Abstract
It has been shown that alternative splicing is especially prevalent in brain and testis when compared to other tissues. To test whether there is a specific propensity of these tissues to generate splicing variants, we used a single source of high-density microarray data to perform both splicing factor and exon expression profiling across 11 normal human tissues. Paired comparisons between tissues and an original exon-based statistical group analysis demonstrated after extensive RT-PCR validation that the cerebellum, testis, and spleen had the largest proportion of differentially expressed alternative exons. Variations at the exon level correlated with a larger number of splicing factors being expressed at a high level in the cerebellum, testis and spleen than in other tissues. However, this splicing factor expression profile was similar to a more global gene expression pattern as a larger number of genes had a high expression level in the cerebellum, testis and spleen. In addition to providing a unique resource on expression profiling of alternative splicing variants and splicing factors across human tissues, this study demonstrates that the higher prevalence of alternative splicing in a subset of tissues originates from the larger number of genes, including splicing factors, being expressed than in other tissues.
INTRODUCTION
The large functional difference between tissues results from complex regulatory machineries that control the tissue-specific expression of genes and yield in turn tissue-specific proteome ensuring tissue-specific functions. Great advances have been made using DNA microarrays to profile gene expression across tissues (1,2). However, large-scale exon expression profiling is necessary to better characterize tissue-specific gene expression regulation. Indeed, the analysis of expressed sequence tags (ESTs), splicing-sensitive microarrays, and high-throughput sequencing data have revealed that most human genes (upto 95% of multiexon genes) generate transcripts having a different exon content, by using alternative promoters [resulting in alternative first exons (AFEs)], alternative polyadenylation sites [resulting in alternative last exons (ALEs)] and alternatively spliced exons (ASEs) (3–9). Different exon combinations then impact on the protein isoforms produced. Indeed, 75% of ASEs result in the removal of protein motifs or domains; AFEs can result in the production of protein isoforms with different N-terminal sequences; and ALEs can result in the production of C-terminal truncated protein isoforms (10–13). Therefore, all these mechanisms play a critical role in increasing the proteome diversity encoded by a limited number of genes. For this reason, we and others developed online resources designed to provide access to reliable annotations of the transcriptome at the exon level (14–18). These resources describe the nature (i.e. exon content) of the transcripts produced by each gene, as well as the potential protein isoforms generated.
In addition to participating in cellular homeostasis, it has been well-established that the differential exon selection process plays a critical role during cellular differentiating programs and development by participating in the production of a tissue-specific proteome (19–26). Therefore, a major challenge is now to develop databases describing the nature of the splicing variants expressed by each gene across normal tissues. Noteworthy, large-scale analyses based on ESTs, splicing sensitive arrays and deep sequencing have demonstrated that some tissues, including brain and testis, expressed more alternatively spliced transcripts than any other tissues, suggesting that brain and testis possess an unusually high level of alternative splicing (3,4,6–8,25–33). However, the mechanisms behind the capability of some tissues to generate a larger number of splicing variants need further investigations. In particular, genome-wide analyses of tissue-specific alternative splicing events and tissue-specific splicing factor expression profile have not been analyzed and compared in the same dataset yet (32,34). Indeed, regulated ASEs require the interplay of cis- and trans-acting factors that repress or activate splice site selection. It has been well-established that variations of the expression level of trans-acting splicing factors play a critical role in tissue-specific alternative splicing. These splicing factors are members of several protein families, including the SR, hnRNP, RBM, MBNL, CELF/CUGBP and KH families (4,19–24,34).
In this work, we performed a genome-wide transcriptomic analysis at the gene and at the exon levels across 11 normal human tissues by using a unique data set that is the Human Exon 1.0 ST Array tissue panel dataset from Affymetrix (8). We provide a list of more than 10 000 exons being differentially expressed across normal human tissues, some of them being validated by Reverse transcription-Polymerase chain reaction (RT-PCR). In addition, a freely available web interface (www.fast-db.com/cgi-bin/easana/index.pl) permits, after registration, to display the tissue dataset from any gene, which provides information on the tissue-specific levels of alternative exons of the query gene. Our analysis confirmed previous observations that tissues like cerebellum and testis express a larger set of transcripts with different exon content when compared to other tissues. We also analyzed and provided the expression profile of 45 splicing regulatory factors across normal tissues. By performing exon and gene expression profiling in the same dataset, we showed that the prevalence of alternative splicing in the cerebellum and testis is likely to originate from a larger number of genes, including genes coding for splicing factors that are more expressed in these tissues.
MATERIALS AND METHODS
Affymetrix exon array data analysis
The publicly available Human Exon 1.0 ST Array tissue dataset (http://www.affymetrix.com/support/technical/sample_data/exon_array_data.affx/) consists in 11 normal human triplicate tissues. The Human Exon 1.0 ST Array tissue panel dataset analysis and visualization were made using EASANA® (GenoSplice technology, www.genosplice.com), which is based on the FAST DB annotation (17,18). The EASANA® visualization module is a web-based interface available after registration at www.fast-db.com/cgi-bin/easana/index.pl.
Data pre-treatment
Exon Array data were normalized by using quantile normalization method. Background correction was made by using the antigenomic probes and probe selection was made as described previously (8). Only probes targeting exons annotated from FAST DB transcripts were selected in order to focus on well-annotated genes, whose mRNA sequences are in public databases (17,18). Among these selected probes, bad-quality probes (e.g. probes labeled by Affymetrix as ‘cross-hybridizing’) and probes with too low intensity signal compared to antigenomic background probes with the same GC content were removed from the analysis. Only probes with a DABG P-value ≤0.05 in at least half of the arrays were considered for statistical analysis (8).
Paired comparisons
Fifty-five paired comparisons were performed by comparing tissues to each other. Differentially expressed exons were identified using the splicing index strategy (8) between triplicate experiment sets from two tissues. Only exons from genes expressed in both compared tissues were analyzed. To be considered as expressed, the Log2 gene signal intensity had to be ≥6.0 and the DABG P-value had to be ≤0.05 for at least half of the gene probes. We performed a paired Student’s t-test to compare the gene-normalized intensity (corresponding to the probe expression level relative to the gene expression level) of each probe in tissue paired comparisons. Therefore, all the probes from an exon had to change similarly to predict the exon as being differentially expressed. Exons were considered significantly differentially regulated when the splicing index fold change was ≥1.5 and the splicing index P-value was ≤0.05.
