Abstract
We have developed a new approach to systematically study post-transcriptional regulation in a small number of cells. Actively translating mRNAs are associated with polysomes and the newly synthesized peptide chains are closely associated with molecular chaperones such as hsp70s, which assist in the proper folding of nascent polypeptides into higher ordered structures. These chaperones provide an anchor with which to separate actively translating mRNAs associated with polysomes from free mRNAs. Affinity capture beads were developed to capture hsp70 chaperones associated with the polysome complexes. The isolated actively translating mRNAs were used for high-throughput expression profiling analysis. Feasibility was demonstrated using an in vitro translation system with known translationally regulated mRNA transcript thymidylate synthase (TS). We further developed the approach using HCT-116 colon cancer cells with both TS and p53 as positive controls. The steady-state levels of TS and p53 mRNAs were unaltered after 5-fluorouracil treatment as assessed by real-time qRT-PCR analysis. In contrast, the protein expression and polysome-associated mRNA levels of both genes were increased. These differences in translational rate were revealed with our new approach from 500 cells. This technology has the potential to make investigation of translational control feasible with limited quantities of clinical specimens.
INTRODUCTION
While most investigators have focused on transcriptional regulation of gene expression in the past, recent studies indicate that post-transcriptional and, especially, translational regulation plays a key role during development, cell cycle control and drug resistance (1–5). Particularly with the recent discovery that non-coding microRNAs are important regulators of translation, it is important to develop a simple and accurate approach to investigate genes regulated at the post-transcriptional and translational levels. We and others have previously developed approaches to systematically investigate genes regulated post-transcriptionally by profiling translationally active mRNAs obtained from isolated polysomes with a high-throughput gene expression analysis platform (6–8). These approaches allowed us to analyze mRNA levels at the final mRNA translation step of protein synthesis. However, the limitation of these approaches is that they all relied on a traditional sucrose gradient density ultracentrifugation procedure to isolate polysome complexes, thus requiring many (up to 109) cells. As a result, current polysome isolation techniques are the major bottleneck for the investigation of post-transcriptionally regulated genes when one is faced with limited quantities of clinical specimens (e.g. needle biopsies of primary and metastatic tumors, isolated circulating tumor cells). There is thus a critical need to develop a novel approach to isolate polysome-loaded mRNAs from very few cells.
The new approach described here stemmed from previous studies indicating that mRNAs being actively translated are associated with multiple units of ribosomes (polyribosomes or polysomes) and the newly synthesized polypeptides are closely associated with molecular chaperones, including members of the hsp70 and hsc70 families (9–11). Chaperones can assist in the efficient folding of newly translated proteins as they are being synthesized on the ribosome and can maintain pre-existing proteins in a stable conformation to avoid premature folding and modifications (12,13). We therefore reasoned that such chaperones can provide a molecular anchor for separating polysome-loaded mRNAs from free mRNAs. In this study, we apply this principle to develop and optimize antibody affinity capture beads for capturing actively translated mRNAs associated with hsp70 family chaperones. The translationally active mRNAs in the polysome complex are pelleted by an antibody recognizing the hsp70/hsc70 chaperone proteins associated with nascent polypeptides of the complex (Figure 1). Bound RNAs are then purified and used for high-throughput expression profiling analysis.
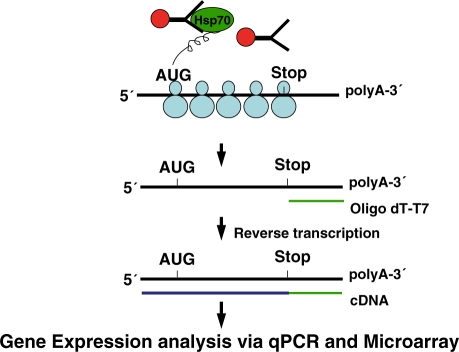
Figure 1.
Schematic diagram of TrIP–Chip approach. Affinity beads with covalently attached anti-Hsp70 antibodies are used to immunoprecipitate a cross-linked complex composed of Hsp70, nascent peptides, polysomes and the actively translating mRNAs. The mRNAs are used to conduct qPCR or converted to a labeled cRNA in a two-step reaction, and hybridized to whole genome microarrays for analysis. If translation is not taking place, there are no nascent peptides with which Hsp70 can associate.
Translational control plays an important role in chemoresistance, such as acute resistance to 5-fluorouracil (5-FU) treatment (14–16). Previous studies have identified that the target enzyme of 5-FU, thymidylate synthase (TS), is regulated at the translational level (15). In addition, as an RNA binding protein, TS not only negatively regulates its own synthesis at the translational level, but also interacts directly with p53 mRNA to downregulate p53 protein expression (17–19). In this study, we developed a novel approach to isolate translationally active mRNAs from a small number (500) of colon cancer HCT-116 cells for gene expression analysis, allowing us to systematically study mRNAs engaged in nascent translation during treatment with the chemotherapeutic drug 5-FU. Two previously identified translationally regulated genes, p53 and TS, were used as positive controls to monitor the process. The translationally active mRNA transcripts were subjected to high-throughput microarray-based expression profiling analysis. Over 500 mRNAs were shown to be differentially regulated in the translationally active mRNA fraction after 5-FU treatment. Genes, such as PP2A, a previously known translationally regulated gene (20), were more abundant in the polysome complex after treatment, and their elevated translation levels were confirmed at the protein level by western immunoblot analysis. Gene Ontology (GO) analysis revealed that genes in translational control are involved in some of the important biological process in response to 5-FU. This novel approach can be used to study translational control mediated by RNA-binding proteins and miRNAs when combined with a parallel expression analysis of steady-state total mRNAs (21–23).
