Abstract
V(D)J recombination entails double-stranded DNA cleavage at the antigen receptor loci by the RAG1/2 proteins, which recognize conserved recombination signal sequences (RSSs) adjoining variable (V), diversity (D) and joining (J) gene segments. After cleavage, RAG1/2 remain associated with the coding and signal ends (SE) in a post-cleavage complex (PCC), which is critical for their proper joining by classical non-homologous end joining (NHEJ). Certain mutations in RAG1/2 destabilize the PCC, allowing DNA ends to access inappropriate repair pathways such as alternative NHEJ, an error-prone pathway implicated in chromosomal translocations. The PCC is thus thought to discourage aberrant rearrangements by controlling repair pathway choice. Since interactions between RAG1/2 and the RSS heptamer element are especially important in forming the RAG-SE complex, we hypothesized that non-consensus heptamer sequences might affect PCC stability. We find that certain non-consensus heptamers, including a cryptic heptamer implicated in oncogenic chromosomal rearrangements, destabilize the PCC, allowing coding and SEs to be repaired by non-standard pathways, including alternative NHEJ. These data suggest that some non-consensus RSS, frequently present at chromosomal translocations in lymphoid neoplasms, may promote genomic instability by a novel mechanism, disabling the PCC’s ability to restrict repair pathway choice.
INTRODUCTION
A hallmark of the adaptive immune system is its ability to recognize a virtually limitless variety of antigens. At the heart of this process lie the B- and T-cell antigen receptors: the immunoglobulins (Ig) and T-cell receptors (TCR). The highly variable antigen binding domains of Ig and TCR molecules are generated by site-specific DNA rearrangements that bring together variable (V), diversity (D) and joining (J) gene segments during lymphocyte differentiation through a process known as V(D)J recombination (1). The biological significance of V(D)J recombination and its proper regulation is profound: it is required for B- and T-cell development (2,3), and errors in receptor gene rearrangements are thought to underlie many of the oncogenic chromosomal translocations found in B- and T-cell lymphomas (4).
V(D)J recombination is initiated by site-specific DNA cleavage mediated by the lymphoid-specific protein products of the recombination activating genes 1 and 2 (RAG1 and RAG2) (5,6). The RAG proteins recognize and bind to specific sequences situated adjacent to the V, D and J coding gene segments, known as recombination signal sequences (RSS). RSS are composed of a highly conserved heptamer sequence (consensus: 5′-CACAGTG-3′) and a less conserved nonamer sequence (consensus: 5′-ACAAAAACC-3′) separated by a less conserved spacer of 12 or 23 base pairs (Figure 1A). The nonamer element is important for initial recognition and binding by the RAG proteins, while the heptamer sequence is important for cleavage (7–9). After cleavage, the heptamer plays a particularly critical role in mediating continued binding of the RAG proteins to the cleaved signal ends (SEs) in the so-called SE complex (SEC) (10).
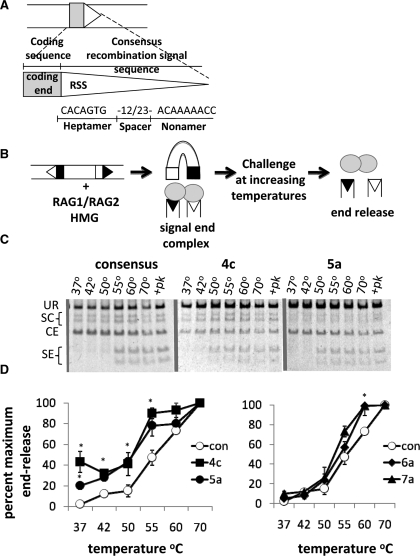
Figure 1.
Alterations to the RSS sequence can destabilize the RAG SE complex. (A) Schematic diagram of a RSS and its adjacent coding sequence. The consensus nucleotide sequence is shown. (B) Biochemical end release assay. Purified GST-tagged core RAG proteins cleave a ∼500 bp PCR product at 37°C. Post-cleavage SE complexes are thermally challenged at increasing temperatures and released SEs detected by electrophoresis. (C) Representative gels for end release assays. UR, unrearranged substrate; SC, single cleavage; CE, coding ends; SE, signal ends. Numbers above each lane indicate the temperatures reactions were heated to before electrophoresis. Samples treated with proteinase K and SDS are indicated with +pk. (D) Quantification of SE release, measured as the combined amount of radioactivity from cleaved products divided by the total amount of radioactivity, from three experiments using two different protein preparations (*P < 0.05). Non-consensus heptamers are abbreviated by the numerical position starting at the site of cleavage and the corresponding nucleotide change (con, consensus). We obtained similar results with GST core RAG1 and GST full length RAG2 (data not shown). Con denotes consensus RSS.
Efficient recombination occurs only between coding gene segments flanked by RSS with spacer lengths differing by one helical turn, a requirement referred to as the ‘12/23 rule’. The RAG proteins initiate recombination by first introducing a nick precisely between the heptamer of the RSS and the adjacent coding sequence, subsequently using the free 3′hydroxyl group to attack a phosphodiester bond on the opposite DNA strand in a direct trans-esterification reaction, thus forming a pair of covalently sealed hairpin coding ends (CE) and a pair of blunt 5′-phosphorylated SEs (11). Hairpin formation requires significant distortion of the DNA at the border between the heptamer and the adjacent coding flank (8,12,13).
CEs and SEs are processed and joined by the classical non-homologous end joining (NHEJ) pathway to form a coding joint and signal joint, respectively (14,15). Interestingly, the RAG proteins appear to play a role in directing the joining steps. After cleavage, the RAG proteins remain tightly bound to the SEs in a SEC that is resistant to competitor DNA, protects SEs from nuclease digestion in vitro (16–18), and appears to be largely dependent upon interactions of the heptamer sequences with the RAG proteins (10). The CEs, however, are much less tightly bound by the RAG proteins (16,17), a feature which has hampered biochemical analysis of RAG-CE associations. The difference in the in vitro stability of these complexes is paralleled by differences in the handling of the two types of DNA ends in vivo: CEs require hairpin opening and are joined imprecisely (19), are much less abundant than SEs in developing lymphocytes (20), and appear to be joined much more quickly than SEs (21). Thus, whereas there is little doubt that a stable RAG-SEC is an important reaction intermediate (see below), in vivo evidence for a four-ended RAG-DNA post-cleavage complex (PCC) involving both CE and SEs remains scant.