Tissue group comparisons
An exon-based statistical group analysis was performed by also using the splicing index strategy. First, the ‘gene-normalized exon intensity’ value in each tissue was calculated for all the exons of all the genes significantly expressed in this tissue. To each exon corresponded three values as the arrays data were generated in triplicate. Second, the average of the ‘gene-normalized exon intensity’ values was calculated for each exon and each tissue (Supplementary Figure S1). Therefore, several numbers (each of them corresponding to the average of the gene-normalized exon intensity values in one tissue) were attributed to each exon. These numbers were then sorted by ascending order for each exon to generate tissue groups. Third, the averages of the gene-normalized exon intensity values were then replaced by the corresponding values obtained in the three experiments in order to perform a statistical analysis on the ascending order. Thanks to this strategy, an unpaired Student’s t-test was performed for each possible group, each group being between three values (when the group contained only one tissue) and 30 values (when the group contained 10 tissues). Finally, the cut point defining two groups of values being statistically significantly different was chosen according to the lower associated p-value of each possible comparison. Groups were considered significantly different when the p-value was ≤0.05 and the fold change was ≥1.5. Constituted groups were then separated in individual tissues. A similar method was used for gene expression-level analysis using the ‘gene signal’ that corresponded to the average of all the selected gene probes. Non-supervised hierarchical clustering (Mev4.0 software from TIGR) using Euclidean distance with complete linkage method was carried out to cluster gene-normalized intensity of differentially regulated exons of genes expressed in at least six tissues.
Splicing and transcription factor gene expression across tissues
The gene signals from 45 splicing factors identified in the literature and 1250 transcription factors identified in the DBD database (http://dbd.mrc-lmb.cam.ac.uk) were retrieved after pre-treatment of the data. Intensity values were displayed with MeV4.0. In addition, the average of the gene signal in the 11 tissues was calculated for each splicing or transcription factor. The distance from the gene signal in a given tissue to the corresponding average in the 11 tissues was calculated. The number of splicing or transcription factors with a tissue-specific gene expression level either above or below (with a 1.2 factor) the gene expression average value in the 11 was calculated.
Global gene expression level
Probe expression analysis was performed with the Expression Console software from Affymetrix (8) to count the number of probesets and genes expressed above DABG across the 11 tissues.
Validation by RT-PCR
One microgram of total RNAs from human breast, cerebellum, heart, liver, muscle, spleen or testis (BioChain) was reverse-transcribed using random primers and the Superscript II® reverse transcriptase (Invitrogen). cDNAs were diluted 400 times and 5 µl of the diluted cDNAs were used for PCR amplification using GoTaq® DNA polymerase (Promega). Primer sequences are provided in Supplementary Table S1.
RESULTS
Identification of differentially expressed exons across 11 normal human tissues
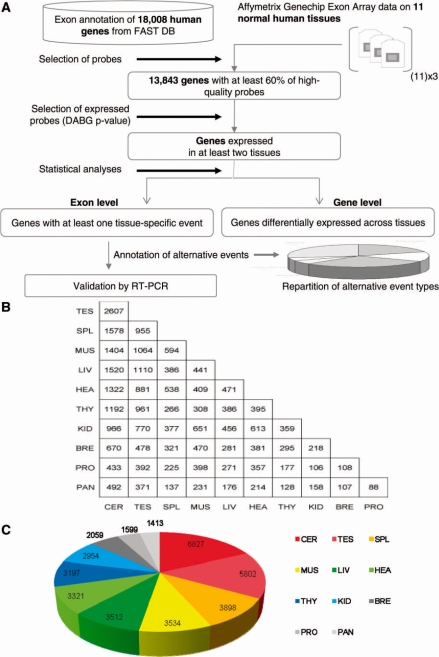
To identify differentially expressed exons across normal human tissues, we analyzed the publicly available dataset from Affymetrix (www.affymetrix.com) in which RNAs from 11 normal human tissues were hybridized on GeneChip® Human Exon 1.0 ST Arrays. Exon arrays contain multiple probes per exon, allowing to analyze gene expression at both transcript and exon levels (8). Using the EASANA® analysis system from GenoSplice technology (www.genosplice.com), 13 843 human genes were analyzed after the selection of ‘good-quality’ probes targeting well-annotated exons of genes with known mRNAs (Figure 1A). Fifty-five paired comparisons of the tissues with each others were performed in order to identify the largest number of differentially expressed exons across tissues. For each paired comparison of tissues, only genes significantly expressed in both tissues were considered for analysis at the exon level. A Student’s t-test was performed to test the difference between ‘splicing index values’ as previously reported (8). Differences between ‘splicing index values’ were considered statistically significant for fold-changes ≥1.5 and P-values ≤0.05.
Figure 1.
Identification of differentially expressed exons by paired comparisons. (A) Workflow. The exon and gene expression profiling across 11 normal tissues was performed using the same dataset from Affymetrix. Stringent criteria were used to select probes from genes expressed in at least two tissues in order to compare the exon content of well-expressed transcripts produced from annotated genes. Among 18 008 human genes annotated in FAST DB based on publicly available mRNA sequences, 13 843 genes were defined by >60% of high-quality probes. (B) Number of exons being differentially expressed when comparing two tissues as indicated. Fifty-five paired comparisons were performed by comparing each tissue to each other. Comparisons between two tissues were performed by considering only genes that were well expressed in both tissues. (C) Number of differentially expressed exons identified for each tissue. Each tissue contained a number of unique differentially expressed exons.
Between 88 and 2607 differentially expressed exons were identified depending on the paired comparison (Figure 1B). After, considering individual tissues, between 1413 (pancreas) and 6827 (cerebellum) differentially expressed exons were identified by taking into account that some exons were simultaneously found in several paired comparisons (Figure 1C). Across the 11 tissues, 11 196 different exons were differentially expressed, which corresponded to 3264 different genes. The cerebellum and testis, followed by the spleen, were the tissues with the largest number of exons being differentially expressed (Figure 1B and C). The list of the differentially expressed exons obtained for each paired comparison is given in Supplementary Table S2, which provides a unique resource of differentially expressed alternative exons.
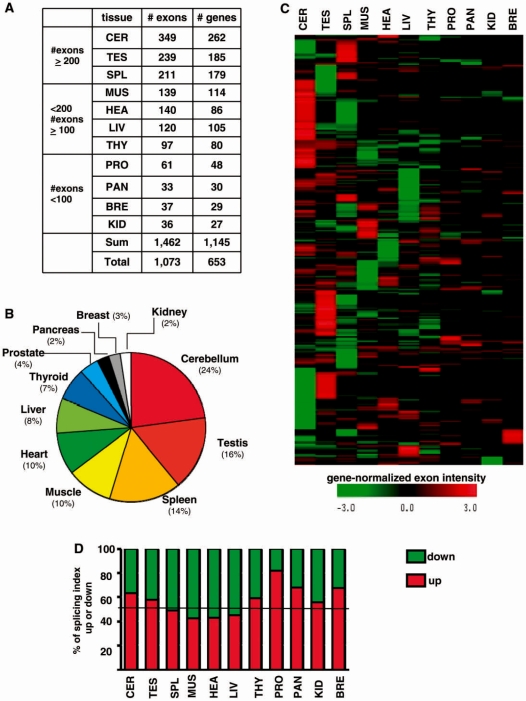
While paired comparisons of tissues identified exons that were differentially expressed between two tissues, they did not allow to determine which tissue or group of tissues specifically expressed a given splicing variant. For that purpose, we compared the exon content of the products of each human gene by considering all the tissues where the gene was expressed in order to identify alternative exons specific to tissues. An exon-based statistical group analysis was performed using the ‘gene-normalized exon intensity’ value that corresponded to the exon expression level relative to the gene expression level as described in ‘Materials and methods’ section and in Supplementary Figure S1. Using this strategy, 1073 unique exons corresponding to 653 unique genes were associated with individual tissues or a group of tissues (including no more than five tissues within a group). Exons found in a group of tissues were then associated with each individual tissue of the group. Between 33 and 349 differentially expressed exons were associated with each tissue as indicated on Figure 2A. The list of the genes containing exons that were differentially expressed in specific tissues is given in Supplementary Table S3, which provides a unique resource of exons being differentially expressed in a tissue-specific manner.
