Abstract
The PTEN-induced putative kinase 1 (PINK1) is a mitochondrially targeted serine–threonine kinase, which is linked to autosomal recessive familial parkinsonism. Current literature implicates PINK1 as a pivotal regulator of mitochondrial quality control, promoting maintenance of respiring mitochondrial networks through cristae stabilization, phosphorylation of chaperones and possibly regulation of mitochondrial transport or autophagy. Pulse—chase studies indicate that PINK1 is rapidly processed into at least two shorter forms, which are distributed in both mitochondrial and cytosolic compartments. Through indirect regulation of mitochondrial proteases and Drp1, PINK1 may act to facilitate localized repair and fusion in response to minor mitochondrial stress. With severe mitochondrial damage, PINK1 facilitates aggregation and clearance of depolarized mitochondria through interactions with Parkin and possibly Beclin1. This switch in function most probably involves altered processing, post-translational modification and/or localization of PINK1, as overexpression of full-length PINK1 is required for mitochondrial Parkin recruitment. Under conditions of PINK1 deficiency, dysregulation of reactive oxygen species, electron transport chain function and calcium homeostasis trigger altered mitochondrial dynamics, indicating compromise of mitochondrial quality control mechanisms. Nevertheless, Parkin- and Beclin1-regulated mitochondrial autophagy remains effective at recycling PINK1-deficient mitochondria; failure of this final tier of mitochondrial quality control contributes to cell death. Thus, PINK1 plays a pivotal, multifactorial role in mitochondrial homeostasis. As autophagic recycling represents the final tier of mitochondrial quality control, whether PINK1 levels are enhanced or reduced, strategies to promote selective mitophagy and mitochondrial biogenesis may prove effective for multiple forms of Parkinson's disease.
GENETIC FACTORS IN PARKINSON DISEASE: CONVERGING ON MITOCHONDRIA
Parkinson disease (PD) and related neurodegenerative disorders are characterized by loss of monoaminergic brainstem neurons and accumulation of aggregated and oxidatively modified proteins in the form of Lewy bodies and Lewy neurites (1,2). While most cases of PD are believed to arise from an unknown combination of environmental factors and genetic susceptibilities, several gene products whose mutations cause familial parkinsonism have been identified. In the last decade, accelerating evidence from modeling these molecular genetic alterations in PD have converged with earlier data from post-mortem and toxic/environmental studies to implicate mitochondrial dysregulation as a central pathogenic mechanism in PD (3–5).
Autosomal dominant PD can be caused by mutations in the leucine-rich repeat kinase 2 (LRRK2) gene (6,7) and by mutation or duplication/triplication of the gene for α-synuclein (8,9), the hallmark component of Lewy pathology. The proteins implicated in autosomal recessive parkinsonism include Parkin, DJ-1, ATP13A2 and PTEN-induced kinase 1 (PINK1) (10–13). Interestingly, dominant forms of familial parkinsonism are often associated with classic Lewy pathology, although other patterns can be observed in some LRRK2 families (6). In contrast, mutations in Parkin, the most common autosomal recessive cause of juvenile parkinsonism, are typically not associated with α-synuclein aggregates. Recently, recessive familial PINK1 mutations were shown to elicit Lewy body pathology (92), whereas the neuropathologies associated with mutations in DJ-1 or ATP13A2 remain to be elucidated (1).
Oxidative stress and mitochondrial electron transport chain dysfunction have long been considered central factors in PD based on human tissue studies (14). Mitochondria are central to the actions of parkinsonian toxins: 1-methyl-4-phenyl-1,2,3,6-tetrahydroxypyridine (15), rotenone (16) and 6-hydroxydopamine (6-OHDA) (17,18). DJ-1 localizes to mitochondria during oxidative stress, where it exhibits peroxiredoxin-like activity (19). PINK1 is a mitochondrially targeted serine/threonine kinase whose loss of function causes striking changes in mitochondrial structure and function [reviewed in (20,21)]. Parkin is an ubiquitin ligase that has recently been shown to play a key role in autophagic clearance of depolarized mitochondria (22). As α-synuclein also affects mitochondrial function (23), these studies collectively implicate a central role for mitochondrial dysregulation in PD pathogenesis.