Perhaps the most compelling evidence for biologically significant associations between the RAG proteins, and CE and SE intermediates after cleavage is provided by analysis of various mutant forms of RAG1 or RAG2, which allow cleavage but hinder the formation of signal joints, coding joints or both, clearly indicating important interactions with both types of DNA intermediates (22–24). Indeed, certain RAG1 and RAG2 mutations destabilize the SEC in vitro and allow coding and SEs to be joined by inappropriate repair pathways, such as homologous recombination and alternative NHEJ, leading to the conclusion that these mutants destabilize the PCC (or the individual RAG-CE and RAG-SECs) in vivo (25,26). These data also strongly suggest that the RAG-PCC normally restricts access of the broken ends to error-prone joining pathways such as alternative NHEJ, which are quite active (26–28), even in cells proficient for classical NHEJ (26,29). Thus, the PCC may play a critical role in preventing genomic instability mediated by alternative NHEJ, a poorly characterized joining pathway that has been implicated in oncogenic chromosomal translocations in lymphoid neoplasms (28,30). This role can be thought of as a restriction on the choice of repair pathway mediated by a continued association between the RAG proteins and the RAG-generated DNA ends, ‘shepherding’ the ends to the high fidelity classical NHEJ pathway.
Given that RAG mutations are not likely to underlie a significant fraction of lymphoid neoplasms, we wondered whether other mechanisms might destabilize the PCC in developing lymphocytes. We reasoned that interactions between the RAG proteins and the SEs might be disrupted by variations in the sequence of the RSS. Indeed, although the RAG proteins primarily interact with the nonamer and heptamer/coding flank border before cleavage (31,32), the importance of the heptamer sequence appears to increase in the SEC. Foot-printing analyses have revealed increased contacts between the RAG proteins and the heptamer after cleavage, and functional analyses show that SEC formation, measured by assembly of precleaved SEs, depends particularly on the heptamer (10). A variety of RSS with non-consensus heptamer sequences are present at antigen receptor loci, many of which are frequently utilized for recombination (33). Furthermore, sequences resembling RSS, some of which bear non-consensus heptamers, are present at certain recurrent chromosomal translocations observed in human lymphoid neoplasms (19,34,35).
Using a variety of in vitro and cell-based assays, we show that certain non-consensus heptamer sequences indeed destabilize the PCC, allowing aberrant repair of both SE and CEs. Our observation that changes in the heptamer sequence can promote aberrant repair of CEs provides firm evidence for a RAG-PCC containing both coding and SEs in cells. We also find that a non-consensus heptamer found at the SIL locus, which is involved in aberrant oncogenic rearrangements in T-cell acute lymphoblastic leukemia (T-ALL), with a fourth and fifth position heptamer mutation causes substantial end release in vitro and allows joining by alternative NHEJ in cells. Based on these results, we propose a novel mechanism for RAG-mediated genomic instability that involves the ability of certain non-consensus RSS to disable the pathway choice restriction normally provided by the RAG-PCC. This model is supported by examination of translocation breakpoints: non-consensus nucleotides at heptamer position 4–5 are often present at both authentic RSS involved in recurrent translocations and RSS-like sequences adjacent to proto-oncogenes.
MATERIALS AND METHODS
Cell culture and protein purification
Chinese hamster ovary fibroblasts (the RMP41 cell line) were grown in DMEM (Invitrogen) supplemented with fetal bovine serum (10%), non-essential amino acids and penicillin–streptomycin. Cells were grown at 37°C in the presence of 5% CO2. GST-tagged truncated RAG proteins were purified from RMP41 fibroblasts by GST affinity chromatography (22) using GST affinity resin (Stratagene). Protein concentrations were determined by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Two independent protein preparations were assayed for each RSS mutation studied and showed consistent results.
Plasmid constructs and mutagenesis
RAG proteins were expressed in the pEBG-1N (RAG1) and pEBG-2C (RAG2) vector (36,37). The DDE mutant (D600A/D708A/E962A) is a catalytically inactive RAG1 mutant used as a control (38,39). A RAG1 ‘nick only’ mutant (R855A/K856A) was used as a control for in vitro nicking and homologous recombination experiments (22). The recombination substrates peCFP-289-NtCFP (25), pGFP-Alt (26) and 290-GFP (26) were used to measure homologous recombination, coding joints made by alternative NHEJ, and all coding joints, respectively. Double-stranded, site-directed mutagenesis (40) was performed on each of these recombination substrates to generate the substrates used in this study, listed in Supplementary Tables S1–3.
Transient transfection
RMP41 fibroblasts were grown to 50% confluency in 24-well plates (Corning). Transfections were performed in single wells. Each transfection contained 800 ng of each plasmid (substrate as indicated), and RAG expression vectors as indicated, 240 µl of serum-free DMEM and 3.6 µl of Fugene 6 transfection reagent (Roche) for a final Fugene 6: DNA ratio of 3 : 2 (26).
Southern blotting
Two-million RMP41 cells were transfected using Fugene 6 reagent (Roche) with 2 µg GST-core RAG1, 2 µg core RAG2 and 4 µg substrate in a 25 cm2 flask (Corning). Cells were harvested 48 h post-transfection. DNA was prepared from cells by the Hirt method used earlier (41). Samples were digested using HpaI and NcoI (New England Biolabs) for 4 h, run on a 2% native agarose gel and then transferred to Genescreen membrane for 5 h in 0.4 N NaOH. The DNA was then crosslinked to the membrane by UV irradiation. Probes were generated with a random priming DNA labeling kit (Invitrogen) using a gel-purified HpaI–NcoI fragment from the pECFP-289-NTCFP vector as a template. Probes were hybridized to blots overnight at 65°C in a solution of 10% Dextran Sulfate, 1 M NaCl and 1% SDS. Blots were washed twice in 2× SSC for 5 min, twice in 2× SSC, 1% SDS for 30 min and twice in 0.1× SSC, 0.1% SDS for 15 min at 65°C and visualized using a Phosphorimager and ImageQuant Software (Molecular Dynamics).