Figure 2.
Identification of differentially expressed exons by group comparison. (A) Number of differentially expressed exons and number of corresponding genes identified by a group comparison. Tissue ranking was done depending on the number of differentially expressed exons associated with each tissue. (B) Proportion of differentially expressed exons identified for each tissue. The number of differentially expressed exons associated with each tissue was divided by the total number of differentially expressed exons identified by a group comparison. (C) Non-supervised hierarchical clustering of gene-normalized exon intensities. Differentially regulated exons across 11 tissues were hierarchically clustered based on their gene-normalized exon intensity. Color scale representing gene-normalized exon intensity is shown below the clustergram. (D) Proportion of ‘splicing index’ fold-changes that were up or down in the tissue group comparison. ‘Splicing index’ values corresponding to the exons identified in the statistical group comparison for each individual tissue were either up (preferentially included) or down (preferentially excluded).
Three major groups of tissues were revealed by this analysis according to the differential expression of exons. Cerebellum, testis and spleen had the largest amount and proportion of differentially expressed exons (i.e. 24, 16 and 14%, respectively) when compared to the other tissues (Figure 2A and B). A second group of tissues was composed of muscle, heart, liver and thyroid that contained between 7 and 10% of the exons being differentially expressed across tissues. A third group was composed of the prostate, pancreas, breast and kidney that contained,<4% of the exons being differentially expressed across tissues.
A non-supervised hierarchical clustering analysis using Mev4.0 software (TIGR, http://www.tm4.org/) based on ‘gene-normalized exon intensity’ values supported this observation. It revealed that each tissue contained alternative exons that were preferentially either included or excluded, as there were exons with both positive and negative ‘gene-normalized exon intensity’ values in each tissue (Figure 2C and D). Therefore, both paired comparisons of tissues and exon-based statistical group analyses revealed that cerebellum, testis and spleen possess a larger proportion of differentially expressed exons than other tissues.
Annotation of the transcriptome at the exon level and validation by RT-PCR
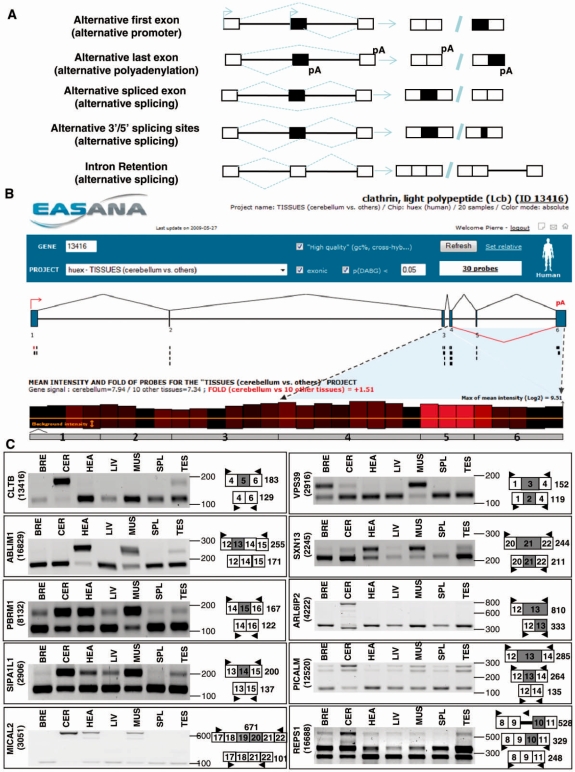
To test by RT-PCR some of the events identified above, we selected exons predicted in the group comparison that were also predicted in several paired comparisons. Within them, we next selected exons with splicing index fold-changes within the range of all the splicing index fold-changes predicting alternative exons (Supplementary Tables S6 and 8). We next developed an approach to classify the differentially expressed exons into AFEs resulting from alternative usage of promoters, ALE resulting from alternative usage of polyadenylation sites, or ASE (Figure 3A). In addition, ASEs can be extended or shortened by the usage of alternative 5′- and 3′-splice sites, and introns can be spliced out or retained (Figure 3A). In an attempt to classify the 1073 differentially expressed exons across tissues identified above (Figure 2A), a manual inspection was performed after uploading the Exon Array data into the EASANA® visualization module, which is based on the FAST DB annotations. FAST DB is a database gathering all the known and well-annotated human alternative transcripts (17,18). By computational comparison of publicly available mRNA sequences with genomic sequences, alternative exons have been annotated in FAST DB as AFEs, ASEs, or ALEs (17,18).
Figure 3.
Annotation and validation of ASEs. (A) Classification of alternative exons. Alternative exons can be classified either as AFEs, ALEs or ASEs. ASEs can be extended or shortened through the use of alternative 5′- and 3′-splice sites and introns can be spliced out or retained. (B) CLTB gene analysis. Genomic structure of the CLTB gene (upper panel). According to FAST DB exon numeration and annotation, the human CLTB gene contains 6 exons and one ASE (indicated by red lines below exon 5). Graphical representation of the exon array probes corresponding to the CLTB gene in the cerebellum compared to the other tissues (lower panel). Each Affymetrix probe corresponding to the CLTB gene is represented by a bar above the numbered grey exons of the gene. The height of each bar is proportional to the mean intensity of the corresponding probe, as calculated from three independent experiments for a given tissue (in this case, the cerebellum). In addition, the color of each bar is proportional to the differences in probe intensities between samples (in this case, cerebellum compared to the mean intensity calculated for the 10 other tissues): a red bar means that the intensity of the corresponding probe was greater in the cerebellum; black corresponds to probes with no intensity variation; and a green bar indicates that the intensity of the corresponding probe was lower in the cerebellum. The bright red bars corresponding to exon 5 indicate that exon 5 may be more included in the cerebellum than in other tissues. Screenshot is from EASANA®. (C) RT-PCR analysis. RT-PCR analyses for several ASEs predicted to be differentially expressed between tissues.
