Abstract
Most of our current knowledge about cellular phenotypes in neurodevelopmental and neurodegenerative diseases in humans was gathered from studies in postmortem brain tissues. These samples often represent the end-stage of the disease and therefore are not always a fair representation of how the disease developed. Moreover, under these circumstances, the pathology observed could be a secondary effect rather than the authentic disease cellular phenotype. Likewise, the rodent models available do not always recapitulate the pathology from human diseases. In this review, we will examine recent literature on the use of induced pluripotent stem cells to model neurodegenerative and neurodevelopmental diseases. We highlight the characteristics of diseases like spinal muscular atrophy and familial dysautonomia that allowed partial modeling of the disease phenotype. We review human stem cell literature on common neurodegenerative late-onset diseases such as Parkinson's disease and amyotrophic lateral sclerosis where patients' cells have been successfully reprogrammed but a disease phenotype has not yet been described. So far, the technique is of great interest for early onset monogenetic neurodevelopmental diseases. We speculate about potential further experimental requirements and settings for reprogrammed neurons for in vitro disease modeling and drug discovery.
INTRODUCTION
In neurology, nerve biopsies are feasible and performed to investigate diseases of the peripheral nervous system. However, owing to the invasiveness of this procedure, neurons in the central nervous system (CNS) are only taken for biopsy under rare conditions. This inability to sample live brain cells limits our knowledge of human neuropathological abnormalities during the course of diseases. Currently, our understanding about disease-related neuronal phenotypes in humans is generated from analyzing postmortem brain tissues that are not always well preserved. In addition, these samples often represent the end-stage of the disease.
Mouse models provide a means to mimic human genetic forms of neurodegenerative diseases, and great insights into mechanisms have been made using transgenic/knockout technologies. However, this approach is limited to monogenetic disorders and thus can only represent a minority of diseases. Owing to technical challenges, species differences and genetic background, even neurologic disorders with defined genes in some cases cannot be adequately modeled by mouse transgenic technology, indicating the need for advancement toward humanized models.
Yamanaka's original reprogramming experiment surprised the scientific community by overturning the dogma that specialized cells in the body retain an immutable identity (1). A set of transcriptional factors has the ability to ‘jump start’ a specific cell fate from a differentiated cell type, in a remarkable demonstration of cell flexibility. In this review, we will examine the recent literature on the use of induced pluripotent stem cells (iPSC) to model neurodegenerative and neurodevelopmental diseases. Although patient-specific iPSC have the promise of being less immunogenic for potential future cell transplantation therapies, our main focus here will be on the future clinical relevance of these cells for in vitro neurologic disease modeling and drug discovery.
IPSC DERIVED FROM HUMAN NEUROLOGIC DISEASES
Diseases that have been modeled for reprogramming can be divided into rare, monocausal genetic diseases and the large group of sporadic and multifactorial diseases. No large-scale disease modeling is currently available for the latter group. It has been more difficult and challenging to obtain conclusive results from this group due to the complexity of the different genetic backgrounds and environmental clues involved in these diseases. However, even patients with monogenetic diseases within families display large genotype–phenotype variability (2), likely due to environmental influence. It will be interesting to determine whether the same variability can be reproduced in iPSC-derived neural cells or if reprogramming in culture eliminates environmental ‘noise’.
Reprogramming of fibroblasts for several neurologic diseases has been reported (Table 1), but few studies have actually recapitulated the phenotype of diseases in the iPSC-derived neuronal population. Successful generation of iPSC-derived neurons has been reported for sporadic middle- or late-age onset neurodegenerative diseases like amyotrophic lateral sclerosis (ALS) and Parkinson's disease (PD) (3–5). From the reprogramming point of view, it is remarkable that aged fibroblasts (up to 85 years old) from ALS and PD patients could still be reset with similar efficiency as fibroblasts from younger patients.
Table 1.
iPSC for modeling human neurological diseases
| Disease | Neuronal pathology | References |
|---|---|---|
| Type 1 SMA | Decrease in motor neuron number after long-term culture | (6) |
| FD | Neuronal differentiation and migration impaired | (7) |
| PD (sporadic) | Not shown | (3,4) |
| ALS (SOD1 mutation) | Not shown | (5) |
| Huntington's disease | Not shown | (3) |
| Rett syndrome (MeCP2 mutation) | Not shown | (31) |
| Down syndrome | Not shown | (3,55) |
Nonetheless, demonstration of disease-related phenotypes in the relevant cell type (i.e. the specific cells that are affected in the disease) has been a major challenge during the past 2 years. It may well be that iPSC-derived neural cells from age-dependent neurodegenerative diseases will not show a phenotypic difference compared with normal control cells with regard to morphology, differentiation and survival. Test assays for challenging these cells for characterization of disease phenotypes could be required. For example, culturing cells under increased oxidative stress may reveal and/or accelerate aberrant neuronal phenotypes in late-onset diseases.
