Abstract
We have developed a high-throughput protein expression and interaction analysis platform that combines cDNA phage display library selection and massive gene sequencing using the 454 platform. A phage display library of open reading frame (ORF) fragments was created from mRNA derived from different tissues. This was used to study the interaction network of the enzyme transglutaminase 2 (TG2), a multifunctional enzyme involved in the regulation of cell growth, differentiation and apoptosis, associated with many different pathologies. After two rounds of panning with TG2 we assayed the frequency of ORFs within the selected phage population using 454 sequencing. Ranking and analysis of more than 120 000 sequences allowed us to identify several potential interactors, which were subsequently confirmed in functional assays. Within the identified clones, three had been previously described as interacting proteins (fibronectin, SMOC1 and GSTO2), while all the others were new. When compared with standard systems, such as microtiter enzyme-linked immunosorbant assay, the method described here is dramatically faster and yields far more information about the interaction under study, allowing better characterization of complex systems. For example, in the case of fibronectin, it was possible to identify the specific domains involved in the interaction.
INTRODUCTION
Generally used methods to screen for protein–protein interactions include tandem affinity purification followed by mass spectrometry, yeast two hybrid system (Y2H), protein complementation assays (PCA), ribosome/RNA display (1–3) and phage display (4). With the exception of tandem affinity purification, which directly assesses proteins that interact with a particular target protein, each of these assays involve time consuming picking and assessment of individual clones after a selection or screening procedure. Consequently although thousands of clones, or even more, may be generated in any of these screening/selection procedures, the number actually analyzed is usually limited to the number of wells on a microtiter plate (96 or multiples thereof) and is therefore significantly smaller. Whether such manual random picking strategies can identify all possible interactions with a given bait-protein depends upon the protein under study and cannot be predicted in advance. In particular, random picking is not successful if a large number of different binders are expected, such as occurs in the screening of a protein with multiple interaction partners, or a few clones are over-represented during the selection process. A novel approach in which the number of screened clones could be increased by several orders of magnitude would therefore represent a major advance in the field.
In this article we applied a deep DNA sequencing strategy, first to the characterization of a cDNA phage display library, and subsequently to the identification of the proteins interacting with the enzyme tissue transglutaminase 2 (TG2), a molecule of increasing interest, in that its biological activity has been associated with a variety of pathologies of primary importance (5). The purpose was to follow the progress of the selection and analyze the whole output in a comprehensive manner, rather than performing random clone picking and enzyme-linked immunosorbant assay (ELISA). Among the available deep-sequencing systems, 454 pyrosequencing (6) is the only one that is suitable for this approach due to the longer read length that can be achieved: up to 450 bp per sequence and a total of over 1 million reads per run with the recently introduced Titanium system.
Phage display has been widely used to select antibodies or short peptides from libraries, however, phage display with cDNA libraries has been rarely used due to the large number of non-functional clones present. This problem has been recently addressed by the concept of filtering DNA for the presence of open reading frames (ORFs), and a number of different approaches have been taken (7–11). In this manuscript, the cloning procedure was based on pPAO2, a display system developed by us (12,13) that directly filters DNA for ORFs within a phage display context, in such a way that they are amenable to subsequent selection or screening. In this vector, DNA fragments are cloned upstream of the β-lactamase gene and clones are selected for ampicillin resistance. After this step, referred to as ORF filtering, the β-lactamase gene is removed by Cre-lox mediated recombination and an in-frame ORF-g3p fusion product is displayed on the phage surface. This vector has a strong bias for ORFs corresponding to real genes rather than ORFs of no biological significance, indicating that the lactamase gene functions as a folding reporter (14), akin to those previously described using either GFP (15), chloramphenicol resistance (16) or beta-galactosidase (17). However, unlike these other systems, functional analysis based on binding activity can also be subsequently carried out. As a target protein we used TG2, a human enzyme showing a number of different biological functions. In the cytosol, it is involved in protein cross-linking (18), signaling activity (19), protein disulfide isomerase activity (20), and even activity as a protein kinase (21). The protein is also found extracellularly where it is involved in the maintenance of tissue stability, cell adhesion and migration. Notably, aberrant functions of TG2 have been associated with a number of pathological conditions, including neurodegenerative diseases and several autoimmune diseases as diabetes, multiple sclerosis and rheumatoid arthritis (5). Antibodies against TG2 are used routinely to diagnose celiac disease. The approach described here was used to identify a set of TG2 interactors. Some of these had been previously described, while others are novel. Furthermore, the sequences of the selected cDNA fragments allowed us to identify the specific domains partaking in the interactions with TG2.
MATERIALS AND METHODS
RNA and cDNA fragments preparation
Three different mRNA sources were used to create the libraries: human colon carcinoma (HCC); human lung fibroblasts (HF) and human pancreatic islets (HPI). The HCC sample was obtained by pooling roughly equivalent amounts of polyA+ RNA obtained from the following cell lines: SW 620, CaCo-2, W.Dr C2Bbe1, CoLo 320 DM, HCT 116, HT29, SW 480. The HF sample was obtained by pooling RNA from Wi 38 and HFL 1. The HPI sample was obtained by purifying polyA+ RNA from a few thousand human pancreatic islets. For each sample, mRNA (1 µg HCC or 1 µg HF or 300 ng HPI) was mildly fragmented by heating for 6 min at 95°C prior to reverse transcription with random hexamers. The single-stranded cDNA was normalized individually with a corresponding mRNA (3 μg of either HCC or HF samples and 900 ng of HPI sample polyA+ RNA) according to Carninci et al. (22). The normalized cDNA was depleted of poly-dT tails by hybridization (3 h at 37°C) with biotinylated poly-dA and separation on streptavidin magnetic beads as described before (22). The unbound material was recovered, purified on Millipore Microcone PCR filter (UFC7PCR50) and used to build the libraries. The directional cloning of the random cDNA fragments in the correct sense orientation was based on a proprietary method developed by Quark Pharmaceuticals Inc (23). Asymmetric adapters (containing BssHII and NheI restriction sites for further cloning) were ligated to the single-stranded cDNA, the cDNA was amplified for 15 cycles with adapter specific primers, separated from short DNA fragments by gravity flow gel filtration on a Chroma Spin—400 column (BD Clontech), reamplified for an additional 13 cycles and 100–600 bp fragments were purified on GeneClean Turbo (Q-Biogene).
