cDNA library. Of these snoR105 and snoR108 were isolated previously as 106 and 110 bp cDNAs (21). The full-length sequences obtained here for snoR105, snoR108 and snoR146 showed these genes to be ∼130 bp long and to be related by virtue of their mode of expression and conserved sequences—particularly in the 30 bp 5′ extension from the C box (Figure 3A). The U13 snoRNAs had two regions of complementarity to the 3′-end of 18S rRNA and human U13 may function in the 3′ cleavage of the 18S rRNA although its excat function remains to be elucidated (51). The three U13 genes have promoter elements (USE and TATA boxes) upstream of their coding sequences normally found in spliceosomal snRNA genes. This gene organization has only been found to date in U3 and MRP snoRNAs which are known to be capped and are required for cleavage of pre-rRNAs. Orthologues of U13 genes are present in many other plant species although there is extensive sequence variation outside of the rRNA-interacting sequences, as seen in snoR105, snoR108 and snoR146.
Orphan snoRNAs have been found in many eukaryotic organisms and recent data suggests that they may target other RNAs such as mRNAs or act as precursor molecules for the production of small regulatory RNAs (miRNAs or siRNAs) (28–31). Here, we have identified eight putative orphan snoRNAs. Our analysis of the orphan snoRNAs identified by Marker et al. (21) suggested that snoR109 and snoR112 are likely to be scaRNAs; and snoR111, a box H/ACA snoRNA. We were unable to find rRNA/snRNA targets for snoR106, snoR107 and snoR110 which therefore remain orphan snoRNA candidates. Some of the Arabidopsis orphan snoRNAs have orthologues in other plant species suggesting that they are bona fide RNA species—for example, snoR28 (18), snoR110 (21), and snoR133, snoR145, snoR149, snoR158, snoR159, snoR163 and snoR164 (this study) (Table 2). Of the 13 currently predicted orphan snoRNAs in Arabidopsis (Table 2), snoR6 (18) and snoR157 were predicted genes due to their sequence containing features of box C/D or H/ACA snoRNAs and their position as part of polycistronic gene clusters but as yet do not have orthologues in other species or any evidence of expression. These may therefore be genes or pseudogenes which have accumulated mutations and lost their ability to interact with target RNAs or generate stable snoRNPs. Mutations resulting in the gradual loss of functional genes is reflected by the presence of gene fragments observed when comparing related polycistronic clusters and accumulation of mutations resulting in loss of complementary sequences and even the evolution of new snoRNAs has also been observed (18,34). Nevertheless, Arabidopsis and other plant species contain orphan snoRNAs which may function in different RNA metabolism pathways to affect gene regulation and ultimately plant growth and development. Orphan snoRNAs in other organisms have novel functions in mRNA alternative splicing or can be processed to snoRNA-derived small RNAs or miRNAs by RNA silencing machinery—it will be particularly interesting to elucidate the function(s) of plant orphan snoRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council (BBSRC) [BBS/B/13519] and [BB/G024979/1]; the Scottish Government Rural and Environment Research and Analysis Directorate (RERAD) [SCR/909/03]; the BioGreen 21 Program of the Rural Administration of the Republic of Korea [20080401034051]; and the EU FP6 Programme Network of Excellence on Alternative Splicing (EURASNET) [LSHG-CT-2005-518238]. Funding for open access charges: Biotechnology and Biological Sciences Research Council.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kiss T. Small nucleolar RNAs: an abundant group of non-coding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 2.Filipowicz W, Pogačić V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 3.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 4.McKeegan KS, Debieux CM, Boulon S, Bertrand E, Watkins NJ. A dynamic scaffold of pre-snoRNP factors facilitates human box C/D snoRNP assembly. Mol. Cell. Biol. 2007;19:6782–6793. doi: 10.1128/MCB.01097-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2’-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richard P, Darzacq X, Bertrand E, Jady BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J. 2003;22:1–11. doi: 10.1093/emboj/cdg394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tycowski KT, Aab A, Stietz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr. Biol. 2004;14:1985–1995. doi: 10.1016/j.cub.