Abstract
We have studied the stimulation of topoisomerase IV (Topo IV) by the C-terminal AAA+ domain of FtsK. These two proteins combine to assure proper chromosome segregation in the cell. Stimulation of Topo IV activity was dependent on the chirality of the DNA substrate: FtsK stimulated decatenation of catenated DNA and relaxation of positively supercoiled [(+)ve sc] DNA, but inhibited relaxation of negatively supercoiled [(−)ve sc] DNA. The DNA translocation activity of FtsK was not required for stimulation, but was required for inhibition. DNA chirality did not affect any of the activities of FtsK, suggesting that FtsK possesses an inherent Topo IV stimulatory activity that is presumably mediated by protein–protein interactions, the stability of Topo IV on the DNA substrate dictated the effect observed. Inhibition occurs because FtsK can strip distributively acting topoisomerase off (−)ve scDNA, but not from either (+)ve scDNA or catenated DNA where the enzyme acts processively. Our analyses suggest that FtsK increases the efficiency of trapping of the transfer segment of DNA during the catalytic cycle of the topoisomerase.
INTRODUCTION
Accurate replication of DNA and faithful segregation of the newly replicated sister chromosomes to the daughter cells are required to ensure stable inheritance of the genetic material. In prokaryotic cells, the DNA replication machinery induces positive supercoiling ahead of the fork. These positive linkages can also diffuse behind the replication machinery to take the form of precatenanes (interwindings of the two partially replicated sister molecules) that are converted to catenanes (interwound, fully replicated sister DNA duplexes) at the end of the replication (1). Accumulation of positive supercoils blocks replication fork progression and catenanes prevent segregation of the genetic information if they are not removed (2). In Escherichia coli, two essential type II DNA topoisomerases, DNA gyrase and topoisomerase IV (Topo IV), manage DNA topology during replication. Gyrase introduces negative supercoils into the DNA to compensate for the accumulation of the positive supercoils ahead of the fork (3); whereas the primary role of Topo IV, which can contribute to replication fork progression by relaxing positive supercoils, is to unlink the catenated chromosomes (4–6).
Type II DNA topoisomerases transiently cleave both strands of a segment of a DNA duplex, termed the gate (G) segment and, in the presence of ATP, pass another segment [the transport (T) segment] of either the same (e.g. during superhelical DNA relaxation) or a different duplex (e.g. during decatenation) through the break followed by religation of the cleaved DNA (7). The enzyme efficiently removes replication catenanes and relaxes positive supercoils, but is considerably less active on negatively supercoiled [(−)ve sc] DNA (4,8,9). Topo IV is composed of a dimer of ParE, which contains the ATPase domain, and a dimer of ParC, which contains the DNA binding, DNA cleavage and religation domains (9). The ParC subunit includes a globular C-terminal domain (CTD) that adopts a unique structural fold termed a β-pinwheel (10,11). Truncation of the ParC CTD ablates the preference of the enzyme for replicative catenanes and positive supercoiled DNA (11).
Several proteins can modulate the activities of Topo IV. SeqA, which prevents overinitiation of chromosome replication, stimulates the relaxing and decatenating activities of Topo IV (12). MreB, an actin-like cytoskeletal element, modulates the decatenation activity of Topo IV in a manner dependent on its polymerization state (13). We have shown previously that the CTD of FtsK interacts with the ParC subunit of Topo IV and stimulates its decatenation activity in vitro (14). FtsK is a bifunctional protein that links cytokinesis and chromosome segregation via its N-terminus that is anchored in the cell division septum and its cytoplasmic CTD (FtsKC), a hexameric ATP-dependent DNA translocase that mobilizes chromosomal DNA trapped at mid-cell during cytokinesis (15). FtsKC is necessary for normal chromosome segregation in part, because it is required for XerCD-catalyzed resolution of chromosomal dimers at dif (16). This resolution reaction, by the action of FtsKC at XerCD-dif, can also serve to unlink replication catenanes if Topo IV activity is impaired (17). Then, the interaction between FtsK, Topo IV and the XerCD-dif recombination system may create a complex of enzymes stationed at mid-cell during cytokinesis that are necessary for the final topological disengagement of the daughter chromosomes and the management of errant segments of DNA, as well as improve the efficiency of these processes by increases in local concentration and enzymatic activity.
In this report, we extend our understanding of the mechanism by which FtsKC modulates Topo IV activity. We show that, in vitro, regulation of Topo IV activities by FtsKC depends on the DNA subsrate: Topo IV activity on positive supercoiled DNA or replicative catenanes is stimulated, whereas its activity on negative supercoiled DNA is inhibited. This modulation depends on the ParC CTD. We suggest that this differential activity relates to the different stability of Topo IV on DNA.