Partial disease modeling with spontaneous induction of a disease phenotype was reported in two young childhood-onset monogenetic diseases: spinal muscular atrophy (SMA) and familial dysautonomia (FD) (6,7). Both diseases are autosomal recessive and share the common trait of rapid disease progression within the first years of life. In addition, both diseases are associated with loss of function of the gene as well as a role in RNA processing.
SMA is a group of autosomal recessive diseases caused by large deletions or point mutations in the survival motor neuron (SMN) genes, leading to loss of function of the survival motor neuron protein. SMN1 gene encodes a 20 kb protein, spans 9 exons and has a role in RNA processing (8–11). SMA type 1 is characterized by mutations in the SMN1 gene and promotes fast progressing degeneration of motor neurons, inducing muscular atrophy and symmetric proximal lower limb weakness. SMA type 1 onset occurs around the sixth month of life, and death often occurs due to respiratory failure before the age of 2. Ebert et al. (6) were able to derive iPSC from fibroblasts from a single SMA patient and showed a decrease in iPSC-derived motor neuron survival after 6 weeks of differentiation, compared with iPSC-derived neurons from the patient's unaffected mother . It remains to be shown whether the neuronal cells derived were indeed functional (i.e. able to fire action potentials or make neuromuscular junctions). Moreover, authors detected an increase in the nuclear number of SMA ‘gems’—protein aggregates that correlate with disease intensity—in fibroblasts and iPSC derived from the SMA patient. Interestingly, this deficiency could be reversed in fibroblasts or SMA-iPSC by increasing wild-type SMA protein levels using non-specific SMN-inducing compounds (6). This work showed for the first time a proof-of-principle for a potential future drug-screening platform using the iPSC technology. However, initial enthusiasm was decreased by the absence of other SMA patients and controls. Incorporation of additional control and patient cells would have reduced the concern that the observed phenotype is a consequence of the intrinsic iPSC variability system (discussed in what follows).
FD is an autosomal recessive disease mostly occurring in persons of Ashkenazi Jewish descent (12). The disease is characterized by degeneration of sensory and autonomic neurons, leading to severe and often lethal autonomic dysfunction. Common clinical features include alacrima, hypoactivity and relative indifference to pain and temperature. A splicing defect in the IkB kinase complex-associated protein (IKBKAP) gene results in a tissue-specific splicing defect, inducing a loss of function or reduced levels of the IKAP protein (13). IPSC derived from three patients with FD revealed that neural crest precursors, specifically, had low levels of IKBKAP expression. In addition, a defect in neuronal differentiation and migration was reported. A drug candidate, kinetin, was able to reduce the levels of mutant IKBKAP splice forms and improved neuronal differentiation, but not cell migration, in iPSC-derived neural crest precursors, suggesting incomplete phenotype complementation (7). Drug screening using kinetin-like variations could be performed using recovery of both of neuronal differentiation and cell migration phenotypes as readout in future studies.
PD is the second most common neurodegenerative disease. Prominent clinical features are motor symptoms (bradykinesia, tremor, rigidity and postural instability) and non-motor-related PD symptoms (olfactory deficits, autonomic dysfunction, depression and sleep disorders). PD is a synucleinopathy, with accumulation of misfolded alpha-synuclein, that forms intracellular inclusions: Lewy bodies and Lewy neurites. Loss of dopaminergic (DA) neurons in the substantia nigra of the midbrain and in other brain regions is a characteristic neuropathological hallmark (14,15). Several different techniques to produce DA neurons in culture from human embryonic stem cells (HESC) are currently available. They include co-culture systems [such as mesencephalic astrocytes (16) and stromal cell-derived inducing activity (17)] and direct differentiation protocols (18). When these cells were transplanted into animal models of PD, functional integration was observed, although technical issues were reported (19). Disease modeling in the dish using embryonic stem cells (ESC) is still limited. Even though specific toxicity and cell death of α-synuclein overexpression were shown in mouse ESC-derived DA neurons (20,21), α-synuclein overexpressed in human neural embryonic cells resulted in patterns of degeneration that recapitulate PD features only in some cases (22). Mouse iPSC-derived precursors were differentiated into DA neurons and transplanted into 6-OHDA-lesioned rats, a rat model of DA depletion. The authors showed that a striatal graft of 1−3 × 105 iPSC-derived neurons expressed midbrain DA markers and functionally integrated after transplantation (23). Primary fibroblasts from sporadic PD patients were successfully reprogrammed and differentiated into DA neurons as efficiently as those from healthy individuals (3,4). These results are promising, as age does not seem to interfere with reprogramming. Phenotypic differences were not reported, indicating that more subtle analysis or even strong stressors or toxins will be necessary to reveal phenotypes of diseases with late onset.
