Abstract
During the past three decades, we have witnessed remarkable advances in our understanding of the molecular etiologies of hereditary neurodegenerative diseases, which have been accomplished by ‘positional cloning’ strategies. The discoveries of the causative genes for hereditary neurodegenerative diseases accelerated not only the studies on the pathophysiologic mechanisms of diseases, but also the studies for the development of disease-modifying therapies. Genome-wide association studies (GWAS) based on the ‘common disease–common variants hypothesis’ are currently undertaken to elucidate disease-relevant alleles. Although GWAS have successfully revealed numerous susceptibility genes for neurodegenerative diseases, odds ratios associated with risk alleles are generally low and account for only a small proportion of estimated heritability. Recent studies have revealed that the effect sizes of the disease-relevant alleles that are identified based on comprehensive resequencing of large data sets of Parkinson disease are substantially larger than those identified by GWAS. These findings strongly argue for the role of the ‘common disease–multiple rare variants hypothesis’ in sporadic neurodegenerative diseases. Given the rapidly improving technologies of next-generation sequencing next-generation sequencing (NGS), we expect that NGS will eventually enable us to identify all the variants in an individual's personal genome, in particular, clinically relevant alleles. Beyond this, whole genome resequencing is expected to bring a paradigm shift in clinical practice, where clinical practice including diagnosis and decision-making for appropriate therapeutic procedures is based on the ‘personal genome’. The personal genome era is expected to be realized in the near future, and society needs to prepare for this new era.
INTRODUCTION
Neurodegenerative diseases are usually characterized by onset in late adulthood, a slowly progressive clinical course and neuronal loss with regional specificity in the central nervous system. In Alzheimer disease, Parkinson disease (PD), spinocerebellar ataxias and amyotrophic lateral sclerosis, neurodegeneration preferentially involves the cerebral cortex, extrapyramidal system, cerebellum and spinal cord, respectively. Although the majority of neurodegenerative diseases are sporadic, Mendelian inheritance patterns have been well documented. Intriguingly, the clinical presentations and neuropathological findings of hereditary forms of these neurodegenerative diseases are often indistinguishable from the sporadic diseases, raising the possibility that common pathophysiologic mechanisms underlie both hereditary and sporadic neurodegenerative diseases.
During the past three decades, there have been remarkable advances in our understanding of the etiologies of hereditary neurodegenerative diseases, which have been accomplished by ‘positional cloning’ efforts (1–4). The identification of the causative genes for hereditary neurodegenerative diseases has accelerated studies on the pathophysiologic mechanisms of diseases and the development of disease-modifying therapies based on these discoveries has now become a reality.
Molecular bases of neurodegenerative disease with Mendelian traits
Establishment of positional cloning strategies (1–4) including high throughput linkage analysis employing microarrays (5–6) has further accelerated the search for the causative genes for diseases with Mendelian traits. Furthermore, the availability of the human genome sequence (7) has tremendously accelerated the discovery of causative genes. Despite this progress, however, the mutations causing a substantial number of hereditary diseases remain to be identified. In familial amyotrophic lateral sclerosis (FALS), in particular, its causative genes have been identified in only 25–30% of FALS cases, suggesting that the majority of FALS genes remain to be identified (8–10). When the pedigree size is limited, it is difficult to narrow the candidate region by linkage analysis; hence, tremendous effort is still required to identify the causative genes based on positional cloning strategies. In diseases such as FALS in which the clinical severity is substantial, the number of living affected individuals is often limited. Thus, for many disorders, we need high throughput comprehensive resequencing capability to identify the causative mutations located in broad candidate regions of 10–100 Mb.
MOLECULAR BASIS OF SPORADIC NEURODEGENERATIVE DISEASES
For sporadic neurodegenerative diseases, which comprise the majority of the cases, we are still far from understanding their molecular etiologies despite the clues obtained on the basis of neuropathological findings. For example, although we know that accumulation of senile plaques in which the β amyloid protein is the major component underlies both sporadic and familial Alzheimer disease, we have little knowledge on the molecular etiologies of sporadic Alzheimer disease. We occasionally observe affected siblings or relatives with neurodegenerative diseases, which raises the possibility of involvement of genetic factors in these diseases. To identify susceptibility genes that account for the heritability seen for complex traits, genome-wide association studies (GWAS) employing common single nucleotide polymorphisms (SNPs) have been conducted. The theoretical framework for GWAS is the ‘common disease–common variant hypothesis’, in which common diseases are attributable in part to allelic variants present in more than 1–5% of the population (11–13).