MATERIALS AND METHODS
Cell culture
The human colon cancer cell line HCT-116 was obtained from ATCC and maintained at 37°C and 5% CO2 in 75-cm2 plastic tissue culture flasks with McCoy’s 5A medium containing 10% fetal bovine serum. Cells at 2 × 105 density were plated in six-well tissue culture plates for 24 h. Cells were then treated with 5 µM 5-FU for 24 h; untreated cells were used as controls.
In vitro transcription/translation
Promega’s TnT Quick Coupled rabbit reticulocyte lysate Transcription/Translation System was utilized to transcribe TS mRNA and subsequently synthesize TS protein. A plasmid, pGEM-pcHTS-1, containing the full-length human TS cDNA (1 µg), was used as the template for the in vitro transcription/translation reaction (15). TS mRNA expression was determined by real time qRT-PCR analysis and the TS protein was analyzed using western immunoblot analysis.
Western blot analysis
Equal amounts of protein (25 µg) extracted from lysed cell lines were separated on 10% sodium dodecyl sulfate-polyacrylamide gels by the method of Laemmli (24). Proteins were probed with mouse anti-TS monoclonal antibody (1:5000 dilution) (Zymed Laboratories, CA, USA), anti-p53 mouse monoclonal antibody (1:1000) (Santa Cruz Biotech Inc., CA, USA), anti-α-tubulin (1:1000) (Santa Cruz Biotech Inc., CA, USA) and anti-PP2A/C (1:1000) (Santa Cruz Biotech Inc., CA, USA). Horseradish peroxidase-conjugated antibodies against mouse or rabbit (1:1000, Santa Cruz Biotechnology) were used as the secondary antibodies. Protein bands were visualized with a chemiluminescence detection system using the Super Signal substrate.
Construction of antibody affinity capture beads
We have systematically evaluated three different types of magnetic beads, Dynabeads®M-280 Sheep anti-Mouse IgG, Dynabeads®M-280 Sheep anti-Rabbit IgG and Dynabeads®Sheep anti-Rat IgG (Invitrogen, Inc). The required amount of thoroughly suspended beads were transferred into an Eppendorf tube. Beads were washed two times with washing buffer (PBS with 0.1% BSA, pH 7.4 for Dynabeads®M-280 Sheep anti-Mouse IgG and Dynabeads®M-280 Sheep anti-Rabbit IgG and PBS with 0.1% BSA and 2 mM EDTA, pH 7.4 for Dynabeads®Sheep anti-Rat IgG). Then 1 µg/µl target immunoglobulin was added (a ratio of 1 µg/107 beads was found to produce sufficient binding). We used three types of antibodies: HSP70 mouse monoclonal antibody, HSC70 (HSP73) rabbit polyclonal antibody (Millipore Inc.) and HSC70 (HSP73) rat monoclonal antibody (BioVision Inc.). Beads with antibody were incubated with slow tilt rotation for 24 h at 2–8°C. The tube was then placed on a magnet for 2 min and supernatant was removed with a pipette. Antibody-coated beads were washed three times with washing buffer and used for immunoprecipitation.
Isolation of polysome-associated mRNA transcripts
To prepare cytoplasmic extracts, cells from six-well tissue culture plates were harvested with 0.05% Trypsin-EDTA (Invitrogen, CA, USA) and washed with ice-cold PBS containing 100 µg/ml cycloheximide (Sigma, MO, USA). Cells were counted and 10 000 cells were incubated with 800 μl of McCoy’s 5A medium containing 10% fetal bovine serum and 100 µg/ml cycloheximide (Sigma, MO, USA) for 5 min at 37°C. After incubation, 200 µl of DSP (1 mM) (Pierce, IL, USA) was introduced as a cross-linking reagent and incubation was carried out for 5 min at 37°C. Excess DSP was quenched with 1 M Tris–HCl (pH 7.4). The cells were washed twice by centrifugation at 1000 r.p.m. for 3 min, the supernatant discarded and the cell pellets were rinsed with ice-cold PBS containing 100 µg/ml cycloheximide (Sigma, MO, USA). The final pellets were swollen for 20 min in 500 µl of low salt buffer (LSB) (20 mM HEPES, pH 7.4, 100 mM KCl, 2 mM MgCl2) containing 1 mM dithiothreitol and lysed by the addition of 200 µl lysis buffer (1× LSB containing 1.2% Triton X-100) (Sigma) followed by brief vortexing. One-tenth (70 µl) of the above lysate was transferred to the Ig-coated beads, and incubation carried out for 1–2 h at 2–8°C.
After incubation with the HSP70/HSP73 antibody-conjugated magnetic beads, the polysome complexes containing translationally active mRNA transcripts were isolated, and the mRNAs eluted from beads conjugated with HSP70/HSP73 using the Array Pure Nanoscale RNA Purification Kit (Epicentre, WI, USA).