ALS or Lou Gehrig's disease is a progressive fatal neurodegenerative disease affecting mainly motor neurons. The most common clinical features of ALS are degeneration of motor neurons producing fasciculation, muscle wasting and hyper-reflexia. Respiratory complications usually develop in patients with advanced disease and the cause of death is generally paralysis of the respiratory muscles and diaphragm. With a projected lifetime risk of 1/2000, ALS is considered one of the most common motor neuron diseases (24,25). ALS is universally fatal, with a median age of onset of 55 years and survival of 2–5 years from symptoms onset. Although the exact pathophysiological mechanisms underlying neurodegeneration in ALS remain uncertain, the presence of a persistent inflammatory reaction prompted researchers to study the involvement of a non-cell-autonomous component in motor neuron death. HESC have been used for modeling both the autonomous and the non-cell-autonomous effects of ALS in vitro, using a gene that is mutated in 20% of the familial cases, superoxide dismutase 1 (SOD1) (26–28). IPSC technology allows for the unprecedented opportunity to also include patient genetic background in the cell-modeling system. Dimos et al. (5) successfully reprogrammed cells from two familial ALS patients. The iPSC generated were able to differentiate in motor neurons (the affected neuronal subtype in ALS pathology), but no phenotype has yet been observed or reported. It remains to be determined whether iPSC-derived neurons have the potential to recapitulate ALS late-onset pathology in vitro and if both familial and sporadic (vast majority of ALS) cases share common phenotypic traits in culture.
MODELING NEURODEVELOPMENTAL DISORDERS
We anticipate that iPSC technology could be used for modeling complex neurodevelopmental disorders such as autism and schizophrenia. However, initial results would probably emerge from modeling monogenetic, early-onset occurrences of these diseases. An example of a neurodevelopmental disease with potential for disease modeling by iPSC technology is Rett syndrome. Rett syndrome is characterized by arrested development in early childhood, regression of acquired skills, loss of speech, stereotypical movements, microcephaly, seizures, autistic characteristics and mental retardation (29). Rett patients have mutations in the X-linked gene, MeCP2, a gene that binds to methylated cytosines in the DNA and is believed to epigenetically regulate global expression of genes (30). In fact, Rett patients' iPSC have been recently generated, but neither their X-inactivation/reactivation status nor their differentiation potential has been extensively studied (31).
Studying the in vitro phenotypic consequences of the mutation in specific genes can help to identify a molecular mechanism responsible for subtle alterations in the nervous system, perhaps pointing to common mechanisms for more complex, multigenetic diseases. A future challenge for neurodevelopmental disorders is the contribution of genetic background and environmental clues. New gene-targeting techniques in human pluripotent cells such as homologous recombination and zinc-finger nucleases may help to eliminate background noise and individual variability (32–37). Effective gene targeting in HESC could disrupt a specific disease-related gene and the resulting neuronal behavior could be compared with the patient's neuron containing the mutation.
As an alternative for revealing complex neuronal phenotypes and niche-specific behaviors, patient-derived cells could be transplanted on the CNS of animal models. In fact, HESC were shown to integrate and form functional connections with host cells when transplanted in the ventricles of embryonic mice (38). Such a chimeric model was recently proposed as an in vivo model to study environmental contributions to complex neurological diseases (39).
USING NEURONAL CELLS FOR DRUG SCREENING
In the past, drug screening was performed in human cell lines and they have represented a major step forward in medical therapy progress. A prime example is the development of vaccines for polio, which were originally made by in vitro studies on cell lines like HeLa cells (40).
iPSC derived from patients seem to offer a significant advantage as they take into consideration the patient's background, the affected cell type and the developmental time. In addition, they allow the generation of both genetic and sporadic forms of the disease.