Although GWAS have successfully revealed numerous susceptibility genes for common diseases such as diabetes as well as neurodegenerative diseases, the odds ratios associated with these risk alleles are generally low and account for only a small proportion of estimated heritability (14–16). It is assumed that risk alleles with large effect size may be rare in frequency and hard to detect by GWAS employing common SNPs. Emerging new technologies of next-generation sequencers will eventually enable the identification of all the variants including ‘rare variants’ in single subjects. In this review, future directions for identifying disease-relevant genetic variations on the basis of comprehensive resequencing of the human genome employing the next-generation sequencers are discussed.
ROLE OF RARE VARIANTS IN NEURODEGENERATIVE DISEASES
The general finding that the odds ratios associated with risk alleles identified for disease susceptibility by GWAS are low indicates that GWAS based on the ‘common disease–common variants hypothesis’ are not effective in identifying genetic risks with large effect sizes. High λs, estimating recurrent risks for siblings of affected individuals have been demonstrated in many diseases with complex traits, but the genetic risk factors identified by GWAS do not account for the high λs. Current experience with GWAS strongly suggests that rarer variants that are hard to detect by GWAS may account for the ‘missing’ heritability. Such rare variants may have large effect sizes as genetic risk factors for diseases. Thus we need a paradigm shift from the ‘common disease–common variants hypothesis’ to a ‘common disease–multiple rare variants hypothesis’ to identify disease-relevant alleles with large effect sizes.
The prominent role of rare variants in neurodegenerative disease is best highlighted by the recent discovery of the glucocerebrosidase gene (GBA) as a robust genetic risk factor for PD (17–18). PD, which is characterized by tremor, rigidity, bradykinesia, and postural instability, is the second most common neurodegenerative disease after Alzheimer disease, with onset typically in late adulthood. The prevalence of PD has been estimated to be 0.3% in the general population and 1% in people over 60 years of age. Although α-synuclein (SNCA), leucine-rich repeat kinase 2 (LRRK2), UCHL-1, Parkin (PARK2), PTEN-induced putative kinase 1 (PINK1) and DJ-1 have been identified as causative genes for familial PD, PD patients with pathogenic mutations in these genes are rare, and most of the PD cases are sporadic, the etiologies of which are poorly understood. A population-based study coupled with genealogy information demonstrated that the estimated risk ratio for PD for siblings of patients with PD was significantly elevated (λs = 6.3), indicating that genetic factors substantially contribute to the development of sporadic PD (18). Recent clinical observations (19) suggested the association of sporadic PD with heterozygous mutations in the glucocerebrosidase gene (GBA) encoding the enzyme that is deficient in patients with Gaucher disease, an autosomal recessive lysosomal storage disease. Furthermore co-morbidity of PD and Gaucher disease had previously been described (20). We conducted an extensive resequencing analysis of GBA in PD patients and controls, and found that GBA variants that are pathogenic for Gaucher disease confer a robust susceptibility to sporadic PD, and, even account for familial clustering of PD (18) (Fig. 1). The combined carrier frequency of the ‘pathogenic variants’ was as high as 9.4% in PD patients and significantly more frequent than in controls (0.37%) with a markedly high odds ratio of 28.0 (95% CI, 7.3 to 238.3) for PD patients compared with controls.
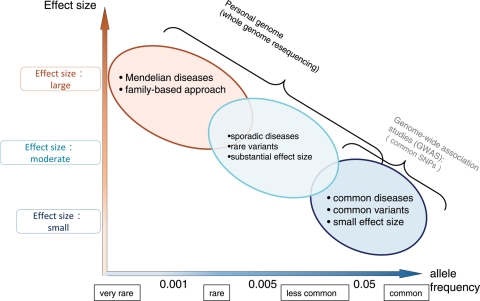
Figure 1.