Flow cytometry
FACS analysis was carried out using a BD LSRII flow cytometer (BD biosciences) equipped with FacsDiva software after cells were trypsinized, pelleted and resuspended in DPBS (Invitrogen) supplemented with 0.5% BSA (Sigma Aldrich) and 5 mM EDTA. FlowJo software was used for data analysis. Cells were harvested for FACS analysis 48 h post-transfection for coding joint formation and alternative NHEJ. Cells were harvested 60 h post-transfection for homologous recombination. Background levels of recombination detected with a catalytically dead DDE RAG1 mutant were subtracted from each sample. Student's T-test assuming equal variance was used to calculate statistical significance.
Thermal challenge assays to measure stability of the SEC
For RAG-mediated cleavage, 100 ng of recombination substrate (PCR product from 289-nt CFP) was incubated for 3 h at 37°C with 200 ng purified RAG protein and 200 ng of purified recombinant HMGB1 in a buffer containing 50 mM Hepes (pH 8.0), 25 mM KCl, 4 mM NaCl, 1 mM DTT, 0.1 μg BSA, 5 mM CaCl2 and 5 mM MgCl2 (22). Reactions were then aliquoted into microfuge tubes and incubated at different temperatures, as indicated, or treated with stop buffer [10 mM Tris (pH 8.0), 10 mM EDTA, 0.2% SDS, 0.35 mg/ml proteinase K (Sigma Aldrich)] for 30 min and then ran out on 4–20% acrylamide TBE gels (Invitrogen). DNA was visualized using SYBR safe DNA gel stain (Invitrogen). To generate the substrate used for the thermal stability experiments pECFP-289-NtCFP constructs with appropriate RSS mutations were digested with HpaI and NdeI (New England Biolabs) and a 1.6 Kb fragment was gel purified and then used for PCR amplification using NtCFP_upPCR 5′-CGCGCCGAGGTGAAGTTCGAGG-3′ and NtCFP_downPCR 5′-TGCCCCAGGATGTTGCCGTCCTCC-3′ to generate a 477 bp PCR product. PCR amplification was performed as follows: 94°C, 2 min; 94°C, 15 s; 65°C, 30 s; 72°C, 30 s for 25 cycles; and 72°C, 7 min. Student's T-test assuming equal variance was used to calculate statistical significance.
In vitro nicking assay
An α-32P body-labeled substrate (800 bp) was generated by PCR from 290-GFP using the HY102 (5′-CTGAACTTGTGGCCGTTTA-3′) and HY98 (5′-AAGCAGAGCTGGTTTAGTGAAC-3′). Reactions were carried out using 100 ng of purified PCR product was incubated with 200 ng GST-coreRAG1/GST-coreRAG2 or GST-coreRAG1(R855A/K856A)/GST-coreRAG2 and 200 ng HMGB1 in buffer containing Mg2+ (25 mM MOPS-pH 7.0, 2 mM DTT, 100 ng BSA, 5 mM CaCl2, 19 mM KOAc, 5 mM MgCl2), as previously reported (22). Reactions mixtures were incubated for 10 min at 37°C. Magnesium was added to a final concentration of 5 mM, and the reactions were incubated to an additional 30 or 60 min before termination with an equal volume of formamide loading dye (95% formamide, 2 mM EDTA). Products were fractionated under denaturing conditions on a 10% polyacrylamide gel (8 M urea, 10% formamide). Gels were analyzed using a Storm phosphoimager and quantified using Quantity One software (Bio-Rad). Percent nicking at each RSS (%12 or 23) and percent hairpin formation (%hp) was calculated as the amount of radioactivity in the nicking and hairpin bands, size adjusted, divided by the total amount of radioactivity in each lane.
RESULTS
Non-consensus nucleotides at the fourth or fifth heptamer positions destabilize the SEC in vitro
To determine whether sequence variations in the heptamer affect the strength of RAG binding to SEs in the SEC, we employed the thermal stability assay schematized in Figure 1B. SEC were formed by cleavage of a radiolabeled 477 bp DNA substrate at 37°C by core RAG1 and core RAG2, which we then challenged by incubation at temperatures ranging from 37°C to 70°C. We measured SE release by native polyacrylamide gel electrophoresis. RAG-SECs do not migrate at defined electrophoretic mobilities in these gels, so that SEs are not visualized as discrete bands unless they are released by treatment with proteinase K/SDS or incubation at elevated temperatures (55°C or above with a consensus RSS sequence) (Figure 1C). With the consensus 12/23 RSS substrate, complete end release was not observed at temperatures below 70°C (Figure 1C), in agreement with previous results using a variation of this assay (25). As expected (8,41), we found that individual alterations of the first three nucleotides of the heptamer virtually abolished cleavage (data not shown). Mutation of the fourth or fifth nucleotide in the heptamer of the 12-RSS significantly diminished SEC stability (Figure 1C and D, left panel; P < 0.05). No statistically significant SEC instability was observed at lower temperatures with substrates bearing alterations in the sixth or seventh positions of the heptamer although more SEs were released at 60°C with these heptamer variations than with a consensus pair of RSS (Figure 1C and D, right panel). These data suggest that changes to the fourth and fifth positions of the heptamer substantially destabilize the SEC in vitro, whereas more distal mutations have a much less severe phenotype.
To further characterize the effect of non-consensus RSS on the stability of the SEC, we altered the fourth and fifth position heptamer sequence to every possible non-consensus nucleotide and examined SEC stability in vitro. We found that any change made to the consensus heptamer sequence at the fourth position destabilized the SEC (Supplementary Figure S1A; *P < 0.05). Unlike changes to the fourth position of the heptamer, the fifth heptamer position was most affected by the 5a mutation, as 5t and 5c showed little end release in our biochemical end release assay (Supplementary Figure S1B). Taken together, these data suggest that RAG proteins are sensitive to all non-consesnsus heptamers bearing fourth position mutations and only some non-consensus heptamers with fifth position mutations (5a).
Non-consensus heptamers influence repair of SEs in cells
To investigate the effects of non-consensus heptamer sequences on the stability of the post-cleavage SEC in cells, we employed an established assay that measures the levels of homologous recombination generated by free SEs (25,26). This assay uses a reporter substrate containing two non-functional copies of the cyan fluorescent protein (CFP) gene: one has an N-terminal truncation and the other is interrupted by an insertion flanked by both a 12RSS and a 23RSS (schematized in Figure 2A). Repair of the interrupted CFP allele by classical NHEJ, as occurs during normal V(D)J recombination, results in a non-functional copy of CFP, but repair by homologous recombination may generate a functional CFP gene, producing fluorescent cells that can be readily quantified by flow cytometry. We previously employed this assay to demonstrate that an intact SEC, in the context of consensus RSS and wild type core RAG1 and core RAG2, allows few SEs to enter the homologous recombination pathway, whereas mutant core RAG proteins that destabilize the SEC stimulate homologous recombination (25).