One of the great benefits of reprogramming cells is the possibility of studying the developmental steps from human neural cells before they are differentiated into mature neurons. Neural progenitor cells in culture can give rise to glial and neuronal populations (41). These populations can further differentiate into subtypes of glia as well as subtypes of neurons with distinct properties (i.e. DA or cholinergic). It is not unlikely that some neurodegenerative diseases have their origins in the neural progenitor population rather than in the mature neuron. In these cases, therapeutic interventions should be tested at an early stage of development. Finally, iPSC technology could help to unveil the changes in connectivity properties between neurons that are affected by a given neurologic disease.
Stem cell biologists can borrow a number of well-established tools from the neurosciences to address neuronal maturation and circuitry integration in vitro. For evaluating single-cell properties in a disease context, one could analyze neuronal morphology, branching, spine density/size/maturation and neuronal polarity. Circuitry integration analysis could be assessed by calcium influx transients, and pre-synaptic connections could be revealed after rabies virus infection. Likewise, paired electrophysiological recordings could address synaptic strength between cells. Recent reports have used such tools in a disease context and have been able to find altered neuronal phenotypes in complex disorders (42–44). In addition, as different types of neural cells can be generated from iPSC, neuron-autonomous and non-autonomous effects (i.e. the interplay between neurons and glia) can be studied individually or in combination.
Technology is already available to generate functionally active, cortical-like mouse neurons from differentiated cells without the need for complete reprogramming to iPSC (45,46). However, it remains to be seen whether this technology can generate the plethora of neuronal subtypes that iPSC-derived neurons can. Moreover, direct conversion from somatic cells to postmitotic neurons, without the step of pluripotency, will limit the amount of cells available to study the disease and will likely not recapitulate disease phenotypes that occur in the neural progenitor state.
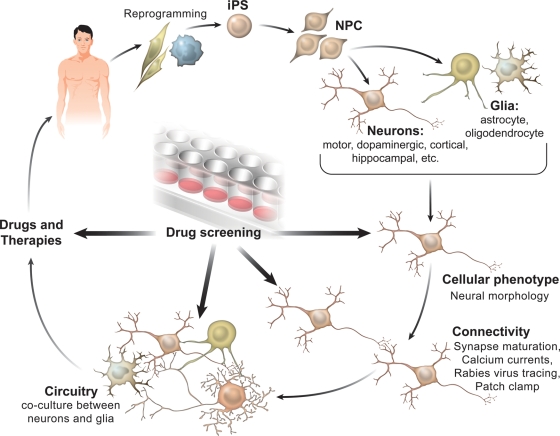
Once a consistent abnormal disease-related phenotype is identified, screening platforms can be developed to test compounds (proteins, small molecules, small hairpin RNAs) that revert or protect the cellular phenotype. After rigorous testing, therapeutic compounds will emerge from the screenings that could potentially benefit a large cohort of patients (Fig. 1).
Figure 1.
iPSC to model neurodegenerative and neurodevelopmental diseases. Human iPSC from neurologic patients and controls are generated after somatic tissue reprogramming (e.g. skin or blood cells). Neural progenitor cells (NPC) are generated and are further differentiated into neurons and/or glial cells. Neurons are then differentiated into subtypes of neurons such as dopaminergic, cholinergic, etc. Cellular phenotype is assessed by measuring neuronal morphology (i.e. process branching, spine density/size/maturation). Next, connectivity and circuitry integration can be analyzed by calcium influx transients, electrophysiology and transneuronal tracing with the rabies virus. In addition, the cross-talk between neurons and glia can be studied to tease out autonomous and non-autonomous aspects of the disease. Once a distinct disease-related phenotype is identified, drug-screening platforms can be developed to test compounds that improve cellular phenotype. Therapeutic compounds could emerge from the screenings, potentially benefiting neurologic patients.