Library cloning and filtering
The creation and validation of the pPAO10 phagemid vector is extensively reported in Supplementary Figure 1 and Supplementary Data. Escherichia coli DH5αF′, F′/endA1 hsd17 (rK− mK+) supE44 thi-1 recA1 gyrA (Nalr) relA1 _ (lacZYA-argF) U169 deoR (F80dlacD-(lacZ)M15) strain was used. The pPAO10 phagemid vector and the purified cDNA fragments (either pooled HCC+HF, or HPI), with BssHII and NheI restriction sites, were digested and ligated using T4 DNA ligase at a molar ratio of 1:5. Totally 5 μg of the ligated DNA were transformed by electroporation into DH5αF′ bacteria, which were then plated on 2xTY agar plates supplemented with chloramphenicol (pPAO10 resistance, 34 μg/ml) and ampicillin (selective marker for ORFs, 15 μg/ml) and allowed to grow for 24 h at 28°C. Dilutions were also plated on either chloramphenicol/ampicillin or chloramphenicol alone to determine the library size. Bacteria were collected, pooled, thoroughly mixed, supplemented with 20% sterile glycerol and stored at –80°C in small aliquots. One aliquot immediately underwent the recombination step yielding a standard phagemid construct for display without β-lactamase as previously described (12).
Panning procedures
Production and rescue of phagemid particles was performed as described elsewhere (24); the phages were resuspended in 2% milk in PBS (MPBS) at a concentration of 1012/ml. Selections on solid phase were performed on Nunc Immunotubes coated with human recombinant TG2 as described before (25). Selections on soluble TG2 were performed as follows: 1012 phages diluted in 4% BSA were mixed with biotinylated human recombinant TG2 and incubated for 30 min in rotation followed by 30 min of static incubation at RT. Dynabeads MyOne Streptavidin beads (Dynal Biotech ASA) were added to human TG2-phages mix and panned in rotation for 45 min at RT; the beads were then washed four and 12 times with PBS added with 0.1% Tween and four and 12 times with PBS for the first and second round of selection, respectively. Bound phages were eluted by adding 1 ml of E.coli DH5αF′, at OD600 0.5, at 37°C, for 45 min, with occasional shaking. Bacteria were plated on agar plates added with chloramphenicol, grown O/N at 30°C, and finally harvested and either grown immediately to produce phages undergoing the following round of selection or stored in small aliquots at –80°C.
454 Deep Sequencing of cDNA Inserts
An amount of 200 ng cDNA fragments cut with BssHII and NheI from the library were purified by MinElute columns (Qiagen) in order to remove shorter fragments. Ligation of the purified samples to specific adaptors and preparation of the single strand libraries (sstDNA) were performed following the manufacturer's; instruction (Roche). The sstDNA libraries were quantitated by RiboGreen RNA Quantitation Kit (Invitrogen) and checked for quality by capillary electrophoresis (Agilent Bioanalyzer 2100 with the RNA Pico 6000 LabChip kit; Agilent Technologies). The sstDNA libraries were then amplified in emulsion as required by the 454 sequencing protocol. The reactions were recovered by isopropanol emulsion breaking and enriched for positive reaction beads. Each enriched sample was separately loaded onto one-eighth of the PicoTiterPlate (PTP) and was sequenced according to the 454 GS-FLX Titanium protocol.
Bioinformatic analysis
Sequences were processed with a custom analysis workflow procedure mainly based on PERL scripts. Both raw and analyzed data were stored in a relational database. Briefly, sequences were mapped onto the human genome (NCBI build 36) using gmap software and matching sequences were compared with annotated genes. Each gene was then ranked according to the number of supporting sequences. The ‘depth index’ for each gene was defined as the maximum number of overlapping sequences (i.e. sequences supporting the same genic region). The ‘focus index’—defined as (depth – 1)/rank—ranges between 0 (indicating a broad distribution of sequences over the gene) and 1 (indicating that all sequences are ‘focused’ on the same region). Data are accessible through a Web based interface (available at http://www.interactomeataglance.org/) implemented in php and java.
Cloning of selected cDNA fragments
After each selection, plasmid DNA was isolated from colonies rescued from the plates. 0.1 ng of each preparation was used as a template for the inverse PCR reaction. A pair of specific primers was designed for each of the top ranking genes, centering on the epitope region identified by the overlapping reads. The forward primer was synthesized with phosphorylated 5′-end in order to allow ligation of blunt ends and PCR was performed with a Phusion High-Fidelity DNA Polymerase (Finnzymes) according to the supplier’s protocol. After gel purification, the PCR product was ligated by T4 DNA ligase O/N at 16°C, and the ligation reaction transformed into DH5αF′ competent cells. Colonies obtained on chloramphenicol plates were sequenced to assess the successful cloning of specific gene fragments.
ELISA
Expression and testing of selected clones, either in the phage format or as soluble polypeptides, for recognition of human TG2 in ELISA was performed according to standard protocols and as previously described (25). Briefly, phage ELISA was performed by coating Costar ELISA strips with human and mouse recombinant TG2 or BSA and WNT4 (as negative control) at 10 μg/ml. Wells were blocked with 2% MPBS and rinsed. Phages of individual clones used in 1:1 dilution with 4% MPBS were added to the wells, followed by HRP-conjugated anti-M13 monoclonal antibody (Amersham Pharmacia). The complexes were revealed with TMB (3,3′,5,5′-tetramethylbenzidine) and read at A450. ELISA on soluble TG2 was performed as follows: 1012 phages of individual clones diluted in 4% BSA were mixed with biotinylated human recombinant TG2 and incubated for 30 min in rotation followed by 30 min of static incubation at RT. Dynabeads MyOne Streptavidin beads (Dynal Biotech ASA) were added to the human TG2-phage mix and incubated in rotation for 45 min at RT. After five washes with PBST, HRP-conjugated anti-M13 monoclonal antibody (Amersham Pharmacia) was added for 45 min. After five additional washes with PBST the complexes were revealed with TMB and read at A450. All experiments were performed in three replicates.