Recent studies of PINK1 and its potential relationships to heat shock protein 75 kDa/TNF-receptor-associated protein 1 (Hsp75/TRAP1), the mitochondrial serine protease high temperature requirement A2 (HtrA2)/Omi and Parkin begin to implicate a network of pathways converging on mitochondrial quality control. Rather than linear pathways, a level of functional redundancy is likely to exist for such an important process, which may explain some apparently disparate observations in different experimental systems. As PINK1 mutations are associated with accelerated disease presenting at younger ages, studying the regulation and function of wild-type PINK1 should yield therapeutically relevant insights towards slowing the progression of PD.
OVERVIEW OF MITOCHONDRIAL QUALITY CONTROL
A simplified overview of mitochondrial quality control includes the following steps: (a) prevention of damage, (b) localized repair and proteolysis with dynamic remodeling and (c) autophagic degradation of entire mitochondrial segments; each of which also depends upon (d) effective and coordinated biogenesis of nuclear and mitochondrial DNA-encoded components (Fig. 1). Mitochondrial fusion facilitates intramitochondrial repair and exchange of mtDNA (24), whereas fission can serve to temporarily isolate defective segments and/or promote their autophagic clearance (25,26). Recent studies indicate that key regulators of mitochondrial health such as PINK1 exhibit coordinated functions at each of these tiers.
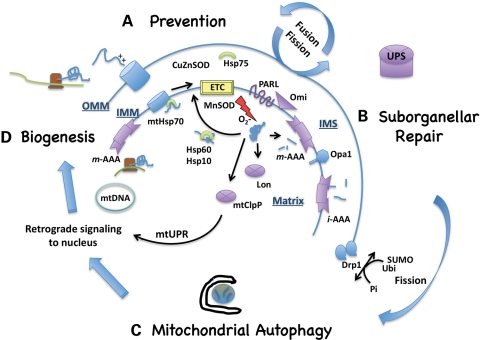
Figure 1.
Mitochondrial quality control mechanisms. (A) Prevention of mitochondrial damage or misfolded proteins represents the first tier of mitochondrial quality control. Chaperones (represented in green) such as mtHsp70 (mortalin) and transmembrane processing peptidases such as the AAA proteases operate in concert with mitochondrial biogenesis to ensure the proper import and folding of electron transport chain (ETC) subunits. Superoxide dismutase (SOD) enzymes in the matrix and intermembrane space (IMS) prevent oxidative damage. Dynamic cycles of fusion–fission and transport allow for interchange of proteins and mtDNA and adjustments of cellular mitochondrial distribution to meet localized cellular demands. (B) The second tier of mitochondrial quality control operates to temporarily sequester and repair misfolded or oxidatively damaged components. Limited proteolysis of Opa1 and post-translational modifications of Drp1 regulate altered mitochondrial dynamics to sequester damaged segments, with re-fusion of reparable segments governed in part by membrane potential. Misfolded proteins that are not successfully disaggregated by heat shock proteins (Hsp) can be degraded by several proteolytic systems (represented in purple) in the matrix, IMS or, in the case of outer mitochondrial membrane (OMM) proteins, the ubiquitin—proteasome system (UPS). The mtClpP system generates retrograde signals to the nucleus to stimulate further transcription of mitochondrial proteases as part of the mitochondrial unfolded protein response (mtUPR). (C) Mitochondrial segments that are too extensively damaged for local repair undergo whole organelle recycling through macroautophagy. Implicit in the success of this final tier of mitochondrial quality control, particularly for neurons that are dependent upon mitochondrial respiration, is (D) upregulation of mitochondrial biogenesis to replace the degraded organelles. IMM, inner mitochondrial membrane; mt, mitochondrial; O2−, superoxide radical; PARL, presenilin-associated rhomboid-like protease; Omi/HtrA2, high temperature requirement A2 mitochondrial protease; Opa1, optic atrophy 1; Drp1, dynamin-related protein 1; Ubi, ubiquitin; SUMO, small ubiquitin-like modifier.