To illustrate the annotation process, the CLTB gene was provided as an example on Figure 3B. A brain-specific CLTB splicing variant containing a supplementary exon (exon 5 on Figure 3B) has been cloned and results in the production of a protein isoform containing a supplementary conserved region of 22 residues near the amino terminus (35). The alternative splicing of exon 5 is indicated by a broken red line below exon 5 (upper panel, Figure 3B). To manually inspect the Exon Array data, each Affymetrix probe corresponding to the CLTB gene is computationally represented by a bar above the numerated gray exons (lower panel, Figure 3B). The color of each bar indicates the variation of the probe intensity across tissues (in that case, the probe intensity in the cerebellum compared to the mean probe intensity obtained in the 10 other tissues). Bright red bars corresponding to exon 5 probes (lower panel, Figure 3B) suggested that CLTB exon 5 was more frequently included in the cerebellum than in the other tissues. This prediction was validated by RT-PCR as exon 5 was specifically included in the cerebellum (CLTB, Figure 3C). Likewise, several cassette exons were identified and validated, as illustrated for the ABLM1, PBRM1, SIPA1L1, MICAL2 and VPS39 genes (Figure 3C). Cases of 5′- and 3′-alternative spliced sites and intron retentions were also identified and validated, as illustrated for the SXN13, ARL6IP2, PICALM and REPS1 genes (Figure 3C).
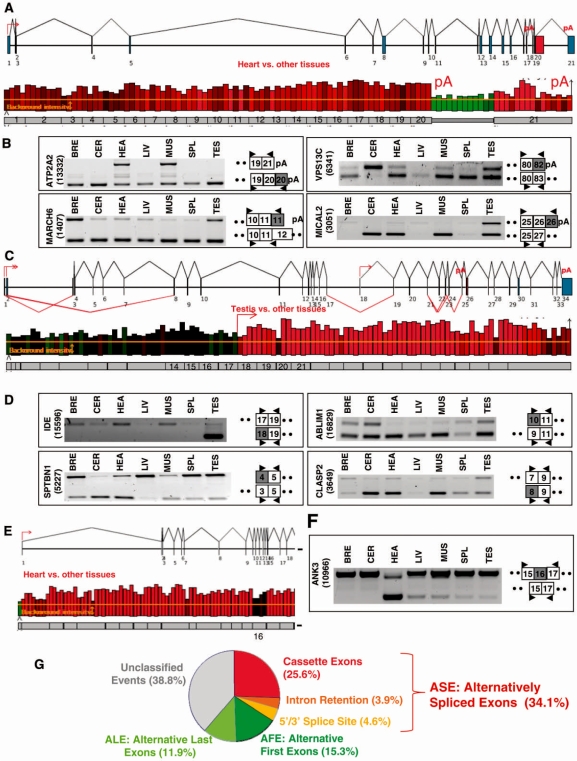
In addition to these alternative splicing events, we identified and validated several cases of alternative polyadenylation sites, as illustrated for the ATP2A2 gene (upper panel, Figure 4A). The transcripts produced by the ATP2A2 gene can end either in intron 20 or in exon 21, as indicated by the ‘pA’ symbol above exons 20 and 21. Exon array data suggested that the ratio of the transcripts ending in intron 20 or exon 21 varied when comparing heart (or muscle, not shown) to the other tissues (lower panel, Figure 4A). This case was validated by RT-PCR analysis (ATPA2, Figure 4B), as well as several other cases of alternative polyadenylation sites, as illustrated on Figure 4B for the MARCH6, VPS13C and MICAL2 genes.
Figure 4.
Annotation and validation of alternative first and last exons. (A) ATP2A2 gene analysis. Genomic structure of the ATP2A2 gene (upper panel). According to FAST DB exon numeration and annotation, the human ATP2A2 gene contains 21 exons and two potential last exons, as indicated by a pA symbol above exons 20 and 21. Graphical representation of the exon array probes corresponding to the ATP2A2 gene in the heart compared to the other tissues (lower panel). Each Affymetrix probe corresponding to the ATP2A2 gene is represented by a bar above the numbered grey exons of the gene. The bright green bars corresponding to 3′-end extended exon 20 and the red bars corresponding to exon 21 indicated that exons 20 and 21 were differentially used between heart and other tissues. Screenshot is from EASANA®. (B) RT-PCR analysis of ALEs. RT-PCR analyses for several ALEs predicted to be differentially expressed between tissues. (C) IDE gene analysis. Genomic structure of the IDE gene (upper panel). According to FAST DB exon numeration and annotation, the human IDE gene contains 34 exons and three potential first exons, as indicated by a red arrow above exons 1, 2 and 18. Graphical representation of the exon array probes corresponding to the IDE gene in the testis compared to the other tissues (lower panel). Each Affymetrix probe corresponding to the IDE gene is represented by a bar above the numbered grey exons of the gene. The bright red bars corresponding to exon 18 and downstream exons indicated that the transcripts starting with exon 18 were more expressed in the testis than in other tissues. Screenshot is from EASANA®. (D) RT-PCR analysis of AFEs. RT-PCR analyses for several AFEs predicted to be differentially expressed across tissues. (E) ANK3 gene analysis. Genomic structure of the ANK3 gene and graphical representation of the exon array probes corresponding to the ANK3 gene in the heart compared to the other tissues (upper and lower panels). Each Affymetrix probe corresponding to the ANK3 gene is represented by a bar above the numbered grey exons of the gene. The black bars corresponding to exon 16 compared to the red bars corresponding to the other exons indicate that exon 16 may be skipped in the heart. Screenshot is from EASANA®. (F) RT-PCR analysis for the ANK3 gene. The RT-PCR analyses for the ANK3 gene demonstrated that ANK3 exon 16 was more frequently skipped in the heart than in other tissues. (G) Classification of the differentially expressed exons between tissues. Sixty-one percent of the differentially expressed exons across tissues were already annotated in FAST DB as ASEs, AFEs or ALE.
Another mechanism leading to variation in mRNA exon content involves alternative promoters (Figure 3A). This was illustrated by the IDE gene that contains an internal promoter indicated by a red arrow above exon 18 (upper panel, Figure 4C). Remarkably, only the intensity of the probes corresponding to exon 18 and downstream exons varied when comparing testis to the other tissues (lower panel, Figure 4C). RT-PCR analysis demonstrated that testis more strongly expressed the forms starting in exon 18 than other tissues (IDE, Figure 4D). Other differentially expressed alternative promoters were identified and validated, as illustrated for the SPTBN1, ABLIM1 and CLASP2 genes (Figure 4D).
Among the 1073 events identified above (Figure 2A), 26, 4 and 5% corresponded to cassette exons, intron retentions and 5′- or 3′-spliced sites, respectively (Figure 4G), and 15 and 12% corresponded to AFE and ALEs, respectively (Figure 4G). This repartition is in agreement with the proportion of these different types of alternative exons in databases (3–9,14–18,36).