Research paradigm to identify disease-related variations based on comparison of effect sizes of variants and allele frequencies of the variants in population. Adapted by permission from Macmillan Publishers Ltd: Nature, 461: 747–53 (2009) (16).
The molecular effects of the ‘pathogenic variants’ in PD remain to be elucidated. Gain of toxic functions of the mutant glucocerebrosidase proteins independent of enzyme activity might be involved in the pathogenesis. Intriguingly, however, all the variants associated with PD are ‘pathogenic variants’ for Gaucher disease, raising the possibility that a decreased glucocerebrosidase activity plays a role in the pathogenesis of PD. Identification of a splice junction mutation, ‘IVS6+1g>a’, which is predicted to lead to a loss of function due to a premature stop codon, in this study may further support this notion (18).
Many GWAS have recently been conducted to identify susceptibility genes for PD. Satake et al. (21) have recently published the results of their GWAS on Japanese PD cases and controls. They found four genetic risk factors including LRRK2 and SNCA. As shown in the Table 1, the odds ratios are relatively low (1.24–1.37) despite the significant P-values. More importantly, the GBA locus on chromosome 1 was not detected in their GWAS, presumably because rare variants such as those in GBA are hard to detect by GWAS using common SNPs (tag SNPs). These results provide the following lessons: (i) risk factors with substantially high odds ratio are present in common diseases such as PD; (ii) rare variants are present at low frequencies; (iii) multiple rare GBA variants were detected only through comprehensive resequencing of the gene; and (iv) GWAS on the same population did not identify the locus for the susceptibility gene harboring multiple rare variants. These data demonstrate the power of resequencing strategies for the identification of rare variants in neurodegenerative disease.
Table 1.
Comparison of allele frequencies and odds ratios of disease-relevant variations
| Variants | Parkinson disease (%) | Controls (%) | Odds ratio (95% confidence interval) |
|---|---|---|---|
| GBAa | 9.4 | 0.4 | 28.0 (7.3–238.3) |
| SNCA (rs11931074)b | 32 | 42 | 1.50 (1.34–1.68) |
| LRRK2 (rs1994090)b | 11 | 8 | 1.43 (1.20–1.70) |
| BST1 (rs11931532)b | 45 | 40 | 1.22 (1.09–1.35) |
| PARK16 (rs947211)b | 43 | 48 | 1.23 (1.11–1.37) |
As Manolio et al. (16) recently described that GWAS have identified hundreds of genetic variants associated with complex human diseases and traits, and have provided valuable insights into their genetic architecture. However, because most variants identified to date confer relatively small increments in risk and explain only a small proportion of familial clustering, a remaining challenge will be to define the genetic basis of the ‘missing’ heritability.
APPLICATION OF HIGH THROUGHPUT RESEQUENCING FOR THE IDENTIFICATION OF RARE GENETIC VARIANTS WITH LARGE EFFECT SIZES
As discussed earlier, GWAS are inefficient in identifying rare variants associated with disease susceptibility and instead whole genome resequencing will be required. For PD, clinical observations suggested an association of PD and Gaucher disease (19–20). Without such clinical observations, however, comprehensive whole genome or exome resequencing will be required to identify rare variants relevant to disease. To accomplish this goal, high throughput resequencing efforts employing next-generation sequencers will be the most promising approach.
Developing next-generation sequencing technologies
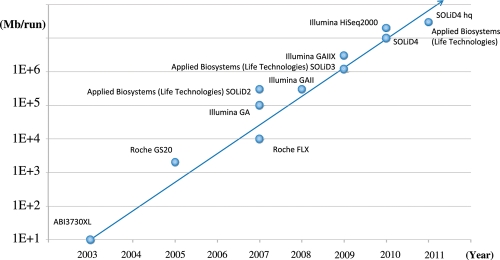
The automated Sanger method is considered a ‘first-generation’ technology, and newer methods are referred to as next-generation sequencing (NGS) (22). As shown in Figure 2, the throughput of NGS is dramatically increasing. As of 2010, the throughput is 100–200 Gb/run. Since the cost for whole genome resequencing for a read depth sufficient to identify variants with a high accuracy is still expensive, it is not easy to resequence the whole genome of a large number of individuals. Thus, we need to develop strategies to efficiently identify disease-relevant variants employing technologies with high accuracy and reasonable cost.
