Abstract
Formation of eukaryotic ribosomes requires more than 150 biogenesis factors which transiently interact with the nascent ribosomal subunits. Previously, many pre-ribosomal intermediates could be distinguished by their protein composition and rRNA precursor (pre-rRNA) content. We purified complexes of ribosome biogenesis factors from yeast cells in which de novo synthesis of rRNA precursors was down-regulated by genetic means. We compared the protein composition of these largely pre-rRNA free assemblies with the one of analogous pre-ribosomal preparations by semi-quantitative mass spectrometry. The experimental setup minimizes the possibility that the analysed pre-rRNA free protein modules were derived from (partially) disrupted pre-ribosomal particles and provides thereby strong evidence for their pre-ribosome independent existence. In support of the validity of this approach (i) the predicted composition of the analysed protein modules was in agreement with previously described rRNA-free complexes and (ii) in most of the cases we could identify new candidate members of reported protein modules. An unexpected outcome of these analyses was that free large ribosomal subunits are associated with a specific set of ribosome biogenesis factors in cells where neo-production of nascent ribosomes was blocked. The data presented strengthen the idea that assembly of eukaryotic pre-ribosomal particles can result from transient association of distinct building blocks.
INTRODUCTION
Eukaryotic ribosomes are composed of four ribosomal RNAs (rRNA) and about 80 ribosomal proteins. In addition to these structural ribosomal components, ribosome biosynthesis requires more than 150 non-ribosomal proteins and many non-coding, small RNAs. Transcription of the DNA coding for rRNA (rDNA) by RNA polymerase I (Pol-I) results in an rRNA precursor (pre-rRNA) which is bound by ribosomal proteins, processed by several endo- and exonucleases and folded into its final conformation. Processing of pre-rRNAs and assembly steps go along with multiple changes in the set of ribosome biogenesis factors associated with pre-rRNAs. Therefore, different ribosomal precursors can be distinguished by their content of pre-rRNAs and associated factors (1–7).
Assembly of about 40 ribosome biogenesis factors, U3 snoRNA, pre-rRNA and ribosomal proteins seems to occur already during synthesis of the primary rRNA transcript and results in the formation of the small subunit (SSU) processome, also referred to as 90S pre-ribosome (8–10). In yeast, SSU-processome components are involved in early pre-rRNA cleavages at processing sites A0 and A1 to remove the 5′external transcribed spacer (5′ETS) and in the cut at A2 within the internal transcribed spacer (ITS1) (8), leading to the two maturation branches in which biogenesis of the SSU and LSU (large subunit) further proceeds (11). Only a few of the SSU-processome associated proteins, like Enp1p, do not dissociate from nuclear SSU pre-rRNA processed at sites A0, A1 and A2. Therefore, Enp1p associates with nuclear RNPs containing the 35S pre-rRNA and SSU processome components as well as with SSU precursors which are exported to the cytoplasm and contain 20S pre-rRNA and factors involved in late steps of SSU biogenesis (1,12). Among them are the serine kinase Rio2p, the putative endonuclease Nob1p and Ltv1p, all of which are required for removal of the 3′ region of 20S pre-rRNA leading to mature 18S rRNA containing SSUs (13–16).
Many of the more than 50 LSU biogenesis factors assemble transiently after cleavage of the ITS1 at site A2. A complex series of pre-rRNA processing events follows, in which interactions of many non-ribosomal proteins with the nascent pre-rRNA are required to generate finally the mature 5S, 5.8S and 25S rRNA containing LSU. First, 27SA2 pre-rRNA containing RNPs are formed. Then, after maturation of the 5′ end of 5.8S rRNA precursors at site B1 involving Nop7p (17), 27SB1 containing particles are generated (2). 27SB pre-rRNA containing complexes include factors which are probably directly involved in cleavage of 27SB pre-rRNA at site C2 in the internal transcribed spacer region 2 (ITS2) resulting in the separation of 5.8S and 25S pre-rRNAs. After cleavage in the ITS2 proteins involved in further nuclear trimming of 5.8 and 25S pre-rRNA, among them Rix1p (18), and proteins which accompany or guide the LSU through the nuclear pore associate with the LSU precursors. Finally, before entry into mRNA translating polysomes the nascent LSUs undergo cytoplasmic maturation steps which include the release of several biogenesis factors, e.g. Arx1p, Tif6p and Nmd3p (19–21).
Indications for the rather complex protein composition of different pre-ribosomal particles came mainly from mass spectrometry based identification of ribosome biogenesis factors which co-purified from yeast cell extracts on affinity matrices (1–4,7,8,22–24). One systematic approach was to apply the tandem affinity purification procedure (TAP) on TAP tagged ribosome biogenesis factors (25) in buffer conditions which are thought to preserve some, but not all aspects of RNP architecture. Some smaller pre-rRNA free subcomplexes of ribosome biogenesis factors could be enriched from yeast extracts in which pre-ribosomes were either depleted from cellular extracts by differential centrifugation (26) or in which they were further destabilized by high salt conditions (27,28), by treatment with phosphatase inhibitors (29) or by inactivation/depletion of ribosome biogenesis factors (3,29,30). Isolation of such pre-rRNA free ribosome biogenesis factor complexes gave important first insights into the modular architecture of eukaryotic pre-ribosomes. Whether submodules of ribosome biogenesis factors form or persist in vivo independent of pre-ribosomal particles, and associate with and subsequently act on pre-ribosomal particles as distinct units remained less clear.
Here, we describe a systematic approach to isolate ribosome assembly factor sub-complexes from pre-rRNA depleted yeast cells. Down-regulation of pre-rRNA synthesis was achieved shifting a mutant expressing defective Pol-I transcription factor Rrn3p to restrictive temperature. Using semi-quantitative mass spectrometry we compared the composition of protein assemblies co-purifying with tagged ribosome biogenesis factors in a one step affinity purification procedure from yeast cells with or without ongoing rRNA de novo synthesis. Our data support and extend the catalogue of protein submodules involved in ribosome biogenesis and they indicate that several of them are not only architectural entities of nascent ribosomes but also exist in the cell independent of pre-ribosomes. In addition, we provide evidence that Arx1p, and several factors believed to play a role in nucleo-cytoplasmic translocation and/or LSU pre-rRNA maturation associate with non-nascent LSUs and therefore might have an additional role in other aspects of the LSU.
MATERIALS AND METHODS
Yeast strains and microbiological procedures
Yeast strains used in this study are listed in Table 1.
Table 1.
Yeast strains used in this study
| Name | Genotype | TOY | Origin |
|---|---|---|---|
| CG379 | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52 | 543 | (31) |
| YCC95 | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8 | – | (31) |
| CG379 Noc1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, NOC1::NOC1-TAP-TRP1 | 576 | this article |
| YCC95 Noc1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8, NOC1::NOC1-TAP-TRP1 | – | this article |
| CG379 Nop7p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, NOP7::NOP7-TAP-TRP1 | 580 | this article |
| YCC95 Nop7p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8, NOP7::NOP7-TAP-TRP1 | 544 | this article |
| CG379 Rio2p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, RIO2::RIO2-TAP-TRP1 | 587 | this article |
| YCC95 Rio2p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8, RIO2::RIO2-TAP-TRP1 | 551 | this article |
| CG379 Rix1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, RIX1::RIX1-TAP-TRP1 | 583 | this article |
| YCC95 Rix1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8, RIX1::RIX1-TAP-TRP1 | 547 | this article |
| CG379 Arx1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, ARX1::ARX1-TAP-TRP1 | 584 | this article |
| YCC95 Arx1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8, ARX1::ARX1-TAP-TRP1 | 548 | this article |
| CG379 Enp1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, ENP1::ENP1-TAP-TRP1 | 588 | this article |
| YCC95 Enp1p-TAP | MATα, ade5, his7-2, leu2-112, trp1-289, ura3-52, rrn3-8, ENP1::ENP1-TAP-TRP1 | 552 | this article |
To construct strains CG379 and YCC95 (31) with endogenously TAP-tagged ribosome biogenesis factors (Arx1p, Enp1p, Noc1p, Nop7p, Rio2p and Rix1p) the TAP-TRP1-cassette on plasmid pBS1479 (54) was PCR-amplified using the respective primers given in Table 2. The purified PCR product was transformed into competent yeast cells (55). The correct genomic integration of the TAP-TRP1 cassette was verified by selection for tryptophan prototrophy and western blot analysis.