EXPERIMENTAL PROCEDURES
FtsK expression and purification
Purification of FLAG-FtsK50CK997A was as described by Aussel et al. (18) and Espeli et al. (14). BL21(λDE3)-pET11a-His-FtsK50C was grown at 37°C in 20 l of LB medium to OD600 = 0.2. The growth temperature was then reduced to 25°C. When the culture reached an OD600 = 0.4, IPTG was added to 0.6 mM and growth was continued for 3 h. Cells were harvested and resuspended in 100 ml of buffer A [50 mM Tris–HCl (pH 7.5 at 4°C), 1 mM DTT, 1 mM EDTA, 10% glycerol] containing 0.06% lysozyme, 150 mM NaCl and 20 mM EDTA. After 10 min of incubation at 0°C, triton X-100 was added to 1.0% and the cell suspension was incubated for 10 min at 25°C, followed by incubation for 10 min at 0°C. The soluble lysate was recovered after centrifugation at 100 000g for 1 h, diluted in buffer A to 50 mM NaCl and loaded onto a SP-Sepharose column (26 ml) equilibrated in buffer A+50 mM NaCl and 1% triton X-100. The column was washed with 75 ml buffer A+50 mM NaCl and 1% triton and the protein eluted using a linear gradient (260 ml) of 50–500 mM NaCl in buffer A+1% triton X-100. FtsK50CK997A eluted at 240 mM NaCl as scored by SDS–PAGE analysis. Peak fractions were pooled (Fraction 2, 32.5 ml, 88 mg protein). After dialysis against buffer B [50 mM Tris–HCl (pH 7.5 at 4°C), 300 mM NaCl, 10% glycerol, 0.8% triton X-100], Fraction 2 was applied to a TALON column (8 ml) that had been previously equilibrated in buffer B. The column was washed with 24 ml buffer B and protein was eluted using a linear gradient (80 ml) of 50–400 mM imidazole in buffer B. Peak fractions were pooled (Fraction 3, 4.5 ml, 4 mg protein). After dialysis against buffer A, Fraction 3 was applied to a Heparin agarose column (1 ml) that had been previously equilibrated with buffer A+50 mM NaCl and 1% triton X-100. The column was washed with 5 ml buffer A+50 mM NaCl and 1% triton X-100 and protein was eluted using a step gradient of 50–400 mM NaCl+1% triton X-100 in buffer A. Peak fractions were pooled (Fraction 4, 1 ml, 2.63 mg protein), dialyzed against storage buffer [Tris–HCl (pH 7.5 at 4°C), 150 mM NaCl, 1 mM EDTA, 1 mM DTT and 38% glycerol] and stored at −80°C.
Immunoblotting
Immunoblotting was performed as described in Espeli et al. (14) with some modifications. An anti-FLAG antibody conjugated to horseradish peroxidase (1: 250 dilution) in 1× PBS was incubated with the blot for 2 h at room temperature. The blot was washed four times quickly, once for 15 min and three times for 5 min each with 60 ml of 1× PBS, 2% milk, and then FtsK50C was detected using ECL western blotting detection reagents, as described by the manufacturer (Amersham Bioscience).
Decatenation of multiply linked DNA dimers
Multiply linked, form II : form II DNA dimers were prepared as described by Marians (19). Assays were performed as described in Espeli et al. (14).
Superhelical DNA relaxation
Positively supercoiled [(+)ve sc] DNA was prepared following the protocol of McClendon et al. (20). Reverse gyrase was a gift of T. S. Hsieh (Duke University Medical Center). Topo IV was reconstituted by mixing ParE in 5% molar excess over either the wild-type or mutant ParC in their storage buffers and incubated at 0°C for 30 min. Reaction mixtures (20 µl) containing 50 mM HEPES–KOH (pH 8.0), 10 mM MgOAc, 10 mM DTT, 100 µg/ml BSA, 20 mM KCl, 2 mM ATP, 5 nM supercoiled plasmid DNA and the indicated amounts of Topo IV and FtsK50C were incubated at 37°C for 10 min. EDTA and NaCl were then added to 22 and 330 mM, respectively, and the incubation continued for 3 min. One-sixth volume of a loading dye mixture was added and the DNA products were analyzed by electrophoresis through 1% agarose gels for 16 h at 22 V using 50 mM Tris–HCl (pH 7.9 at 23°C), 40 mM NaOAc and 1 mM EDTA as the electrophoresis buffer. The gels were stained with ethidium bromide and the images were recorded using a Kodak imaging system.
Assay for constrained supercoils
The indicated amount of FtsK50C was incubated with 5 nM partially relaxed (+)ve scDNA for 30 min at 37°C in 20 µl of 50 mM Tris–HCl (pH 7.5 at 25°C), 10 mM MgCl2, 10 mM DTT, 50 mM KCl, 100 µg/ml BSA and 2 mM AMP-PNP. Vaccinia virus Topo I (0.5 pg; the gift of S. Shuman, Memorial Sloan-Kettering Cancer Center) was then added to the reaction mixture and the incubation continued for 30 min at 37°C. SDS and proteinase K were then added to 1.0% and 1 mg/ml, respectively, and the incubation continued for 30 min at 37°C. One-sixth volume of a loading dye mixture was then added and the DNA products were analyzed by electrophoresis as described above.
DNA cleavage assay
Reaction mixtures (20 µl) containing 50 mM HEPES–KOH (pH 8.0), 10 mM MgOAc, 10 mM DTT, 100 µg/ml BSA, 20 mM KCl, 2 mM AMP-PNP, 5 nM (+)ve sc plasmid DNA, 4 nM Topo IV and either 60 nM FtsK50C or FtsK50CK997A were incubated 10 min at 37°C. SDS was then added to 1.0% and incubation continued for 4 min. EDTA and proteinase K were then added to 23 mM and 0.9 mg/ml, respectively, and the incubation continued for an additional 15 min. One-sixth volume of a loading dye mixture was then added and the DNA products were analyzed by electrophoresis as described above.