Quantitative gene expression analysis via real-time quantitative reverse transcription-polymerase chain reaction
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis was performed on the control mRNAs. PCR primers and probes (eIF4E, TS, p53, PP2A) were purchased from Applied Biosystems (Foster City, CA, USA). qRT-PCR was performed on an ABI 7500HT instrument under the following conditions: 25°C for 10 min and 37°C for 30 min of reverse transcription; 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 35 s (n = 3). Signal was collected at the endpoint of every cycle. The expression values of genes from different samples were calculated by normalizing with equal RNA input and relative quantitation values were plotted.
Two-round RNA amplification
To systematically discover post-transcriptionally and translationally regulated genes affected by 5-FU treatment, isolated translationally active mRNAs were used to profile gene expression using human whole-genome array analysis. Due to the limited quantities of cellular RNA isolated from 500 control HCT116 cells and cells treated with 5-FU, we employed two rounds of RNA amplification using the Agilent Low RNA Input Linear Amplification Kit (Agilent, CA, USA), which was also used to amplify mRNAs isolated from polysomes. The T7 promoter primer (1.2 µl) was added and the total reaction volume was adjusted to 11.5 µl with nuclease-free water. The primer and template were incubated at 65°C for 10 min and then placed on ice for 5 min. cDNA Master Mix (8.5 µl) was added to each sample and incubation carried out at 40°C for 2 h, 65°C for 15 min and on ice for 5 min. Then 60 µl of amplification reaction buffer was added to 20 µl of cDNA product and incubation proceeded at 40°C for 4 h. After purification, cRNA products were dried in a Speed Vacuum apparatus and the total volume adjusted to 10.5 µl. Random Hexamers (1.0 µl) were added to the cRNA products, incubation performed at 65°C for 10 min and the tube chilled to 4°C for 5 min. To each sample, 9.5 µl of cDNA Master Mix supplemented with Cy3-labeled NTPs was added and incubated at 40°C for 2 h, 65°C for 15 min and on ice for 5 min. Then 59 µl of amplification reaction buffer was added to 21 µl of cDNA product and incubation proceeded at 40°C for 4 h. The mini elute clean up kits (Qiagen, CA, USA) were utilized to purify the amplified samples after the first round amplification, and the RNeasy mini kits (Qiagen, CA, USA) were used to purify the labeled RNA after the second round in preparation for expression analysis.
Array hybridization and gene expression analysis
All reagents were provided in the Gene expression Hybridization Kit (Agilent, CA, USA). Eleven microliters of 10 × Blocking Agent and nuclease-free water was added to 1.65 µg of Cy3-labeled linearly amplified cRNA to bring the volume to 53 µl. Then 2.2 µl of 25 × Fragmentation buffer was added. After vortexing, the RNAs were fragmented by incubating at 60°C for 30 min. To prepare hybridization mix, 55 µl of fragmented cRNA and 55 µl of 2 × GE Hybridization Buffer HI-RPM were mixed and carefully loaded onto the array gasket well to avoid bubbles. Hybridization was carried out at 10 r.p.m. at 65°C for 17 h in a hybridization oven. After hybridization, the arrays were washed with Gene Expression Wash Buffer 1 (Agilent, CA, USA). The slides were then dried and kept in the dark until scanning. Images were captured on an Axon GenePix 4200A scanner.
The resulting image was quantified and the intensity of each spot divided by the median spot intensity to provide a scaled and comparable number across multiple arrays. After dot-grid and quality-control analysis, gene expression analysis was conducted using GeneSpring software 7.2 (Agilent, CA, USA), which allows multi-filter comparisons using data from different experiments to perform the normalization, generation of restriction lists and functional classification of differentially expressed genes. Under the Cross-Gene Error Model, normalization was applied in two steps: (i) ‘per chip normalization’ in which each measurement was divided by the 50th percentile of all measurements in its array; and (ii) ‘per gene normalization’ in which all the samples were normalized against specific control samples. Then data were filtered for ≥4-fold changes. The expression profiles of the different groups were compared using one-way analysis of variance (ANOVA) with cut-off P < 0.05. Comparisons of gene lists across different groups were performed using Venn diagrams and clustering was performed with the Condition Tree algorithm. The GO groupings and Gen Maps 2.0 program were used in conjunction with GeneSpring to identify pathways and functional groups of genes.
RESULTS
Preparation of antibody affinity capture beads
The principle of our polysome isolation process is illustrated in Figure 1. Translationally active mRNAs are associated with multiple units of ribosomes and the newly synthesized polypeptides are closely associated with molecular chaperones, such as hsp70s. These molecular chaperones, which assist in the proper folding of nascent polypeptides into higher ordered structures, provide an anchor for separating translationally active mRNAs associated with polysomes from free mRNAs. Affinity antibody capture beads will capture hsp70s chaperones associated with the polysome complexes so that polysomes can be separated from monosomes and free mRNAs. The isolated translationally active mRNAs are used for array based or real-time qRT-PCR gene expression analysis. To prepare and characterize the affinity capture beads, a number of different hsp70/hsc70 antibodies were evaluated to optimize the conditions for binding with the polysome complex. A schematic diagram of the use of these affinity capture beads for polysome isolation is presented in Supplementary Figure S1.