Table 2.
Oligonucleotides used in this study
| Name | Sequence | Purpose | Gene/Locus |
|---|---|---|---|
| 177 | ATCTGCCGACGATTATGCTCAATATTTAGATCA AGATTCAGACTCCATGGAAAAGAGAAG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | NOC1 |
| 178 | TAATTTACAACACCGAAGTGTTTAGTTAATGTA TTATTATTTTTACGACTCACTATAGGG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | NOC1 |
| 638 | GCCAAACAAAAAGCTAAACTGAATAAACTAGA TTCCAAGAAATCCATGGAAAAGAGAAG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | NOP7 |
| 639 | AGACAAAATTTTTGAGAGGCTATTGGAAAAGA AGAGAAAATACGACTCACTATAGGG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | NOP7 |
| – | AAATCTAAAAATGGATAAACTAGGAAACTATA TACTAGAGTCCATGGAAAAGAGAAG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | RIO2 |
| – | TTGATTATTTGCGGCCATTTATGCAGTCGTCTA AACTAAATACGACTCACTATAGGG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | RIO2 |
| 646 | CGAATTAAGTGATGACGAAGAGGAGGAGGAA GAAGGAGAATCCATGGAAAAGAGAAG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | RIX1 |
| 647 | TCTAGTCGAAATATAACCAAACAAAATCTGGTT GATATTATACGACTCACTATAGGG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | RIX1 |
| 648 | TGAGACATCAAATGGCGGAGTTGAAGAAACCA TGAAAATGTCCATGGAAAAGAGAAG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | ARX1 |
| 649 | TATATTATTTATATACTAGCTTTAGAAATGATG AAGTTTCTACGACTCACTATAGGG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | ARX1 |
| – | TCCACAGGAAGCTAATGATGATTTAATGATTG ATGTCAATTCCATGGAAAAGAGAAG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | ENP1 |
| – | CCGAGCGATATAAAATTGATGAAAAATTGATA TTACAGCATACGACTCACTATAGGG | Primer to obtain amplicon of pBS1479 for genomic integration of TAP-Tag | ENP1 |
| 205 | CATGGCTTAATCTTTGAGAC | 32P-labelled probe for northern blot hybridization (detection of 18S rRNA) | – |
| 212 | CTCCGCTTATTGATATGC | 32P-labelled probe for northern blot hybridization (detection of 25S rRNA) | – |
| 210 | GGCCAGCAATTTCAAGTTA | 32P-labelled probe for northern blot hybridization in ITS2 region (detection of 35/32S, 27SA2, 27SB, 7S pre-rRNAs) | – |
| 1819 | GTAAAAGCTCTCATGCTCTTGCC | 32P-labelled probe for northern blot hybridization in ITS1 region (detection of 35/32S, 23S, 20S pre-rRNAs) | – |
All used yeast strains were cultivated at 24°C in YPD (1% yeast extract, 2% bacto peptone, 2% glucose); temperature shifts were performed at starting OD600 of 0.5 for 3 h at 37°C.
Steady-state analysis of (pre-) rRNA
To analyse steady-state levels of different (pre-) rRNA species of yeast strains CG379 and YCC95 at permissive and non-permissive temperature, 1 ml of cell culture with OD600 of 1 was harvested at 24°C or after 3 h at 37°C. RNA was extracted by hot phenol–chloroform treatment (56), loaded on denaturing agarose gels and analysed by northern blot. Hybridization was performed in 50% formamide; 5 × SSC; 0,5% SDS; 5 × Denhards solution at 30°C with the following 32P-labelled probes: # 205 (18S): 5′-CATGGCTTAATCTTTGAGAC-3′; # 212 (25S): 5′-CTCCGCTTATTGATATGC-3′; # 210 (E-C2): 5′-GGCCAGCAATTTCAAGTTA-3′; # 1819 (D-A2): 5′-GTAAAAGCTCTCATGCTCTTGCC-3′. The blots were washed twice for 15 min with 2 × SSC at 30°C. Labelled rRNA signals were detected using a Phosphor Imager FLA3000 (Fujifilm). Data were quantified using MultiGauge V3.0 (Fujifilm).
Protein detection by western blot analysis
Expression levels of TAP-tagged biogenesis factors in yeast strains CG379 and YCC95 at 37°C were determined by western blot analysis. Same amounts of whole cell extracts, prepared by cell lysis using glass beads, were analysed using PAP visualization reagent (DakoCytomation, Z 0113) in a dilution of 1:3000 for detection of the TAP-Tag. For detection of rpS8 a rabbit anti-rpS8 antibody was diluted 1:2000 (G. Dieci). Protein signals were visualized by chemiluminescence using a Fluorescence Image Reader LAS3000 (Fujifilm). Data was quantified using MultiGauge V3.0 (Fujifilm).
Fluorescence microscopy
Temperature shifts of logarithmically growing yeast cells and in situ detection of tagged proteins were performed as described (35). Totally 0.1 × volume of 37% formaldehyde (methanol stabilized) was added to cultures and fixation of cells was performed for 1 h at 37°C. Spheroblasting of cells was done in 0.1 M potassium phosphate buffer pH 7.5 for 45 min at 30°C using 50 µg/ml zymolyase T100 (Seikagaku Corporation). Fixed spheroblasts were put on poly-l-lysine treated three-well diagnostic slides (Menzel–Glaeser), blocked with 2% BSA in 1xTBS/0,1%NP40 and treated with the following antibodies in 1 × TBS/0,1%BSA: rabbit anti-Protein A (Sigma, P-3775) in a dilution of 1:50 000 and mouse anti-Nop1p (Abcam, ab4575) in a dilution of 1:1000. For fluorescence detection the secondary antibodies Alexa Fluor 594 goat anti-rabbit (Molecular Probes, A-11 012) and Alexa Fluor 488 goat anti-mouse (Molecular Probes, A-11 017) were used in dilutions of 1:500 in 1 × TBS/0,1%BSA. DNA staining was achieved by subsequent treatment with 1 µg/ml DAPI in Moviol-solution [0.1 µg/ml Moviol (Hoechst) in 25% Glycerin/0.1 M Tris pH 8.5].
Images were captured with an AxioCam MR CCD camera mounted on an Axiovert 200M Zeiss microscope and processed with Axiovision V 4.7.1.0 and Adobe Photoshop.
Protein affinity purification experiments
Growth and temperature shifts of yeast strains (Table 1) were performed as indicated. Two to six litres of cell culture (400 ml culture for small scale RNA analysis) were harvested and whole cell extracts were prepared in 1 volume of lysis buffer (50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 0.15% NP-40, 1 mM PMSF, 2 mM benzamidine). An equal volume of glass beads (Ø 0.75–1 mm) was added and cells were lysed at 4°C in a bead mill (pulverisette 6/Fritsch, for small scale Vibrax/IKA). After centrifugation in a Ti45 rotor for 30′ with 35 000 rpm (small scale: 2 × 10′ in a table top at 14.000rpm) the cleared lysate was incubated with 300 µl (small scale: 50ul) of equilibrated IgG–Sepharose (Amersham) for 2 h at 4°C. Beads were washed five times with 2 ml and once with 10 ml of lysis buffer. Affinity purification experiments using Dynabeads® Pan Mouse IgG (Invitrogen, Cat. No.11 041) were performed essentially the same as with IgG-Sepharose, using buffer A200 (20 mM Tris–HCl, pH 8, 200 mM KCl, 5 mM MgAc, 1 mM DTT, 0.5% Triton X-100, 0.1% Tween 20).
RNA was extracted from beads and extracts as described (36). For immunological detection of Protein A tagged proteins samples from beads and extracts were taken and analysed by western blot using PAP visualization reagent (DakoCytomation, Z 0113) in a dilution of 1:3000.
For comparative quantifications of proteins contained in affinity purified complexes TAP-tagged proteins bound to beads were eluted using AcTEV™ Protease (Invitrogen) for 2 h at 16°C and precipitated with methanol/chloroform.