ATPase assay
Reaction mixtures (20 µl) containing 50 mM HEPES–KOH (pH 8.0), 10 mM MgOAc, 10 mM DTT, 100 µg/ml BSA, 20 mM KCl, 5 nM supercoiled plasmid DNA, 320 nM [α-32P]ATP, 1 mM ATP and 10 nM FtsK50C were incubated for the indicated time at 37°C. When testing Topo IV activity, 4 nM of (+)ve scDNA, 50 nM [α-32P]ATP, 2 mM ATP, 60 nM of FtsK50CK997A and the indicated amount of Topo IV were incubated for 10 min at 37°C. Reactions were stopped by adding ADP and cold ATP, 3 mM each. The ratio of ATP to ADP was analyzed by ascending thin layer chromatography on PEI plates, developed with a mixture of 0.5 M LiCl and 1 M HCOOH.
ParC (ΔCTD) purification
BL21 (λDE3)-pET11a-parCΔ483-752 was grown at 37°C in 20 l of LB medium to OD600 = 0.2. The growth temperature was then reduced to 25°C. When the culture reached an OD600 = 0.4, IPTG was added to 0.6 mM and growth was continued for 3 h. Cells were harvested and resuspended in 100 ml of buffer C [50 mM Tris–HCl (pH 7.5 at 4°C), 5 mM DTT, 1 mM EDTA, 10% glycerol] containing 0.06% lysozyme, 150 mM NaCl, 20 mM EDTA and 0.5% triton X-100. After 20 min of incubation at 0°C, the soluble lysate was recovered after centrifugation at 100 000g for 1 h, and loaded onto a Q-Sepharose column (42 ml) equilibrated in buffer C+50 mM NaCl. The column was washed with 210 ml buffer C+50 mM NaCl and protein was eluted using a linear gradient (420 ml) of 50–600 mM NaCl in buffer C. Peak fractions were pooled (Fraction 2, 35 ml, 157 mg protein), diluted to 50 mM NaCl with buffer C and applied to a Heparin agarose column (17 ml) equilibrated in buffer C+50 mM NaCl. The column was washed with 51 ml buffer C+50 mM NaCl and protein was eluted using a linear gradient (170 ml) of 50–500 mM NaCl in buffer C. Peak fractions were pooled (Fraction 3, 37.5 ml, 50 mg protein), adjusted to 1 M NaCl and applied to a 10 ml hydroxylapatite: cellulose (60 : 17) column equilibrated in buffer C+1 M NaCl. The column was washed with 30 ml buffer C+1 M NaCl and protein was eluted using a linear gradient (100 ml) of 0–600 mM (NH4)2SO4 in buffer C+1 M NaCl. Peak fractions were pooled (Fraction 4, 6 ml, 17 mg protein) and dialyzed against storage buffer [50 mM Tris–HCl (pH 7.5 at 4°C), 150 mM NaCl, 1 mM EDTA, 5 mM DTT, 40% glycerol].
RESULTS
FtsK stimulates Topo IV-catalyzed relaxation of (+)ve scDNA
Previously, we had observed stimulation of the decatenation activity of Topo IV, using a purified FtsK CTD (Domain 3) tagged with the FLAG epitope at the N-terminus (FtsKC) (14). Because this protein lacked amino acids 179–230, which are required for hexamerization (18), high concentrations were necessary to achieve oligomerization and stimulation of Topo IV. In this report, we have used a version of FtsK that includes amino acids 179–230 linked to the N-terminus of Domain 3 that is also tagged with the FLAG epitope (FtsK50C). Although the amino acids 179–230 are necessary to promote the assembly of the hexameric ring structure on the DNA, they can also cause aggregation of the protein. However, our protein preparation was subjected to analytical gel filtration and no protein aggregates were observed (data not shown). As we have shown for FtsKC (14), FtsK50C also interacts with the ParC subunit of Topo IV (Figure 1A).
Figure 1.
FtsK50C stimulates both Topo IV-catalyzed decatenation and (+)ve scDNA relaxation. (A) FtsK50C interacts with Topo IV. Two and one half pmol of FtsK50C protein and 180 pmol of the indicated proteins were spotted on a nitrocellulose membrane. Immunoblotting was then performed as in Espeli et al. (14). FtsK50C was visualized by ECL western analysis using an anti-FLAG–HRP conjugate antibody. (B and C) Reaction mixtures containing either no Topo IV or 0.05 nM Topo IV, the indicated amounts of FtsK50C, and either form II : form II DNA dimers (B) or (+)ve scDNA (C) were incubated and analyzed as described under ‘Experimental Procedures’ section. Addition of either 15 or 60 nM of FtsK50C stimulated decatenation by 2.2-fold (B) and relaxation of (+)ve scDNA by 3- and 3.3-fold, respectively (C). F I, form I DNA; F II, form II DNA.