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- 10.Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;7:344–351. doi: 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 11.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase centre. Mol. Cell. 2003;11:425–435. doi: 10.1016/s1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 12.Badis G, Fromont-Racine M, Jacquier A. A snoRNA that guides the two most conserved pseudouridine modifications within rRNA confers a growth advantage in yeast. RNA. 2003;9:771–779. doi: 10.1261/rna.5240503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu B, Fournier MJ. Interference probing of rRNA with snoRNPs: a novel approach for functional mapping of RNA in vivo. RNA. 2004;10:1130–1141. doi: 10.1261/rna.7190104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B, Liang X.-H, Piekna-Przybylska D, Liu Q, Fournier MJ. Mis-targeted methylation in rRNA can severely impair ribosome synthesis and activity. RNA Biol. 2008;5:249–254. doi: 10.4161/rna.6916. [DOI] [PubMed] [Google Scholar]
- 15.Cavaillé J, Butting K, Kiefmann M, lalande M, Brannen CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavaillé J, Vitali P, Basyuk E, Hüttenhofer A, Bachellerie J.-P. A novel brain-specific box C/D small nucleolar RNA processed from tandemly repeated introns of a noncoding RNA gene in rats. J. Biol. Chem. 2001;276:26374–26383. doi: 10.1074/jbc.M103544200. [DOI] [PubMed] [Google Scholar]
- 17.Barneche F, Gaspin C, Guyot R, Echeverria M. Identification of 66 box C/D snoRNAs in Arabidopsis thaliana: extensive gene duplications generated multiple isoforms predicting new ribosomal RNA 2′-O-methylation sites. J. Mol. Biol. 2001;311:57–73. doi: 10.1006/jmbi.2001.4851. [DOI] [PubMed] [Google Scholar]
- 18.Brown JWS, Clark GP, Simpson CG, Leader DJ, Lowe TM. Multiple snoRNA gene clusters from Arabidopsis. RNA. 2001;7:5718–5732. [PMC free article] [PubMed] [Google Scholar]
- 19.Hüttenhofer A, Kiefmann M, Meier-Ewart S, O’Brien J, Lehrach H, Bachellerie JP, Brosius J. RNomics: an experimental approach that identifes 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J. 2001;20:2943–2953. doi: 10.1093/emboj/20.11.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie. 2002;84:775–790. doi: 10.1016/s0300-9084(02)01402-5. [DOI] [PubMed] [Google Scholar]
- 21.Marker C, Zemann A, Terhorst T, Kiefmann M, Kastenmayer JP, Green P, Bachellerie JP, Brosius J, Huttenhofer A. Experimental Rnomics: Identification of 140 candidates for small non-messenger RNAs in the plant Arabidopsis thaliana. Curr. Biol. 2002;12:2002–2013. doi: 10.1016/s0960-9822(02)01304-0. [DOI] [PubMed] [Google Scholar]
- 22.Chen CL, Liang D, Zhou H, Zhou M, Chen YQ, Qu LH. The high diversity of snoRNAs in plants: identification and comparative study of 120 snoRNA genes from Oryza sativa. Nucleic Acids Res. 2003;31:2610–2613. doi: 10.1093/nar/gkg373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen C.-L, Chen C.-J, Vallon O, Huang Z.-P, Zhou H, Qu L.-H. Genome-wide analysis of box C/D and Box H/ACA snoRNAs in Chlamydomonas reinhardtii reveals extensive organisation into intronic gene clusters. Genetics. 2008;179:21–30. doi: 10.1534/genetics.107.086025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, Chen YQ, Qu LH. snoSeeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34:5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bazeley PS, Shepelev V, Talebizadeh Z, Butler MG, Fedorova L, Filatov V, Fedorov A. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene. 2008;408:172–179. doi: 10.1016/j.gene.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hertel J, Hofacker IL, Stadler PF. SnoReport: computational identification of snoRNAs with unknown targets. Bioinformatics. 2008;24:158–164. doi: 10.1093/bioinformatics/btm464. [DOI] [PubMed] [Google Scholar]