We also identified a large set of predicted differentially expressed exons that were not annotated as alternative exons in FAST DB (Figure 4G, unclassified events). For example, ANK3 exon 16 was predicted to be differentially expressed when comparing heart to the other tissues (Figure 4E). RT-PCR analysis demonstrated that ANK3 exon 16 was more frequently skipped in heart than in the other tissues (Figure 4F). The list of events and their annotation is given in Supplementary Data for each tissue (Supplementary Table S3), which provides a resource for alternative exons not being yet annotated in databases. It is also possible to use the EASANA® visualization module (Supplementary Figure S2) to retrieve information on the tissue-specific levels of annotated or new alternative exons of queried genes (www.fast-db.com/cgi-bin/easana/index.pl).
Expression profile of splicing factors across tissues
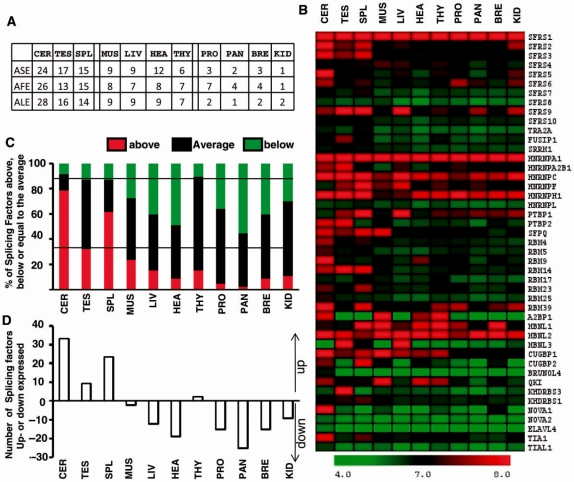
The classification of the differentially expressed exons across the 11 tissues by alternative event categories, that is, ASEs, AFEs and ALEs, revealed a similar rank as that observed on Figure 2B. In particular, the cerebellum, testis and spleen were enriched in ASEs being differentially expressed when compared to the other tissues (ASE, Figure 5A).
Figure 5.
Profile of splicing factors gene expression across 11 tissues. (A) Tissue ranking of classified and differentially expressed exons across tissues. A similar tissue ranking was obtained for ASEs, AFEs and ALE that were differentially expressed across tissues. (B) HeatMap of 45 splicing factor gene intensity. The color scale representing gene normalized intensity is shown below the HeatMap. (C) Proportion of splicing factors being up- or down-expressed in each tissue compared to their average expression level in the 11 tissues. The mean of the gene signal in the 11 tissues was calculated for each splicing factor. The distance between the gene signal in a given tissue and the corresponding mean in the 11 tissues was calculated. The number of splicing factors with a gene expression level above or below the gene expression average in the 11 tissues was calculated. (D) Number of splicing factors being up- or down-expressed in each tissue. The number of splicing factors being less expressed in a given tissue was subtracted from the number of splicing factors being more expressed in the same tissue.
Because ASEs are controlled by splicing factors, we next investigated the expression profile of splicing factors across tissues in the same dataset to be able to compare genome-wide alternative splicing events and splicing factor expression profile. In the literature, we selected 45 well-characterized splicing regulatory factors that have been shown to participate in the selection of ASE. These include members of the SR, hnRNP, RBM, MBNL, CELF and KH families (4,19–24,34). All these factors are known to recognize and bind to RNA regulatory sequences located in introns and/or exons and to control the use of splicing sites selected by the spliceosome. The normalized gene expression values (gene signals) of the 45 splicing regulatory factors across the 11 tissues were retrieved. Intensity values were then displayed with MeV4.0. This analysis revealed that, first, each tissue was characterized by a specific splicing factor expression profile (columns, Figure 5B). Second, each splicing factor presented a specific expression profile across tissues (lines, Figure 5B). Many features of this analysis have been previously reported, as it will be underlined in the ‘Discussion’ section.
This analysis also suggested that cerebellum, testis and spleen expressed the largest number of splicing factors at a relatively high level. To test this hypothesis, we quantified, for each tissue, the number of splicing factors having a gene expression value above or below the corresponding mean gene expression value calculated for the 11 tissues (Supplementary Table S4). As shown on Figure 5C and D, a larger proportion of splicing factors was highly expressed (i.e. gene expression value above the mean) in the cerebellum, testis and spleen than in the other tissues, and a smaller proportion of splicing factors was poorly expressed (i.e. gene expression value below the mean) in the cerebellum, testis and spleen than in the other tissues. These data indicated that tissues with the largest number of differentially expressed ASEs (Figure 5A) also had the largest number of highly expressed splicing factors (Figure 5B–D). The expression pattern of splicing factors can be retrieved either in Supplementary Table S4 and Figure S4 or by using the EASANA® visualization module as described in Supplementary Figure S5.
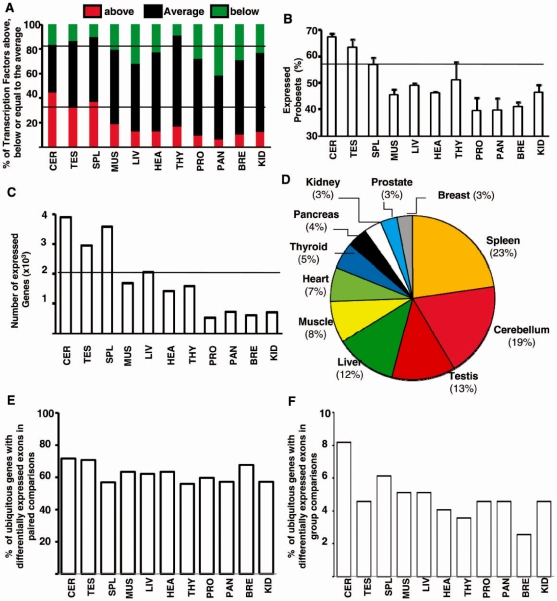
Global gene expression level
To test whether there was a propensity of cerebellum, testis and spleen to generate transcripts with a different exon content from that of other tissues (Figure 5A), we investigated whether a high expression level of splicing factors was a specific feature of these tissues. First, similar results were obtained by performing transcription factor gene expression profiling, as a larger proportion of transcription factors were more expressed in the cerebellum, testis and spleen than in the other tissues and a smaller proportion of transcription factors were less expressed in the cerebellum, testis and spleen than in the other tissues, as observed for splicing factors (comparing Figures 5C and 6A). Second, we observed that a larger proportion of probesets were above the DABG in the cerebellum, testis and spleen than in the other tissues (Figure 6B). Third, there were almost twice more genes expressed in the cerebellum, testis, and spleen than in the other tissues (Figure 6C). Finally, a statistical group analysis based on gene expression level (gene signal) revealed that the tissues expressing a larger proportion of differentially expressed genes were those expressing a larger proportion of differentially expressed exons (comparing Figures 2B and 6D). Therefore, these analyses revealed that the larger number of splicing factors being highly expressed in the cerebellum, testis and spleen (Figure 5C and D) is likely to be part of a more global profile of gene expression (Figure 6). This was strengthened by the observation of a similar ranking by analyzing ASEs, AFE and ALEs (Figure 5A). Altogether, the apparent propensity of cerebellum, testis and spleen to express more alternative transcripts than other tissues is likely to originate from their ability to express more genes.