FtsK50C was active in stimulating Topo IV decatenation of multiply linked DNA dimers at concentrations one-eightieth of that required for FtsKC (Figure 1B). These DNA dimers are purified from oriC plasmid DNA replication reactions in vitro, where the only topoisomerase present is DNA gyrase, which is very inefficient at unlinking replication catenanes. The dimers are two sister form II DNA molecules linked up to 35 times. Each rung on the ladder observed in Figure 1B represents a difference in the intermolecular linking number (Lki) of 1; more highly linked dimers have a greater Rf. As Topo IV unlinks the catenanes, the Lki distribution shifts to lower values, with the final product being the completely unlinked form II molecules.
Although the main role of Topo IV in the cell is to remove any links between replicated chromosomes, it can also relax (+)ve supercoils generated during replication fork progression and can replace gyrase completely in vitro and partially in vivo (21,22). We therefore asked whether stimulation of Topo IV by FtsKC extended to relaxation of (+)ve scDNA. This substrate was prepared by treating (−)ve scDNA with reverse gyrase, the only topoisomerase known that can introduce positive superhelical twists into DNA (23). Reaction mixtures containing Topo IV, FtsK50C and (+)ve scDNA were incubated for 10 min and quenched with EDTA and NaCl. The DNA products were separated by agarose gel electrophoresis (Figure 1C). Only 0.05 nM Topo IV was required to establish an observable (+)ve scDNA relaxation reaction. As in the decatenation reaction, FtsK50C stimulated Topo IV-catalyzed (+)ve scDNA relaxation.
Neither DNA translocation nor DNA topology modification by FtsK is responsible for stimulation of Topo IV activity
FtsK is an exceptionally fast DNA translocase [∼6 kb/s; (24,25)]. Positive supercoils accumulate ahead of the translocating enzyme, whereas negative supercoils accumulate behind it (18). We have suggested previously that the observed stimulation of Topo IV activity might result from FtsK translocation, generating a preferred substrate in the vicinity of the topoisomerase (14). We reasoned that if this were the case, removing the motor activity of FtsK50C should abolish the observed stimulation. Because FtsK50C uses the energy of ATP hydrolysis to translocate along the DNA (18,25), we purified a Walker A (K997A) mutant of FtsK50C tagged with six His residues at the N-terminus. This variant FtsK has no detectable ATPase activity (Figure 2A) and thus, based on previous studies (18,25), cannot translocate on DNA. Surprisingly, both the decatenation and (+)ve scDNA relaxation activities of Topo IV were stimulated by the presence of FtsK50CK997A to the same extent as by the wild-type protein (Figure 2B and C). We conclude that the observed stimulation is likely to be independent of FtsK translocation.
Figure 2.
Stimulation of Topo IV activity is not a result of either DNA translocation or alteration of DNA topology by FtsK50C. (A) The FtsK50C K997A variant has no detectable ATPase activity. Reaction mixtures containing [α-32P]ATP, 5 nM (−)ve sc DNA, the indicated amount of FtsK50C or FtsK50C K997A were incubated for 3 min at 37°C and analyzed as described under ‘Experimental Procedures’ section. The results of three independent experiments were averaged. (B and C) An ATPase mutant of FtsK50C stimulates both Topo IV-catalyzed decatenation and (+)ve scDNA relaxation. Reaction mixtures containing either no Topo IV or 0.05 nM Topo IV, the indicated amounts of FtsK50CK997A, and either form II : form II DNA dimers (B) or (+)ve scDNA (C) were incubated and analyzed as described under ‘Experimental Procedures’ section. Addition of 3.8, 7.5, 15 and 60 nM FtsK50CK997A stimulated decatenation by 2.5-, 2.8-, 3.5- and 3.2-fold, respectively (B). Addition of 15 and 60 nM of FtsK50CK997A stimulated relaxation of (+)ve scDNA by 3- and 2.4-fold, respectively (C). (D) Neither the binding of FtsK50C nor of FtsK50CK997A affects either the writhe or the twist of DNA. Reaction mixtures containing either no or the indicated amounts of either FtsK50C or FtsK50CK997A were incubated with relaxed (+)ve scDNA in the presence of AMP-PNP for 30 min as described under ‘Experimental Procedures’ section. Vaccinia virus Topo I was then added to remove unconstrained supercoils. DNA products were analyzed as described under ‘Experimental Procedures’ section. The DNA substrate is shown in the left-most lane of the gel.
Due to the fact that the concentration of FtsK50C in the reaction mixtures is in considerable excess compared with that of the DNA, we addressed the possibility that the binding of FtsK50C to the DNA was effecting a change in topology that either stimulated Topo IV activity or accounted for the observed stimulation directly. We used vaccinia virus topoisomerase as a probe to assess whether the binding of FtsK50C to DNA caused any alterations in topology. The vaccinia virus enzyme is a type IB topoisomerase that relaxes both negative and positive supercoils. After binding of a protein to the DNA, treatment with vaccinia virus Topo I will relax all supercoils in the portion of the molecule that is not constrained by bound protein; alterations in DNA topology induced by bound protein remain and are subsequently detectable by agarose gel electrophoresis (26). FtsK50C and FtsK50CK997A were bound to partially relaxed (+)ve scDNA in the presence of the non-hydrolyzable ATP analogue AMP-PNP (to prevent translocation), and the protein–DNA complex was then treated with vaccinia virus Topo I (Figure 2D). The DNA topology was the same in the absence or presence of FtsK50C. Under the conditions used, these FtsK variants bound DNA well (Supplementary Figure 1); we therefore conclude that FtsK50C does not constrain the DNA.