Translationally regulated genes can be captured by affinity capture beads
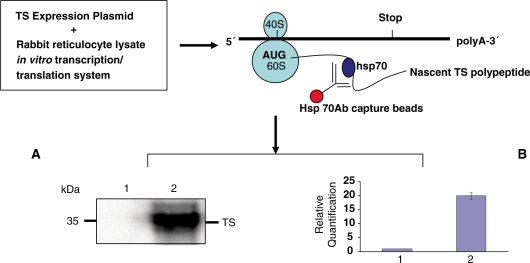
To determine the feasibility of isolating the hsp70-associated translational complex, we utilized an in vitro rabbit reticulocyte lysate coupled transcription/translation system with an expression construct pGEM-pcHTS-1 providing expression of full-length TS mRNA (15). Our results clearly indicate that we can immunoprecipitate the TS translation complex with hsp70 antibody affinity capture beads as shown in Figure 2. The absence of TS protein with control beads coupled to an anti-tubulin antibody (lane 1) indicates the specificity of our hsp70 antibody affinity capture beads (Figure 2A). We can detect the presence of newly synthesized TS protein in the isolated complex using western immunoblot analysis (lane 2) (Figure 2A). The associated TS mRNA template was also detected using real-time qRT-PCR analysis (lane 2, Figure 2B).
Figure 2.
Western immunoblot analysis of in vitro translated TS protein expression isolated using hsp70/hsc70 antibody affinity capture beads (lane 2); unrelated α-tubulin antibody beads were used as negative control (lane 1) (A). Real-time qRT-PCR analysis of in vitro transcribed TS mRNA expression (lane 1, control; lane 2, TS mRNA) (B).
Translationally regulated genes can be captured by affinity capture beads from HCT-116 colon cancer cells
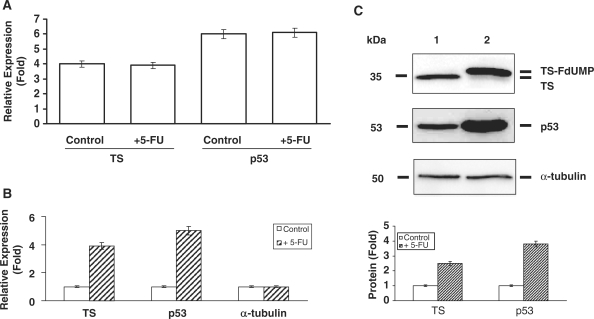
To directly demonstrate that we are capturing polysomes with affinity capture beads, HCT-16 colon cancer cells was incubated with 400 µM puromycin, a drug that triggers premature polypeptide termination and polysome disassembly. HCT-116 colon cancer cells treated with 400-µM puromycin reduced the total RNA levels associated with the hsp70 RNP complex compared to control cells (Supplementary Figure S2A). Real-time qRT-PCR analysis further revealed decreased level of translational initiation factor eIF4E with puromycin treatment (Supplementary Figure S2B). To further demonstrate directly in vivo that we can isolate translationally active mRNA complexes associated with hsp70 proteins, we utilized the HCT-116 human colon cancer cell line model with or without 5-FU treatment. We used two genes, p53 and TS, previously shown to be translationally regulated to monitor the isolation and quantification process (1,18). We were able to isolate actively translating mRNAs with our affinity capture approach as determined by both the presence of p53 and TS mRNAs. As shown in Figure 3A, the steady-state total cellular mRNA levels of TS and p53 was unchanged after 5-FU exposure (n = 3, P < 0.05) (15,18). By contrast, both TS and p53 mRNA transcripts isolated from the polysome complex were elevated by 5-FU treatment (n = 3, P < 0.01, CV < 5%) (Figure 3B). These results were supported by the corresponding increases of TS and p53 proteins determined by western blot analysis (Figure 3C).
Figure 3.
Real-time qRT-PCR analysis of TS and p53 mRNA expression from total RNA isolated from control and 5-FU-treated HCT116 cells (n = 3, P < 0.05) (A); Real-time qRT-PCR analysis of TS, p53 and α-tubulin mRNA expression from hsp70/hsc70 antibody affinity capture bead isolated RNAs from control and 5-FU-treated HCT116 cells (n = 3, P < 0.05) (B); Western immunoblot analysis of the expression of TS and p53 proteins isolated using hsp70/hsc70s antibody affinity capture beads (lane 1, control; lane 2, treated with 5-FU) (C). The lower panel shows the quantified protein expression levels for TS and p53.
We have previously reported that as an RNA-binding protein TS can suppress the expression of p53 at the translational level by direct interaction with p53 mRNA (18,19). To further demonstrate that we can capture endogenous translationally regulated events independent of 5-FU treatment, we used previously developed colon cancer cell lines HCT-C (TS–) and HCT-C (TS+) to show that the suppression of p53 mRNA translation by RNA-binding protein TS can be delineated with the hsp-70 affinity bead capture approach. Our experimental results indicated that the decreased p53 protein expression due to translational arrest by the TS protein can be revealed by hsp-70 antibody affinity capture beads (Supplementary Figure S3A, lane 2). The reduced level of actively translated p53 mRNA in the hsp-70 nascent translation complex was consistent with the reduction of p53 protein levels while total levels of p53 mRNA were unchanged (Supplementary Figure S3B).