Comparative quantification of proteins purified with TAP–tagged biogenesis factors
Protein samples were lyophilized, resuspended in 20 µl dissolution buffer (iTRAQTM labelling kit, Invitrogen) and reduced with 5 mM Tris–(2-carboxyethyl)phosphine at 60°C for 1 h. Cysteins were blocked with 10 mM methyl-methanethiosulfonate (MMTS) at room temperature for 10 min as described previously (37,38). After trypsine digest for 20 h at 37°C, tryptic peptides of the purifications of interest were labelled with different combinations of the four iTRAQTM reagents according to the manufacturer (Invitrogen). Pairs of labelling reactions were combined and lyophilized.
The combined differently labelled peptides were dissolved for 2 h in 0.1%TFA and loaded on a nano-flow HPLC-system (Dionex) harbouring a C18-Pep-Mep column (LC-Packings). The peptides were separated by a gradient of 5% to 95% of buffer B (80% acetonitrile/0.05% TFA), fractions were mixed with five volumes of CHCA (alpha-cyano-4-hydroxy cinnamic acid; Sigma) matrix (2 mg/ml in 70% acetonitrile/0.1%TFA) and spotted on a MALDI-target. Spotted fractions were analysed by a 4700 series MALDI-TOF/TOF-system (Applied Biosystems). The six most intense peptide peaks per spot detected in the MS mode were further fragmented yielding the respective MSMS spectra. Measured m/z ratios were assigned to peptides and respective proteins by MASCOT database search in a yeast protein database. Only proteins identified by at least two non-redundant peptides with a Confidence Interval >95% were included in the analysis; except for the Noc1–TAP analysis. Known contaminants in IgG-Sepharose affinity purification experiments (ribosomal proteins, translation factors, heat-shock proteins) were excluded from further quantitation analysis. Otherwise, the peak area for iTRAQTM reporter ions were interpreted and corrected by the GPS-Explorer software (Applied Biosystems) and Excel (Microsoft). Average of all peptides of a given protein was calculated and outliers were deleted by manual evaluation. For determination of protein co-purification depending on rRNA de novo synthesis ratios of the signal intensities of the respective reporter ions were calculated. The ratio found for bait proteins was normalized to 1.
Sucrose gradient analysis
Cycloheximide was added to yeast cultures grown for 2 h at 37°C of OD600 0.5–0.8 to a final concentration of 100 μg/ml. After 15 min incubation at the given temperature and 15 min incubation on ice, the cells were harvested and lysed in lysis buffer (20 mM HEPES pH 7.5, 10 mM KCl, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 100 μg/ml cycloheximide) using glass beads. An amount of 600 μg whole-cell extract was loaded on a 10–50% sucrose gradient and centrifugation was performed for 2.5 h at 39 000 rpm and 4°C in a SW40 rotor. Gradients were fractionated using a BioLogic LP chromatography system from Bio-Rad and analysed by western blot using PAP visualization reagent (DakoCytomation, Z 0113) in a dilution of 1:3000 for detection of the TAP-Tag. The distribution of ribosomal particles in the gradient was determined by analyzing the blot with rabbit anti-rpS8 antibody in a dilution of 1:2000.
RESULTS
Analysis of expression levels and intracellular localization of ribosome biogenesis factors after shut down of rRNA de novo synthesis
We developed a general strategy to identify proteinaceous (sub-) complexes incorporated in pre-ribosomal RNPs which can form in vivo independent of the presence of (pre-) rRNA and might represent thereby functional building blocks of the RNPs.
To this end we used yeast genetics to conditionally inactivate Pol-I dependent rRNA synthesis and analysed the resulting effects on expression levels and intracellular localization of a selection of ribosome biogenesis factors. We compared then the protein composition of assemblies of biogenesis factors affinity purified from cells with or without ongoing rRNA synthesis by semi-quantitative mass spectrometry.
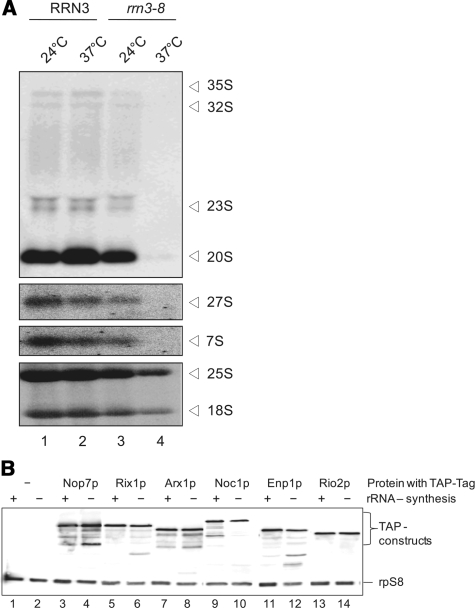
First, we analysed how the steady state levels of rRNA precursors were affected after specific shut down of the Pol-I machinery in a conditional temperature-sensitive mutant of the essential Pol-I transcription factor Rrn3p. Therefore, (pre-) rRNA levels in the rrn3-8 mutant strain YCC95 (31) and in the corresponding RRN3 wild-type strain CG379 were analysed at permissive (24°C) and restrictive temperature (37°C) by northern blot hybridization (Figure 1A). Pol-I transcribes a region of the rDNA-locus to yield the 35S rRNA precursor, which is processed in various endo- and exonucleolytic cleavage steps to finally result in the mature 18S, 5.8S and 25S rRNA. rRNA precursor levels of both large and small ribosomal subunit (respectively 27S and 20S pre-rRNA) were reduced after 3h incubation at 37°C to a residual amount of ∼5% in the rrn3-8 mutant when compared to the level in wild-type cells (Figure 1A, compare lane 2 with lane 4).
Figure 1.
Analysis of cellular pre-rRNA and ribosome biogenesis factor levels after shut down of rRNA de novo synthesis. (A) Northern hybridization analysis of precursor and mature rRNA species of both ribosomal subunits was performed on RNA extracts from whole cells. Yeast strains with RRN3 and with rrn3-8 background were analysed at permissive (24°C) and restrictive (3 h 37°C) temperature. RNA from equal number of cells was loaded and different oligonucleotides (‘Materials and methods’ section) were used for detection of the different indicated (pre-) rRNA species (B) Protein levels of TAP–tagged ribosome biogenesis factors in RRN3 (rRNA synthesis +) and in rrn3-8 background (rRNA synthesis –) were analysed at restrictive (3h, 37°C) temperature by western blotting using PAP visualization reagent. Equal loading was controlled by determination of the protein level of rpS8.
Next, we created derivates of the strain YCC95 carrying the rrn3-8 allele, and of the corresponding RRN3 wild-type strain CG379. The coding sequence of the TAP-tag was integrated by homologous recombination in front of the stop codon of a selection of genes coding for different ribosome biogenesis factors. In this way, these genes remained under the control of their endogenous promoters but coded for C-terminal TAP-tagged fusion proteins. We chose for this approach proteins which have been described to be involved in specific biogenesis stages of either the small (Enp1p, Rio2p) or the large (Noc1p, Nop7p, Rix1p, Arx1p) ribosomal subunit.
Immunodetection of the ProteinA moiety of their TAP-tag by western blotting showed that in almost all cases the expression level of the tagged proteins was comparable in wild-type and rrn3-8 background at 37°C (Figure 1B, compare lanes 3 and 4, 5 and 6, 7 and 8, 11 and 12, 13 and 14). The protein level of Noc1p-TAP showed a slight reduction to ∼40% (Figure 1B, lanes 9 and 10) after shift to the restrictive temperature in the rrn3-8 background in comparison to wild-type background. This result indicates that some proteinaceous constituents of pre-ribosomes are still present when rRNA de novo synthesis is shut down, although—in general—pre-rRNA-containing particles are strongly reduced.