Cloning of interactors for PCA
Details for pα and pω vector construction are described in (26). The DNA fragments of interactors were cut from pPAO10 with BssHII and NheI restriction enzymes, and ligated into the pω vector. Human TG2 was PCR amplified from pTrcHis-htTG (25) with the forward primer htTG-EcoRI (5′-AGTCGGATCCGAATTCATGGCCGAGGAGCTGGTC-3′) and the reverse primer htTG-HindIII (5′-AGCTAAGCTTTTAGGCGGGGCCAATGATG-3′). The TG2 gene was digested with EcoRI and HindIII, and ligated into the pα vector. As positive control, co-transformed bacteria pα-RON2/pω-sc7 were used according to Secco et al. (26). pω-ΔG2 was used as negative control for the interaction with TG2: ΔG2 is a scFv recognizing a portion of the Cholera toxin (DG).
PCA validation of interactors
The DH5αF′ were co-transformed with both pα-TG2 and pω-interactor vectors. After incubation for 1 h at 37°C without selective pressure, the co-transformed cells were plated onto 2xTY agar plates, supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol, and incubated O/N at 30°C.
A single clone of each co-transformed bacteria was grown in 2xTY broth, supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol, at 37°C to OD600 0.5. Bacteria were plated either onto agar plate supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol for titration, or onto plates supplemented with increasing ampicillin concentration (15–20–30 µg/ml) and 1 mM IPTG. The plates were incubated at 28°C for 48 h and bacterial growth was scored from negative (–) to highly positive (+++).
RESULTS
Strategy
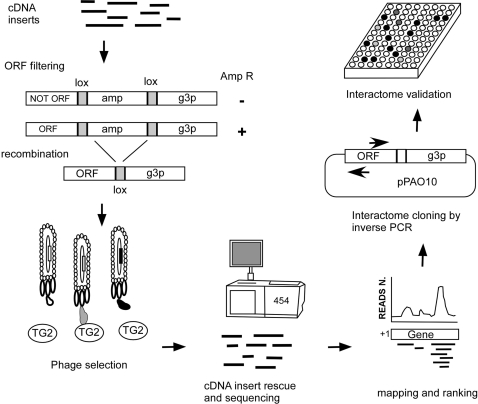
The overall strategy we have developed to identify the TG2 interactome consists of four key steps as outlined in Figure 1: (i) Generation of an ORF filtered phage display library. mRNA is fragmented into calibrated lengths, reverse transcribed into cDNA, normalized, cloned into the filtering vector and clones encoding ORFs are filtered out using ampicillin selection. (ii) Selection of interacting ORFs on a target protein. After elimination of the β-lactamase-coding region by Cre-lox mediated recombination, ORF displaying phages are challenged with purified recombinant TG2 through two cycles of selection and amplification. (iii) Massive sequencing of selected inserts and ranking of reads. cDNA insert fragments are recovered from the selected libraries, sequenced with 454 pyrosequencing and ranked according to their frequency. (iv) Recovery and validation of the most frequent ORFs. Highly scored ORFs are recloned from the library by inverse PCR (27) and their interactions validated by ELISA and the PCA .
Figure 1.
Interactome discovery pipeline. Poly A+ RNA is fragmented and retro-transcribed to cDNA by random priming. After normalization and linker ligation cDNAs are cloned into pPAO10 ORF-filtering vector and the library is filtered on chloramphenicol/ampicillin containing plates. After CRE-mediated recombination, ORF-displaying phages are challenged with TG2 throughout two cycles of selection and amplification. The ORF inserts of selected phages are obtained by digestion with restriction fragments and massively sequenced. Reads are mapped to the human genome and identified genomic regions are ranked on the basis of read coverage. Clones containing candidate interacting ORFs are rescued from selected libraries by inverse PCR using primers matching the contig sequence core. Recovered clones are expressed and validated by ELISA.
Construction of phage display ORFs cDNA library
PolyA+ RNA was obtained from human colon carcinoma cells (HCC), human lung fibroblasts (HF) and purified human pancreatic islets (HPI). mRNA samples were fragmented with a calibrated length of 100–600 bases in order to improve the uniformity of sequence coverage across transcripts (28). Orientated cDNA fragments (23) were prepared by random primer reverse transcription and normalization (22). Finally a cDNA display library was created within the context of an improved version of the pPAO2 phagemid vector (pPAO10, see Supplementary Figure 1 and Supplementary Data). After cDNA fragment ligation and transformation two libraries of 9 × 106 and 2.1 × 107 clones were obtained for pooled HCC+HF or HPI cDNAs, respectively. The size of the libraries was reduced to 4 × 105 and 1.2 × 106, respectively after ORF filtering by ampicillin selection. This corresponds to a reduction in clone numbers of ∼95% (95.6% for HCC+LF and 94.3% for HPI) which is in line with theoretical expectations as well as with previous observations (12), and indicates that ORF filtration was successful. On the basis of the observed numbers, each library represents a potential of at least 40 ORF fragments per gene, assuming that all 24 000 human genes are equally represented in the normalized cDNA.
Library characterization by massive sequencing
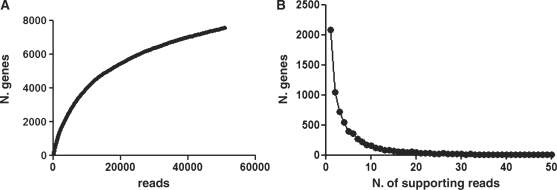
The two libraries were pooled (termed NS, for ‘not selected’ library), cDNA fragments were recovered after pooled phagemid digestion with BssHII and NheI restriction enzymes and were subjected to 454 sequencing. We obtained 67 587 reads from the NS library (Table 1). These were aligned to the human genome sequence (NCBI build 36) using gmap software. Totally 51 071 sequences had at least 95% identity and 90% overlap. 7576 genes were identified by at least 1 read (Figure 2A) (complete data at http://www.interactomeataglance.org) confirming the high diversity of the NS library. The efficacy of cDNA normalization was shown by the fact that 6259 (corresponding to 83%) of the identified genes were represented by no more than 10 reads in the library (Figure 2B). In order to analyze the efficacy of the ORF filtration in more detail we created a subset of so called ‘perfect sequences’. These had no sequence differences compared with the annotated genome sequence, and contained vector encoded restriction sites at both ends, indicating that they represented complete phage inserts. This was considerably facilitated by the long reads we could obtain with the 454 GS-FLX Titanium. Totally 12 119 reads fulfilling the ‘perfect sequences’ requirements were obtained. The length of 11 060 of these (91.2 %) sequences were multiples of 3 bp, corresponding to ORFs in our filtering system and 99% of these contained no stop codons, confirming the quality of the filtering procedure. Of those containing stop codons, 67% were amber codons that are suppressed in the DH5αF′ E.coli strain. Furthermore, 85% of these sequences mapped to the correct frame of the gene, and consequently represented real genes rather than spurious ORFs.