FtsK does not affect the DNA cleavage–religation equilibrium of Topo IV
The catalytic mechanism of type II DNA topoisomerases can be separated into distinct steps: binding of the G segment, binding/hydrolysis of ATP and trapping of the T segment, transient cleavage of the G segment and passage of the T segment through the break, resealing of the G segment and exit of the T segment via the protein gate (27). We reasoned that the stimulation of Topo IV activities could be the result of a stimulation of one of these steps by the presence of FtsK.
To determine whether the presence of FtsK50C affected the Topo IV cleavage–religation equilibrium, we analyzed the effect of FtsK50C on the ability of the Topo IV to bind DNA and make a double-strand DNA break. Topo IV was incubated with DNA and AMP-PNP in either the presence or absence of FtsK50C, SDS was added to denature cleavable complexes, the DNA products were treated with proteinase K and then analyzed by gel electrophoresis (Figure 3B). Neither FtsK50C nor FtsK50CK997A affected Topo IV-mediated DNA cleavage.
Figure 3.
Effect of FtsK50C on the catalytic mechanism of Topo IV. (A) Reaction mixtures containing either no NTP or 2 mM AMP-PNP, 7.5 nM Topo IV and 5 nM (+)ve scDNA were incubated and analyzed as described under ‘Experimental Procedures’ section. The fraction of DNA cleaved is presented (average of three independent experiments). (B) FtsK does not alter the DNA cleavage–religation equilibrium of Topo IV. Reaction mixtures containing 2 mM AMP-PNP, 4 nM Topo IV, 5 nM (+)ve scDNA and 60 nM of either FtsK50C or FtsK50CK997A were incubated and analyzed as described under ‘Experimental Procedures’ section. The fraction of DNA cleaved is presented (the average of three independent experiments). (C) FtsK50CK997A does not affect the ATPase activity of Topo IV. Reaction mixtures containing the indicated amounts of Topo IV, 5 nM (+)ve sc DNA, [α-32P]ATP and either no FtsK50CK997A or 60 nM FtsK50CK997A were incubated and analyzed as described under ‘Experimental Procedures’ section. The results of three independent experiments were averaged.
Nucleotide binding by Drosophila melanogaster or yeast Topo II stimulates the cleavage of DNA (28–30). ATP binding or hydrolysis by Topo IV also shifts the DNA cleavage–religation equilibrium, increasing the extent of the cleavage [Figure 3A and (31)]. Increasing the ATPase activity of Topo IV should therefore result in an increase in DNA cleavage. As FtsK50C did not affect DNA cleavage by Topo IV, we predicted that FtsK50C would not affect the ATPase activity of Topo IV. As FtsK50C is itself a powerful ATPase (18), we could only test the effect of the Walker A FtsK50C variant on the ATPase activity of Topo IV. As expected, the rate of ATP hydrolyzed was unchanged in the presence of FtsK50CK997A mutant (Figure 3C). Thus, stimulation of Topo IV activity by FtsK50C is unlikely to be the result of a shift in the cleavage–religation equilibrium because of an increase in ATP turnover by Topo IV.
Stimulation of Topo IV by FtsK50C is dictated by the binding of the topoisomerase to the substrate
To explore the possibility that FtsK affects Topo IV activity in a manner dependent on the chirality of the crossings of the DNA, we determined the effect of FtsK50C on relaxation of (−)ve sc DNA. Topo IV is 10- to 20-fold more efficient at relaxing (+)ve sc than (−)ve sc (8,32). Thus, whereas only 0.05 nM Topo IV is required to establish (+)ve scDNA relaxation (Figure 1C), 2 nM Topo IV is required to relax (−)ve sc DNA (Figure 4A). Surprisingly, the presence of FtsK50C inhibited the (−)ve sc DNA relaxation activity of Topo IV (Figure 4A). Furthermore, unlike the stimulation of (+)ve sc relaxation, which did not require the translocation activity of FtsK50C, inhibition of (−)ve sc relaxation did require FtsK50C translocation; FtsK50CK997A had no effect (Figure 4B).
Figure 4.
Translocation of FtsK50C inhibits Topo IV-catalyzed relaxation of (−)ve sc DNA. Reaction mixtures containing either no Topo IV, 2 nM Topo IV, (−)ve sc DNA and the indicated amounts of either FtsK50C (A) or FtsK50CK997A (B) were incubated and analyzed as described under ‘Experimental Procedures’ section. Fifteen and 60 nM FtsK50C inhibited (−)ve sc DNA relaxation by 66 and 96%, respectively, whereas 15 and 60 nM FtsK50CK997A stimulated (−)ve sc DNA relaxation by 8 and 25%, respectively (B).