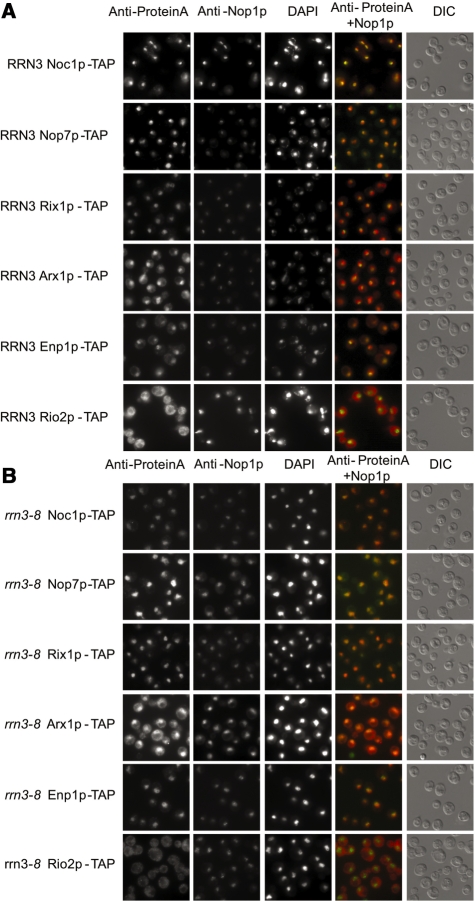
Next we analysed the localization of the tagged proteins in both wild-type and rrn3-8 mutant background by immunocytochemistry. To detect subcellular compartments we used an antibody raised against the nucleolar protein Nop1p, and DAPI staining of the DNA for visualization of the nucleus (Figure 2).
Figure 2.
Subcellular localization of TAP–tagged ribosome biogenesis factors after shut down of Pol I transcription. The localization of the indicated tagged proteins in yeast strains with (A) RRN3 and with (B) rrn3-8 background after 3 h shift to 37°C was analysed by fluorescence microscopy using an antibody directed against the Protein A moiety of the TAP-tag. Nucleolar structures were visualized by an anti–Nop1p antibody, yeast nuclei were stained with DAPI. Additionally, the ProteinA-signal (red) and the staining for Nop1p (green) were overlaid for better visualization of subcellular distribution. Whole yeast cells morphology was visualized by differential contrast (DIC). For better comparison the chosen exposition time for detection of the Protein A was not changed during analysis of the different tagged proteins. This results in slightly overexposed pictures for Nop7p-TAP and Arx1p-TAP.
As expected (27,39), in the wild-type background Noc1p-TAP and Nop7p-TAP showed co-localization with the nucleolar marker protein Nop1p (Figure 2A). The staining for Rix1p-TAP overlapped with the DAPI signal, arguing for nucleoplasmic localization. A very faint cytoplasmic signal as it was reported in the literature was less evident (4). Arx1p-TAP was also detected in both the nucleoplasm and cytoplasm (4). The SSU biogenesis factor Enp1p-TAP showed both nuclear and weak cytoplasmic staining (1), whereas Rio2p-TAP showed strong cytoplasmic signals (14). When rRNA synthesis was shut down in the rrn3-8 mutant background by applying restrictive conditions (3 h at 37°C), the signal detected by the anti Nop1p antibody appeared to be less defined when compared to the clear crescent shaped signals seen in the wild-type strain background (Figure 2B) and overlapped significantly with DAPI stained nucleoplasmic regions. This kind of intra-nuclear redistribution is most probably due to putative changes in the structure of the yeast nucleolus as a result of inhibition of Pol-I transcription (40). We observed a similar redistribution for Noc1p-TAP and Nop7p-TAP which showed co-staining with Nop1p in the wild-type strain background. On the other hand, shut down of rRNA de novo synthesis did apparently not lead to major nucleo-cytoplasmic redistribution of any of the investigated proteins, except Enp1-TAP which showed a more pronounced nuclear accumulation under these conditions, also when factor distribution was investigated by semi-quantitative profile analysis (data not shown). Accordingly, the nucleo-cytoplasmic distribution of most of the herein analysed factors is not exclusively dependent on ongoing ribosome biogenesis.
Analysis of ribosome biogenesis factors association with (pre-) rRNA after shut down of rRNA de novo synthesis
Next, we wanted to know with which and with how much residual (pre-) rRNA the tagged ribosome biogenesis factors are still associated after rRNA de novo synthesis is shut down. Therefore, we affinity purified the tagged ribosome biogenesis factors from wild type and rrn3-8 mutant cells after 3 h shift to restrictive conditions and analysed (pre-) rRNA contained in extracts and affinity purified fractions by northern blot hybridization. In parallel, the relative amounts of affinity purified, tagged ribosome biogenesis factors were determined by western blotting.
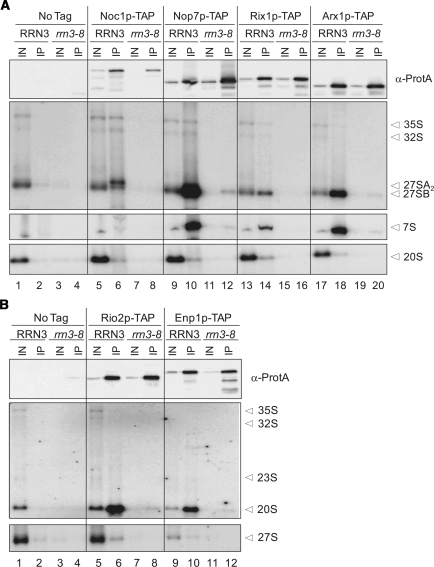
Pre-rRNA species which copurified with the different tagged ribosome biogenesis factors were as described from previous experiments (1,4,7,14,29). In wild-type background, Noc1p-TAP precipitated predominantly 27SA2 pre-rRNA, smaller quantities of 35S, 32S and 27SB pre-rRNA (Figure 3A, lane 6). Nop7p-TAP associated mainly with 27SB and 7S pre-rRNA. Early precursors like 35S and 32S pre-rRNA co-purified to a lower extent (Figure 3A, lane 10). Both Rix1p-TAP and Arx1p-TAP showed incorporation in pre-ribosomal particles containing 27SB and 7S pre-rRNA (Figure 3A, lanes 14 and 18). Both studied ribosome biogenesis factors of the small ribosomal subunit, Enp1p and Rio2p, showed association with 40S precursor particles containing the 20S pre-rRNA (Figure 3B, lanes 6 and 10). Only small amounts of 35S and 32S pre-rRNA were detected to co-purify with Enp1p-TAP. For all the analysed factors the co-purification efficiency of pre-rRNAs contained in precursors of the corresponding ribosomal subunit for whose maturation they are not required for was comparably low, indicating the reasonable specifity of the analysis (Figure 3A and B, lanes 6, 10, 14 and 18).
Figure 3.
Co-purification of pre-rRNA with different TAP-tagged biogenesis factors of the (A) large and (B) small ribosomal subunit after shut down of Pol-I transcription. Northern hybridization analysis of precursor rRNA species of both ribosomal subunits was performed on RNA extracted from whole cell extracts (IN) and from affinity purified TAP-tagged (A) Noc1p, Nop7p, Rix1p, Arx1p, (B) Rio2p and Enp1p (IP). Yeast strains carrying the RRN3 wild-type allele or the rrn3-8 allele were analysed at restrictive (3 h, 37°C) temperature. Different oligonucleotides (‘Materials and methods’ section) were used for detection of the different indicated (pre-) rRNA species. In parallel the amounts of tagged protein in input and affinity purified fractions were determined by western blotting using PAP visualization reagent. For each precipitation sample same signal intensities in IN and IP lanes reflect a purification recovery of 1%.
Western blot analysis indicated that for all studied tagged factors the efficiencies of their affinity purification were comparable no matter whether rDNA transcription was shut down or not. In contrast, the amount of co-purifying pre-rRNA was clearly reduced when rDNA transcription was impaired (Figure 3A and B, compare lanes 6 and 8, 10 and 12, 14 and 16, 18 and 20). The ratio of purified tagged protein versus co-purified pre-rRNA was strongly increased in this situation indicating that pre-rRNA free protein complexes could be enriched. Interestingly, in cells where rDNA transcription was not impaired, comparison of the amounts of purified Noc1p-TAP, Rix1p-TAP and Nop7p-TAP with the corresponding amounts of copurified pre-rRNA (wild-type, Figure 3A, lanes 6, 10 and 14) indicated that already under these conditions purified Noc1p-TAP and Rix1p-TAP fractions contained significant amounts of pre-rRNA-free protein (complexes). Accordingly, assemblies of ribosome biogenesis factors that are affinity purified under standard conditions contain varying amounts of pre-ribosomal RNPs and pre-rRNA free protein (complexes).