Figure 2.
Increased throughput of next-generation sequencers.
To reduce the cost of high throughput resequencing, enrichment of exons or target regions using oligonucleotide arrays or oligonucleotide ‘bait’ in solution have been preferentially employed. With this strategy, all exons in the genome or selected regions implicated in disease can be efficiently enriched (23–25). With this approach, more than 90% of target regions can be enriched, and these enriched genomic regions can be subjected to massive resequencing using NGS. This approach is currently being intensively used for the identification of disease-relevant variants, cancer profiling and applications to genetic diagnosis (26–34). Dihydroorotate dehydrogenase, the causative gene for Miller syndrome, has recently been discovered by exome resequencing employing NGS with a mean read depth of 40× (30). Limitations of this approach are that capture efficiency may not be complete and that additional resequencing may be required to fully cover the target regions.
Given the ever increasing throughput of NGS and the dramatically decreasing costs, it will soon be a realistic approach to conduct whole genome resequencing employing NGS. To date, whole genome sequences of at least eight individuals have been described (35–39). These studies have shown that there are more than 3 million SNPs in the human genome. In one study, among the 3.3 million SNPs, 8996 known non-synonymous SNPs and 1573 novel non-synonymous SNPs were identified. Interestingly, 32 alleles exactly matched mutations previously registered in the Human Gene Mutation Database. In addition, 345 insertions/deletions were observed to overlap the coding sequence and had the potential to alter protein function (39). These results indicate that it will be challenging to determine the variations that are relevant to diseases among the numerous variations.
The throughput of NGS is increasing at a rapid rate and several hundred Gb can now be generated in just one ‘run’. The enormous amount of data will result in significant challenges in appropriately interpreting the data. Given the enormous numbers of short-read sequences (∼100 bp), informatics analyses including mapping to reference sequences and identification of variations require a huge computational power (40–44). Furthermore, mutations can be variable including single base substitutions, insertions/deletions and structural variations. It is difficult to efficiently identify all the variations using currently available software. Functional annotation of variants identified in NGS will be important, and availability of databases containing variations and the functional annotation will be needed.
With current NGS, it is important to realize that there are error reads inherently associated with these technologies. Although the average rates of error are less than 0.1–0.2%, it is essential to minimize errors before attempting to identify disease-relevant variations. With increasing sequencing depth, error rate substantially decreases, but some errors remain and the error rates may depend on sequencing cycles, sequence context and other factors (45,46).
Application of NGS for diseases with Mendelian traits
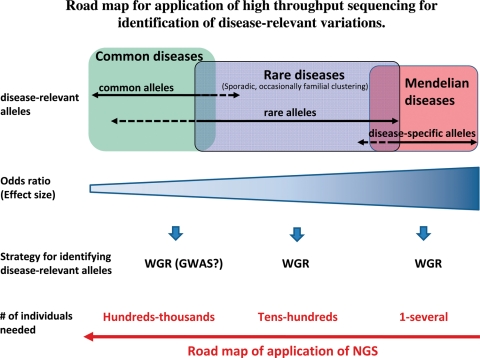
Lupski et al. (47) has recently applied whole genome resequencing to identify the causative mutation for a family with a recessive form of Charcot–Marie–Tooth disease. They sequenced the whole genome of the proband, identified all potential functional variants in genes likely to be related to the disease, and identified and validated compound, heterozygous, causative alleles in SH3TC2 (the SH3 domain and tetratricopeptide repeats 2 gene), involving two mutations, in the proband and the other affected family members. This study strongly encourages future applications of NGS to identify causative genes for Mendelian diseases (Fig. 3).
Figure 3.
Road map for application of high throughput sequencing for the identification of disease-relevant alleles. WGR, whole genome resequencing; GWAS, genome-wide association studies.
Application of NGS to sporadic diseases
To identify disease-relevant variants in sporadic diseases, it is essential to analyze larger sample sizes of cases and controls than those needed to identify causative genes for diseases with Mendelian traits. The number of individuals needed to identify disease-relevant variants will depend on the odds ratio of the disease-related alleles and the allele frequencies in the population. In the case of GBA as the risk factor for PD (described above), initial screening of 100 PD patients and 100 controls was sufficient to identify this gene as a risk factor for PD (18). In addition, we occasionally observe patients with familial aggregation (48,49). Because disease-relevant alleles with large effect sizes seem to underlie familial clustering, such cases should be good candidates to apply comprehensive resequencing with NGS.