Expression analysis of 5-FU responsive translationally active genes isolated from polysome complexes in HCT-116 cells
After we confirmed and optimized our isolation process, we sought to determine novel post-transcriptionally regulated genes affected by 5-FU treatment. To do so, the mRNAs isolated with the antibody affinity capture beads were used to quantify mRNA expression using human CodelinkTM whole-genome microarray analysis. We identified over 500 genes that were differentially regulated by 5-FU treatment, including our positive control genes p53 and TS. A selected gene list is shown in Table 1 (complete list in Supplementary Table S1). Many genes involved in translational control were upregulated. This includes genes such as TS (15), p53 (18), PP2A (20), hsp70 (25), thymidine kinase (26) and ornithine decarboxylase (27), which have previously been shown to be regulated by 5-FU at the post-transcriptional or translational level. We also confirmed the microarray expression analysis data using real-time qRT-PCR analysis with 10 randomly selected genes. Our results showed that the expression levels of selected genes quantified by microarray analysis are in good correlation (P < 0.02) with real-time qRT-PCR results (Table 2). The downregulated genes include ATG5, Claudin18, FGFR4, GPCR162, KIF21A and PDE1C, and some of these genes (ATG5, Claudin18 and FGFR4) are reported to be associated with chemoresistance (28–30).
Table 1.
Post-transcriptionally regulated genes mediated by 5-FU treatment in HCT116 colon cancer cells isolated by TrIP–Chip analysis (partial list)
| Gene ID | Fold change | Gene name |
|---|---|---|
| NM_001416 | 4.515 | Eukaryotic translation initiation factor 4A, isoform 1 (EIF4A1) |
| NM_022170 | 9.517 | Eukaryotic translation initiation factor 4H (EIF4H), transcript variant 1 |
| NM_006597 | 11.35 | Heat-shock 70-kDa protein 8 (HSPA8), transcript variant 1 |
| NM_182640 | 4.873 | Mitochondrial ribosomal protein S9 (MRPS9), nuclear gene encoding mitochondrial protein |
| NM_020360 | 5.118 | Phospholipid scramblase 3 (PLSCR3) |
| NM_000937 | 22.14 | Polymerase (RNA) II (DNA directed) polypeptide A, 220 kDa (POLR2A) |
| NM_006232 | 6.268 | Polymerase (RNA) II (DNA directed) polypeptide H (POLR2H) |
| NM_032959 | 5.668 | DNA-directed RNA polymerase II polypeptide J-related gene (POLR2J2) |
| NM_021128 | 6.834 | Polymerase (RNA) II (DNA directed) polypeptide L, 7.6 kDa (POLR2L) |
| NM_005837 | 6.522 | Processing of precursor 7, ribonuclease P subunit (S. cerevisiae) (POP7) |
| NM_021129 | 18.13 | Pyrophosphatase (inorganic) 1 (PPA1) |
| NM_177983 | 4.641 | Protein phosphatase 1G (formerly 2C), magnesium-dependent, gamma isoform (PPM1G) |
| NM_138689 | 7.384 | Protein phosphatase 1, regulatory (inhibitor) subunit 14B (PPP1R14B) |
| NM_024607 | 11.47 | Protein phosphatase 1, regulatory (inhibitor) subunit 3B (PPP1R3B) |
| NM_138558 | 8.723 | Protein phosphatase 1, regulatory (inhibitor) subunit 8 (PPP1R8), transcript variant 2 |
| NM_002715 | 5.813 | Protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform (PPP2CA) |
| NM_001009552 | 10.09 | Protein phosphatase 2 (formerly 2A), catalytic subunit, beta isoform (PPP2CB), transcript variant 2 |
| NM_033625 | 59.35 | Ribosomal protein L34 (RPL34) |
| NM_000996 | 21.23 | Ribosomal protein L35a (RPL35A) |
| NM_015414 | 5.287 | Ribosomal protein L36 (RPL36) |
| NM_021029 | 17.53 | Ribosomal protein L36a (RPL36A) |
| NM_000998 | 8.972 | Ribosomal protein L37a (RPL37A) |
| NM_000999 | 100.7 | Ribosomal protein L38 (RPL38) |
| NM_001000 | 7.078 | Ribosomal protein L39 (RPL39) |
| NM_000972 | 10.35 | Ribosomal protein L7a (RPL7A) |
| NM_001030 | 9.202 | Ribosomal protein S27 (metallopanstimulin 1) (RPS27) |
| NM_001032 | 22.91 | Ribosomal protein S29 (RPS29), transcript variant 1 |
| NM_001005 | 5.221 | Ribosomal protein S3 (RPS3) |
| NM_001010 | 136.6 | Ribosomal protein S6 (RPS6) |
| NM_001011 | 21.93 | Ribosomal protein S7 (RPS7) |
| NM_001033 | 4.609 | Ribonucleotide reductase M1 polypeptide (RRM1) |
| NM_001034 | 30.43 | Ribonucleotide reductase M2 polypeptide (RRM2) |
| NM_006819 | 7.226 | Stress-induced phosphoprotein 1 (Hsp70/Hsp90-organizing protein) (STIP1) |
| NM_003258 | 48.01 | Thymidine kinase 1, soluble (TK1) |
| NM_016399 | 16.27 | TP53 regulated inhibitor of apoptosis 1 (TRIAP1) |
| NM_001071 | 4.999 | Thymidylate synthetase (TYMS) |
| NM_003400 | 5.32 | Exportin 1 (CRM1 homolog, yeast) (XPO1) |
| NM_002789 | 18.14 | Proteasome (prosome, macropain) subunit, alpha type, 4 (PSMA4) |
| NM_002791 | 9.663 | Proteasome (prosome, macropain) subunit, alpha type, 6 (PSMA6) |
| NM_002793 | 36.45 | Proteasome (prosome, macropain) subunit, beta type, 1 (PSMB1) |
| NM_002802 | 5.809 | Proteasome (prosome, macropain) 26S subunit, ATPase, 1 (PSMC1) |
| NM_005805 | 4.907 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 (PSMD14) |
| NM_007273 | 7.634 | Prohibitin 2 (PHB2) |
| NM_00546 | 5.75 | Tumor protein p53 (TP53) |
| NM_004152 | 26.8 | Ornithine decarboxylase antizyme 1 (OAZ1) |
| NM_004394 | 4.549 | Death-associated protein (DAP) |
Table 2.