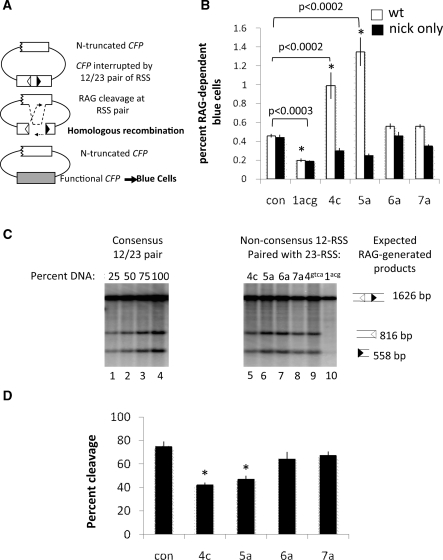
Figure 2.
Certain non-consensus RSS affect handling of SEs after cleavage. (A) Homologous recombination assay. Repair of a non-functional CFP gene by homologous recombination after RAG cleavage of plasmid 289-ntCFP leads to the expression of the CFP gene resulting in the detection of blue fluorescent cells, measurable by FACS analysis. (B) Quantification of FACS data (n = 6). Background, as measured by CFP expression in cells transfected with a catalytically inactive RAG1 allele, DDE, was subtracted from each sample. Con denotes consensus RSS. (C) Southern blot showing products of RAG cleavage of 289-ntCFP by cores RAG1 and RAG2. Hirt preps were digested with HpaI and NcoI to generate a 1626 bp fragment. Cleavage by the V(D)J recombinase generates a pair of SEs that are 816 bp (12RSS) and 558 bp (23RSS) and can be visualized using an internally radiolabeled probe. First four lanes show a titration of Hirt DNA from a transfection with a consensus pair of RSS. Percent of DNA loaded is found above each lane. Lanes 4–10 show levels of SEs in vivo at various non-consensus RSSs, labeled above each lane. (D) Quantification of in vitro cleavage by purified RAG proteins, measured as the combined amount of radioactivity from cleaved products divided by the total amount of radioactivity. Examples of gels are shown in Figure 1C in the proteinase K treated lane. * denotes P < 0.05.
We generated a series of homologous recombination substrates containing point mutations at each of the last four positions of the 12RSS heptamer paired with a consensus 23RSS (Supplementary Table S1). Catalytically inactive RAG proteins gave low background levels of fluorescence when paired with a consensus 12RSS, which was increased by active RAG1/2 (Supplementary Figure S2A), as shown previously (25). This stimulation depended upon the presence of an RSS and a homologous template for repair (Supplementary Figure S2B). As expected, an RSS bearing substitutions at the first three heptamer positions (1acg) reduced homologous recombination to levels below those observed with consensus RSS (Figure 2B) and abolished cleavage, as measured by Southern blotting of DNA purified from transfected cells (Figure 2C, lane 10). Mutations in the fourth and fifth heptamer positions, however, significantly and reproducibly increased homologous recombination 2–3-fold (Figure 2B, Supplementary Figure S2A) (P < 0.0002), even though they reduced levels of SEs by 25–50% in cells (Figure 2C) and in vitro (quantified in Figure 2D; examples of gels shown in Figure 1C). Mutations more distal to the cleavage site (6a, 7a) did not significantly affect cleavage (Figure 2C; quantified in vitro cleavage shown in Figure 2D) or levels of homologous recombination (Figure 2A–C, lanes 7–9). Changing the fourth heptamer position to another nucleotide (4t and 4g) gave similar results, as did placing the 4c and 5a mutations in the context of a 23RSS, while fifth position changes to 5t and 5c only slightly increased the levels of homologous recombination (data not shown). These data indicate that non-consensus nucleotides at the fourth and fifth position in the heptamer, though they reduce the efficiency of cleavage, destabilize the RAG-SEC in cells, making ends available to alternative repair pathways.
A previous study detected an accumulation of nicks at certain cryptic RSSs (42). Because we previously demonstrated that RAG-mediated nicks initiate homologous recombination by wild-type RAG proteins at a consensus RSS pair (25), we wondered whether the increase in homologous recombination at non-consensus RSSs was a result of increased nicking at these sites. Therefore, we examined RAG-mediated nicking of substrates bearing non-consensus heptamers in vitro. None of the RSS variations tested significantly increased nicking compared to a consensus pair of RSS after 30 min incubation (Supplementary Figure S3). Similar results were obtained with a longer incubation of 60 min (data not shown). To directly examine the role of nicks in initiating homologous recombination at non-consensus RSS, we tested the non-consensus RSS substrates with a ‘nick-only’ RAG1 mutant (R855A/K856A) that nicks at wild-type levels, but is severely impaired for DSB formation (22). We first assessed this mutant’s ability to nick at non-consensus RSS in vitro and found that the nick only RAG1 mutant exhibited similar levels of nicking at all RSS variations compared to wild-type RAG1 (Supplementary Figure S3). Subsequently, we tested the nick-only mutant’s ability to stimulate homologous recombination in substrates containing non-consensus heptamers. Although both the nick-only RAG1 and the wild-type RAG1 proteins produced virtually identical levels of homologous recombination with a consensus 12/23RSS pair (Figure 2B), in agreement with previous results (25), the nick-only mutant did not increase levels of homologous recombination on substrates with fourth and fifth heptamer mutations (Figure 2B). Together, these data strongly suggest that RAG-mediated double-strand breaks, not nicks, are the primary lesions that stimulate homologous recombination in substrates containing 4c and 5a heptamer variations.