As shown in the road map in Figure 3, we may need to wait until the cost for resequencing goes down substantially to apply whole genome resequencing to identify disease-relevant variants for sporadic diseases. To lessen the cost burden, whole exome analysis of a large number of samples is an alternative approach. Analysis of pooled DNAs may also be an alternate approach (45).
CONCLUSION
As discussed earlier, whole genome resequencing is a promising strategy for identifying causative genes and clinically relevant variations. Beyond this, whole genome resequencing is expected to bring a paradigm shift in clinical practice, where the diagnosis and decision-making for appropriate therapeutic procedures is based on the ‘personal genome’. The realization of ‘personal genome’ era is expected to come soon, and the genetics community needs to prepare for this exciting new era in genetics research.
FUNDING
This work was supported in part by KAKENHI (Grant-in-Aid for Scientific Research) on Priority Areas, Applied Genomics, Global Center for Education and Research for Chemical Biology of the Diseases, and Scientific Research (A) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and a Grant-in-Aid for ‘the Research Committee for Ataxic Diseases’ of the Research on Measures for Intractable Diseases from the Ministry of Health, Welfare and Labour, Japan. Funding to pay the Open Access publication charges for this article was provided by University of Tokyo.
ACKNOWLEDGEMENTS
The author appreciates Dr Laura P. W. Ranum for critical reading of the manuscript and valuable suggestions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.The Huntington's Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. doi:10.1016/0092-8674(93)90585-E. [DOI] [PubMed] [Google Scholar]
- 2.Koenig M., Hoffman E.P., Bertelson C.J., Monaco A.P., Feener C., Kunkel L.M. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. doi:10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 3.Monaco A.P., Neve R.L., Colletti-Feener C., Bertelson C.J., Kurnit D.M., Kunkel L.M. Isolation of candidate cDNAs for portions of the Duchenne muscular dystrophy gene. Nature. 1986;323:646–650. doi: 10.1038/323646a0. doi:10.1038/323646a0. [DOI] [PubMed] [Google Scholar]
- 4.Collins F.S. Positional cloning: let's not call it reverse anymore. Nat. Genet. 1992;1:3–6. doi: 10.1038/ng0492-3. doi:10.1038/ng0492-3. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda Y., Nakahara Y., Date H., Takahashi Y., Goto J., Miyashita A., Kuwano R., Adachi H., Nakamura E., Tsuji S. SNP HiTLink: a high-throughput linkage analysis system employing dense SNP data. BMC Bioinformat. 2009;10:121. doi: 10.1186/1471-2105-10-121. doi:10.1186/1471-2105-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krueger K.A., Tsuji S., Fukuda Y., Takahashi Y., Goto J., Mitsui J., Ishiura H., Dalton J.C., Miller M.B., Day J.W., et al. SNP haplotype mapping in a small ALS family. PLoS ONE. 2009;4:e5687. doi: 10.1371/journal.pone.0005687. doi:10.1371/journal.pone.0005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. doi:10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi Y., Seki N., Ishiura H., Mitsui J., Matsukawa T., Kishino A., Onodera O., Aoki M., Shimozawa N., Murayama S., et al. Development of a high-throughput microarray-based resequencing system for neurological disorders and its application to molecular genetics of amyotrophic lateral sclerosis. Arch. Neurol. 2008;65:1326–1332. doi: 10.1001/archneur.65.10.1326. doi:10.1001/archneur.65.10.1326. [DOI] [PubMed] [Google Scholar]
- 9.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. doi:10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wijesekera L.C., Leigh P.N. Amyotrophic lateral sclerosis. Orphanet. J. Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. doi:10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reich D.E., Lander E.S. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. doi:10.1016/S0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 12.Collins F.S., Guyer M.S., Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. doi: 10.1126/science.278.5343.1580. doi:10.1126/science.278.5343.1580. [DOI] [PubMed] [Google Scholar]