Figure 6.
Profile of gene expression across 11 tissues. (A) Number of transcription factors being up- or down-expressed in each tissue compared to their average expression level in the 11 tissues. The mean of the gene signal in the 11 tissues was calculated for each transcription factor. The distance between the gene signal in a given tissue and the corresponding mean in the 11 tissues was calculated. The number of transcription factors with a gene expression level above or below the gene expression average in the 11 tissues was calculated. (B) Percentage of selected probesets expressed above DABG in each tissue. (C) Number of genes expressed above DABG in each tissue. (D) Statistical group analysis of gene expression levels. The tissue ranking using gene expression level (gene signal) values in parallel to those of alternative exons (compared with Figures 2B); that is, the same types of samples with the largest number of differentially expressed exon (spleen, cerebellum and testes, see Figure 2B) were among those with the most differentially expressed genes. (E) Percentage in each tissue of genes that were significantly expressed in 11 tissues and that contained differentially expressed exons in paired comparisons. The proportion of genes with differentially expressed exons in each tissue was calculated among 524 genes (corresponding to 100%) that were significantly expressed in the 11 tissues and that were predicted to contain differentially expressed alternative exons in all paired comparisons. (F) Percentage in each tissue of genes that were significantly expressed in 11 tissues and that contained differentially expressed exons in group comparisons. The proportion of genes with differentially expressed exons in each tissue was calculated among 196 genes (corresponding to 100%) that were significantly expressed in the 11 tissues and that were predicted to contain differentially expressed alternative exons in all group comparisons.
This conclusion was further supported by the analysis of the splicing pattern of genes expressed in all analyzed tissues. As shown on Figure 6E and F, all tissues contained a similar proportion of genes producing alternative exons by using only the set of ubiquitous genes in contrast to what we observed by using all genes (Figures 1 and 2). However, each tissue expressesed different splicing isoforms produced from the ubiquitous genes, as illustrated on Supplementary Figure S9. This was expected as the different tissues did not express the same set of splicing factors (Figure 5).
Biological impact of tissue-specific exons
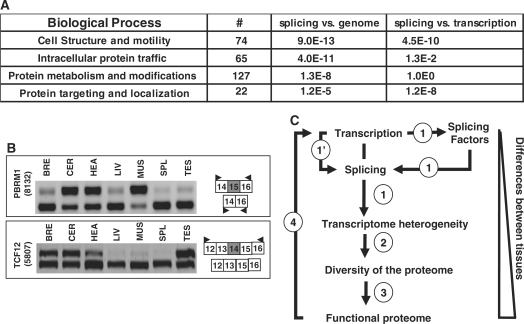
To test whether the genes bearing differentially expressed exons across tissues were involved in specific biological processes, we performed a functional analysis using the PANTHER software (www.pantherdb.org). The 653 analyzed genes containing differentially expressed exons across tissues (Figure 2A) were enriched for Gene Ontology functional categories, including ‘cell structure and motility’, ‘intracellular protein traffic’, ‘protein targeting and localization’ and ‘protein metabolism and modification’ (Figure 7A). Therefore, the functions of genes products that control the fate and post-translational modifications of proteins may be particularly affected by differential exon selection in a tissue-specific manner. Remarkably, by comparing the genes having a differential expression across tissues (Figure 6D), we observed that ‘cell structure and motility’ and’protein targeting and localization’ were processes enriched in the ‘splicing’ list compared to the ‘transcription’ list. Noteworthy, many alternative splicing events affect protein domains that control the intracellular localization of proteins by deletion/insertion of exons coding for subcellular localization signals (10). Therefore, tissue-specific alternative splicing events may impact the proteome, first by affecting protein domains and second by affecting gene products involved in the control of protein modifications and fate.
Figure 7.
Functional consequences of differential exon selection. (A) Biological process. Number of genes presenting differentially regulated exons across tissues and associated with specific biological processes as defined by PANTHER (www.pantherdb.org). Biological process analysis was performed using Bonferroni correction. Statistical significance calculated by comparing splicing-regulated genes to the genome or splicing-regulated genes to transcriptional-regulated genes. (B) RT-PCR analysis. The RT-PCR analyses for the PBRM1 and TCF12 transcriptional factors demonstrated that different spliced isoforms were differentially expressed across analyzed tissues. (C) Successive layers of regulation drive an increasing divergence between tissues. Key transcriptional regulators determine the nature and the number of genes being expressed during cell specialization. Tissues expressing a larger number of genes express a larger number of splicing factors that in turn impact the exon content of the gene products (1). Transcriptional regulators may also affect transcript exon content (1’). As a consequence, the transcriptome differs in terms of both transcript expression level and transcript exon content, resulting in a more diversified proteome (2). Outcomes of differentially expressed exons impinge on a third layer of regulation [(3), the ‘functional proteome’] as there was an enrichment of splicing-regulated genes involved in ‘intracellular protein traffic’ and ‘protein metabolism and modification’. This process may be maintained by impacting on transcriptional regulators (4).
Furthermore, we observed a large set of transcriptional regulators bearing tissue-specific differentially expressed exons (Supplementary Table 5), as previously reported (13). This was illustrated with the PBRM1 and TCF12 genes (Figure 7B). Therefore, the transcriptome diversity at the exon level originates from tissue-specific gene expression level and in turn impacts on gene transcriptional control. In conclusion, tissues expressing a larger set of genes, including splicing factors, express different splicing variants when compared to other tissues (Figure 7C). Divergences with other tissues increase consequently, as protein isoforms translated from these splicing variants differentially impact protein fate, as well as gene expression regulation (Figure 7C).
DISCUSSION
In this study, we developed bioinformatics tools to profile gene and exon expression across 11 normal human tissues. In particular, we provided a list of more than 10 000 exons being differentially expressed across normal human tissues (Supplementary Tables S2 and 3). In addition, a freely available web interface (www.fast-db.com/cgi-bin/easana/index.pl) permits after registration to display tissue dataset from any gene, which provides access to splicing variant expression profile across normal tissues of query genes (see Supplementary Figures S2 and 9 for details regarding the use of the web interface). We also provided the expression profile of 45 splicing regulatory factors across normal tissues (Figure 5, Supplementary Table S4 and Figure S4), which can be extended to any queried gene thanks to the freely available web interface, as described in Supplementary Figure S5.
It has been well-established that some tissues, including brain and testis, express a larger set of splicing variants than other tissues (3–9,21–26,36). In this study, we demonstrated for the first time that spleen is also a tissue expressing a high proportion of alternative transcripts (Figure 2). Remarkably, several recent reports have indicated that alternative splicing plays a critical role in the activation of lymphocytes during the immune response that occurs in part in spleen (37). A critical role for the HNRNPL splicing factor, which is more expressed in spleen than in other tissues (Supplementary Figure S3) as already reported, has been shown in this process (38–40).