Table 1.
Summary of reads obtained from 454 sequencing
| NS | II-BIO | II-SP | |
|---|---|---|---|
| Total numer of sequences | 67 587 | 60 275 | 61 863 |
| Total with 5′ | 45 127 | 43 097 | 37 340 |
| Total with 3′ | 40 663 | 39 985 | 35 224 |
| Total with 5′ and 3′ | 27 946 | 30 541 | 24 049 |
| Average length | 245 | 246 | 211 |
| Reads mapping | 51 071 | 47 393 | 43 623 |
| Genes | 7576 | 7091 | 7524 |
Summary of reads obtained from 454 sequencing of not selected (NS) and selected (BIO, SP) libraries. Number of total sequences, and of sequences with vector encoded restriction site at one or both ends are reported. Reads were defined as mapping if aligned to the genome with at least 95% identity and 90% overlap. Genes identified by at least one read are reported.
Figure 2.
(A) Rank abundance curve obtained by plotting the total number of mapped informative reads (presence of both 454 primers) versus the total number of identified genes. (B) Chart shows how many genes are supported by different number of reads. As a result of cDNA normalization the vast majority of genes are represented by up to 10 reads while only very few genes show more reads (with a maximum of 680). X-axis has been limited to 50.
Library selection
ORF displaying phages were used for the selection of interacting proteins using purified recombinant human TG2 as the target. TG2 is a structurally complex multi-domain enzyme, undergoing extensive conformational changes under different physiological conditions (29,30). In order to account for the possibility that TG2 may be denatured by solid-phase absorption (perhaps leading to the binding of different targets), selection was carried out using two different methods: in the first the library was challenged with solid phase immobilized human TG2 (SP selection); in the second, soluble biotinylated TG2 was used (BIO Selection). In the latter case, after incubation with the phage library, the biotinylated TG2 along with potential associated phages was recovered after incubation with the phage library using streptavidin-conjugated magnetic beads. Bound phages were subsequently eluted with conventional methods. In order to avoid a significant restriction in output diversity (31) only two cycles of selection were performed for both the SP and BIO proteins. Twenty random colonies were checked after each selection: electrophoretic analysis of insert sizes and subsequent restriction fingerprinting showed a substantial diversity of selected clones, indicating that no single clone(s) dominated after the selection (not shown).
Massive sequencing of selected inserts and ranking of reads
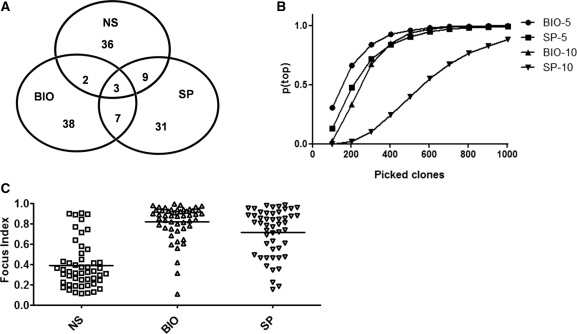
After two selection rounds using TG2 that was either directly immobilized (SP), or biotinylated (BIO), the selected cDNA fragments were recovered by digesting phagemid DNA with BssHII and NheI. The purified fragments were used for 454 based sequencing, yielding 61 863 (II-SP) and 60 275 (II-BIO) different reads (Table 1). Compared with the not selected (NS) library, the diversity after selection did not appear to be significantly reduced, in that sequences corresponding to over 7000 different genes for the two selections were matched by at least 1 read. However, this included a total of 3232 genes not listed in the original non-selected data set, indicating that the library diversity was even greater than that described by the first sequencing data set. Reads corresponding to the not selected and the selected libraries were mapped to the human genome and matched regions were ranked on the basis of the total number of reads obtained for every gene. The 50 genes found most frequently in the not selected and the two selected libraries (Supplementary Table S1) were further analyzed and validated. Among these genes (Figure 3A) only three were common to all three libraries and seven more to the two selected ones, indicating that the two TG2 forms used for selection appear to differ in their binding properties. Moreover, the differing composition of the two selections confirms that selection did not appear to be biased towards non-specific high frequency phage clones. Interestingly, the top five genes for the immobilized TG2 selection comprised only 4.9% of the total number of reads, while for the biotinylated TG2, the top five genes comprised 15.4% of reads, indicating that if 96 clones were picked at random for sequencing and/or ELISA, according to standard techniques, the liklihood of identifying all 10 of these clones would be very low. This is represented in Figure 3B where the theoretical chance [p(top)] of identifying the top five (or top 10) clones identified using massive sequencing is plotted versus the number of random picked clones that would have to be assayed in order to identify the same top ranking genes in a traditional system: to identify the top five genes with 95% certainty, over 500 random clones would have to be picked for each selection. The fact that the two different forms of TG2 selected different interactors may seem counterintuitive at first sight. However, this confirms results we published using phage display antibody libraries, in which we showed that the same antigen presented in different formats could result in the selection of different antibodies (31).
Figure 3.
(A) Venn diagram of the first 50 genes on the ranking list of non selected (NS) and selected (BIO, SP) libraries. (B) Probability of identifying all top five or top 10 genes by random selecting sample of clones of increasing size. Probability (pTop) has been approximated by a 10 000 cycle simulation for each sample size (picked clones). Simulation was performed using a custom developed PERL script. (C) Focus indexes for the first 50 genes of each library ranked on the basis of the number of supporting reads (coverage). The horizontal bars represent the mean value.