A series of heat shock proteins and mitochondrial proteases regulate: (1) normal biosynthetic processing and assembly of mitochondrial electron transport chain (ETC) complexes and mitochondrial ribosomes, (2) refolding or degradation of misfolded, oxidized and aggregated proteins and (3) limited proteolysis as a signaling mechanism to regulate mitochondrial and cellular responses to stress (Fig. 1). Chaperones such as mitochondrial Hsp70 assist with the import and assembly of ETC components in the inner membrane (27). Together with antioxidant enzymes of the intermembrane and matrix spaces, these chaperones serve to prevent mitochondrial protein aggregation and cristae damage, promoting efficient respiratory function. Oxidative damage and aggregation triggers chaperone-mediated refolding or proteolysis by membrane-bound AAA proteases or Lon, which shows particular selectivity for oxidized proteins. In addition, unfolded proteins can trigger a mitochondrial unfolded protein response with retrograde signaling to the nucleus mediated by products of the mitochondrial ClpP protease. Other regulatory modifications that are triggered include liberation of inner membrane-tethered HtrA2/Omi by the rhomboid protease PARL and limited proteolysis of the fusion protein Opa1 (28). Combined with altered phosphorylation, ubiquitination or SUMOylation of the fission protein Drp1 (29), these post-translational modifications modulate fission–fusion dynamics and potentially cell death in response to mitochondrial stress.
With more severe mitochondrial injury, the fission–fusion balance tips toward accumulation of isolated mitochondrial fragments, which may paradoxically show enlarged fluorescent diameters due to swelling or perinuclear aggregation. Thus, ultrastructural, three-dimensional fluorescence reconstruction and live imaging studies each provide complementary information for analysis of fission–fusion dynamics. The decreased probability that depolarized segments of mitochondria will re-fuse with the mitochondrial reticulum has been proposed as a mechanism that permits selective autophagic clearance (25), although other studies indicate that autophagy is an active participant in fragmenting PINK1-deficient mitochondrial networks (26).
Mitochondrial autophagy can occur under several circumstances. The first occurs during non-selective bulk autophagy in response to nutrient deprivation. The second is selective developmental removal of mitochondria in erythrocytes or lens cells. Selective mitophagy in yeast may also occur in response to growth conditions where aerobic respiration is not needed. In these situations, the mitochondria are likely to have been functional at the time of removal (30,31). In contrast, a growing number of recent studies highlight a role for autophagy in selective removal of damaged mitochondria (22,26). As discussed below, the regulation of selective autophagy for damaged or depolarized mitochondria represents an exciting new frontier in basic and PD research.
THE AUTOPHAGY MACHINERY: AN OVERVIEW
Macroautophagy is a major form of autophagy that involves membrane rearrangements to sequester and deliver cargo through fusion with lysosomes. Other forms of autophagy include chaperone-mediated autophagy, in which proteins are unfolded to enter the lysosome through Lamp2a receptors, and microautophagy in which cytosolic proteins enter through lysosomal membrane invaginations. The major form of autophagy responsible for degrading organelles or protein aggregates is macroautophagy, which is hereafter referred to as autophagy.
A growing number of signaling pathways contribute to the regulation of autophagy in mammalian cells [reviewed in (32)]. These include the mammalian target of rapamycin (mTOR) and insulin/Akt signaling, which suppress nutrient-regulated autophagy, and AMP kinase and mitogen-activated protein kinases (33,34), which promote autophagy under conditions of energy crisis or mitochondrial injury. Classic starvation-induced autophagy involves inactivation of mTOR, de-repression of Autophagy-related gene 1 (Atg1) protein function and activation of the Beclin1/Atg6 complex containing the class III phosphatidylinositol 3-kinase (PIK3C3/Vps34). Local alteration in membrane PI(3)P composition recruits proteins that lead to the deposition of small ubiquitin-like proteins Atg12 and microtubule-associated protein 1 light chain 3 (LC3/Atg8), which mediates the extension and curvature of the nascent autophagosome membrane. The covalent conjugation of LC3 to autophagosome membranes forms a useful marker that can be followed by its altered mobility in immunoblots or by (immuno)fluorescent detection of puncta in neuritic and somatic compartments (35). Moreover, LC3 interacts with adaptor proteins such as p62 and NBR1, which mediate the enrichment of ubiquitinated cargo inside autophagosomes (36).
The majority of autophagy regulation studies have been conducted using yeast cells, hepatocytes or fibroblasts, predominantly under conditions of nutrient stress. Unlike hepatocytes or mesenchymal cells, however, neurons are not programmed to digest cytoplasm for the purpose of maintaining blood nutrient levels. Moreover, neurons show a highly distinct physiology, including the large distances covered by neuronal processes and their high mitochondrial metabolic demand (37,38). For these reasons, transport-related cytoskeletal processes are likely to play a particularly important role (39), and much remains to be learned concerning the regulation of neuronal autophagy under both physiological and disease states.