Comparison of microarray and qRT-PCR results (two sided Pearson correlation analysis P = 0.02)
| Gene name | Microarray (fold) | qRT-PCR (fold) |
|---|---|---|
| RRM1 | 4.60 | 5.80 |
| OAZ1 | 26.1 | 11.5 |
| TK1 | 48.2 | 8.90 |
| EIF4A | 4.50 | 5.50 |
| PSMC1 | 5.80 | 7.50 |
| RPS3 | 5.20 | 3.85 |
| STIP1 | 7.22 | 5.60 |
| HSPA5 | 7.29 | 6.14 |
| STX3 | 7.21 | 5.78 |
| NEDD8 | 4.72 | 6.30 |
It should be noted that although technically changes in mRNA levels are being measured in these hybridization assays, increased mRNA abundance in this polysome fraction would be expected to indicate increased levels of translation. However, some of which will also reflect in the levels of increased transcription or mRNA turn over. Changes in transcription would only be revealed when total steady-state mRNA abundance is measured if there is no translation control. We performed GO analysis using the DAVID Bioinformatics Resource to discover important gene pathways involved in post-transcriptional regulation (http://david.abcc.ncifcrf.gov/) (31) (Figure 4). DAVID Gene Functional Classification algorithm allows us to condense our gene list into organized classes based on cellular functions. The genes are grouped and clustered based on their cellular and molecular functions, which help us to identify functional related genes presented as a fuzzy heat map graphic view (Figure 4A). A zoomed-in view of cluster one is shown in Figure 4B. Most of the genes in cluster one are associated with protein translational initiation process. The fuzziness capability is a unique feature to maximally preserve biological patterns and to discover fine differences for a given gene. More than 20 clusters were represented with unique functions, such as protein synthesis, cell cycle control and RNA binding. The distribution of major classified gene clusters was presented as a pie chart (Figure 4C).
Figure 4.
GO analysis was performed using the DAVID bioinformatics suite. The DAVID Gene Functional Classification algorithm allows us to condense our gene list into organized classes based on cellular functions. The genes are grouped and clustered based on their cellular and molecular functions, which helps to identify functionally related genes, presented as a fuzzy heat map graphic view. The annotation terms are ordered based on the enrichment scores associated with the groups. Green represents a positive association between the gene terms; conversely, black represents an unknown relationship. The scattered pattern indicates functional differences. More than 20 clusters were represented with unique functions such as protein synthesis, cell cycle control and RNA binding (A). A zoomed-in view of the upper left cluster representing genes involved in translational initiation (B). The distribution of each set of classified genes is presented as a pie chart (C).
Confirmation of post-transcriptionally regulated genes
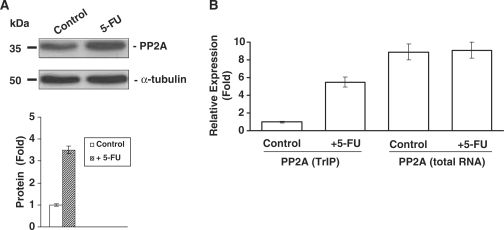
To confirm directly that some of the newly identified genes were indeed regulated post-transcriptionally, we measured protein expression by western blot and quantified mRNA by real-time qRT-PCR analysis. The results indicated that proteins such as PP2A was regulated most likely at the translational level (Figure 5A), as the total mRNA level of PP2A was not changed, whereas the polysome bound PP2A mRNA level was increased (Figure 5B).
Figure 5.
Western immunoblot analysis of PP2A protein expression isolated using hsp70/hsc70 antibody affinity capture beads from control and 5-FU-treated HCT116 colon cancer cells (lane 1, control; lane 2, plus 5-FU). Quantification of the protein levels for PP2A is provided in the lower panel (A). Real-time qRT-PCR analysis of total PP2A mRNA and polysome associated mRNA levels in control and 5-FU-treated HCT-116 colon cancer cells (B).