In order to determine whether the sequencing data could be used to provide information beyond gene identification, and identify domains within proteins that were responsible for binding to TG2, we developed a ‘focus index’ for each ranked gene. This represents the ratio between the depth of read coverage at the deepest site and the total number of reads per gene. The closer this ratio is to one, the more ‘focused’ the reads are to a single site or domain within the gene, while the lower the index, the more widely distributed the reads are on each gene. Figure 3C shows the focus index for the first 50 genes of each of the three libraries (NS, SP and BIO). It can be clearly seen that reads from the unselected library have a low focus index, indicating that these reads are distributed throughout each of the identified genes, while reads from the selected libraries have far higher focus indices, indicating that specific interacting domains have been selected (an example of such focussing is shown for genes MYO18A and LAP3 in Supplementary Figure S2).
Interacting clones validation
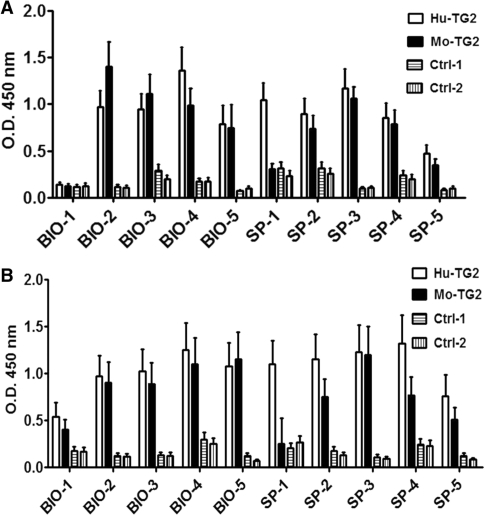
In order to confirm that the selection procedure really identified TG2 protein-binding partners, clones corresponding to the top five genes from each selection (Table 2) were selected for further analysis. All sequences were in the correct mRNA orientation and 7 out of 10 were in the correct reading frame, whereas two (SP-5 and BIO-4) were in reading frames 2 and 3, respectively, thus completely altering the actual amino acid sequence. These amino acid sequences were tested by protein BLAST aligment and did not give any significant homology to known proteins. The tenth clone (SP-4) is a non-protein-coding transcript which however in this context encodes for an ORF. The corresponding gene is reported in Table 2 only for reference. Clones were recovered from the corresponding selected library by inverse PCR (27) using a pair of specific primers centering on the region identified by the overlapping reads. After cloning, one to three different sequences were recovered for each of the 10 top scoring clones. After validation by sequencing, one clone for each interacting protein was selected for further analysis. cDNA fragments were expressed on the phage surface and analyzed for TG2 recognition by ELISA. All ten clones were tested by ELISA using both human and mouse TG2 as well as two unrelated control proteins (BSA and WNT4) in both solid phase and as soluble antigen. As shown in Figure 4A 9 of the 10 clones showed a positive signal with human TG2 (but not with the control proteins) when tested in solid phase ELISA. Interestingly, the specificity of the binding was also confirmed using recombinant mouse TG2; all cDNA fragments tested, except SP-1, gave the same results as with the human protein. The same clones were also positive when tested on the soluble antigens (Figure 4B), further confirming the specificity of the interaction.
Table 2.
Description of the ELISA-validated top five ranking clones from the selected libraries
| Code | Gene name and frame | Hugo | Accession number | First base | Length (bp) |
|---|---|---|---|---|---|
| SP selected clones | |||||
| SP-1 | Leucine aminopeptidase 3, frame 1 | LAP3 | NM_015907.2 | 1078 | 186 |
| SP-2 | Glutathione S-transferase omega 2, frame 1 | GSTO2 | NM_183239.1 | 623 | 177 |
| SP-3 | Fibronectin 1, transcript variant 1, frame 1 | FN1 | NM_212482.1 | 1581 | 390 |
| SP-4 | H19, imprinted maternally expressed transcript (non-protein coding) | H19 | NR_002196.1 | 50 | 114 |
| SP-5 | Heat shock 70 kDa protein 1 A, frame 2 | HSPA1A | NM_005345.4 | 1343 | 396 |
| BIO selected clones | |||||
| BIO-1 | Aldolase B, fructose-bisphosphate, frame 1 | ALDOB | NM_000035.2 | 126 | 354 |
| BIO-2 | Tetratricopeptide repeat domain 31, frame 1 | TTC31 | NM_022492.4 | 311 | 330 |
| BIO-3 | SPARC related modular calcium binding 1, frame 1 | SMOC1 | NM_022137.4 | 1139 | 408 |
| BIO-4 | Zinc finger protein 23 (KOX 16), frame 3 | ZNF23 | NM_145911.1 | 1728 | 498 |
| BIO-5 | Myosin XVIIIA, frame 1 | MYO18A | NM_078471.3 | 4956 | 291 |
Top five ranking clones from each selection. Ranking is based on the number of reads. Name, frame, abbreviation, accession number, first base of the gene fragment, as well as length of the clone is reported.
Figure 4.
ELISA reactivity of the top five clones from BIO and SP selection. Reactivity was tested on recombinant human and mouse TG2 proteins and on negative control proteins (BSA and WNT4) coated on solid surface (A) and as soluble biotinylated proteins (B).
Phage display of polypeptides is a reliable method for the identification of antigens and binding structures. Although it is considered to be highly specific, it generally requires confirmation of the binding using alternative methods. In our case, we used the PCA, a method previously used in our laboratory (26). All the selected TG2 interactors were cloned into the pω vector, in frame with the C-terminal fragment (aa 196–286) of TEM-1 β-lactamase, while the human TG2 gene was cloned into the pα vector, fused to the α fragment (aa 1–195, N-terminal). The rationale of the system is that when the two partners interact, β-lactamase activity is reconstituted and the clone gains resistance to ampicillin. Once a stable bacterial clone expressing TG2 in the pα context was obtained, it was transformed with plasmids encoding the individual interactors expressed in the pω vector. The resulting clones, with resistance to both chloramphenicol and kanamycin (encoded in the backbones of the two plasmids), were challenged with increasing amounts of ampicillin, ranging from 15 µg/ml to 30 µg/ml. The growth at each concentration was scored and reported on a semiquantitative scale ranging from negative (–) to very positive (+++). The results are summarized in Table 3. Specificity of interaction for TG2 was confirmed as most of the clones grew well even at the highest ampicillin concentration, and did not grow when tested against an unrelated target at the lowest ampicillin concentration. Two of the clones (SP-4 and BIO-3) grew slowly at higher ampicillin concentrations, while two (BIO-1 and BIO-4) did not grow at all on ampicillin. Of these four clones, SP-4 and BIO-4 were derived from ORFs that were in a different frame to that of the annotated gene.