Mechanistic variations from classic starvation-induced autophagy have been reported in several models of cell injury (40). For instance, Beclin1/PIK3C3-independent autophagy or mitophagy is observed in neuronal cells treated with the mitochondrial complex I inhibitor MPP+ (41), in resveratrol-treated breast carcinoma cells (42), in PUMA/Bax-induced autophagy (43) and in autophagic neurite retraction elicited by the G2019S mutant of leucine-rich repeat kinase 2 (LRRK2) (44), the most common genetic cause of PD. These studies suggest that mitochondrial injury or oxidative damage may elicit alternative mechanisms of nucleation. Moreover, a loss of dependence on Beclin1 also implies an escape from other regulatory mechanisms, such Bcl-2 suppression of Beclin 1-dependent autophagy (45). While additional studies are needed to determine the alternative mechanisms involved, the few examples of Beclin1-independent autophagy described thus far each implicate autophagy in a detrimental role, perhaps due to overactivation.
Mechanisms that regulate selective mitophagy are just beginning to be elucidated. In yeast, Uth1 and Atg32 are necessary for selective clearance of mitochondria, but potential mammalian homologs remain to be discovered (46,47). The BH3-only protein NIX is required for selective mitophagy during erythrocyte maturation, and BNIP3 regulates hypoxia-induced mitophagy (31). Interestingly, membrane depolarization can bypass the requirement for NIX in NIX−/− fibroblasts (31), implicating alternative mechanisms leading to mitochondrial autophagy that depend on the cellular context. The most recent mammalian candidate for Atg5-dependent mitophagy of depolarized mitochondria is the ubiquitin ligase Parkin (22), which may act to recruit adaptor proteins such as p62 (48).
ROLE OF ENDOGENOUS PINK1 IN MAINTAINING FUNCTIONAL MITOCHONDRIAL NETWORKS
Mitochondrial dysfunction, as evidenced by decreased mitochondrial membrane potential and oxygen consumption, has consistently been observed in mammalian PINK1 loss-of-function studies [summarized in tabular form in (20)]. Given that mitochondria isolated from human brain tissues and peripheral cells of sporadic PD patients exhibit reduced mitochondrial complex I activity (14), it is interesting to note that decreased complex I activity can be observed in cellular models of PINK1 deficiency (49,50) and in striatal mitochondria of PINK1 knockout mice (51), although this is not consistently observed (52). In our experience, chronic low-dose treatment with the mitochondrial complex I inhibitor MPP+ results in marked alterations in complex I and complex IV subunit expression (J. Zhu and C.T. Chu, unpublished data). Thus, it can be difficult to distinguish primary from secondary deficits. On the other hand, both direct and indirect downstream effects that contribute to cellular dysfunction could represent useful targets for therapeutic intervention. Interestingly, substrate limitation as a result of mitochondrial calcium dysregulation has been proposed as an upstream mechanism influencing electron transport chain activity (53).
In addition to functional changes, PINK1 expression levels influence cristae density in SH-SY5Y cells, with the knockdown lines showing decreased cristae and the overexpressing lines showing increased cristae density (26). Co-immunoprecipitation studies using tagged PINK1 demonstrate an interaction with mitofilin (54), a mitochondrial inner membrane protein that regulates cristae morphology. Moreover, PINK1 exists as a dimer that may migrate with several respiratory chain complexes (55). The discovery of the putative molecular chaperone TNF-receptor associated protein 1 (TRAP1)/Hsp75 as a substrate for PINK1 (56) also suggests a possible role regulating the proper folding and insertion of newly synthesized or imported proteins. As maintenance of a proper redox balance is important for protein folding and misfolding, it is striking that multiple model systems of PINK1 deficiency also implicate PINK1 in limiting oxidative stress (26,51,57,58). Additionally, PINK1 transcription in lymphocytes is enhanced by Forkhead box subgroup O transcription factors that are classically involved in protecting cells from oxidative stress-related cell death (59).