CAUTIONARY NOTES
The available lines of HESC are notoriously variable with regard to epigenetic marks, expression profile and differentiation propensity (47,48). Even though the initial hope was that iPSC technology would generate pluripotent cells that were less variable than HESC cell lines, recent reports suggest that significant intrinsic variability remains in the iPSC lines generated to date (49). Pick et al. (49) detected abnormal expression of imprinted genes in a significant number of iPSC lines. Those differences have been generally attributed to the introduction of reprogramming factors, using randomly integrating viral vectors and/or by persistent donor cell gene expression (4,50). It remains to be seen if iPSC generated in the absence of integrating factors still recapitulate intrinsic variability (51–53). Moreover, expression profile analysis on integration-free human iPSC has shown an expression signature in iPSC that is distinct from both the original population and standard HESC (51). An intriguing study recently performed by Hu et al. (54) showed that the neuronal differentiation competence of iPSC was highly variable when compared with HESC differentiation and surprisingly independent of transgene expression. More data are necessary to uncover the levels of variability between HESC and iPSC lines (generated with different methods) in both undifferentiated and differentiated states. Determining the variability levels between lines and clones will allow researchers to elucidate more robust phenotypes on cells derived from diseased iPSC. Moreover, dissecting the iPSC-intrinsic variability may provide clues as to which wild-type iPSC would be the most suitable experimental control and how many control lines should be derived for each experiment. For example, it is currently unclear whether the best controls for diseased iPSC lines should be iPSC derived from age/gender-matched donors or from a family member regardless of age/gender.
CONCLUDING REMARKS
Scientists are now using the powerful iPSC technology to investigate early stages of human development and to model diseases. So far, this technique has been shown to be of specific interest for monogenetic neurodevelopmental diseases, providing an innovative way to understand disease pathology. Modeling late-onset neurodegenerative diseases and multifactorial neurodevelopmental diseases will require additional advances. For example, in the future, the switch from one cell type to another might help the direct conversion of astrocytes to motor neurons in spinal cord trauma patients or other neurodegenerative diseases, perhaps combining gene and cell therapy in vivo. On the other hand, the recapitulation of all stages of neural development by iPSC is an invaluable tool to depict the exact moment of the disease onset, optimizing therapeutical interventions. The potential of cellular reprogramming is limited only by human creativity and ethical guidelines. Neuroscientists in the past could not have imagined a scenario in which patient-derived neural cell types would be readily accessible to thousands of laboratories around the world, and researchers in the future will never imagine neuroscience without it.
FUNDING
The authors are funded by the California Institute of Regenerative Medicine (CIRM) and National Institutes of Health (NIH). F.H.G. is supported by The Lookout Fund and the Mathers Foundation. M.C.N.M. is supported by Christopher and Dana Reeve Foundation (CDRF); B.W. is supported by Tom Wahlig Foundation and is a Feodor Lynen fellow of the Alexander von Humboldt Foundation.
ACKNOWLEDGEMENTS
The authors would like to thank J. Simon for illustrations and M.L. Gage for editorial comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. doi:10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 2.Sieberts S.K., Schadt E.E. Moving toward a system genetics view of disease. Mamm. Genome. 2007;18:389–401. doi: 10.1007/s00335-007-9040-6. doi:10.1007/s00335-007-9040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park I.H., Arora N., Huo H., Maherali N., Ahfeldt T., Shimamura A., Lensch M.W., Cowan C., Hochedlinger K., Daley G.Q. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. doi:10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G.W., Cook E.G., Hargus G., Blak A., Cooper O., Mitalipova M., et al. Parkinson's disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. doi:10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dimos J.T., Rodolfa K.T., Niakan K.K., Weisenthal L.M., Mitsumoto H., Chung W., Croft G.F., Saphier G., Leibel R., Goland R., et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. doi:10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- 6.Ebert A.D., Yu J., Rose F.F., Jr, Mattis V.B., Lorson C.L., Thomson J.A., Svendsen C.N. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457:277–280. doi: 10.1038/nature07677. doi:10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee G., Papapetrou E.P., Kim H., Chambers S.M., Tomishima M.J., Fasano C.A., Ganat Y.M., Menon J., Shimizu F., Viale A., et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature. 2009;461:402–406. doi: 10.1038/nature08320. doi:10.1038/nature08320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burglen L., Lefebvre S., Clermont O., Burlet P., Viollet L., Cruaud C., Munnich A., Melki J. Structure and organization of the human survival motor neuron (SMN) gene. Genomics. 1996;32:479–482. doi: 10.1006/geno.1996.0147. doi:10.1006/geno.1996.0147. [DOI] [PubMed] [Google Scholar]
- 9.Lorson C.L., Androphy E.J. The domain encoded by exon 2 of the survival motor neuron protein mediates nucleic acid binding. Hum. Mol. Genet. 1998;7:1269–1275. doi: 10.1093/hmg/7.8.1269. doi:10.1093/hmg/7.8.1269. [DOI] [PubMed] [Google Scholar]
- 10.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. doi:10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 11.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. doi:10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunt P.W., McKusick V.A. Familial dysautonomia. A report of genetic and clinical studies, with a review of the literature. Medicine (Baltimore) 1970;49:343–374. [PubMed] [Google Scholar]
- 13.Slaugenhaupt S.A., Blumenfeld A., Gill S.P., Leyne M., Mull J., Cuajungco M.P., Liebert C.B., Chadwick B., Idelson M., Reznik L., et al. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 2001;68:598–605. doi: 10.1086/318810. doi:10.1086/318810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braak H., Sastre M., Del Tredici K. Development of alpha-synuclein immunoreactive astrocytes in the forebrain parallels stages of intraneuronal pathology in sporadic Parkinson's disease. Acta Neuropathol. 2007;114:231–241. doi: 10.1007/s00401-007-0244-3. doi:10.1007/s00401-007-0244-3. [DOI] [PubMed] [Google Scholar]
- 15.Goedert M. Alpha-synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2001;2:492–501. doi: 10.1038/35081564. doi:10.1038/35081564. [DOI] [PubMed] [Google Scholar]
- 16.Roy N.S., Cleren C., Singh S.K., Yang L., Beal M.F., Goldman S.A. Functional engraftment of human ES cell-derived dopaminergic neurons enriched by coculture with telomerase-immortalized midbrain astrocytes. Nat. Med. 2006;12:1259–1268. doi: 10.1038/nm1495. doi:10.1038/nm1495. [DOI] [PubMed] [Google Scholar]
- 17.Kawasaki H., Mizuseki K., Nishikawa S., Kaneko S., Kuwana Y., Nakanishi S., Nishikawa S.I., Sasai Y. Induction of midbrain dopaminergic neurons from ES cells by stromal cell-derived inducing activity. Neuron. 2000;28:31–40. doi: 10.1016/s0896-6273(00)00083-0. doi:10.1016/S0896-6273(00)00083-0. [DOI] [PubMed] [Google Scholar]
- 18.Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. doi:10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho M.S., Lee Y.E., Kim J.Y., Chung S., Cho Y.H., Kim D.S., Kang S.M., Lee H., Kim M.H., Kim J.H., et al. Highly efficient and large-scale generation of functional dopamine neurons from human embryonic stem cells. Proc. Natl Acad. Sci. USA. 2008;105:3392–3397. doi: 10.1073/pnas.0712359105. doi:10.1073/pnas.0712359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamashita H., Nakamura T., Takahashi T., Nagano Y., Hiji M., Hirabayashi T., Amano T., Yagi T., Sakai N., Kohriyama T., et al. Embryonic stem cell-derived neuron models of Parkinson's disease exhibit delayed neuronal death. J. Neurochem. 2006;98:45–56. doi: 10.1111/j.1471-4159.2006.03815.x. doi:10.1111/j.1471-4159.2006.03815.x. [DOI] [PubMed] [Google Scholar]
- 21.Crews L., Mizuno H., Desplats P., Rockenstein E., Adame A., Patrick C., Winner B., Winkler J., Masliah E. Alpha-synuclein alters Notch-1 expression and neurogenesis in mouse embryonic stem cells and in the hippocampus of transgenic mice. J. Neurosci. 2008;28:4250–4260. doi: 10.1523/JNEUROSCI.0066-08.2008. doi:10.1523/JNEUROSCI.0066-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider B.L., Seehus C.R., Capowski E.E., Aebischer P., Zhang S.C., Svendsen C.N. Over-expression of alpha-synuclein in human neural progenitors leads to specific changes in fate and differentiation. Hum. Mol. Genet. 2007;16:651–666. doi: 10.1093/hmg/ddm008. doi:10.1093/hmg/ddm008. [DOI] [PubMed] [Google Scholar]