- 13.Pritchard J.K. Are rare variants responsible for susceptibility to complex diseases? Am. J. Hum. Genet. 2001;69:124–137. doi: 10.1086/321272. doi:10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. doi:10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Visscher P.M. Sizing up human height variation. Nat. Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. doi:10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 16.Manolio T.A., Collins F.S., Cox N.J., Goldstein D.B., Hindorff L.A., Hunter D.J., McCarthy M.I., Ramos E.M., Cardon L.R., Chakravarti A., et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. doi:10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidransky E., Nalls M.A., Aasly J.O., Aharon-Peretz J., Annesi G., Barbosa E.R., Bar-Shira A., Berg D., Bras J., Brice A., et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson's disease. N. Engl. J. Med. 2009;361:1651–1661. doi: 10.1056/NEJMoa0901281. doi:10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitsui J., Mizuta I., Toyoda A., Ashida R., Takahashi Y., Goto J., Fukuda Y., Date H., Iwata A., Yamamoto M., et al. Mutations for Gaucher disease confer high susceptibility to Parkinson disease. Arch. Neurol. 2009;66:571–576. doi: 10.1001/archneurol.2009.72. doi:10.1001/archneurol.2009.72. [DOI] [PubMed] [Google Scholar]
- 19.Goker-Alpan O., Schiffmann R., LaMarca M.E., Nussbaum R.L., McInerney-Leo A., Sidransky E. Parkinsonism among Gaucher disease carriers. J. Med. Genet. 2004;41:937–940. doi: 10.1136/jmg.2004.024455. doi:10.1136/jmg.2004.024455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neudorfer O., Giladi N., Elstein D., Abrahamov A., Turezkite T., Aghai E., Reches A., Bembi B., Zimran A. Occurrence of Parkinson's syndrome in type I Gaucher disease. QJM. 1996;89:691–694. doi: 10.1093/qjmed/89.9.691. [DOI] [PubMed] [Google Scholar]
- 21.Satake W., Nakabayashi Y., Mizuta I., Hirota Y., Ito C., Kubo M., Kawaguchi T., Tsunoda T., Watanabe M., Takeda A., et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat. Genet. 2009;41:1303–1307. doi: 10.1038/ng.485. doi:10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 22.Metzker M.L. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. doi:10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 23.Okou D.T., Steinberg K.M., Middle C., Cutler D.J., Albert T.J., Zwick M.E. Microarray-based genomic selection for high-throughput resequencing. Nat. Methods. 2007;4:907–909. doi: 10.1038/nmeth1109. doi:10.1038/nmeth1109. [DOI] [PubMed] [Google Scholar]
- 24.Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. doi: 10.1038/nmeth1111. doi:10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 25.Hodges E., Xuan Z., Balija V., Kramer M., Molla M.N., Smith S.W., Middle C.M., Rodesch M.J., Albert T.J., Hannon G.J., et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–1527. doi: 10.1038/ng.2007.42. doi:10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 26.Volpi L., Roversi G., Colombo E.A., Leijsten N., Concolino D., Calabria A., Mencarelli M.A., Fimiani M., Macciardi F., Pfundt R., et al. Targeted next-generation sequencing appoints c16orf57 as clericuzio-type poikiloderma with neutropenia gene. Am. J. Hum. Genet. 2010;86:72–76. doi: 10.1016/j.ajhg.2009.11.014. doi:10.1016/j.ajhg.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedges D.J., Burges D., Powell E., Almonte C., Huang J., Young S., Boese B., Schmidt M., Pericak-Vance M.A., Martin E., et al. Exome sequencing of a multigenerational human pedigree. PLoS ONE. 2009;4:e8232. doi: 10.1371/journal.pone.0008232. doi:10.1371/journal.pone.0008232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zaghloul N.A., Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. 2010;26:168–176. doi: 10.1016/j.tig.2010.01.006. doi:10.1016/j.tig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Biesecker L.G. Exome sequencing makes medical genomics a reality. Nat. Genet. 2010;42:13–14. doi: 10.1038/ng0110-13. doi:10.1038/ng0110-13. [DOI] [PubMed] [Google Scholar]