To better understand the prevalence of alternative splicing in a set of tissues, we analyzed the expression level of splicing factors in the same dataset. Genome-wide analyses of tissue-specific alternative splicing events and tissue-specific splicing factor expression profile had not been performed in the same dataset yet (32,34). Because tissue ranking, which is based either on the number of differentially expressed exons or on the number of differentially expressed genes, is likely to depend on the compared tissues, an important improvement of our analysis was to draw conclusions that derived from the same dataset to survey exon and gene profiling. Thanks to this strategy, we demonstrated for the first time that tissues expressing the largest proportion of splicing variants (Figures 1 and 2) were tissues that expressed the largest number of splicing factors at a high level (Figure 5). High level of a larger number of splicing factors is expected to improve the chance of exons to be differentially selected: two tissues expressing the same set of splicing factors are more likely to express the same splicing variants compared to a tissue that would express a different set of splicing factors (3,4,19–24). In addition, as each tissue was characterized by a specific splicing factor expression profile (Figure 5B) and as a larger number of splicing factors being expressed in a tissue increases the number of possible combinations for the ratio between splicing factor levels, the possibility of alternative splicing regulation in this tissue may in turn increase, given the combinatorial mode of splicing regulation. A limit to this analysis is that DNA microarray data measure mRNA expression levels. However, among the splicing factors that were highly expressed in the set of analyzed tissues (Figure 5B, Supplementary Table S4 and Figure S3), several cases have been well documented at both RNA and protein levels. For examples, NOVA1 and PTB2, but not PTB1, are highly expressed at the mRNA and protein levels in the cerebellum (4,23,24,26); CUGBP2 (ETR-3) protein has been shown to be more expressed in brain and spleen than other tissues (4,22); MBNL3 protein has been shown to be expressed in testis, spleen and liver, but not in brain, muscle and heart (21); a restricted and high expression level of A2BP1 (FOX1) protein in cerebellum, muscle and heart has been reported (26). Although some mRNAs encoding splicing factors may not be translated, there is probably a strong relationship between the large number of splicing factors being more expressed in the cerebellum, testis and spleen, and the capability of these tissues to express more splicing variants than other tissues.
The large number of splicing factors being express in cerebellum, testis and spleen (Figure 5) is part of a more global gene expression profile because these tissues express a larger number of genes, including transcription factors, at higher levels (Figure 6). This large number of expressed genes in the cerebellum has already been reported and it was estimated that >50% of the mouse genome would be expressed during the development of male germ cells (1,25). This propensity of a set of tissues to express a larger number of genes is likely to have two major impacts in terms of alternative splicing. First, splicing factors are among the genes that are more expressed in these tissues. Second, increasing the number of genes that are expressed in a given tissue increases the probability of generating splicing variants. Therefore, it can be concluded that higher levels of alternative splicing events in a set of human tissues originate from their ability to express a larger set of genes. These tissues are likely to express highly specific splicing factors, as recently shown in the nervous system (41), in the same way as they express other tissue-restricted genes like transcriptional factors. A third mechanism by which tissue-specific transcriptional programs may impact on splicing is based on the functional coupling between transcription and splicing. Although, we observed no correlation between gene expression regulation or gene expression level and splicing (Supplementary Figures S6 and 7) as previously reported (42), a rather qualitative than quantitative change in gene transcriptional activity may impact gene product splicing owing to alternative promoter usage and/or chromatin marks. Indeed, it has been shown that an alternative promoter switch can impact on the exon content of the gene products (43) and increasing evidences indicate a link between chromatin marks and splicing (44). Therefore, tissue-specific transcriptional programs may impact on the transcriptome at the exon level, either directly or indirectly through splicing factor expression level. In both cases, the apparent propensity of tissues like cerebellum, testis and spleen to generate more splicing variants is likely to be a consequence of a global gene expression pattern. This conclusion is supported by our observation that all three types of alternative exons controlled by alternative promoters, splicing and polyadenylation, respectively, were more prominent in the cerebellum, testis and spleen (Figure 5A).
Our analysis supports a simple model where the divergence between tissues is expanding through successive layers of regulation starting from the number (and obviously the identity) of expressed genes. The first layer of regulation that controls the number of genes expressed in a tissue impinges on a second layer of regulation controlling splicing variants expression. Indeed, more genes expressed in a tissue will increase the ability to generate splicing variants that results in proteome diversity (Figure 7C). Outcomes of differentially expressed exons impinge on a third layer of regulation (the ‘functional proteome’) as there was an enrichment in splicing-regulated genes involved in ‘intracellular protein traffic’ and ‘protein metabolism and modification’ (Figure 7A). This process may be maintained by impacting on transcriptional regulators (Figure 7B and Supplementary Table S5), whose functions are known to be regulated by alternative splicing (13). Moreover, gene expression level and alternative splicing may act complementary because some biological processes were preferentially affected by alternative splicing (Figure 7A), as already reported in mouse (42).
Finally, the ability of some tissues to express more genes correlated with a larger number of transcription factors being expressed (Figure 6) and the number of differentially expressed exons/genes in tissues may reflect the relative abundance of cell types in these tissues. Indeed, the tissues (cerebellum, testis and spleen) that express the largest set of genes and splicing variants have a large diversity of cell types. Therefore, the larger numbers of genes and splicing variants being expressed in these tissues could represent the cellular heterogeneity of these tissues: if each cell type expresses a specific splicing variant, cellular heterogeneity will increase the number of splicing variants when compared to a homogenous cell population. Accordingly, a recent study has revealed a large number of alternative splicing variations by comparing different brain regions (45). Cell specialization is a key driving force in organism complexity and recent evidences have indicated that alternative splicing is more frequent in organisms with increased cellular and functional specialization (46). Our data support the notion that exonic regulations play a role in cell specialization of complex tissues in addition to the regulation of gene expression levels.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union FP6 (NoE EURASNET), INSERM AVENIR program, ANR and Institut National du Cancer; Association Française contre les Myopathies (to P.dl G.); European Union FP6 (NoE EURASNET to P.dl G.); INCa (to L.G.); Canceropole Ile-de-France (to M.D.); INSERM (to M.D.). Funding for open access charge: INSERM.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Shyamsundar R, Kim YH, Higgins JP, Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D, Brown PO, Pollack JR. A DNA microarray survey of gene expression in normal human tissues. Genome Biol. 2005;6:R22. doi: 10.1186/gb-2005-6-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saito-Hisaminato A, Katagiri T, Kakiuchi S, Nakamura T, Tsunoda T, Nakamura Y. Genome-wide profiling of gene expression in 29 normal human tissues with a cDNA microarray. DNA Res. 2002;9:35–45. doi: 10.1093/dnares/9.2.35. [DOI] [PubMed] [Google Scholar]