Loss of PINK1 and Parkin result in similar steady-state morphological changes in mitochondria, although conclusions drawn from Drosophila and mammalian models of PINK1 deficiency have implicated opposite effects on mitochondrial fission and fusion. The role of PINK1 in influencing mitochondrial dynamics has been extensively reviewed (20,21,60). PINK1 overexpression increases Drp1 phosphorylation (S.J. Cherra and C.T. Chu, unpublished data), and in PINK1-deficient cells, calcineurin-mediated dephosphorylation of Drp1 triggers mitochondrial fission (61), as observed by several groups (26,61,62). Because dynamic changes in mitochondrial fission, fusion and transport are all likely to occur as part of the processes governing local repair or autophagic degradation, it is perhaps not surprising that static estimates of mitochondrial morphology may yield differing results among model systems (20,63). Alternatively, it is possible that PINK1 will show different effects depending upon differences in post-translational regulation, allowing it to act as a sensor for changing mitochondrial and cellular needs.
POST-TRANSLATIONAL REGULATION OF PINK1
While knockdown studies of endogenous mammalian PINK1 indicate a key role for PINK1 in maintaining functioning mitochondrial networks, preventing mitochondrial ROS generation, calcium dysregulation, membrane depolarization and fragmentation, not all neuroprotective activities of PINK1 depend upon its N-terminal mitochondrial localization sequence [(64) R.K. Dagda, S.J. Cherra III and C.T. Chu, unpublished data]. Processed, and presumably active, forms of PINK1 are found distributed in both mitochondrial and cytoplasmic compartments (65,66), and there is evidence to support PINK1 activities in both intramitochondrial (56,67,68) and cytosolic compartments (69,70).
One of the difficulties studying PINK1 is that many commercially available antibodies show insufficient avidity to recognize endogenous levels of PINK1. Thus, the degree of overexpression in many transfection studies is unknown, representing an important caveat for studies where PINK1 goes from undetectable to robustly detectable. In one study, tagged PINK1 was subjected to metabolic pulse—chase studies, demonstrating rapid processing of full-length PINK1 (∼67 kDa) to at least two distinct forms termed Δ1 (∼58 kDa) and Δ2 (∼50 kDa). While the Δ1 form is sensitive to proteasome degradation, the Δ2 form is long lived (66). The levels of PINK1 are also regulated by heat shock protein chaperones (65) and by Parkin (71,72), although whether or not either is a direct substrate of the other remains controversial (73). In addition to N-terminal proteolysis by mitochondrial signal peptidases, additional processing is likely given the apparent molecular weight changes and the observation that C-terminal truncation regulates PINK1 kinase activity (67). The possibility of processing from both ends of the protein presents unique challenges for interpreting studies of overexpressed PINK1, as only forms that retain the tag are detected, and additional experimental reagents or strategies are needed.
Using an antibody raised to a central region of human PINK1, the major endogenous bands are processed (26). Overexpression of C-terminally tagged PINK1-3xFlag reveals that proteasome inhibitors stabilize an ∼58 kDa form of N-terminally processed PINK1-3xFlag (26), consistent with Δ1. However, some endogenous bands migrate lower than this Flag-tagged band, implicating PINK1 processing from both ends. While these forms of PINK1 appear to be long-lived, with little evidence of stabilization by proteasome inhibitors, chronic mitochondrial injury induces a striking loss of these processed endogenous bands (J. Zhu and C.T. Chu, unpublished data). Differences in stability or localization of overexpressed versus endogenous PINK1 may reflect saturation of chaperone, mitochondrial import or processing systems. Given that mitochondrial membrane potential drives the import of many mitochondrial proteins (74), it is likely that import and processing of PINK1 are altered by mitochondrial depolarization, favoring accumulation of full-length PINK1 (66,75). The effects of membrane potential on long-lived processed forms of PINK1 are unknown, but it is conceivable that the different forms of PINK1 mediate different functions within the cell, allowing PINK1 to serve as a sensor for mitochondrial status.
PINK1 AND MITOCHONDRIAL AUTOPHAGY
Ultrastructural studies of PINK1-deficient cells often display increased lysosomal content (26,58,76), shown to be due to an increase in mitochondrial autophagy [(26), Fig. 2]. Knockdown of essential autophagy proteins exacerbates cell death in stable PINK1 shRNA lines, indicating that autophagy plays a compensatory role in PINK1-deficient cells (26). Mitochondrial autophagy induced by PINK1 deficiency is regulated by canonical Beclin 1-dependent mechanisms (77). Moreover, endogenous Parkin levels increase in some stable PINK1-deficient lines (78), suggesting a role for Parkin-mediated mitophagy as a compensatory response. Although it remains to be determined whether or not Parkin is necessary for mitophagy in PINK1-deficient cells, cell survival is enhanced by transient overexpression of Parkin (26). As Parkin may show multiple neuroprotective mechanisms, we employed siRNA to Atg7, the E1-like activating protein for Atg12 and LC3, to study the contribution of autophagy. Inhibition of autophagy substantially reduced the ability of Parkin to confer protection (78), indicating that a major neuroprotective effect of Parkin in this context involves autophagy.