Non-consensus heptamers influence repair of CEs in cells
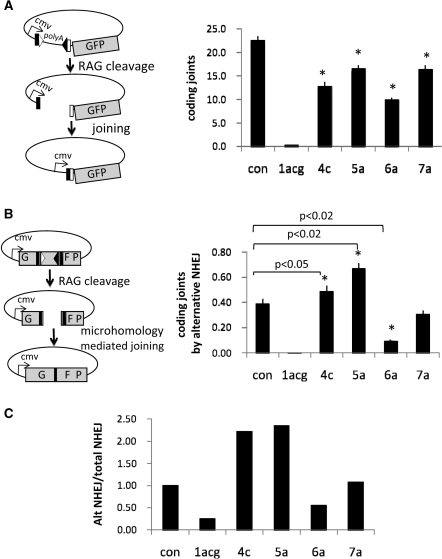
Our homologous recombination assay measures only SEs (in previous work we were unable to detect initiation of recombination from CEs, presumably because they are covalently sealed) (25). We, therefore, examined the fate of CEs formed by cleavage of consensus and non-consensus RSS using two different assays, both measuring recombination catalyzed by full-length RAG1/2. Figure 3A shows results of assays for coding joint formation. Alterations at the fourth–seventh heptamer positions reduced coding joint formation, likely due to the reduction in cleavage (Figure 2C). To gauge whether these RSS alterations led to aberrant release of CEs from the PCC, we measured repair of CEs by alternative NHEJ using a substrate that requires excessive deletion of nucleotides from both ends (20 nts total deletion) and relies on a 9 nt microhomology to create a functional GFP gene (26,29) (schematized in Figure 3B). Consensus RSS gave extremely low levels of GFP-positive cells (Figure 3B), reflecting inefficient use of alternative NHEJ in the presence of wild-type RAG proteins, consistent with our previous results (26,29). In sharp contrast, RSS bearing a fifth position heptamer mutation (5a) yielded significantly higher levels of alternative NHEJ compared to the consensus RSS (P < 0.02) (Figure 3B), even though coding joint formation (measured using a conventional substrate) was reduced (Figure 3A). We observed similar results with a fourth position heptamer variant (4C), which, despite a 50% reduction in normal coding joint formation (Figure 3A), yielded a small but statistically significant (P < 0.05) increase in alternative NHEJ (Figure 3B). The negative effects of heptamer alterations on cleavage lead to an underestimate of their effects on alternative NHEJ. To take this into account, we calculated the ratio of alternative NHEJ products to standard coding joints. Both the 4c and 5a heptamer mutations increased this ratio over 2-fold, but changes to the sixth and seventh position of the heptamer did not (Figure 3C). These data suggest that certain changes to the consensus RSS sequence can affect handling of CEs, increasing their availability to alternative NHEJ.
Figure 3.
Certain non-consensus RSS alter handling of CEs after cleavage. (A) Schematic diagram of GFP reporter construct. GFP is under the control of the CMV promoter. RAG cleavage and subsequent coding joint formation deletes a poly-adenylation site located upstream of the GFP, thereby allowing GFP expression. Right panel shows quantification of FACS data for coding joint formation (n = 5, *P < 0.02). RMP41 cells were transiently transfected with full-length RAG proteins and a fluorescent reporter for coding joint formation, 290-GFP containing either a consensus pair of RSS or a non-consensus 12RSS, as indicated. (B) Alternative NHEJ assay (n = 5). Cleavage by the RAG proteins followed by resection of 10 nts from each CE reveals a nine-nucleotide microhomology used to direct the precise repair of the GFP gene. Highly deleted coding joints that use microhomologies are characteristic of joining by the alternative NHEJ pathway. Right panel shows coding joint formation by alternative NHEJ assay as measured by transient transfection of RMP41 cell with full-length RAG proteins using ALT-GFP reporter substrate containing either a consensus pair of RSS or a non-consensus 12RSS, as indicated. (C) Ratio of alternative NHEJ to total NHEJ, normalized to one for a consensus pair of RSS. Similar results were obtained with core RAG2 (data not shown).
A non-consensus heptamer found at the SIL locus destabilizes the SEC in vitro and allows aberrant repair of CEs in cells
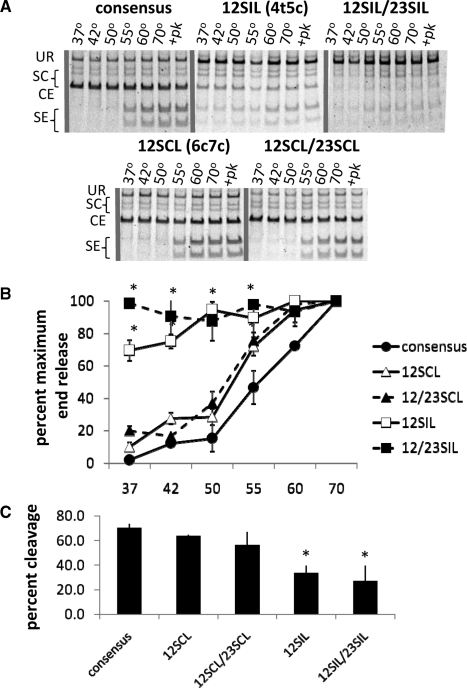
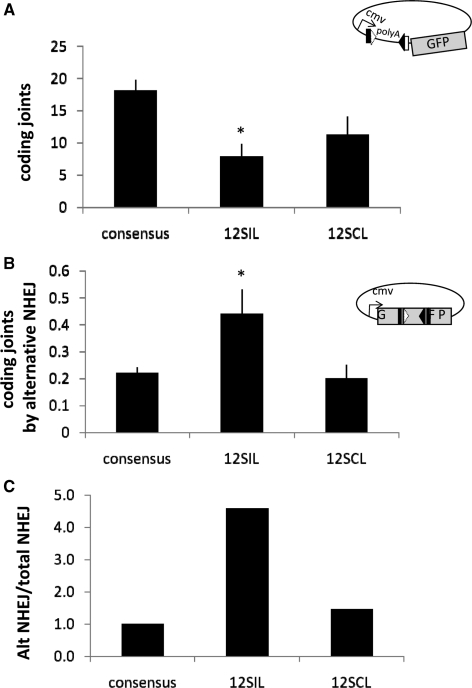
The cryptic RSS found at the SIL and SCL loci contain multiple changes to the consensus heptamer and nonamer sequences and are thus very poor substrates for V(D)J recombination (35,42). We wondered whether the heptamer variations found at these cryptic RSS, including a fourth and fifth position mutation at the SIL locus (4t5c) and a sixth and seventh position mutation at the SCL locus (6c7c), could destabilize the RAG-SEC and allow repair of breaks at these sites by alternative NHEJ (a consensus nonamer was provided to facilitate cleavage). We found that the 4t5c cryptic heptamer found at the SIL locus almost completely destabilizes the post-cleavage SEC, allowing 70% end release at 37°C (Figure 4A and quantified in Figure 4B, P < 0.02), while modestly decreasing cleavage (to 50% of that observed with a consensus pair of RSS; quantified in Figure 4C) when paired to a consensus 23-RSS. Pairing two 4t5c mutations, one at the 12RSS and one at the 23RSS, resulted in complete end release even at 37°C, allowing cleavage at 40% of wild-type levels (quantified in Figure 4C). A 6c7c heptamer mutation, found at the SCL locus, did not significantly destabilize the SEC (Figure 4A and quantified in Figure 4B) and did not affect cleavage in vitro (Figure 4A and quantified in Figure 4C). As expected, 4t5c and 6c7c both significantly decreased coding joint formation (Figure 5A) (35). Interestingly, the 4t5c, but not the 6c7c cryptic heptamer, increased the level of alternative end joining 4–5-fold (Figure 5B and C). These data suggest that cryptic RSS found the SIL locus may promote joining by error-prone alternative DNA repair pathways.