DISCUSSION
We have described here a novel approach to systematically study post-transcriptionally regulated genes from as few as 500 cells with hsp70/hsc70 antibody affinity capture beads. The results obtained from puromycin treatment further confirmed that the RNAs captured by hsp70/hsc70 antibody affinity capture beads are indeed associated with active translation events (Supplementary Figure S4). In this study, we have developed a method to capture mRNAs engaged in nascent translation, and we used this approach to identify a number of genes potentially post-transcriptionally regulated by 5-FU treatment when combined with a parallel expression analysis of total steady-state mRNA transcripts (6). We expect that some of these transcripts will be useful in understanding 5-FU related chemosensitivity. Many of these genes would have been missed if we had just profiled total RNAs, since translational control does not alter steady-state mRNA levels but rather translational efficiency. The minimal overlap of our gene list with previously reported genes affected by 5-FU treatment in analyzing steady-state mRNAs suggests that we predominantly capture and reflect changes of mRNAs involved in nascent translation (32). It is not surprising to see there are a number of stress-induced proteins that were upregulated by 5-FU exposure. GO analysis by DAVID provides an overview of genes associated with different biological processes (Figure 4). There are over 40% of genes that are related to protein biosynthesis (ribosomal proteins, translational initiation factors andRNA-binding proteins). Other genes involved are protein chaperons, RNA processing proteins, cell cycle control genes and phosphoproteins. Known drug-resistance-related genes such as calmodulin, EIFs related to mTOR pathway, and proteasome 26S subunit were all upregulated. Our results clearly showed that translational control played a major role in response to 5-FU treatment.
We are able to monitor the polysome isolation process using known translationally regulated genes, such as TS and p53, to ensure that the isolation process is robust and reproducible (Figure3B and C). We also noticed that the TS and p53 bands represent mostly full-length protein as analyzed by western immunoblot. This could be because the antibodies only recognize full-length proteins and not the shorter polypeptides. We randomly selected 10 genes from the gene expression array analysis to confirm the results by real-time qRT-PCR analysis (Table 2). Our results showed that the two sets of data are well correlated. We also demonstrated that our new approach can capture endogenous translationally regulated genes independent of drug response (Supplementary Figure S3). Furthermore, the large degree of overlap with expression profiles obtained using the high-density sucrose gradient ultracentrifugation approach suggests that we have indeed captured most of the mRNAs in the polysomal translational complex (Supplementary Figure S4 and Supplementary Data). The non-overlapping genes may be accounted for by biases attendant in each of the approaches, neither of which may be able to recapitulate completely the repertoire of polysome-associated mRNAs. The amount of hsp70 proteins in the polysome complex are enriched (Supplementary Figure S4), which is highly consistent with previous studies indicating that hsp70s are one of the major family of chaperones involved in translation; other chaperone proteins (e.g. hsp40 and hsp90) are also part of the hsp70 complex (9,11,33).
The new approach we have developed here adds a key tool necessary for investigating post-transcriptionally regulated genes from a small number of cells. The combination of this methodology with gene expression analysis represents a powerful approach for understanding genes translationally regulated by RNA-binding proteins or miRNAs in a variety of important biological processes. The current limit of isolating polysomes for expression analysis was the RNA quantity requirement of the expression platform. By coupling our approach with the digital expression analysis offered by the next generation DNA sequencing, we should be able to systematically investigate translational control from single cell in the future.
This study establishes the principle for integrating translational control by isolating actively translated mRNAs associated with protein chaperones such as hsp70. The data we obtained in this study indicated that the capture approach using hsp70s isolated most of the mRNAs involved in nascent translation since hsp70s are major contributors to efficient protein folding globally (34). We are aware that there are other types of protein chaperones such as hsp40 and hsp90. However, as both hsp40 and hsp90 are part of the hsp70 molecular chaperone complex associated with nascent polypeptides (33), we believe that it is possible to capture most of the translational complex using the hsp70 antibody affinity beads. Nonetheless, the method we developed here may be further improved by combining antibody capture beads consist of all major protein chaperones.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Stony-Brook Translational Research Laboratory Start-up fund and NIH/NCI CA114043 (to J.J.); and MH075020 (to J.J.). Funding for open access charge: Translational Research Laboratory Start-up Fund.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Chu E, Allegra CJ. The role of thymidylate synthase as an RNA binding protein. Bioessays. 1996;18:191–198. doi: 10.1002/bies.950180306. [DOI] [PubMed] [Google Scholar]
- 2.Chu E, Cogliati T, Copur SM, Borre A, Voeller DM, Allegra CJ, Segal S. Identification of in vivo target RNA sequences bound by thymidylate synthase. Nucleic Acids Res. 1996;24:3222–3228. doi: 10.1093/nar/24.16.3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz JA, Mullner EW. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 2000;14:1641–1652. doi: 10.1096/fj.14.11.1641. [DOI] [PubMed] [Google Scholar]