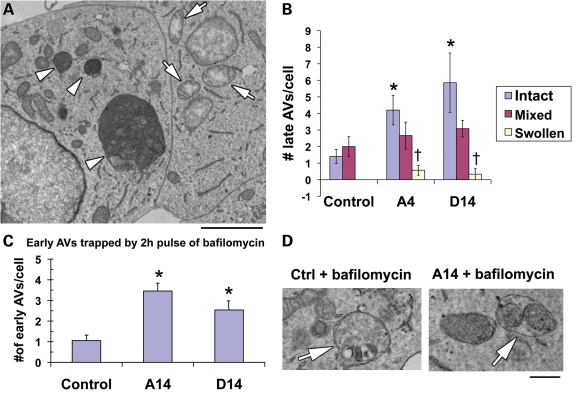
Figure 2.
Selective mitophagy in PINK1-deficient cells. (A) The majority of SH-SY5Y cells stably expressing PINK1 shRNA exhibit autophagic vacuoles (AVs; arrowheads) and at least some morphologically preserved mitochondria. As observed in other pathological states (41), occasional giant AVs are seen. Less than 20% of cells, however, exhibit swelling of all mitochondria without increased AVs (arrows). Scale bar = 2 µm. (B) The number of AVs per cell was analyzed by separating cells into three groups: those that exhibited relatively preserved mitochondrial ultrastructure, those with a mixture of intact and swollen mitochondria and those with exclusively swollen mitochondria. When compared with control cells with intact mitochondria, PINK1 shRNA cells with intact mitochondria exhibited significantly increased numbers of AVs/cell. In contrast, PINK1 shRNA cells with swollen mitochondria showed significantly fewer AVs than shRNA cells exhibiting intact or a mixture of intact and swollen mitochondria. *P < 0.05 versus control/intact; †P < 0.05 versus respective shRNA line/intact and versus respective shRNA line/mixed; 25–30 cells analyzed per group. These data show that mitochondrial quality in PINK1-deficient cells is positively correlated with the ability to induce autophagy. (C) Early autophagosomes with recognizable cargo are rare in both control and PINK1 shRNA cells. To assess relative rates of autophagosome formation, the fusion inhibitor bafilomycin A1 (10 nm) was applied for 2 h prior to fixation with 2% glutaraldehyde in order to trap newly formed autophagosomes. Both A series and D series PINK1 shRNA lines exhibit increased AV formation over 2 h. *P > 0.05 versus control. (D) Early AVs in bafilomycin-treated control cells contain a variety of cytoplasmic structures (left, arrow), while mitochondria comprise a prominent component of early AVs in the PINK1 shRNA line (right, arrow). Scale bar = 500 nm. These data support a selective process of mitophagy in PINK1-deficient cells.
In our ultrastructural studies, a minor fraction of PINK1-deficient cells exhibit swollen, pale mitochondria reminiscent of those observed in toxin-treated cells (Fig. 2A, right cell). Quantitative analysis indicates that these cells do not show upregulation of lysosomes/late autophagic vacuoles (Fig. 2B), and most probably account for the low level of basal cell death observed in the PINK1 shRNA cells (26). In contrast, cells with relatively intact mitochondrial morphologies show the highest content of late autophagic vacuoles (Fig. 2B), further supporting a role for autophagy in mitochondrial quality control and cytoprotection among individual cells in these stable lines.
As steady-state levels of autophagic vacuoles are not necessarily reflective of autophagic flux or activity, we performed several studies to verify enhanced mitochondrial autophagy. PINK1 knockdown cells show reduced mitochondrial mass as assessed through western blot analysis for either matrix or membrane proteins, or by analysis of the cytoplasmic area occupied by mitochondria (26). Flux studies using the autophagosome–lysosome fusion inhibitor bafilomycin demonstrate intact lysosomal turnover of both early and total LC3 puncta, as well as lysosome-dependent loss of GFP-tagged mitochondria (26). Further ultrastructural studies revealed that a 2 h pulse of bafilomycin trapped approximately three times as many early autophagosomes in PINK1 shRNA cells compared with control cells (Fig. 2C), indicating an increased rate of autophagosome formation. In contrast to control cells that showed a random mixture of cytoplasmic constituents, mitochondria were enriched within early autophagosomes of PINK1-deficient cells (Fig. 2D), consistent with selective mitophagy.