- 23.Wernig M., Zhao J.P., Pruszak J., Hedlund E., Fu D., Soldner F., Broccoli V., Constantine-Paton M., Isacson O., Jaenisch R. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson's disease. Proc. Natl Acad. Sci. USA. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. doi:10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vucic S., Kiernan M.C. Pathophysiology of neurodegeneration in familial amyotrophic lateral sclerosis. Curr. Mol. Med. 2009;9:255–272. doi: 10.2174/156652409787847173. doi:10.2174/156652409787847173. [DOI] [PubMed] [Google Scholar]
- 25.Eisen A. Amyotrophic lateral sclerosis: a 40-year personal perspective. J. Clin. Neurosci. 2009;16:505–512. doi: 10.1016/j.jocn.2008.07.072. doi:10.1016/j.jocn.2008.07.072. [DOI] [PubMed] [Google Scholar]
- 26.Karumbayaram S., Kelly T.K., Paucar A.A., Roe A.J., Umbach J.A., Charles A., Goldman S.A., Kornblum H.I., Wiedau-Pazos M. Human embryonic stem cell-derived motor neurons expressing SOD1 mutants exhibit typical signs of motor neuron degeneration linked to ALS. Dis. Model Mech. 2009;2:189–195. doi: 10.1242/dmm.002113. doi:10.1242/dmm.002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchetto M.C., Muotri A.R., Mu Y., Smith A.M., Cezar G.G., Gage F.H. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. doi:10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 29.Samaco R.C., Hogart A., LaSalle J.M. Epigenetic overlap in autism-spectrum neurodevelopmental disorders: MECP2 deficiency causes reduced expression of UBE3A and GABRB3. Hum. Mol. Genet. 2005;14:483–492. doi: 10.1093/hmg/ddi045. doi:10.1093/hmg/ddi045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chahrour M., Zoghbi H.Y. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. doi:10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Hotta A., Cheung A.Y., Farra N., Vijayaragavan K., Seguin C.A., Draper J.S., Pasceri P., Maksakova I.A., Mager D.L., Rossant J., et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 2009;6:370–376. doi: 10.1038/nmeth.1325. doi:10.1038/nmeth.1325. [DOI] [PubMed] [Google Scholar]
- 32.Zwaka T.P., Thomson J.A. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. doi:10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 33.Davis R.P., Grandela C., Sourris K., Hatzistavrou T., Dottori M., Elefanty A.G., Stanley E.G., Costa M. Generation of human embryonic stem cell reporter knock-in lines by homologous recombination. Curr. Protoc. Stem Cell Biol. 2009;11:5B.1.1–5B.1.34. doi: 10.1002/9780470151808.sc05b01s11. [DOI] [PubMed] [Google Scholar]
- 34.Davis R.P., Costa M., Grandela C., Holland A.M., Hatzistavrou T., Micallef S.J., Li X., Goulburn A.L., Azzola L., Elefanty A.G., et al. A protocol for removal of antibiotic resistance cassettes from human embryonic stem cells genetically modified by homologous recombination or transgenesis. Nat. Protoc. 2008;3:1550–1558. doi: 10.1038/nprot.2008.146. doi:10.1038/nprot.2008.146. [DOI] [PubMed] [Google Scholar]
- 35.Di Domenico A.I., Christodoulou I., Pells S.C., McWhir J., Thomson A.J. Sequential genetic modification of the hprt locus in human ESCs combining gene targeting and recombinase-mediated cassette exchange. Cloning Stem Cells. 2008;10:217–230. doi: 10.1089/clo.2008.0016. doi:10.1089/clo.2008.0016. [DOI] [PubMed] [Google Scholar]
- 36.Zou J., Maeder M.L., Mali P., Pruett-Miller S.M., Thibodeau-Beganny S., Chou B.K., Chen G., Ye Z., Park I.H., Daley G.Q., et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. doi:10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hockemeyer D., Soldner F., Beard C., Gao Q., Mitalipova M., DeKelver R.C., Katibah G.E., Amora R., Boydston E.A., Zeitler B., et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. doi:10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muotri A.R., Nakashima K., Toni N., Sandler V.M., Gage F.H. Development of functional human embryonic stem cell-derived neurons in mouse brain. Proc. Natl Acad. Sci. USA. 2005;102:18644–18648. doi: 10.1073/pnas.0509315102. doi:10.1073/pnas.0509315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muotri A.R. Modeling epilepsy with pluripotent human cells. Epilepsy Behav. 2009;14(Suppl. 1):81–85. doi: 10.1016/j.yebeh.2008.09.021. [DOI] [PubMed] [Google Scholar]