- 3.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 5.Zavolan M, Kondo S, Schonbach C, Adachi J, Hume DA, Hayashizaki Y, Gaasterland T. Impact of alternative initiation, splicing, and termination on the diversity of the mRNA transcripts encoded by the mouse transcriptome. Genome Res. 2003;13:1290–1300. doi: 10.1101/gr.1017303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Clark TA, Schweitzer AC, Chen TX, Staples MK, Lu G, Wang H, Williams A, Blume JE. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugnet CW, Srinivasan K, Clark TA, O'B;rien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Yura K, Shionyu M, Hagino K, Hijikata A, Hirashima Y, Nakahara T, Eguchi T, Shinoda K, Yamaguchi A, Takahashi K, et al. Alternative splicing in human transcriptome: functional and structural influence on proteins. Gene. 2006;380:63–71. doi: 10.1016/j.gene.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 12.Xing Y, Lee CJ. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taneri B, Snyder B, Novoradovsky A, Gaasterland T. Alternative splicing of mouse transcription factors affects their DNA-binding domain architecture and is tissue specific. Genome Biol. 2004;5:R75. doi: 10.1186/gb-2004-5-10-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shionyu M, Yamaguchi A, Shinoda K, Takahashi K, Go M. AS-ALPS: a database for analyzing the effects of alternative splicing on protein structure, interaction and network in human and mouse. Nucleic Acids Res. 2009;37:D305–D309. doi: 10.1093/nar/gkn869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koscielny G, Le Texier V, Gopalakrishnan C, Kumanduri V, Riethoven JJ, Nardone F, Stanley E, Fallsehr C, Hofmann O, Kull M, et al. ASTD: the alternative splicing and transcript diversity database. Genomics. 2009;93:213–220. doi: 10.1016/j.ygeno.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Bhasi A, Philip P, Sreedharan VT, Senapathy P. AspAlt: a tool for inter-database, inter-genomic and user-specific comparative analysis of alternative transcription and alternative splicing in 46 eukaryotes. Genomics. 2009;94:48–54. doi: 10.1016/j.ygeno.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 17.de la Grange P, Dutertre M, Correa M, Auboeuf D. A new advance in alternative splicing databases: from catalogue to detailed analysis of regulation of expression and function of human alternative splicing variants. BMC Bioinformatics. 2007;8:180. doi: 10.1186/1471-2105-8-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de la Grange P, Dutertre M, Martin N, Auboeuf D. FAST DB: a website resource for the study of the expression regulation of human gene products. Nucleic Acids Res. 2005;33:4276–4284. doi: 10.1093/nar/gki738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee KS, Squillace RM, Wang EH. Expression pattern of muscleblind-like proteins differs in differentiating myoblasts. Biochem. Biophys. Res. Commun. 2007;361:151–155. doi: 10.1016/j.bbrc.2007.06.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ladd AN, Stenberg MG, Swanson MS, Cooper TA. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 2005;233:783–793. doi: 10.1002/dvdy.20382. [DOI] [PubMed] [Google Scholar]
- 23.Boutz PL, Chawla G, Stoilov P, Black DL. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2007;21:71–84. doi: 10.1101/gad.1500707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boutz PL, Stoilov P, Li Q, Lin CH, Chawla G, Ostrow K, Shiue L, Ares M., Jr, Black DL. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee TL, Pang AL, Rennert OM, Chan WY. Genomic landscape of developing male germ cells. Birth Defects Res. C Embryo Today. 2009;87:43–63. doi: 10.1002/bdrc.20147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 2007;8:819–831. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 27.Fagnani M, Barash Y, Ip JY, Misquitta C, Pan Q, Saltzman AL, Shai O, Lee L, Rozenhek A, Mohammad N, et al. Functional coordination of alternative splicing in the mammalian central nervous system. Genome Biol. 2007;8:R108. doi: 10.1186/gb-2007-8-6-r108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venables JP. Alternative splicing in the testes. Curr. Opin. Genet. Dev. 2002;12:615–619. doi: 10.1016/s0959-437x(02)00347-7. [DOI] [PubMed] [Google Scholar]
- 29.Elliott DJ, Grellscheid SN. Alternative RNA splicing regulation in the testis. Reproduction. 2006;132:811–819. doi: 10.1530/REP-06-0147. [DOI] [PubMed] [Google Scholar]
- 30.Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Res. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noh SJ, Lee K, Paik H, Hur CG. TISA: tissue-specific alternative splicing in human and mouse genes. DNA Res. 2006;13:229–243. doi: 10.1093/dnares/dsl011. [DOI] [PubMed] [Google Scholar]
- 32.Grosso AR, Gomes AQ, Barbosa-Morais NL, Caldeira S, Thorne NP, Grech G, von Lindern M, Carmo-Fonseca M. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res. 2008;36:4823–4832. doi: 10.1093/nar/gkn463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol. 2004;5:R74. doi: 10.1186/gb-2004-5-10-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gabut M, Chaudhry S, Blencowe BJ. SnapShot: The splicing regulatory machinery. Cell. 2008;133:192–192.e1. doi: 10.1016/j.cell.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Jackson AP, Parham P. Structure of human clathrin light chains. Conservation of light chain polymorphism in three mammalian species. J. Biol. Chem. 1988;263:16688–16695. [PubMed] [Google Scholar]
- 36.Beaudoing E, Gautheret D. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 2001;11:1520–1526. doi: 10.1101/gr.190501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller SN, Ahmed R. Lymphoid stroma in the initiation and control of immune responses. Immunol. Rev. 2008;224:284–294. doi: 10.1111/j.1600-065X.2008.00657.x. [DOI] [PubMed] [Google Scholar]
- 38.Melton AA, Jackson J, Wang J, Lynch KW. Combinatorial control of signal-induced exon repression by hnRNP L and PSF. Mol. Cell Biol. 2007;27:6972–6984. doi: 10.1128/MCB.00419-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zikherman J, Weiss A. Alternative splicing of CD45: the tip of the iceberg. Immunity. 2008;29:839–841. doi: 10.1016/j.immuni.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 40.Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calarco JA, Superina S, O'H;anlon D, Gabut M, Raj B, Pan Q, Skalska U, Clarke L, Gelinas D, van der Kooy D, et al. Regulation of vertebrate nervous system alternative splicing and development by an SR-related protein. Cell. 2009;138:898–910. doi: 10.1016/j.cell.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Kornblihtt AR. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 44.Kornblihtt AR, Schor IE, Allo M, Blencowe BJ. When chromatin meets splicing. Nat. Struct. Mol. Biol. 2009;16:902–903. doi: 10.1038/nsmb0909-902. [DOI] [PubMed] [Google Scholar]
- 45.Johnson MB, Kawasawa YI, Mason CE, Krsnik Z, Coppola G, Bogdanovic D, Geschwind DH, Mane SM, State MW, Sestan N. Functional and evolutionary insights into human brain development through global transcriptome analysis. Neuron. 2009;62:494–509. doi: 10.1016/j.neuron.2009.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinogradov AE, Anatskaya OV. Organismal complexity, cell differentiation and gene expression: human over mouse. Nucleic Acids Res. 2007;35:6350–6356. doi: 10.1093/nar/gkm723. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.