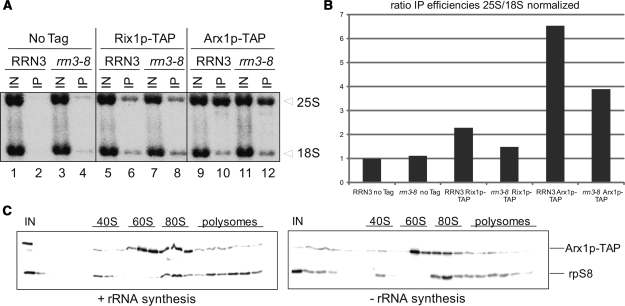
In contrast to early 60S biogenesis factors like Noc1p and Nop7p (data not shown), previous analyses suggested that Arx1p and Rix1p associate with nascent ribosomal subunits containing mature 25S rRNA. However Arx1p and Rix1p seem to be largely excluded from 60S ribosomal subunits which are able to associate with 40S ribosomal subunits and initiate translation (4,41). Therefore, we compared the relative amounts of mature 25S rRNA and 18S rRNA which co-purify with Rix1p-TAP and Arx1p-TAP on an IgG-Dynabead matrix after in vivo inactivation of the rDNA transcription machinery (Figure 4A). When extracts were prepared from cells where rDNA transcription took place, the ratio of 25S rRNA to 18S rRNA retained on IgG-Dynabeads was six times higher in experiments with strain Y584 (Arx1p-TAP) and about two times higher in the experiment with strain Y583 (Rix1p-TAP) than with strain Y543 expressing no TAP tagged protein. Interestingly, when the same analysis was performed with cells where rDNA transcription was shut down, the ratio of 25S rRNA to 18S rRNA retained on IgG-Dynabeads was still about four times higher for the strain expressing Arx1p-TAP in comparison to the strain expressing no tagged protein (Figure 4A and B). In addition, sedimentation analysis of whole cellular extracts on sucrose gradients showed that after shut down of rRNA de novo synthesis the major population of Arx1p-TAP co-sediments with 60S ribosomal subunits (Figure 4C). Accordingly, in agreement with previous analyses, Arx1p associates with LSUs containing mature 25S rRNA. Among them is, most likely, a population of nascent 60S ribosomal subunits, but Arx1p seems also to be attached to non-nascent free large ribosomal subunits which are not engaged in translation.
Figure 4.
Co-purification of mature 25S and 18S rRNA with TAP-tagged Rix1p and Arx1p after shut down of Pol-I transcription. (A) Northern hybridization analysis of mature rRNA species of both ribosomal subunits was performed on RNA extracted from whole cell extracts (IN) and from affinity purified TAP-tagged Rix1p and Arx1p (IP). Yeast strains carrying the RRN3 wild-type allele or the rrn3-8 allele were analysed at restrictive (3 h, 37°C) temperature. Different oligonucleotides ‘Materials and methods’ section) were used for detection of the indicated 18S and 25S rRNA species. Same signal intensities in IN and IP lanes reflect a purification recovery of 1%. (B) The ratio of affinity precipitation efficiencies for 25S and 18S rRNAs was calculated and normalized to the one found in the untagged wild-type yeast strain using MultiGauge V3.0 (Fujifilm). (C) Sedimentation behaviour of TAP–tagged Arx1p was analysed on sucrose density gradients with cellular extracts of strains carrying the RRN3 wild-type allele (rRNA synthesis +) or the rrn3-8 allele (rRNA synthesis −) after 3 h shift to restrictive temperature. Distribution of ribosomal particles (40S, 60S, 80S, polysomes) in the gradient was determined by OD254 measurement (data not shown) and western blot analysis of the gradient fractions using an anti-rpS8 antibody. The amount of TAP-tagged Arx1p in each fraction and in the input-sample (IN) was also visualized by western blot analysis.
Changes in the protein composition of purified ribosome biogenesis factor assemblies after shut down of rRNA de novo synthesis
In summary, these analyses indicated that ribosome biogenesis factor assemblies purified from cells, in which rRNA de novo synthesis was shut down, were largely devoid of pre-rRNA and, except for Arx1p-TAP purified assemblies, also of mature rRNA molecules. Next, we compared by semi-quantitative mass spectrometry how the protein composition of these assemblies changes after in vivo depletion of nascent pre-rRNA.
One-step affinity purification of ProteinA tagged ribosome biogenesis factors from cells with or without ongoing rDNA transcription was performed using IgG–sepharose. Tryptic peptides from the purified assemblies of these two pools were separately labelled using two different iTRAQ reagents (38). Then, the two differentially labelled samples were combined, the mixture of peptides was fractionated by nano-flow reversed-phase-chromatography and analysed by MALDI-TOF/TOF (matrix assisted laser desorption ionization-time of flight) mass spectrometry.
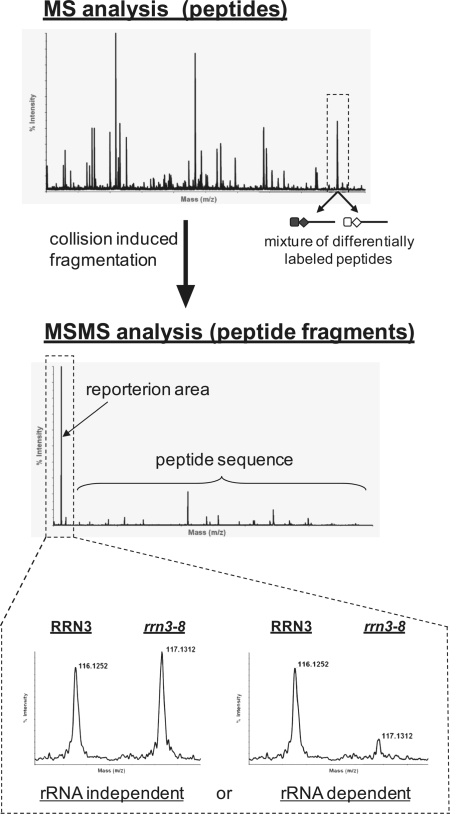
In the fragmentation chamber of the mass spectrometer different reporter groups (114, 115, 116, 117 Da) are released from the specific iTRAQ labels and can be used to determine how much of the analysed peptide was linked to the respective iTRAQ reagent (Figure 5). Furthermore, the peptide itself is fragmented and analysis of the resulting fragment masses allows identification of the protein from which it was derived (38).
Figure 5.
Schematic view of relative protein quantitation set up. A typical MS spectrum is shown, where each peak represents a mixture of sequence-identical but differentially iTRAQ-labelled peptides from affinity purifications of wild-type and rrn3-8 mutant strains. In the MSMS–mode peptides are selected for fragmentation, yielding in peptide fragments with sequence specific m/z ratios used for identification of the respective protein by database search. In addition the iTRAQ reporter ions of different masses are released and are used for relative quantitation and subsequent determination of pre-rRNA-dependent or pre-rRNA-independent co-purifications.