Table 3.
Validation of selected clones by PCA assay
| TG2 |
Negative Control | ||||
|---|---|---|---|---|---|
| Cam/ Kan | 15 Amp | 20 Amp | 30 Amp | 15 Amp | |
| BIO-1 | + | – | – | – | – |
| BIO-2 | + | +++ | +++ | +++ | – |
| BIO-3 | + | ++ | + | + | – |
| BIO-4 | + | +/– | – | – | – |
| BIO-5 | + | +++ | +++ | +++ | – |
| SP-1 | + | +++ | +++ | +++ | – |
| SP-2 | + | +++ | +++ | +++ | – |
| SP-3 | + | +++ | +++ | +++ | – |
| SP-4 | + | ++ | + | + | – |
| SP-5 | + | +++ | +++ | +++ | –– |
| Positive control | + | +++ | +++ | +++ | + |
| Negative control | + | – | – | – | – |
In vivo validation of selected interactors. E. coli cells containing the pα-TG2 vector were transformed with the pω vector carrying the cloned interactors. Bacteria were plated on plates supplemented with kanamicin/chloramphenicol to check the presence of both vectors, and on plates supplemented with increasing ampicillin concentration (15 to 20 to 30 μg/ml) and IPTG 1 mM to assay interaction. Positive and negative controls are described in the ‘Materials and Methods’ section.
DISCUSSION
Over one thousand scientific reports related to TG2 have been published in the past five years, attesting to the growing interest in this enzyme. Although belonging to a well-known family of enzymes and being itself an extensively studied molecule, the biological significance of TG2 and its enzymatic functions are still far from completely elucidated, mainly due to the many different biological functions in which it is thought to be involved. The enzyme substrate specificity, and as many as 138 targets substrates, have been identified to date using a number of different techniques including phage display (33–35; listed in the TRANSDAB online database http://genomics.dote.hu/wiki/; 36). In contrast, only a limited number of interaction partners has been identified and experimentally validated: only nine partners are present in the same TRANSDAB database. It is therefore clear that an approach attempting to systematically identify TG2 protein-binding partners is very much needed, and the present study provides a first step in this direction. Within a more general context, TG2 is only one of many proteins with very complex interaction networks, the characterization of which requires appropriate tools that allow the screening of a large number of potential interacting partners. In the present work, we show that by using massive sequencing in combination with phage display cDNA libraries, we are able to increase the number of screened clones by at least two orders of magnitude compared with a typical ELISA-based screen employing microtiter plates. In such a conventional strategy only a few hundred clones are assayed, whereas here we report more than 60 000 sequences, with the theoretical upper limit restricted only by the capacity of the sequencing system. Despite replacing a relatively cheap assay (ELISA) with a more expensive one (massive sequencing) may seem a major drawback, several important details should be considered. First, the amount of information obtained by massive sequencing far exceeds that which can be obtained by traditional screening. Second, positive clones identified by ELISA still need identification by standard sequencing, and the number of required sequences can be very high when the binders of interest are many and a few clones are over-represented. Thirdly, massive sequencing provides a ranking of the most frequently selected clones, with the number of times each is selected. In Figure 3B, we have carried out a simulation of the chance of identifying the top five (or 10) clones identified using massive sequencing for a specific number of picked clones. For example, if 100 clones for each selection were picked at random, the chance that the top ranked five clones for each selection would be picked is 30% for the BIO selection and 13% for the SP selection (and essentially zero for the top ten clones). In fact, one would have to pick 1000 clones to have a 99% chance of picking the top five clones in both selections. In this article we carried out analysis after two rounds of selection, which gives the best balance between high numbers of positive clones and broad diversity. However, given the great analytical potential of this method, it may be possible to carry out analysis after a single selection round, comparing the net enrichment of clones by direct comparison to the unselected library. This is likely to identify significantly more potential interacting partners than could ever be analyzed using the traditional approach.
Finally, by virtue of the extremely deep sequencing that is carried out, in addition to providing information on interacting proteins, this method also identifies the domains responsible for the interactions directly in a first screen. Such detailed information on the interaction domains would not be possible using traditional methods, where little more than the identification of the interacting gene would be expected to be obtained, and following such identification, new libraries would have to be created for each protein in order to identify the responsible interacting domain.
Although the costs of 454 sequencing are relatively high at the moment, greater availability is likely to lead to a reduction in price, and as barcoding becomes more sophisticated, multiplexed analysis will become more straightforward, allowing broader screening, and the profiling of the interactomes of several players involved in a biological process simultaneously. In this study we confirmed the reliability of the β-lactamase based selection system to select ORFs. We have shown that over 91% of clones filtered using this system comprise ORFs representing multiples of 3 nt that lack non-suppressible stop codons.
Seven out of the ten top ranking sequences identified were derived from the correct frame. In general, cloned sequences not expressed in the correct frame but nevertheless recognized by a reactant (e.g. in immunorecognition) are described as mimotopes; i.e. random polypeptides whose structures resemble those of other proteins normally recognized by the selector. These two clones were confirmed as being mimotopes after they were recloned with the addition of an extra base to allow translation in frame 1. The subsequent ELISA analysis (not shown) revealed a loss in recognition by both human and mouse TG2, indicating that the polypeptide translated in the annotated frame does not interact with TG2.