We considered that the differential effects of FtsK on (+)ve and (−)ve sc DNA relaxation derived from the possibility that FtsK did not translocate as well on (+)ve scDNA compared with (−)ve sc DNA. If this were the case, there should be an observable difference in FtsK50C ATPase activity, using the two differentially supercoiled DNA as effectors. FtsK50C ATPase assays were therefore performed at limiting concentration of FtsK50C and ATP using (+)ve and (−)ve sc DNA as effectors (Figure 5). The rate of ATP hydrolysis in the presence of the two DNA effectors was indistinguishable. These data suggest that the chirality-dependent difference in the effect of FtsK50C on Topo IV activity is probably a manifestation of differential modes of Topo IV binding to the DNA, affecting the probability of forming a productive stimulatory complex with FtsK50C, rather than being related to an effect of the topology differences on FtsK. The processivity of Topo IV is much greater with (+)ve scDNA than (−)ve sc DNA, which implies that Topo IV–(+)ve scDNA complexes persist for longer time than Topo IV–(−)ve sc DNA complexes (8,33).
Figure 5.
The ATPase activity of FtsK50C is insensitive to DNA chirality. Reaction mixtures containing [α-32P]ATP, either no FtsK50C or 10 nM of FtsK50C, and either (−)ve sc or (+)ve scDNA were incubated for the indicated times and analyzed as described under ‘Experimental Procedures’ section. The results of six independent experiments were averaged.
Much of the discrimination shown by Topo IV between DNA substrates can be attributed to the CTD of ParC. Removal of this region of ParC results in a slight overall reduction of the affinity of the variant Topo IV for all DNA substrates (binding to the G segment) and a dramatic loss of topological discrimination (binding to the T segment) (11). CTD-truncated (residues 1–482) ParC was purified. The reconstituted ParC(ΔCTD) Topo IV exhibited reduced activity compared with wild-type Topo IV and lost discrimination between DNAs of opposite chirality, as shown previously by Corbett et al. (11). Given that the activity of ParC(ΔCTD) Topo IV is independent of the substrate, we predicted that the FtsK50C derivatives would have equivalent effects on the relaxation of (+)ve and (−)ve sc DNA by Topo IV. This proved to be the case. FtsK50C now inhibited, rather than stimulated, ParC(ΔCTD) Topo IV relaxation of (+)ve scDNA (Figure 6A) while retaining its ability to inhibit relaxation of (−)ve sc DNA (Figure 6B). FtsK50CK997A no longer stimulated relaxation of (+)ve scDNA, but also had no effect on either the (+)ve or (−)ve sc DNA relaxation activity of ParC(ΔCTD) Topo IV (Figure 6C and D, respectively). The increased concentrations of ParC(ΔCTD) Topo IV required to establish relaxation of the DNA substrates suggest a roughly 80- to 120-fold decrease in activity. This observation is consonant with the demonstration by Stone et al. (33) that relaxation of positive supercoils by Topo IV has an inherent processivity of ∼80 strand passage events, as well as with the observation of Neuman et al. (33) that relaxation of negative supercoils is perfectly distributive. Because ParC(ΔCTD) Topo IV can no longer discriminate topology, its activity is expected to be distributive [this is apparent in the data of Corbett et al. (11)]. We suggest that because of this change in activity, ParC(ΔCTD) Topo IV can no longer establish a stimulatory complex with either FtsK50C or FtsK50CK997A and that this destabilized ParC(ΔCTD) Topo IV can be displaced by translocating wild-type FtsK50C.
Figure 6.
FtsK50C, but not FtsK50CK997A, inhibits the DNA relaxation activity of ParC(ΔCTD) Topo IV. Reaction mixtures containing the indicated amounts of ParC(ΔCTD)Topo IV, either FtsK50C or FtsK50CK997A, and either (+)ve scDNA (A and C, respectively) or (−)ve sc DNA (B and D, respectively) were incubated and analyzed as described under ‘Experimental Procedures’ section. ParC(ΔCTD) Topo IV, Topo IV reconstituted with wild-type ParE and ParC(ΔCTD).
DISCUSSION
We have investigated further the mechanism by which FtsK50C modulates Topo IV activities. This modulation was dependent on the DNA substrate. When Topo IV was bound to multiply linked DNA dimers and (+)ve scDNA, the CTD of FtsK stimulated the unlinking of DNA strands; however, when Topo IV was bound to (−)ve sc, FtsK50C inhibited DNA unlinking. Inhibition of Topo IV activity required the DNA translocation activity of FtsK50C, whereas stimulation of Topo IV activity did not, suggesting that the differential effects might be attributable to preferential substrate utilization by FtsK. However, the DNA translocation activity of FtsK50C was insensitive to the chirality of the DNA crossing segments, which is not the case for Topo IV activity.
It has been proposed that recognition by Topo IV of the intrinsic orientation of the T and G segments (chiral-sensing model) results in differential activity (32,36). As Topo IV relaxes (+)ve scDNA much faster than (−)ve sc DNA, it assumes that Topo IV preferentially binds the left-handed nodes formed by (+)ve scDNA (32,36). However, the differential regulation of Topo IV by FtsK50C cannot be explained only by the handedness of the DNA, because FtsK50C did not have the same effect on (−)ve sc DNA and catenated DNA, which both contain right-handed nodes (the intermolecular helical crossing in replicative catenanes are nominally right-handed, they arise by denaturation of right-handed duplex turns). However, recently Neuman et al. (33) have shown that the apparent chiral discrimination of Topo IV is a function of its processivity. Unlinking of right-hand crossings in single molecule experiments was shown to be perfectly distributive, whereas unlinking of left-hand crossings was previously shown to have an average processivity of ∼80 strand passage events (36). Topo IV is thus significantly more processive on (+)ve scDNAs compared with (−)ve scDNAs (8,32,33,36), implying increased stability of Topo IV on those substrates that support greater processivity. Similarly, decatenation of multiply linked DNA dimers by Topo IV is highly processive (5).