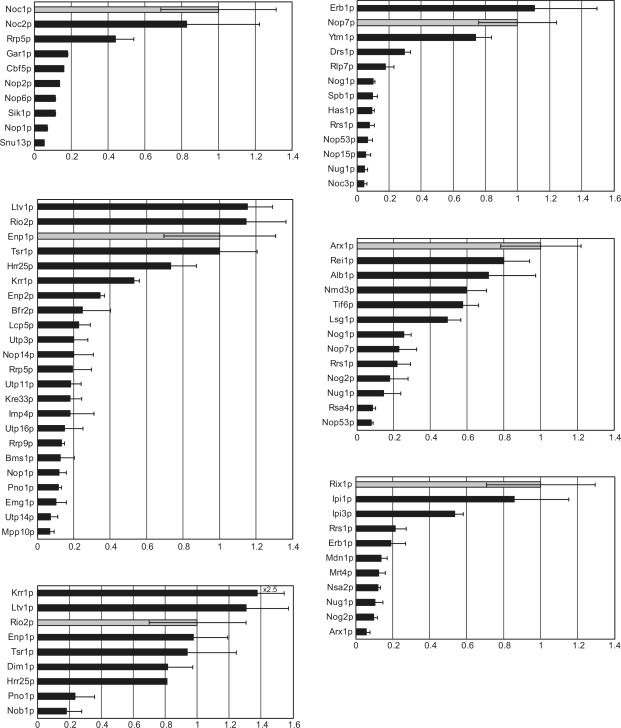
This methodology allowed us to compare the relative abundance of established ribosome biogenesis factors co-purifying with Noc1p-TAP, Nop7p-TAP, Rio2p-TAP, Enp1p-TAP, Rix1p-TAP or Arx1p-TAP from cells carrying either the temperature sensitive rrn3-8 or the corresponding wild-type RRN3 allele (Figure 6). As expected from the literature (1), Enp1p-TAP purified from RRN3 wild-type cells co-purified with a large set of proteins which are believed to be part of the SSU-processome, an assembly of factors which are required for nuclear steps of eukaryotic SSU maturation and which associate in vivo with an U3-snoRNP. Association of most of these factors with Enp1p-TAP was largely reduced (rrn-8:RRN3 ratio <0.2) when rDNA transcription was shut down, indicating that co-purification of Enp1p-TAP with SSU-processome components is mediated through pre-rRNA. On the other hand, co-purification of several other factors with Enp1p-TAP remained largely unaffected by inactivation of the rDNA transcription factor Rrn3p (rrn3-8:RRN3 ratio for Ltv1p 1.15, for Tsr1p 1.0, for Krr1p 0.53, for Hrr25p 0.73, for Rio2p 1.15). Apparently, these proteins are part of a, most likely proteinaceaous, module which can interact with SSU precursors but whose in vivo existence does not require the presence or ongoing production of pre-rRNA. This interpretation is in general agreement with the recent observation, that an Enp1p-Ltv1p-rpS3 complex can be extracted from pre-ribosomes in high salt conditions (28). Comparison of the protein compositions of Rio2p-TAP affinity-purifications from cells with or without ongoing rRNA synthesis confirmed the existence of this protein module. Association of Enp1p (rrn3-8:RRN3 ratio 0.97), Ltv1p (rrn3-8:RRN3 ratio 1.3), Tsr1p (rrn3-8:RRN3 ratio 0.94), Krr1p (rrn3-8:RRN3 ratio 3.44), Hrr25 (rrn3-8:RRN3 ratio 0.81, based on one peptide), and additionally of Dim1p (rrn3-8:RRN3 ratio 0.82) with Rio2p-TAP remained largely unaffected when rDNA transcription was shut down. Interestingly, two other ribosome biogenesis factors involved in pre-SSU maturation, Nob1p and Pno1p/Dim2p co-purified as expected from earlier experiments (1) with Rio2p-TAP in the RRN3 wild-type strain, but their association was largely reduced after inactivation of rrn3-8 indicating that the Nob1p and Pno1p/Dim2p co-purification with Rio2p-TAP is pre-rRNA mediated.
Figure 6.
Semi-quantitative comparison of co-purifying ribosome biogenesis factors from cells with or without ongoing rRNA synthesis. The diagrams show the ratio of identified proteins co-purifying with TAP-tagged bait proteins from cells carrying the RRN3 wild-type and the rrn3-8 mutant allele. The bars indicate the average value of the calculated RRN3:rrn3-8 ratios for all identified peptides of the indicated protein. Error bars represent the standard deviation of these ratios (P < 0.05). The ratio of the bait protein (highlighted by a grey bar) is set to one. For co-purified proteins, ratios >0.5 were considered to reflect associations with bait proteins barely influenced by the absence of de novo rRNA synthesis. Association of proteins with intermediate ratios between 0.25 and 0.5 were classified to be stronger affected by the shutoff of rRNA synthesis. Nevertheless they are still significantly co-purified in the rrn3-8 mutant. Associations of proteins with ratios below 0.25 seem to be strongly dependent on rRNA de novo synthesis. Two independent purifications for the indicated bait proteins were performed and for each purification mass spectrometry analysis was repeated once. Quantitation of one representative experiment is shown.
Using the same approach we analysed with which proteins the LSU biogenesis factors Noc1p, Nop7p and Rix1p associate in vivo in the absence of rRNA de novo synthesis. Previous studies showed that in conditions leading to in vivo or ex vivo disruption of pre-ribosomes, Noc1p could be purified in a complex with Noc2p (27), Nop7p in a complex with Erb1p and Ytm1p (29) and Rix1p in a complex with Ipi1p and Ipi3p (18). The experimental approach used in the present study now indicated that these three protein complexes can form or persist in vivo independent of the presence and de novo synthesis of pre-ribosomes (Figure 6, see also Supplementary Figure S1). In addition, some significant residual association of Rrp5p with the Noc1p/Noc2p complex (rrn3-8:RRN3 ratio 0.44) and of the nucleolar DEAD box helicase Drs1p with the Nop7p complex (rrn3-8:RRN3 ratio 0.29) was observed when rRNA de novo synthesis was shut down, suggesting that these proteins are part of the corresponding protein modules. [Co-purification of Rrp5p with Noc1p was also observed in the Hurt laboratory (E. Hurt and J. Bassler, personal communication)].
Semi-quantitative mass spectrometry analysis of Arx1p-TAP affinity purifications from extracts of cells with or without ongoing rRNA synthesis showed that significant amounts of Rei1p (rrn3-8:RRN3 ratio 0.81), Alb1p (rrn3-8:RRN3 ratio 0.67) Lsg1p (rrn3-8:RRN3 ratio 0.5), Tif6p (rrn3-8:RRN3 ratio 0.63) and Nmd3p (rrn3-8:RRN3 ratio 0.55) co-purify with Arx1p-TAP independent of the presence of nascent large ribosomal subunits. Our observation that Arx1p-TAP significantly associated with mature 25S rRNA when rRNA de novo synthesis was shut down (see above, Figure 4A) indicated that it associates not only with nascent subunits but also with free, non-nascent 60S ribosomal subunits. Accordingly, copurification of Rei1p, Alb1p, Lsg1p, Tif6p and Nmd3p with Arx1p-TAP in conditions where neo-production of large ribosomal subunits is inhibited might be due to their (pre-) ribosome independent association with Arx1p or due to their Arx1p independent association with mature, free non-nascent 60S ribosomal subunits.
DISCUSSION
A large variety of ribosome biogenesis factors is needed for efficient production of eukaryotic ribosomes. Many of them interact in a coordinated transient way with nascent ribosomal subunits leading to the formation of specific pre-ribosomal intermediates. It was shown that in conditions leading to (partial) disruption of pre-ribosomes (3,17–30) or after differential centrifugation (26) (pre-) rRNA free protein modules of ribosome biogenesis factors can be isolated from yeast cellular extracts by affinity purification. The experimental approach chosen in this article, namely in vivo down-regulation of rRNA synthesis followed by one step affinity purification and comparative protein analysis of the purified assemblies by semi-quantitative mass spectrometry, turned out to reveal several new aspects about protein modules involved in eukaryotic ribosome maturation. First it allowed to formally analyse which protein modules can form or persist in vivo independent of the presence and de novo synthesis of pre-ribosomes. The strong argument provided in this article for pre-ribosome independent in vivo existence of protein modules (Rio2p/Enp1p-, Rix1p-, Noc1p- and Nop7p modules) involved in ribosome biogenesis implies that these modules represent functional entities which can transiently interact with nascent ribosomal subunits en bloc. Recently, Watkins and colleagues observed the accumulation of a 50S U3 sno-RNP which contained Rrp5p and Dbp4p and other unidentified factors in mammalian cells in which RNA polymerase I was inactivated (10). Altogether, these analyses indicate the existence of various pre-ribosome independent ribosome biogenesis factors modules in eukaryotic cells.