- 30.Ng S.B., Buckingham K.J., Lee C., Bigham A.W., Tabor H.K., Dent K.M., Huff C.D., Shannon P.T., Jabs E.W., Nickerson D.A., et al. Exome sequencing identifies the cause of a mendelian disorder. Nat. Genet. 2010;42:30–35. doi: 10.1038/ng.499. doi:10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi M., Scholl U.I., Ji W., Liu T., Tikhonova I.R., Zumbo P., Nayir A., Bakkaloglu A., Ozen S., Sanjad S., et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc. Natl Acad. Sci. USA. 2009;106:19096–19101. doi: 10.1073/pnas.0910672106. doi:10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. doi:10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maher B. Exome sequencing takes centre stage in cancer profiling. Nature. 2009;459:146–147. doi: 10.1038/459146b. doi:10.1038/459146b. [DOI] [PubMed] [Google Scholar]
- 34.Ng P.C., Levy S., Huang J., Stockwell T.B., Walenz B.P., Li K., Axelrod N., Busam D.A., Strausberg R.L., Venter J.C. Genetic variation in an individual human exome. PLoS Genet. 2008;4:e1000160. doi: 10.1371/journal.pgen.1000160. doi:10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster S.C., Miller W., Ratan A., Tomsho L.P., Giardine B., Kasson L.R., Harris R.S., Petersen D.C., Zhao F., Qi J., et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. doi:10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G., Hall K.P., Evers D.J., Barnes C.L., Bignell H.R., et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. doi:10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., Wang W., Li R., Li Y., Tian G., Goodman L., Fan W., Zhang J., Li J., Guo Y., et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. doi:10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahn S.M., Kim T.H., Lee S., Kim D., Ghang H., Kim D.S., Kim B.C., Kim S.Y., Kim W.Y., Kim C., et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. doi:10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler D.A., Srinivasan M., Egholm M., Shen Y., Chen L., McGuire A., He W., Chen Y.J., Makhijani V., Roth G.T., et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. doi:10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 40.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. doi:10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li R., Li Y., Kristiansen K., Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. doi:10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Mu Q. Soap-HT-BLAST: high throughput BLAST based on Web services. Bioinformatics. 2003;19:1863–1864. doi: 10.1093/bioinformatics/btg244. doi:10.1093/bioinformatics/btg244. [DOI] [PubMed] [Google Scholar]
- 43.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. doi:10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. doi:10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mitsui J., Fukuda Y., Azuma K., Tozaki H., Ishiura H., Takahashi Y., Goto J., Tsuji S. Multiplexed resequencing analysis to identify rare variants in pooled DNA with barcode indexing employing next-generation sequencer. J. Hum. Genet. 2009 doi: 10.1038/jhg.2010.46. [DOI] [PubMed] [Google Scholar]
- 46.Druley T.E., Vallania F.L., Wegner D.J., Varley K.E., Knowles O.L., Bonds J.A., Robison S.W., Doniger S.W., Hamvas A., Cole F.S., et al. Quantification of rare allelic variants from pooled genomic DNA. Nat. Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. doi:10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lupski J.R., Reid J.G., Gonzaga-Jauregui C., Rio Deiros D., Chen D.C., Nazareth L., Bainbridge M., Dinh H., Jing C., Wheeler D.A., et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. N. Engl. J. Med. 2010;362:1181–1191. doi: 10.1056/NEJMoa0908094. doi:10.1056/NEJMoa0908094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sveinbjornsdottir S., Hicks A.A., Jonsson T., Petursson H., Gugmundsson G., Frigge M.L., Kong A., Gulcher J.R., Stefansson K. Familial aggregation of Parkinson's disease in Iceland. N. Engl. J. Med. 2000;343:1765–1770. doi: 10.1056/NEJM200012143432404. doi:10.1056/NEJM200012143432404. [DOI] [PubMed] [Google Scholar]
- 49.Fang F., Kamel F., Lichtenstein P., Bellocco R., Sparen P., Sandler D.P., Ye W. Familial aggregation of amyotrophic lateral sclerosis. Ann. Neurol. 2009;66:94–99. doi: 10.1002/ana.21580. doi:10.1002/ana.21580. [DOI] [PMC free article] [PubMed] [Google Scholar]