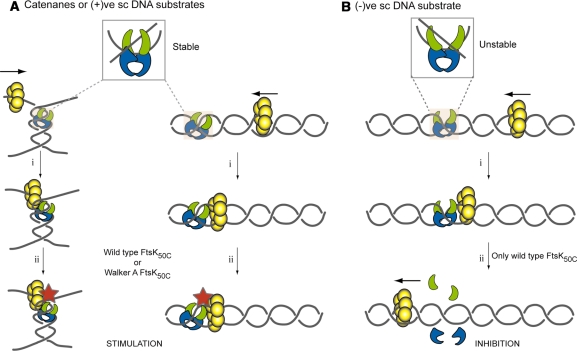
The presence of the CTD of ParC influences Topo IV substrate selectivity. There is some decrease in generalized DNA binding of ParC(ΔCTD) Topo IV compared with wild-type. This measurement is thought to probe affinity for the G segment of DNA (11); however, the difference in affinity is insufficient to account for the dramatic change in activity of the truncated enzyme compared with the wild-type, and the authors proposed that this implied that the CTD presumably bound the T segment of DNA. Thus, in the absence of the ParC CTD, preferential relaxation of DNA substrates by Topo IV is lost. Concomitant with the decreased activity of the ParC(ΔCTD) Topo IV, stimulation by FtsK50C was no longer observed, only inhibition was observed with DNA substrates of both chiralities. Thus, the observed differential effect of FtsK likely relates directly to the differential stability of Topo IV on DNA (Figure 7). Either FtsK50C or FtsK50CK997A could form a productive stimulatory complex with Topo IV on DNA substrates on which the latter enzyme acted processively, (+)ve scDNA and replication catenanes. As DNA translocation is not required for the stimulation, FtsK50CK997A can still stimulate Topo IV activity by virtue of binding to a DNA site adjacent to the topoisomerase, perhaps even being attracted to such a site by the affinity between the two enzymes (Figure 7A.i). Whereas with (−)ve sc DNA, where Topo IV acts distributively, a productive complex cannot be formed and translocating FtsK50C is able to displace the bound Topo IV (Figure 7B). Under these circumstances, FtsK50CK997A would be expected to have no effect, as we observed.
Figure 7.
Modulation of Topo IV activity by FtsK50C. Schematics describing the effect of FtsK50C on Topo IV action on either replication catenanes and (+)ve scDNA (A) or (−)ve sc DNA (B). Details are given in the text. The subunits of Topo IV are colored green (ParE) and blue (ParC). Hexamers of FtsK50C are colored yellow. A red star represents stimulation of Topo IV by FtsK50C.
Because supercoiling stimulates decatenation (19), we first thought that FtsKC might stimulate Topo IV by generating supercoiled DNA in a topological domain in the vicinity of Topo IV (14). We proposed that the interaction between these proteins might serve as an anchor of a topological domain, wherein FtsKC pumped supercoiled DNA toward Topo IV. However, the finding in the present study that an ATPase mutant of FtsK still stimulates the decatenation activity of Topo IV indicates that our previous model cannot be correct. This FtsK variant cannot pump the DNA and thus no supercoiled DNA can be created as a preferential substrate for Topo IV. Moreover, it is now known that the binding of FtsKC to DNA does not change the conformation of DNA (37) (and as we report herein). So, what is the molecular basis of the observed stimulation of Topo IV activity by FtsK50C?