- 4.Derrigo M, Cestelli A, Savettieri G, Di Liegro I. RNA-protein interactions in the control of stability and localization of messenger RNA (review) Int. J. Mol. Med. 2000;5:111–123. [PubMed] [Google Scholar]
- 5.Sheikh MS, Fornace A.J., Jr Regulation of translation initiation following stress. Oncogene. 1999;18:6121–6128. doi: 10.1038/sj.onc.1203131. [DOI] [PubMed] [Google Scholar]
- 6.Ju J, Huang C, Minskoff SA, Mayotte JE, Taillon BE, Simons JF. Simultaneous gene expression analysis of steady-state and actively translated mRNA populations from osteosarcoma MG-63 cells in response to IL-1alpha via an open expression analysis platform. Nucleic Acids Res. 2003;31:5157–5166. doi: 10.1093/nar/gkg702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc. Natl Acad. Sci. USA. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morris DR. Growth control of translation in mammalian cells. Prog. Nucleic Acid Res. Mol. Biol. 1995;51:339–363. doi: 10.1016/s0079-6603(08)60883-1. [DOI] [PubMed] [Google Scholar]
- 9.Beckmann RP, Mizzen LE, Welch WJ. Interaction of Hsp 70 with newly synthesized proteins: implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 10.Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 11.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-i. [DOI] [PubMed] [Google Scholar]
- 12.Hansen WJ, Lingappa VR, Welch WJ. Complex environment of nascent polypeptide chains. J. Biol. Chem. 1994;269:26610–26613. [PubMed] [Google Scholar]
- 13.Houry WA. Chaperone-assisted protein folding in the cell cytoplasm. Curr. Protein Pept. Sci. 2001;2:227–244. doi: 10.2174/1389203013381134. [DOI] [PubMed] [Google Scholar]
- 14.Chu E, Drake JC, Koeller DM, Zinn S, Jamis-Dow CA, Yeh GC, Allegra CJ. Induction of thymidylate synthase associated with multidrug resistance in human breast and colon cancer cell lines. Mol. Pharmacol. 1991;39:136–143. [PubMed] [Google Scholar]
- 15.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, Zinn S, Allegra CJ. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl Acad. Sci. USA. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grem JL, Chabner BA, Chu E, Johnson P, Yeh GC, Allegra CJ. Antimetabolites. Cancer Chemother. Biol. Response Modif. 1991;12:1–25. [PubMed] [Google Scholar]
- 17.Chu E, Schmitz JC, Ju J, Copur SM. An immunoprecipitation-RNA:rPCR method for the in vivo isolation of ribonucleoprotein complexes. Methods Mol. Biol. 1999;118:265–274. doi: 10.1385/1-59259-676-2:265. [DOI] [PubMed] [Google Scholar]
- 18.Ju J, Pedersen-Lane J, Maley F, Chu E. Regulation of p53 expression by thymidylate synthase. Proc. Natl Acad. Sci. USA. 1999;96:3769–3774. doi: 10.1073/pnas.96.7.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu E, Copur SM, Ju J, Chen TM, Khleif S, Voeller DM, Mizunuma N, Patel M, Maley GF, Maley F, et al. Thymidylate synthase protein and p53 mRNA form an in vivo ribonucleoprotein complex. Mol. Cell. Biol. 1999;19:1582–1594. doi: 10.1128/mcb.19.2.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baharians Z, Schonthal AH. Autoregulation of protein phosphatase type 2A expression. J. Biol. Chem. 1998;273:19019–19024. doi: 10.1074/jbc.273.30.19019. [DOI] [PubMed] [Google Scholar]
- 21.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 22.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 23.Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression? Biochem. Soc. Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Theodorakis NG, Morimoto RI. Posttranscriptional regulation of hsp70 expression in human cells: effects of heat shock, inhibition of protein synthesis, and adenovirus infection on translation and mRNA stability. Mol. Cell. Biol. 1987;7:4357–4368. doi: 10.1128/mcb.7.12.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gross MK, Merrill GF. Thymidine kinase synthesis is repressed in nonreplicating muscle cells by a translational mechanism that does not affect the polysomal distribution of thymidine kinase mRNA. Proc. Natl Acad. Sci. USA. 1989;86:4987–4991. doi: 10.1073/pnas.86.13.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rousseau D, Kaspar R, Rosenwald I, Gehrke L, Sonenberg N. Translation initiation of ornithine decarboxylase and nucleocytoplasmic transport of cyclin D1 mRNA are increased in cells overexpressing eukaryotic initiation factor 4E. Proc. Natl Acad. Sci. USA. 1996;93:1065–1070. doi: 10.1073/pnas.93.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansell A, Farnebo L, Grenman R, Roberg K, Thunell LK. Polymorphism of FGFR4 in cancer development and sensitivity to cisplatin and radiation in head and neck cancer. Oral Oncol. 2009;45:23–29. doi: 10.1016/j.oraloncology.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Yasui W, Oue N, Sentani K, Sakamoto N, Motoshita J. Transcriptome dissection of gastric cancer: identification of novel diagnostic and therapeutic targets from pathology specimens. Pathol. Int. 2009;59:121–136. doi: 10.1111/j.1440-1827.2009.02329.x. [DOI] [PubMed] [Google Scholar]
- 30.Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, Brunner T, Simon HU. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat. Cell. Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- 31.Huang da W, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, Lempicki RA. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8:R183. doi: 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyer J, Allen WL, McLean EG, Wilson PM, McCulla A, Moore S, Longley DB, Caldas C, Johnston PG. Pharmacogenomic identification of novel determinants of response to chemotherapy in colon cancer. Cancer Res. 2006;66:2765–2777. doi: 10.1158/0008-5472.CAN-05-2693. [DOI] [PubMed] [Google Scholar]
- 33.Frydman J, Nimmesgern E, Ohtsuka K, Hartl FU. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994;370:111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- 34.Naylor DJ, Hartl FU. Contribution of molecular chaperones to protein folding in the cytoplasm of prokaryotic and eukaryotic cells. Biochem. Soc. Symp. 2001:45–68. doi: 10.1042/bss0680045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.