Of the identified genes, three have been previously reported as either directly interacting with TG2, or as belonging to a family of proteins, a member of which has been identified as interacting with TG2. Fibronectin (FN1) (Table 2, clone SP-3) has been described as interacting with TG2 (36), and has been extensively studied for its implication in TG2 mediated cell adhesion to the extracellular matrix (37). Fibronectin was third in the ranking of the solid phase selection with 685 sequences, and 23 rd in the BIO selection with 181 sequences. When all these sequences were aligned we were not only able to confirm the region previously proposed for TG2 interaction (37), but to further restrict the size from a relatively large 42-kDa fragment to one of only 130 aa, thus confirming the value of obtaining multiple sequences for each interacting protein. Secreted modular calcium-binding (SMOC1) (Table 2, clone BIO-3) is a member of the BM-40/SPARC family of matricellular proteins thought to influence growth factor signaling, migration, proliferation, and angiogenesis (38). SPARC was identified as a TG2 interacting partner using Y2H in a proteome-scale map of the human protein–protein interaction network (39). Finally glutathione S-transferase omega 2 (GSTO2) (Table 2, clone SP2), is part of a family of enzymes that play an important role in detoxification by catalyzing the conjugation of many hydrophobic and electrophilic compounds with reduced glutathione. Glutathione S-transferase P was identified as a TG2-binding partner in immunoprecipitation experiments (40). Although TG2 is found mainly in the cytoplasm, it is also found in the nucleus, plasma membrane, and the extracellular matrix, making it difficult to use cellular co-localization as confirmatory information in this specific case.
On this basis, the other proteins may be considered to belong to a novel class of putative TG2 interacting partners that could be added to the fast-growing TG2 TRANSDAB database (35). Furthermore, a more detailed analysis of many of the other selected gene fragments will likely identify many other potential interaction partners.
In conclusion, by using this cDNA fragment phage display library, we have shown the power that the application of massive 454 deep sequencing can bring to library analysis, and in particular, as it relates to the analysis of protein–protein interactions. In fact, further confirmation of the results described here may allow the replacement of protein expression and ELISA testing for protein–protein interactions with massive deep sequencing. We anticipate that this approach to library and selection analysis can also be extended to the other methods traditionally used to study protein–protein interactions (i.e. PCA and Y2H) as well as to the study of the selection of peptides and antibodies by phage display. Furthermore, beyond the identification of proteins involved in interactions, the use of libraries of fragmented genes as described here also localizes the regions of interactions to domains, rendering the information more useful.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fondazione CRT Progetto Alfieri; Fondazione Cariplo Bando Ricerca Biomedica 2009; National Institutes of Health [NIH RFA-DK-06-002] to D.S.; Compagnia Sanpaolo to C.S.; EC Marie Curie Research Training Network [contract n. MRTN-CT-2006-036032] to R.M. Funding for open access charge: Fondazione CRT Progetto Alfieri.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Huang BC, Liu R. Comparison of mRNA-display-based selections using synthetic peptide and natural protein libraries. Biochemistry. 2007;46:10102–10112. doi: 10.1021/bi700220x. [DOI] [PubMed] [Google Scholar]
- 2.Shen X, Valencia CA, Gao W, Cotten SW, Dong B, Huang BC, Liu R. Ca(2+)/Calmodulin-binding proteins from the C. elegans proteome. Cell Calcium. 2008;43:444–456. doi: 10.1016/j.ceca.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 3.He M, Liu H, Turner M, Taussig MJ. Detection of protein-protein interactions by ribosome display and protein in situ immobilisation. N. Biotechnol. 2009;26:277–281. doi: 10.1016/j.nbt.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Suter B, Kittanakom S, Stagljar I. Interactive proteomics: what lies ahead? Biotechniques. 2008;44:681–691. doi: 10.2144/000112799. [DOI] [PubMed] [Google Scholar]
- 5.Facchiano F, Facchiano A, Facchiano AM. The role of transglutaminase-2 and its substrates in human diseases. Front Biosci. 2006;11:1758–1773. doi: 10.2741/1921. [DOI] [PubMed] [Google Scholar]
- 6.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caberoy NB, Zhou Y, Alvarado G, Fan X, Li W. Efficient identification of phosphatidylserine-binding proteins by ORF phage display. Biochem. Biophys. Res. Commun. 2009;386:197–201. doi: 10.1016/j.bbrc.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caberoy NB, Zhou Y, Jiang X, Alvarado G, Li W. Efficient identification of tubby-binding proteins by an improved system of T7 phage display. J. Mol. Recognit. 2010;23:74–83. doi: 10.1002/jmr.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hust M, Meysing M, Schirrmann T, Selke M, Meens J, Gerlach GF, Dubel S. Enrichment of open reading frames presented on bacteriophage M13 using hyperphage. Biotechniques. 2006;41:335–342. doi: 10.2144/000112225. [DOI] [PubMed] [Google Scholar]
- 10.Faix PH, Burg MA, Gonzales M, Ravey EP, Baird A, Larocca D. Phage display of cDNA libraries: enrichment of cDNA expression using open reading frame selection. Biotechniques. 2004;36:1018–1022, 1024, 1026–1019. doi: 10.2144/04366RR03. [DOI] [PubMed] [Google Scholar]