Figure 4.
A non-consensus heptamer found at the SIL locus destabilizes the RAG SE complex. (A) Representative gels for end release assays, depicted in Figure 1B, using substrates with non-consensus heptamers found at the SCL (6c7c) and SIL (4t5c) locus, note that a consensus nonamer is used to examine post-cleavage effects of heptamer mutations. (B) Quantification of SE release from three experiments (*P < 0.02). (C) Quantification of in vitro cleavage by purified RAG proteins, measured as the combined amount of radioactivity from cleaved products divided by the total amount of radioactivity. Examples of gels are shown in Figure 4A in the proteinase K treated lanes. 12SIL: CACtcTG_ACAAAAACC paired with consensus 23RSS. 12SCL: CACAGcc_ACAAAAACC paired with consensus 23RSS. 12/23SIL: CACtcTG_ACAAAAACC at both RSS. 12/23SCL: CACAGcc_ACAAAAACC at both RSS.
Figure 5.
A non-consensus heptamer found at the SIL locus allows aberrant repair of CEs. (A) Quantification of FACS data for coding joint formation (n = 5, *P < 0.02). RMP41 cells were transiently transfected with full-length RAG proteins and a fluorescent reporter for coding joint formation, 290-GFP containing either a consensus pair of RSS or substrates with non-consensus heptamers, as indicated. (B) Alternative NHEJ assay (n = 5). RMP41 cells were transiently transfected with full-length RAG proteins using ALT-GFP reporter substrate containing either a consensus pair of RSS or a non-consensus 12RSS, as indicated. Coding joint formation by alternative NHEJ was measured by FACS. (C) Ratio of alternative NHEJ to total NHEJ, normalized to one for a consensus pair of RSS. 12SIL: CACtcTG_ACAAAAACC paired with consensus 23RSS. 12SCL: CACAGcc_ACAAAAACC paired with consensus 23RSS. 12/23SIL: CACtcTG_ACAAAAACC at both RSS. 12/23SCL: CACAGcc_ACAAAAACC at both RSS.
DISCUSSION
A PCC containing both CE and SEs is important for restricting repair pathway choice and is disrupted by heptamer alterations
Previous studies have examined the influence of RSS sequence on RAG binding, nicking and hairpin formation (8,9,12,42). In this work we specifically examined whether non-consensus heptamers affect the stability of the PCC and influence joining of CE and SEs. We found that, as expected, mutations in the first three heptamer positions adjacent to the cleavage site impaired cleavage to such an extent that we were unable to study their effects on joining. Alterations in the fourth and fifth positions diminished cleavage but also changed the way both SE and CEs are handled post-cleavage. In vitro, purified RAG proteins prematurely released SEs containing these non-consensus heptamers from the SEC. These results confirm and extend previous results suggesting that RAG-heptamer interactions are important for assembly of the post-cleavage SEC from pre-cleaved SEs (10). We found that the same heptamer alterations that destabilize the SEC complex in vitro affect handling of both CE and SEs in cells, making SEs available to inappropriate repair by homologous recombination and allowing CEs to access alternative NHEJ, effects previously observed only with mutant RAG proteins that destabilize the PCC (25,26). Interestingly, we found that any deviation from the consensus nucleotide at the fourth position destabilizes the SEC, while only one fifth position heptamer variation (5a) displays significant end release in our biochemical assays and allows aberrant joining of V(D)J breaks in cells.
Our data are in close agreement with those of Agard and Lewis, who showed (using a pre-B cell line that expresses endogenous full-length RAG proteins) that non-canonical RSS can selectively impair joining events that require participation of all four ends (inversional recombination) compared with two-ended joining events (coding or signal joint formation by deletion, as measured in our study) (44). Their results indicate that changes to the RSS, particularly in the positions 5–7 of the heptamer, preferentially affect four-ended joining events, consistent with our direct demonstration that similar heptamer alterations (at positions 4 and 5) destabilize the RAG-SEC in vitro and make both CE and SEs available for alternative joining pathways in cells. These results are consistent with our recent demonstration that RAG mutants which destabilize the SEC in vitro also selectively diminish inversional recombination events in cells and in developing lymphocytes, again indicating a destabilization of the PCC (Roth lab, unpublished data), similar to effects of ATM-deficiency (44) and hypomorphic mutations in NBS1 (29,45). Together, these data firmly implicate the heptamer sequence in maintenance of both SE and CEs in a four-ended RAG PCC, which plays an important role in restricting access of the ends to alternative joining pathways. How long this complex persists, and how it might mature into separate coding and SECs (to account for the differential stability and processing of these ends) remain important questions for future investigation.
Our results also extend previous studies showing that RAG mutations, which destabilize the SEC in vitro, allow SE and CEs to inappropriately access alternative repair pathways (homologous recombination and alternative NHEJ) in cells (25,26). Given the lack of biochemical assays for the roles of the RAG proteins in the joining steps of V(D)J recombination, we have been unable to determine whether our RAG mutants affect the PCC, the separate SE and presumptive CE complexes, or all three. Non-consensus heptamers now provide another tool with which to perturb the system.