In the type II topoisomerase catalytic cycle (34,35), the first step is the binding of the catalytic domain (ParC subunits in Topo IV) to the G segment. ATP-induced dimerization of the ATPase domains (ParE in Topo IV) captures the T segment. This capture leads to G segment cleavage and opening, to let the T segment pass through the G segment gap into the interior of the enzyme. The G segment is then religated after this strand passage event. We considered that FtsK50C might affect one of the first steps in this cycle. We could not test the effect of FtsK50C on Topo IV binding to the G segment as FtsK50C itself binds DNA. However, increasing the binding affinity of Topo IV for DNA should also increase the extent of the G segment cleavage. However, FtsK50C had no effect on the formation of covalent cleavable complexes in the presence of AMP-PNP. This result suggested us that FtsK50C did not affect the binding of Topo IV to DNA. Furthermore, FtsK50C did not affect the rate of Topo IV ATP hydrolysis. Therefore, to explain the increased gain in Topo IV activity in the presence of FtsK50C, we suggest an increased efficiency in the trapping of the T segment. During any one reaction cycle for Topo IV, the dimerization of ParE after binding of ATP does not always successfully trap a T segment. If the presence of FtsK50C results in more efficient trapping of the T segment, perhaps because of a slightly deformed path of the bound DNA, more productive catalytic cycles will result, generating the observed stimulation, yet without an effect on the rate of ATP hydrolysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health grant (GM34558).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol. Microbiol. 2004;52:925–931. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 3.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 4.Hiasa H, Marians KJ. Topoisomerase IV can support oriC DNA replication in vitro. J. Biol. Chem. 1994;269:16371–16375. [PubMed] [Google Scholar]
- 5.Hiasa H, Marians KJ. Two distinct modes of strand unlinking during theta-type DNA replication. J. Biol. Chem. 1996;271:21529–21535. doi: 10.1074/jbc.271.35.21529. [DOI] [PubMed] [Google Scholar]
- 6.Khodursky AB, Peter BJ, Schmid MB, DeRisi J, Botstein D, Brown PO, Cozzarelli NR. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc. Natl Acad. Sci. USA. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 8.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E.coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng H, Marians KJ. Escherichia coli topoisomerase IV. Purification, characterization, subunit structure, and subunit interactions. J. Biol. Chem. 1993;268:24481–24490. [PubMed] [Google Scholar]
- 10.Corbett KD, Berger JM. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 11.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 12.Kang S, Han JS, Park JH, Skarstad K, Hwang DS. SeqA protein stimulates the relaxing and decatenating activities of topoisomerase IV. J. Biol. Chem. 2003;278:48779–48785. doi: 10.1074/jbc.M308843200. [DOI] [PubMed] [Google Scholar]
- 13.Madabhushi R, Marians KJ. Actin homolog MreB affects chromosome segregation by regulating topoisomerase IV in Escherichia coli. Mol. Cell. 2009;33:171–180. doi: 10.1016/j.molcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espeli O, Lee C, Marians KJ. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 2003b;278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
- 15.Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 2007;64:1434–1441. doi: 10.1111/j.1365-2958.2007.05755.x. [DOI] [PubMed] [Google Scholar]
- 16.Lesterlin C, Barre FX, Cornet F. Genetic recombination and the cell cycle: what we have learned from chromosome dimers. Mol. Microbiol. 2004;54:1151–1160. doi: 10.1111/j.1365-2958.2004.04356.x. [DOI] [PubMed] [Google Scholar]
- 17.Grainge I, Bregu M, Vazquez M, Sivanathan V, Ip SC, Sherratt DJ. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26:4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108:195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- 19.Marians KJ. DNA gyrase-catalyzed decatenation of multiply linked DNA dimers. J. Biol. Chem. 1987;262:10362–10368. [PubMed] [Google Scholar]
- 20.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase II alpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 21.Peng H, Marians KJ. Decatenation activity of topoisomerase IV during oriC and pBR322 DNA replication in vitro. Proc. Natl Acad. Sci. USA. 1993;90:8571–8575. doi: 10.1073/pnas.90.18.8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 23.Forterre P, Bergerat A, Lopez-Garcia P. The unique DNA topology and DNA topoisomerases of hyperthermophilic archaea. FEMS Microbiol. Rev. 1996;18:237–248. doi: 10.1111/j.1574-6976.1996.tb00240.x. [DOI] [PubMed] [Google Scholar]
- 24.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–590. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 25.Saleh OA, Perals C, Barre FX, Allemand JF. Fast, DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23:2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kampranis SC, Bates AD, Maxwell A. A model for the mechanism of strand passage by DNA gyrase. Proc. Natl Acad. Sci. USA. 1999;96:8414–8419. doi: 10.1073/pnas.96.15.8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schoeffler AJ, Berger JM. Recent advances in understanding structure-function relationships in the type II topoisomerase mechanism. Biochem. Soc. Trans. 2005;33:1465–1470. doi: 10.1042/BST0331465. [DOI] [PubMed] [Google Scholar]
- 28.Lindsley JE, Wang JC. On the coupling between ATP usage and DNA transport by yeast DNA topoisomerase II. J. Biol. Chem. 1993;268:8096–8104. [PubMed] [Google Scholar]
- 29.Mueller-Planitz F, Herschlag D. Coupling between ATP binding and DNA cleavage by DNA topoisomerase II: a unifying kinetic and structural mechanism. J. Biol. Chem. 2008;283:17463–17476. doi: 10.1074/jbc.M710014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Osheroff N. Eukaryotic topoisomerase II. Characterization of enzyme turnover. J. Biol. Chem. 1986;261:9944–9950. [PubMed] [Google Scholar]
- 31.Nurse P, Bahng S, Mossessova E, Marians KJ. Mutational analysis of Escherichia coli topoisomerase IV. II. ATPase negative mutants of parE induce hyper-DNA cleavage. J. Biol. Chem. 2000;275:4104–4111. doi: 10.1074/jbc.275.6.4104. [DOI] [PubMed] [Google Scholar]
- 32.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roca J, Wang JC. DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell. 1994;77:609–616. doi: 10.1016/0092-8674(94)90222-4. [DOI] [PubMed] [Google Scholar]
- 36.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lowe J, Ellonen A, Allen MD, Atkinson C, Sherratt DJ, Grainge I. Molecular mechanism of sequence-directed DNA loading and translocation by FtsK. Mol. Cell. 2008;31:498–509. doi: 10.1016/j.molcel.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.