The strategy applied in this article allowed usage of mild conditions (physiological MgCl2 and salt concentrations) during protein module isolation and semi-quantitative, comparative interpretation of the protein composition of pre-rRNA dependent and independent assemblies. Thereby, several new members of pre-rRNA free ribosome biogenesis factor modules could be identified. Our data suggest that Rio2p forms (a) module(s) with Enp1p, Ltv1p, Tsr1p, Krr1p, Hrr25p and Dim1p; that Rrp5 is a member of a Noc1-Noc2 module, and that Drs1p association with a Nop7p-Erb1p-Ytm1p module is enhanced by pre-rRNA, but does not strictly depend on it. The notion that Drs1p is part of Nop7p-Erb1p-Ytm1p modules is further supported by results of Woolford and colleagues (39). They found allele specific synthetic lethal phenotypes in nop7/drs1 double mutants and observed underproduction of 60S ribosomal subunits in drs1 mutant strains.
The appearance of pre-rRNA-free complexes containing both, putative early assembly biogenesis factors like Enp1p, and those which apparently bind late, like Hrr25p and Rio2p, does not necessarily mean that they have to assemble always as one complex with nascent subunits. The following scenarios are also possible. (i) Enp1p could bind at least at two different sites to the pre-ribosome. The formation of the two-binding sites is timely separated. The first occurring site binds Enp1p, but not late associating factors, whereas the other binding site interacts with a complex containing Enp1p, Rio2p and other late factors. Binding at different sites of the pre-ribosome was recently suggested for Prp43 (42). (ii) Enp1p assembles first on the pre-ribosome and is then replaced by a complex containing both the late factors and Enp1p, and (iii) Enp1 and late factors assemble independently, but are released together.
Interestingly, Nob1p and its putative interaction partner Pno1p/Dim2p (13,43), while consistently found in Rio2p containing pre-ribosomal assemblies (1), seem not to be part of the pre-rRNA free Rio2p protein modules (Figure 6). Beside Nob1p, a putative endonuclease containing a PIN domain (44), and Pno1p/Dim2p, Rio2p itself and several of the Rio2p module components are required for cytoplasmic 3′ processing of 18S pre-rRNA at site D (13,15,16,45,46). Apparently, the interaction of at least two discrete protein modules, the Nob1p-Pno1p/Dim2p module and the Rio2p module(s), with nascent 40S ribosomal subunits at most likely distinct sites seems to be necessary to allow efficient conversion of 3′ extended 18S rRNA into mature 18S rRNA by endonucleolytic cleavage at site D.
An in part unexpected outcome of these analyses was that free large ribosomal subunits are associated with a specific set of ribosome biogenesis factors, including Arx1p, in cells where neo-production of nascent ribosomes was blocked for prolonged times (3 h). We propose that Arx1p associated subunits represent free, non-nascent LSUs. De novo synthesis of ribosomes in yeast requires about 15 min and all analysed pre-rRNAs were turned over, either through degradation or productive processing, during the 3 h of rrn3-8 inactivation applied in the experimental setup (Figures 1A, 3A and B). Thereby, if 25S rRNA containing nascent subunits persisted during 3 h, they behaved in clear contrast to the other pre-ribosomal particles. A possible explanation in favour of this would be that release of Arx1p and other factors from nascent LSUs required active ongoing translation which might be disturbed by an unknown feedback mechanism when Pol-I activity is decreased in cells. On the other hand it was demonstrated by Nomura and colleagues that inactivation of the Pol-I machinery has no impact on ribosomal activity (47). In agreement with this, sucrose gradient analyses argue that small ribosomal subunits stay associated with polysomal fractions when Pol-I transcription is shut down (Figure 4C and our unpublished polysome profile data). Altogether we think it is worth to consider a regulatory role of Arx1p in both the synthesis and function of LSUs. Beside its suggested auxiliary role in nucleo-cytoplasmic translocation of pre 60S subunits (48–50) it might protect both nascent and non-nascent LSUs from degradation and/or regulate their interaction with 40S ribosomal subunits. Interestingly, the human homolog Ebp1, which was suggested to play a role in translational control (51–53), seems to lack the characteristics of yeast Arx1p mediating its interaction with nucleoporines and its efficient nucleo-cytoplasmic shuttling in in vitro assays (48). Possibly Arx1p plays a conserved role in eukaryotic translation while it gained additional non-essential function in nucleo-cytoplasmic transport of the LSU in certain organisms like the fast dividing S. cerevisiae.
In yeast cells lacking the ARX1 gene changes in pre-rRNA maturation or underaccumulation of de novo synthesized LSUs were not detected [(20), our own unpublished data], and polysome profiles revealing an increase in both the free 40S and 60S subunits and the appearance of halfmeres, could be indicative of a role of Arx1p in subunit joining [see for example (49), our unpublished data].
When de novo synthesis of rRNA was shut down several LSU biogenesis factors, as Nmd3p, Tif6p/eIF6, Rei1p, Alb1p and Lsg1p, continued to co-purify with Arx1p while others like Nug1p, Nog2p and Rrs1p ceased in doing so (Figure 6).
Nmd3p was already shown to interact with both nascent and non-nascent free LSUs (54,55), apparently in the LSU central protrusion region near rpL10 (19,56). Tif6p/eIF6 from archaea and yeast binds in vitro specifically to mature large ribosomal subunits, most likely in the subunit interface via rpL23, and prevents their association with SSUs (57,58). Since the suggested Tif6p/eIF6 and Nmd3p LSU binding sites are not in direct neighbourhood, it is conceivable that co-purification of Nmd3p and Tif6p/eIF6 with Arx1p in the absence of pre-ribosomes is due to their interaction with mature free LSUs and not due to an association with a ribosome-free Arx1p protein module.
Altogether, these data suggest that free 60S ribosomal subunits, neo-synthesized or released from 80S ribosomes during translation termination, are decorated with a common set of factors including Arx1p, Tif6p/eIF6, Nmd3p and, eventually, in addition Rei1p, Alb1p and Lsg1p. A dual function in both ribosome synthesis and regulation of LSU recycling during translation, as already suggested for mammalian eIF6/Tif6p (59, 60), would explain the unexpected association of these factors with mature LSUs.
FUNDING
Funding for open access charge: Deutsche Forschungsgemeinschaft (DFG).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the Lehrstuhl Biochemie III for critical discussion and Kristin Hergert and Eduard Hochmuth for technical support. They are grateful to Ed Hurt and Jochen Baßler for sharing unpublished results.