A post-cleavage role for the distal portion of the heptamer sequence in preserving genomic stability
Alterations at positions 4 and 5 of the heptamer allow relatively efficient double-strand break formation but promote use of alternative, potentially dangerous, pathways for resolution of the broken DNA intermediates. Considerable evidence suggests that such heptamer variants participate in oncogenic chromosomal translocations, as they have been identified at both translocation partners: at the authentic RSS in the antigen receptor locus and adjacent to proto-oncogenes, in the form of RSS-like sequences termed ‘cryptic’ RSS. For example at the Ig locus, JH6 (CACAaTG), a 5a variant, frequently appears as a translocation partner joined to BCL2 in B- acute lymphoblastic leukemia (B-ALL) (46–48). Similarly, at the TCR locus, Dbeta1 (CACAaTG), another 5a variant, is frequently used in translocations involving cryptic RSS in the vicinity of proto-oncogenes in T-ALL (49). Our findings suggest that this non-consensus heptamer (5a), which allows efficient cleavage by the V(D)J recombinase, nevertheless perturbs the RAG PCC enough to promote joining by the error prone alternative NHEJ pathway.
We found that non-consensus heptamer changes at the sixth and seventh position have a subtle end release defect in vitro, manifested only at higher temperatures. We speculate that these (and perhaps other) heptamer variants might destabilize the PCC in conjunction with other alterations, such as non-consensus nonamers that are frequently involved in chromosomal translocations (LMO2 CACAGTa_gCAAtAAtt, TAL1 CACAccG_cgAAAAAGG, HOX 11 CACAGcc_cagtcctAaa). Unfortunately, the substantial cleavage defects associated with these multiply altered RSS (42) precludes direct analysis with our end release assay.
Approximately 25% of cases of human T-ALL harbor an oncogenic interstitial deletion between the SIL and Scl genes (50), which appears to be mediated by RAG cleavage at non-consensus RSS bearing changes at positions 4 and 5 (adjacent to SIL) and at positions 6 and 7 (adjacent to Scl), that results in the inappropriate expression of the Tal-1 gene, which is not normally expressed in T cells (51). These cryptic RSS, which bear multiple non-consensus nucleotides at the presumptive nonamer elements, recombine quite inefficiently when tested on extrachromosomal substrates (∼0.1% or less) (35,52), are cleaved poorly by purified RAG proteins in vitro (∼1% of consensus) and also in transfected cells (43). Indeed, recombination events between the two cryptic RSS (SIL/SCL) placed on an extrachromosomal substrate were so infrequent as to be undetectable in transient transfection assays (35). Why, then, is recombination between these two elements seen so (relatively) frequently in T-ALL? Factors such as local chromatin structure and selective growth advantage conferred by the translocation may play important roles. Indeed, recent work has shown that cryptic RSS commonly involved in oncogenic translocations or aberrant RAG-mediated rearrangements are associated with histone H3 trimethylated at lysine 4 (H3K4me3) hot spots (53). The V(D)J recombinase is thought to recognize H3K4me3 histone variants via the PHD domain of RAG2 (54,55), which may be important for targeting the RAG proteins to rearranging antigen receptor loci, known to be in an ‘open’ chromatin configuration with acetylated H3 and H4 as well as H3K4me3 histone variants. Interestingly, although the SIL and SCL cryptic RSS show very little recombination activity in transiently transfected cells, H3K4me3 (in the form of a peptide) stimulates both nicking at hairpin formation at the SIL and SCL cryptic RSS in vitro (53). The RAG proteins may thus be targeted and recombination stimulated at these cryptic sites by H3K4me3.
RAG-mediated nicks accumulate at the SIL and SCL cryptic RSS in vitro (43). Single-stranded nicks could potentially contribute to aberrant rearrangements at these sites, perhaps after conversion to a double-strand break. Aberrant nicking on both strands, which occurs at the SIL cryptic RSS (43), could lead to a double-strand break directly. Such RAG-mediated double-strand breaks formed outside of the canonical cleavage reaction (nick to hairpin conversion within a synaptic complex), may not be protected by the RAG-PCC and could thus be available to alternative repair pathways. We found that a non-consensus heptamer bearing a 4t5c mutation found at the SIL locus destabilizes the RAG-PCC in vitro and allows aberrant joining by alternative NHEJ. These data suggest another possibility: some cryptic RSS may destabilize the PCC and increase the availability of DNA ends to the error-prone alternative NHEJ pathway, which has been implicated in chromosomal translocations in a variety of situations, including in developing B cells (28,30,56).
In sum, end release promoted by non-consensus heptamer sequences at either authentic or cryptic RSS could favor oncogenic rearrangements via two mechanisms: making the ends available for error-prone alternative end joining or by promoting ‘end donation’ (19), which could proceed by either alternative or classical NHEJ and could be facilitated simply by untethering the ends from the normal PCC.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (CA104588). Funding for open access charge: CA104588.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Ludovic Deriano, Bette Pancake and Marc Coussens for their helpful comments on the manuscript.