- 11.Ansuini H, Cicchini C, Nicosia A, Tripodi M, Cortese R, Luzzago A. Biotin-tagged cDNA expression libraries displayed on lambda phage: a new tool for the selection of natural protein ligands. Nucleic Acids Res. 2002;30:e78. doi: 10.1093/nar/gnf077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zacchi P, Sblattero D, Florian F, Marzari R, Bradbury AR. Selecting open reading frames from DNA. Genome Res. 2003;13:980–990. doi: 10.1101/gr.861503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Niro R, Ferrara F, Not T, Bradbury AR, Chirdo F, Marzari R, Sblattero D. Characterizing monoclonal antibody epitopes by filtered gene fragment phage display. Biochem. J. 2005;388:889–894. doi: 10.1042/BJ20041983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waldo GS. Genetic screens and directed evolution for protein solubility. Curr Opin Chem Biol. 2003;7:33–38. doi: 10.1016/s1367-5931(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 15.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 1999;17:691–695. doi: 10.1038/10904. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell KL, Mittermaier AK, Forman-Kay JD, Davidson AR. A simple in vivo assay for increased protein solubility. Protein Sci. 1999;8:1908–1911. doi: 10.1110/ps.8.9.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wigley WC, Stidham RD, Smith NM, Hunt JF, Thomas PJ. Protein solubility and folding monitored in vivo by structural complementation of a genetic marker protein. Nat. Biotechnol. 2001;19:131–136. doi: 10.1038/84389. [DOI] [PubMed] [Google Scholar]
- 18.Griffin M, Casadio R, Bergamini CM. Transglutaminases: nature's; biological glues. Biochem. J. 2002;368:377–396. doi: 10.1042/BJ20021234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fesus L, Piacentini M. Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem. Sci. 2002;27:534–539. doi: 10.1016/s0968-0004(02)02182-5. [DOI] [PubMed] [Google Scholar]
- 20.Hasegawa G, Suwa M, Ichikawa Y, Ohtsuka T, Kumagai S, Kikuchi M, Sato Y, Saito Y. A novel function of tissue-type transglutaminase: protein disulphide isomerase. Biochem. J. 2003;373:793–803. doi: 10.1042/BJ20021084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mishra S, Murphy LJ. Tissue transglutaminase has intrinsic kinase activity: identification of transglutaminase 2 as an insulin-like growth factor-binding protein-3 kinase. J. Biol. Chem. 2004;279:23863–23868. doi: 10.1074/jbc.M311919200. [DOI] [PubMed] [Google Scholar]
- 22.Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y. Normalization and subtraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res. 2000;10:1617–1630. doi: 10.1101/gr.145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Einat P, Zevin-Sonkin D, Gilad S. PCT Patent Publication No. WO 2004/111182. 2008. [Google Scholar]
- 24.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 25.Marzari R, Sblattero D, Florian F, Tongiorgi E, Not T, Tommasini A, Ventura A, Bradbury A. Molecular dissection of the tissue transglutaminase autoantibody response in celiac disease. J Immunol. 2001;166:4170–4176. doi: 10.4049/jimmunol.166.6.4170. [DOI] [PubMed] [Google Scholar]
- 26.Secco P, D'A;gostini E, Marzari R, Licciulli M, Di Niro R, D'A;ngelo S, Bradbury AR, Dianzani U, Santoro C, Sblattero D. Antibody library selection by the {beta}-lactamase protein fragment complementation assay. Protein Eng. Des. Sel. 2009;22:149–158. doi: 10.1093/protein/gzn053. [DOI] [PubMed] [Google Scholar]
- 27.Hoskins RA, Stapleton M, George RA, Yu C, Wan KH, Carlson JW, Celniker SE. Rapid and efficient cDNA library screening by self-ligation of inverse PCR products (SLIP) Nucleic Acids Res. 2005;33:e185. doi: 10.1093/nar/gni184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 29.Mariani P, Carsughi F, Spinozzi F, Romanzetti S, Meier G, Casadio R, Bergamini CM. Ligand-induced conformational changes in tissue transglutaminase: Monte Carlo analysis of small-angle scattering data. Biophys. J. 2000;78:3240–3251. doi: 10.1016/S0006-3495(00)76860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pinkas DM, Strop P, Brunger AT, Khosla C. Transglutaminase 2 undergoes a large conformational change upon activation. PLoS Biol. 2007;5:e327. doi: 10.1371/journal.pbio.0050327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou J, Marzari R, Verzillo V, Ferrero F, Pak D, Sheng M, Yang C, Sblattero D, Bradbury A. Antibodies in haystacks: how selection strategy influences the outcome of selection from molecular diversity libraries. J. Immunol. Methods. 2001;253:233–242. doi: 10.1016/s0022-1759(01)00385-4. [DOI] [PubMed] [Google Scholar]
- 32.Sugimura Y, Hosono M, Wada F, Yoshimura T, Maki M, Hitomi K. Screening for the preferred substrate sequence of transglutaminase using a phage-displayed peptide library: identification of peptide substrates for TGASE 2 and Factor XIIIA. J. Biol. Chem. 2006;281:17699–17706. doi: 10.1074/jbc.M513538200. [DOI] [PubMed] [Google Scholar]
- 33.Keresztessy Z, Csosz E, Harsfalvi J, Csomos K, Gray J, Lightowlers RN, Lakey JH, Balajthy Z, Fesus L. Phage display selection of efficient glutamine-donor substrate peptides for transglutaminase 2. Protein Sci. 2006;15:2466–2480. doi: 10.1110/ps.051818406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Esposito C, Caputo I. Mammalian transglutaminases. Identification of substrates as a key to physiological function and physiopathological relevance. FEBS J. 2005;272:615–631. doi: 10.1111/j.1742-4658.2004.04476.x. [DOI] [PubMed] [Google Scholar]
- 35.Csosz E, Mesko B, Fesus L. Transdab wiki: the interactive transglutaminase substrate database on web 2.0 surface. Amino Acids. 2009;36:615–617. doi: 10.1007/s00726-008-0121-y. [DOI] [PubMed] [Google Scholar]
- 36.Jones RA, Nicholas B, Mian S, Davies PJ, Griffin M. Reduced expression of tissue transglutaminase in a human endothelial cell line leads to changes in cell spreading, cell adhesion and reduced polymerisation of fibronectin. J. Cell. Sci. 1997;110:2461–2472. doi: 10.1242/jcs.110.19.2461. [DOI] [PubMed] [Google Scholar]
- 37.Akimov SS, Krylov D, Fleischman LF, Belkin AM. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell. Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Podhajcer OL, Benedetti L, Girotti MR, Prada F, Salvatierra E, Llera AS. The role of the matricellular protein SPARC in the dynamic interaction between the tumor and the host. Cancer Metastasis Rev. 2008;27:523–537. doi: 10.1007/s10555-008-9135-x. [DOI] [PubMed] [Google Scholar]
- 39.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 40.Piredda L, Farrace MG, Lo Bello M, Malorni W, Melino G, Petruzzelli R, Piacentini M. Identification of ‘tissue’ transglutaminase-binding proteins in neural cells committed to apoptosis. FASEB J. 1999;13:355–364. doi: 10.1096/fasebj.13.2.355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.