We also found that overexpression of PINK1 is capable of suppressing toxin-induced increases in autophagic vacuoles (26), and the ability of RNAi-resistant PINK1 to reverse the autophagic phenotype in stable PINK1 shRNA lines was not dependent upon the N-terminal mitochondrial-targeting sequence. We propose that one effect of PINK1 in maintaining stable mitochondrial networks includes suppression of autophagy. This could be through either direct or indirect mechanisms, such as reducing redox signals for autophagy induction (79). The possibility that PINK1 overexpression could promote maturation/clearance of toxin-induced autophagosomes also remains to be determined.
Interestingly, several recent papers indicate that PINK1 promotes Parkin-associated mitophagy (48,73,75), potentially with direct effects on Beclin 1 (80). As depolarized mitochondria are less capable of processing PINK1 to the proteasomally degraded Δ1 form, it is proposed to accumulate on the surface of mitochondria to recruit Parkin (75). Another study showed that overexpression of PINK1 and Parkin caused trafficking of mitochondria to perinuclear aggresome-assembly areas (73), although this effect was only observed when both proteins were concurrently overexpressed. The trafficking effect could relate to the association of PINK1 with the Miro/Milton-trafficking adaptors (54), as Miro is implicated in both anterograde and retrograde trafficking. As Milton mediates kinesin-dependent anterograde transport, it is interesting to note that the Miro/Milton complex can associate with truncated PINK1, while the effects of overexpressed PINK1 on Parkin recruitment and retrograde mitochondrial aggregation are dependent upon its full-length sequence (75,81). Overexpressed PINK1 has been reported to exhibit a cytosolic orientation of its kinase domain (69), and arrested import/processing of overexpressed PINK1 by depolarized mitochondria appears to be sufficient to recruit Parkin to mitochondria (75).
In these studies, either acute PINK1 knockdown or PINK1−/− MEFs showed impaired ability to recruit Parkin to chemically depolarized mitochondria. On the other hand, increased Parkin promotes compensatory mitophagy in PINK1-deficient SH-SY5Y cells (26). Thus, it is unclear whether or not stable physical association of Parkin with mitochondria is necessary for mitophagy or if transient enzymatic interaction is sufficient. Although Parkin recruitment to mitochondria is associated with its ability to cause loss of fluorescently labeled mitochondria, Parkin recruitment and mitochondrial clearance can be experimentally dissociated (75). Furthermore, studies in neurons may reveal additional regulatory mechanisms, as chemical uncouplers appear to induce complete loss of mitochondrial fluorescence from the affected cells. While glycolysis-competent cell types can survive mitochondrial depletion, excessive mitochondrial degradation is detrimental in neuronal cells (41,82,83).
Another caveat to consider is whether or not loss of mitochondrial fluorescence could reflect other mechanisms. As discussed above, there are several proteases within mitochondria that could contribute to loss of mitochondrial constituents or quenching of mitochondrial fluorescence in chemically depolarized cells. Mitochondrially targeted proteins are subject to proteasome degradation under depolarizing conditions where import is impaired (84), and mitochondrial depolarization or permeability transition dissipates intermembrane space proteins. Quantitative ultrastructural analysis to demonstrate increased mitochondria in autophagosomes at early time points, with EM confirmation of absent mitochondrial structures at later time points, would help resolve these possibilities. Alterations in translational regulation, biosynthesis and import/assembly of mitochondrial constituents could also contribute to depletion of mitochondrial content within cells, affecting quality control and the outcome of autophagic responses to injury. As autophagic recycling represents the final tier of mitochondrial quality control in the presence or absence of sufficient PINK1 function, further strategies to enhance selective mitophagy (38) while promoting mitochondrial biogenesis (85) may prove effective for multiple forms of Parkinson's disease.