REFERENCES
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 3.Shinkai Y, Rathbun G, Lam K.-P, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, et al. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 4.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 6.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 7.Akamatsu Y, Tsurushita N, Nagawa F, Matsuoka M, Okazaki K, Imai M, Sakano H. Essential residues in V(D)J recombination signals. J. Immunol. 1994;153:4520–4529. [PubMed] [Google Scholar]
- 8.Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- 9.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 10.Nagawa F, Kodama M, Nishihara T, Ishiguro K, Sakano H. Footprint analysis of recombination signal sequences in the 12/23 synaptic complex of V(D)J recombination. Mol. Cell Biol. 2002;22:7217–7225. doi: 10.1128/MCB.22.20.7217-7225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 12.Cuomo CA, Mundy CL, Oettinger MA. DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol. Cell. Biol. 1996;16:5683–5690. doi: 10.1128/mcb.16.10.5683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu CP, Sandoval H, Weller GR, Rice PA, Roth DB. Identification of active site amino acids in RAG-1 critical for DNA hairpin formation. Nat. Struct. Mol. Biol. 2006;13:1010–1015. doi: 10.1038/nsmb1154. [DOI] [PubMed] [Google Scholar]
- 14.Lieber MR, Ma Y, Pannicke U, Schwarz K. The mechanism of vertebrate nonhomologous DNA end joining and its role in V(D)J recombination. DNA Repair. 2004;3:817–826. doi: 10.1016/j.dnarep.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Weterings E, Chen DJ. The endless tale of non-homologous end-joining. Cell Res. 2008;18:114–124. doi: 10.1038/cr.2008.3. [DOI] [PubMed] [Google Scholar]
- 16.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 17.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol. Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 18.Jones JM, Gellert M. Intermediates in V(D)J recombination: a stable RAG1/2 complex sequesters cleaved RSS ends. Proc. Natl Acad. Sci. USA. 2001;98:12926–12931. doi: 10.1073/pnas.221471198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological and comparative analyses. Adv. Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 20.Roth DB, Nakajima PB, Menetski JP, Bosma MJ, Gellert M. V(D)J recombination in mouse thymocytes: double-strand breaks near T cell receptor delta rearrangement signals. Cell. 1992;69:41–53. doi: 10.1016/0092-8674(92)90117-u. [DOI] [PubMed] [Google Scholar]
- 21.Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- 22.Huye LE, Purugganan MM, Jiang MM, Roth DB. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell Biol. 2002;22:3460–3473. doi: 10.1128/MCB.22.10.3460-3473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 24.Tsai CL, Drejer AH, Schatz DG. Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev. 2002;16:1934–1949. doi: 10.1101/gad.984502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 26.Corneo B, Wendland RL, Deriano L, Cui X, Klein IA, Wong SY, Arnal S, Holub AJ, Weller GR, Pancake BA, et al. Rag mutations reveal robust alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 27.Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 28.Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol. Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109:811–821. doi: 10.1016/s0092-8674(02)00770-5. [DOI] [PubMed] [Google Scholar]
- 31.Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 32.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hesse JE, Lieber MR, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 34.Aplan PD, Lombardi DP, Kirsch IR. Structural characterization of SIL, a gene frequently disrupted in T-cell acute lymphoblastic leukemia. Mol. Cell Biol. 1991;11:5462–5469. doi: 10.1128/mcb.11.11.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raghavan SC, Kirsch IR, Lieber MR. Analysis of the V(D)J recombination efficiency at lymphoid chromosomal translocation breakpoints. J. Biol. Chem. 2001;276:29126–29133. doi: 10.1074/jbc.M103797200. [DOI] [PubMed] [Google Scholar]
- 36.Spanopoulou E, Roman C.AJ, Corcoran L, Schlissel M, Silver DP, Nemazee D, Nussenzweig M, Shinton SA, Hardy RR, Baltimore D. Functional immunoglobulin transgenes guide ordered B cell differentiation in RAG-1-deficient mice. Genes Dev. 1994;8:1030–1042. doi: 10.1101/gad.8.9.1030. [DOI] [PubMed] [Google Scholar]
- 37.Spanopoulou E, Zaitseva F, Wang F.-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 38.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG-1 and RAG-2 identifies three active site amino acids in RAG-1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deng WP, Nickoloff JA. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 41.Steen SB, Gomelsky L, Speidel SL, Roth DB. Initiation of V(D)J recombination in vivo: role of recombination signal sequences in formation of single and paired double-strand breaks. EMBO J. 1997;16:2656–2664. doi: 10.1093/emboj/16.10.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M, Swanson PC. V(D)J recombinase binding and cleavage of cryptic recombination signal sequences identified from lymphoid malignancies. J. Biol. Chem. 2008;283:6717–6727. doi: 10.1074/jbc.M710301200. [DOI] [PubMed] [Google Scholar]
- 43.Agard EA, Lewis SM. Postcleavage sequence specificity in V(D)J recombination. Mol. Cell Biol. 2000;20:5032–5040. doi: 10.1128/mcb.20.14.5032-5040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, Nuskey B, Sullivan KE, Pandita TK, Bassing CH, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 2006;442:466–470. doi: 10.1038/nature04866. [DOI] [PubMed] [Google Scholar]
- 45.Helmink BA, Bredemeyer AL, Lee BS, Huang CY, Sharma GG, Walker LM, Bednarski JJ, Lee WL, Pandita TK, Bassing CH, et al. MRN complex function in the repair of chromosomal Rag-mediated DNA double-strand breaks. J. Exp. Med. 2009;16:669–79. doi: 10.1084/jem.20081326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nadel B, Marculescu R, Le T, Rudnicki M, Bocskor S, Jager U. Novel insights into the mechanism of t(14;18)(q32;q21) translocation in follicular lymphoma. Leuk. Lymphoma. 2001;42:1181–1194. doi: 10.3109/10428190109097743. [DOI] [PubMed] [Google Scholar]
- 47.Marculescu R, Le T, Bocskor S, Mitterbauer G, Chott A, Mannhalter C, Jaeger U, Nadel B. Alternative end-joining in follicular lymphomas' t(14;18) translocation. Leukemia. 2002;16:120–126. doi: 10.1038/sj.leu.2402324. [DOI] [PubMed] [Google Scholar]
- 48.Li A, Rue M, Zhou J, Wang H, Goldwasser MA, Neuberg D, Dalton V, Zuckerman D, Lyons C, Silverman LB, et al. Utilization of Ig heavy chain variable, diversity, and joining gene segments in children with B-lineage acute lymphoblastic leukemia: implications for the mechanisms of VDJ recombination and for pathogenesis. Blood. 2004;103:4602–4609. doi: 10.1182/blood-2003-11-3857. [DOI] [PubMed] [Google Scholar]
- 49.Tycko B, Palmer JD, Sklar J. T cell receptor gene trans-rearrangements: chimeric gamma delta genes in normal lymphoid tissues. Science. 1989;245:1242–1246. doi: 10.1126/science.2551037. [DOI] [PubMed] [Google Scholar]
- 50.Aplan PD, Lombardi DP, Ginsberg AM, Cossman J, Bertness VL, Kirsch IL. Disruption of the human SCL locus by “illegitimate” V(D)J recombinase activity. Science. 1990;250:1426–1429. doi: 10.1126/science.2255914. [DOI] [PubMed] [Google Scholar]
- 51.Pulford K, Lecointe N, Leroy-Viard K, Jones M, Mathieu-Mahul D, Mason DY. Expression of TAL-1 proteins in human tissues. Blood. 1995;85:675–684. [PubMed] [Google Scholar]
- 52.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol. Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol. Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Matthews AG, Kuo AJ, Ramon-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharpless NE, Ferguson DO, O'H;agan RC, Castrillon DH, Lee C, Farazi PA, Alson S, Fleming J, Morton CC, Frank K, et al. Impaired nonhomologous end-joining provokes soft tissue sarcomas harboring chromosomal translocations, amplifications, and deletions. Mol. Cell. 2001;8:1187–1196. doi: 10.1016/s1097-2765(01)00425-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.