- 40.Syverton J.T., Scherer W.F., Elwood P.M. Studies on the propagation in vitro of poliomyelitis viruses. V. The application of strain HeLa human epithelial cells for isolation and typing. J. Lab. Clin. Med. 1954;43:286–302. [PubMed] [Google Scholar]
- 41.Yeo G.W., Coufal N., Aigner S., Winner B., Scolnick J.A., Marchetto M.C., Muotri A.R., Carson C., Gage F.H. Multiple layers of molecular controls modulate self-renewal and neuronal lineage specification of embryonic stem cells. Hum. Mol. Genet. 2008;17:R67–R75. doi: 10.1093/hmg/ddn065. doi:10.1093/hmg/ddn065. [DOI] [PubMed] [Google Scholar]
- 42.Gutierrez R.C., Hung J., Zhang Y., Kertesz A.C., Espina F.J., Colicos M.A. Altered synchrony and connectivity in neuronal networks expressing an autism-related mutation of neuroligin 3. Neuroscience. 2009;162:208–221. doi: 10.1016/j.neuroscience.2009.04.062. doi:10.1016/j.neuroscience.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 43.Pavlowsky A., Gianfelice A., Pallotto M., Zanchi A., Vara H., Khelfaoui M., Valnegri P., Rezai X., Bassani S., Brambilla D., et al. A postsynaptic signaling pathway that may account for the cognitive defect due to IL1RAPL1 mutation. Curr. Biol. 2010;20:103–115. doi: 10.1016/j.cub.2009.12.030. doi:10.1016/j.cub.2009.12.030. [DOI] [PubMed] [Google Scholar]
- 44.Xu L., Ryugo D.K., Pongstaporn T., Johe K., Koliatsos V.E. Human neural stem cell grafts in the spinal cord of SOD1 transgenic rats: differentiation and structural integration into the segmental motor circuitry. J. Comp. Neurol. 2009;514:297–309. doi: 10.1002/cne.22022. doi:10.1002/cne.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vierbuchen T., Ostermeier A., Pang Z.P., Kokubu Y., Sudhof T.C., Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berninger B., Costa M.R., Koch U., Schroeder T., Sutor B., Grothe B., Gotz M. Functional properties of neurons derived from in vitro reprogrammed postnatal astroglia. J. Neurosci. 2007;27:8654–8664. doi: 10.1523/JNEUROSCI.1615-07.2007. doi:10.1523/JNEUROSCI.1615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rugg-Gunn P.J., Ferguson-Smith A.C., Pedersen R.A. Status of genomic imprinting in human embryonic stem cells as revealed by a large cohort of independently derived and maintained lines. Hum. Mol. Genet. 2007;16(Spec no. 2):R243–R251. doi: 10.1093/hmg/ddm245. [DOI] [PubMed] [Google Scholar]
- 48.Osafune K., Caron L., Borowiak M., Martinez R.J., Fitz-Gerald C.S., Sato Y., Cowan C.A., Chien K.R., Melton D.A. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat. Biotechnol. 2008;26:313–315. doi: 10.1038/nbt1383. doi:10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 49.Pick M., Stelzer Y., Bar-Nur O., Mayshar Y., Eden A., Benvenisty N. Clone- and gene-specific aberrations of parental imprinting in human induced pluripotent stem cells. Stem Cells. 2009;27:2686–2690. doi: 10.1002/stem.205. doi:10.1002/stem.205. [DOI] [PubMed] [Google Scholar]
- 50.Ghosh Z., Wilson K.D., Wu Y., Hu S., Quertermous T., Wu J.C. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS ONE. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. doi:10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marchetto M.C., Yeo G.W., Kainohana O., Marsala M., Gage F.H., Muotri A.R. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. doi:10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin I.I., Thomson J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. doi:10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H., Wu S., Joo J.Y., Zhu S., Han D.W., Lin T., Trauger S., Bien G., Yao S., Zhu Y., et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. doi:10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu B.Y., Weick J.P., Yu J., Ma L.X., Zhang X.Q., Thomson J.A., Zhang S.C. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl Acad. Sci. USA. 2010;107:4335–4340. doi: 10.1073/pnas.0910012107. doi:10.1073/pnas.0910012107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baek K.H., Zaslavsky A., Lynch R.C., Britt C., Okada Y., Siarey R.J., Lensch M.W., Park I.H., Yoon S.S., Minami T., et al. Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature. 2009;459:1126–1130. doi: 10.1038/nature08062. doi:10.1038/nature08062. [DOI] [PMC free article] [PubMed] [Google Scholar]