- 27.Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaille J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 29.Ender C, Krek A, Friedländer M, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathogens. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taft RJ, Glazov EA, Lassmann T, Hayashizake Y, Carnici P, Mattick JS. Small RNAs derived from snoRNAs. RNA. 2009;15:1233–1240. doi: 10.1261/rna.1528909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu LH, Meng Q, Zhou L, Chen Y.-Q. Identification of 10 novel snoRNA gene clusters from Arabidopsis thaliana. Nucleic Acids Res. 2001;29:1623–1630. doi: 10.1093/nar/29.7.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JWS, Echeverria M, Qu LH, Lowe TM, Bachellerie JP, Huttenhofer A, Kastenmayer JP, Green PJ, Shaw P, Marshall DF. Plant snoRNA database. Nucleic Acids Res. 2003b;31:432–435. doi: 10.1093/nar/gkg009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown JWS, Echeverria M, Qu LH. Plant snoRNAs: functional evolution and new modes of gene expression. Trends Plant Sci. 2003a;8:42–49. doi: 10.1016/s1360-1385(02)00007-9. [DOI] [PubMed] [Google Scholar]
- 35.Chen H.-M, Wu SH. Mining of small RNA sequencing data: a new approach to identify small nucleolar RNAs in Arabidopsis. Nucleic Acids Res. 2009;37:e69. doi: 10.1093/nar/gkp225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leader DJ, Clark GP, Watters JA, Beven AF, Shaw PJ, Brown J.WS. Clusters of multiple different small nucleolar RNA genes in plants are expressed as and processed from polycistronic pre-snoRNAs. EMBO J. 1997;16:5742–5751. doi: 10.1093/emboj/16.18.5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shaw PJ, Beven AF, Leader DJ, Brown JWS. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J. Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- 38.Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown J.WS, Shaw PJ. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol. Biol. Cell. 2005;16:260–269. doi: 10.1091/mbc.E04-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown J.WS. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell. 2009;21:2045–2057. doi: 10.1105/tpc.109.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuker M. MFOLD web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waterhouse AM, Procter JB, Martin D.MA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drea S, Leader DJ, Arnold BC, Shaw P, Dolan L, Doonan JH. Systematic spatial analysis of gene expression during wheat caryopsis development. Plant Cell. 2005;17:2172–2185. doi: 10.1105/tpc.105.034058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiss T, Marshallsay C, Filipowicz W. Alteration of the RNA polymerase specificity of U3 sn RNAs during evolution and in vitro. Cell. 1991;65:517–526. doi: 10.1016/0092-8674(91)90469-f. [DOI] [PubMed] [Google Scholar]
- 45.Kruszka K, Barneche F, Guyot R, Ailhas J, meneau I, Schiffer S, Marchfelder A, Echeverria M. Plany dicistronic tRNA-snoRNA genes: a new mode of expression of the small nucleolar RNAs processed by RNase Z. EMBO J. 2003;22:621–632. doi: 10.1093/emboj/cdg040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbezier N, Canino G, Rodor J, Jobet E, Saez-Vasquez J, marchfelder A, Echeverria M. Processing of a dicistronic tRNA-snoRNA precursor: combined analysis in vitro and in vivo revels alternate pathways and coupling to assembly of snoRNP. Plant Physiol. 2009;150:1598–1610. doi: 10.1104/pp.109.137968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watkins NJ, Dickmanns A, Lührmann R. Conserved stem II of the box C/D motif is essential for nucleolar localisation and is required, along with the 15.5K protein, for the hierarcichal assembly of the box C/D snoRNP. Mol. Cell Biol. 2002;22:8342–8352. doi: 10.1128/MCB.22.23.8342-8352.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachellerie JP, Michot B, Nicoloso M, Balakin A, Ni J, Fournier MJ. Anti-sense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem. Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 49.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koroleva OA, Calder G, Pendle AF, Kim SH, Lewandowska D, Brown J.WS, Shaw PJ. Dynamic behaviour of the eIF4A-III, putative core protein of Exon Junction Complex: fast relocation to nucleolus and splicing speckles under hypoxia. Plant Cell. 2009;21:1592–1606. doi: 10.1105/tpc.108.060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavaillé J, Hadjiolov AA, Bachellerie J.-P. processing of mammalian rRNA precursors at the 3’ end of 18S rRNA: identification of cis-acting signals suggests the involvement of U13 small nucleolar RNA. Eur. J. Biochem. 1996;242:206–213. doi: 10.1111/j.1432-1033.1996.0206r.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.