REFERENCES
- 1.Schäfer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 2003;22:1370–1380. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saveanu C, Namane A, Gleizes P, Lebreton A, Rousselle J, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol. Cell. Biol. 2003;23:4449–4460. doi: 10.1128/MCB.23.13.4449-4460.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pérez-Fernández J, Román A, De Las Rivas J, Bustelo XR, Dosil M. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol. Cell. Biol. 2007;27:5414–5429. doi: 10.1128/MCB.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milkereit P, Kühn H, Gas N, Tschochner H. The pre-ribosomal network. Nucleic Acids Res. 2003;31:799–804. doi: 10.1093/nar/gkg165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein KA, Gallagher J.EG, Mitchell BM, Granneman S, Baserga SJ. The small-subunit processome is a ribosome assembly intermediate. Eukaryotic Cell. 2004;3:1619–1626. doi: 10.1128/EC.3.6.1619-1626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harnpicharnchai P, Jakovljevic J, Horsey E, Miles T, Roman J, Rout M, Meagher D, Imai B, Guo Y, Brame CJ, et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- 8.Dragon F, Gallagher J.EG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wery M, Ruidant S, Schillewaert S, Leporé N, Lafontaine DL. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA. 2009;15:406–419. doi: 10.1261/rna.1402709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner AJ, Knox AA, Prieto J, McStay B, Watkins NJ. A novel small-subunit processome assembly intermediate that contains the U3 snoRNP, nucleolin, RRP5, and DBP4. Mol. Cell. Biol. 2009;29:3007–3017. doi: 10.1128/MCB.00029-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trapman J, Retèl J, Planta RJ. Ribosomal precursor particles from yeast. Exp. Cell. Res. 1975;90:95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- 12.Chen W, Bucaria J, Band DA, Sutton A, Sternglanz R. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 2003;31:690–699. doi: 10.1093/nar/gkg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fatica A, Oeffinger M, Dlakić M, Tollervey D. Nob1p is required for cleavage of the 3′ end of 18S rRNA. Mol. Cell. Biol. 2003;23:1798–1807. doi: 10.1128/MCB.23.5.1798-1807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanrobays E, Gelugne J, Gleizes P, Caizergues-Ferrer M. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:2083–2095. doi: 10.1128/MCB.23.6.2083-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seiser RM, Sundberg AE, Wollam BJ, Zobel-Thropp P, Baldwin K, Spector MD, Lycan DE. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 2006;174:679–691. doi: 10.1534/genetics.106.062117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geerlings TH, Faber AW, Bister MD, Vos JC, Raué HA. Rio2p, an evolutionarily conserved, low abundant protein kinase essential for processing of 20 S Pre-rRNA in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:22537–22545. doi: 10.1074/jbc.M300759200. [DOI] [PubMed] [Google Scholar]
- 17.Oeffinger M, Leung A, Lamond A, Tollervey D, Lueng A. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002;8:626–636. doi: 10.1017/s1355838202020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galani K, Nissan TA, Petfalski E, Tollervey D, Hurt E. Rea1, a dynein-related nuclear AAA-ATPase, is involved in late rRNA processing and nuclear export of 60 S subunits. J. Biol. Chem. 2004;279:55411–55418. doi: 10.1074/jbc.M406876200. [DOI] [PubMed] [Google Scholar]
- 19.Hedges J, West M, Johnson AW. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 2005;24:567–579. doi: 10.1038/sj.emboj.7600547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebreton A, Saveanu C, Decourty L, Rain J, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J. Cell. Biol. 2006;173:349–360. doi: 10.1083/jcb.200510080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Senger B, Lafontaine DL, Graindorge JS, Gadal O, Camasses A, Sanni A, Garnier JM, Breitenbach M, Hurt E, Fasiolo F. The nucle(ol)ar Tif6p and Efl1p are required for a late cytoplasmic step of ribosome synthesis. Mol. Cell. 2001;8:1363–1373. doi: 10.1016/s1097-2765(01)00403-8. [DOI] [PubMed] [Google Scholar]
- 22.Gavin A, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 23.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 24.Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- 25.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Séraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 26.Krogan NJ, Peng W, Cagney G, Robinson MD, Haw R, Zhong G, Guo X, Zhang X, Canadien V, Richards DP, et al. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell. 2004;13:225–239. doi: 10.1016/s1097-2765(04)00003-6. [DOI] [PubMed] [Google Scholar]
- 27.Milkereit P, Gadal O, Podtelejnikov A, Trumtel S, Gas N, Petfalski E, Tollervey D, Mann M, Hurt E, Tschochner H. Maturation and intranuclear transport of pre-ribosomes requires Noc proteins. Cell. 2001;105:499–509. doi: 10.1016/s0092-8674(01)00358-0. [DOI] [PubMed] [Google Scholar]
- 28.Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 2006;441:651–655. doi: 10.1038/nature04840. [DOI] [PubMed] [Google Scholar]
- 29.Miles TD, Jakovljevic J, Horsey EW, Harnpicharnchai P, Tang L, Woolford JL. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 2005;25:10419–10432. doi: 10.1128/MCB.25.23.10419-10432.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lebreton A, Rousselle J, Lenormand P, Namane A, Jacquier A, Fromont-Racine M, Saveanu C. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res. 2008;36:4988–4999. doi: 10.1093/nar/gkn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cadwell C, Yoon HJ, Zebarjadian Y, Carbon J. The yeast nucleolar protein Cbf5p is involved in rRNA biosynthesis and interacts genetically with the RNA polymerase I transcription factor RRN3. Mol. Cell. Biol. 1997;17:6175–6183. doi: 10.1128/mcb.17.10.6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Séraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 33.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 34.Milkereit P, Strauss D, Bassler J, Gadal O, Kühn H, Schütz S, Gas N, Lechner J, Hurt E, Tschochner H. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2003;278:4072–4081. doi: 10.1074/jbc.M208898200. [DOI] [PubMed] [Google Scholar]
- 35.Léger-Silvestre I, Milkereit P, Ferreira-Cerca S, Saveanu C, Rousselle J, Choesmel V, Guinefoleau C, Gas N, Gleizes P. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 2004;23:2336–2347. doi: 10.1038/sj.emboj.7600252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferreira-Cerca S, Pöll G, Gleizes P, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Chen X, Andrews PC. Quantitative proteomics analysis of pancreatic zymogen granule membrane proteins. Meth. Mol. Biol. 2009;528:327–338. doi: 10.1007/978-1-60327-310-7_23. [DOI] [PubMed] [Google Scholar]
- 38.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford JL. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA. 2002;8:150–165. doi: 10.1017/s1355838202010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trumtel S, Léger-Silvestre I, Gleizes PE, Teulières F, Gas N. Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell. 2000;11:2175–2189. doi: 10.1091/mbc.11.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hung N, Johnson AW. Nuclear recycling of the pre-60S ribosomal subunit-associated factor Arx1 depends on Rei1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:3718–3727. doi: 10.1128/MCB.26.10.3718-3727.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol. Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tone Y, Toh-E A. Nob1p is required for biogenesis of the 26S proteasome and degraded upon its maturation in Saccharomyces cerevisiae. Genes Dev. 2002;16:3142–3157. doi: 10.1101/gad.1025602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fatica A, Tollervey D, Dlakić M. PIN domain of Nob1p is required for D-site cleavage in 20S pre-rRNA. RNA. 2004;10:1698–1701. doi: 10.1261/rna.7123504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK. Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA. 2001;7:1268–1283. doi: 10.1017/s1355838201013073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanrobays E, Gélugne J, Caizergues-Ferrer M, Lafontaine D.LJ. Dim2p, a KH-domain protein required for small ribosomal subunit synthesis. RNA. 2004;10:645–656. doi: 10.1261/rna.5162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wittekind M, Kolb JM, Dodd J, Yamagishi M, Mémet S, Buhler JM, Nomura M. Conditional expression of RPA190, the gene encoding the largest subunit of yeast RNA polymerase I: effects of decreased rRNA synthesis on ribosomal protein synthesis. Mol. Cell. Biol. 1990;10:2049–2059. doi: 10.1128/mcb.10.5.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgärtel V, Boese G, Bassler J, Wild K, et al. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell. 2007;27:767–779. doi: 10.1016/j.molcel.2007.06.034. [DOI] [PubMed] [Google Scholar]
- 49.Hung N, Lo K, Patel SS, Helmke K, Johnson AW. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell. 2008;19:735–744. doi: 10.1091/mbc.E07-09-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lo K, Johnson AW. Reengineering ribosome export. Mol. Biol. Cell. 2009;20:1545–1554. doi: 10.1091/mbc.E08-10-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 53.Squatrito M, Mancino M, Sala L, Draetta GF. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem. Biophys. Res. Commun. 2006;344:859–868. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 54.Hedges J, Chen Y, West M, Bussiere C, Johnson AW. Mapping the functional domains of yeast NMD3, the nuclear export adapter for the 60 S ribosomal subunit. J. Biol. Chem. 2006;281:36579–36587. doi: 10.1074/jbc.M606798200. [DOI] [PubMed] [Google Scholar]
- 55.Ho JH, Kallstrom G, Johnson AW. Nascent 60S ribosomal subunits enter the free pool bound by Nmd3p. RNA. 2000;6:1625–1634. doi: 10.1017/s1355838200001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 2001;21:3405–3415. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benelli D, Marzi S, Mancone C, Alonzi T, la Teana A, Londei P. Function and ribosomal localization of aIF6, a translational regulator shared by archaea and eukarya. Nucleic Acids Res. 2009;37:256–267. doi: 10.1093/nar/gkn959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Si K, Maitra U. The Saccharomyces cerevisiae homologue of mammalian translation initiation factor 6 does not function as a translation initiation factor. Mol. Cell. Biol. 1999;19:1416–1426. doi: 10.1128/mcb.19.2.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–688. doi: 10.1038/nature07267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miluzio A, Beugnet A, Volta V, Biffo S. Eukaryotic initiation factor 6 mediates a continuum between 60S ribosome biogenesis and translation. EMBO Rep. 2009;10:459–465. doi: 10.1038/embor.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.