HYPOTHESIS: PINK1 AS A SENSOR FOR MITOCHONDRIAL QUALITY CONTROL
As discussed above, PINK1 has been implicated in all tiers of mitochondrial quality control (Fig. 3). Moreover, it has become clear that PINK1 functions converge with those of other PD-linked genes such as Parkin, HtrA2/Omi and DJ-1, acting in both shared and distinct steps (78).
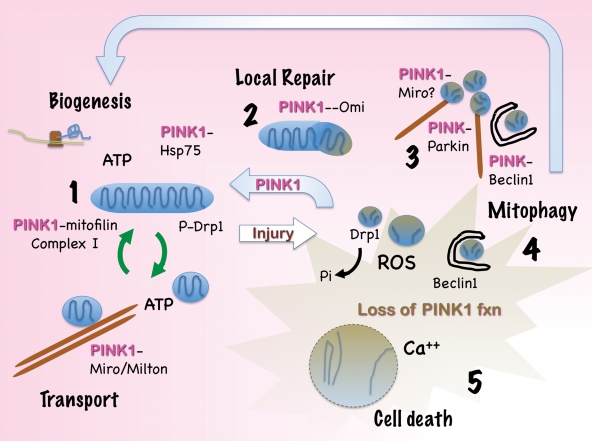
Figure 3.
A pivotal role for PINK1 in mitochondrial quality control. Emerging data indicate that PINK1 acts at multiple levels to promote mitochondrial health. (1) PINK1 deficiency studies suggest that endogenous PINK1 serves to maintain mitochondrial complex I activity, potentially through stabilization of cristae structures and electron transport chain components through interactions with mitofilin or the chaperone Hsp75/TRAP1. PINK1 also suppresses basal and 6-hydroxydopamine-induced mitochondrial ROS production and regulates mitochondrial calcium homeostasis. Interactions involving Miro/Milton suggest a role in regulating microtubule-dependent mitochondrial transport. (2) Maintenance of mitochondrial integrity may also involve interactions with chaperones or indirect regulation of the mitochondrial protease Omi/HTrA2 to effect localized repair. (3) With severe mitochondrial injury, Parkin is recruited to mitochondria, resulting in mitochondrial ubiquitination and formation of large perinuclear aggregates with features of aggresomes. These are cleared by Beclin 1-dependent autophagy, and expression of full-length PINK1 facilitates Parkin translocation, mitochondrial aggregation and eventual clearance. Interactions of PINK1 with Parkin, Beclin 1 and potentially Miro/Milton may contribute to this clearance. (4) Under conditions of PINK1 deficiency, the earlier tiers of mitochondrial quality control are compromised. Increased ROS and calcium signals trigger dephosphorylation of Drp1, mitochondrial fission and mitophagy, which plays a neuroprotective role in this setting. Interestingly, Parkin and Beclin 1 also function to clear PINK1-deficient mitochondria, suggesting that autophagy functions as one of the final lines of defense against mitochondrial injury. (5) Dysregulation of autophagy involving either insufficient, or excessive mitochondrial clearance, contributes to neuronal cell death.
PINK1 regulates normal mitochondrial function and possibly mitochondrial transport, which is critical to normal functioning of neurons (86). HtrA2/Omi and Parkin have each been implicated downstream of PINK1 (87–89), yet HtrA2 may not be essential for all PINK1-related functions (90). HtrA2 is an intermembrane space protease, whose phosphorylation is indirectly regulated by PINK1. Although its exact function is unknown, HtrA2 may mediate localized degradation of misfolded proteins or activation of signaling pathways in response to localized stress.
If damage is more substantial, mitochondrial fission and autophagy of depolarized mitochondria are triggered as another line of defense regulated by Parkin (22), as observed both in PINK1-deficient cells (26) and as a PINK1-regulated mitophagy pathway (75,80). Interestingly, Parkin and PINK1 have also been implicated in regulating mitochondrial biogenesis (52,91) or protein import (63) to complete the cycle. Furthermore, PINK1 functions may change depending upon mitochondrial and cellular signals for post-translational regulation, allowing it to act as a sensor for mitochondrial health. Although this model requires experimental testing, it offers an interesting conceptual framework for building our understanding of normal PINK1 function(s) and what may go wrong in Parkinson's and related diseases.
To summarize, there is exciting evidence for a key role for PINK1 in several pathways of mitochondrial quality control and mitochondrial autophagy (Fig. 3). On the basis of the existing information, PINK1 is hypothesized to show differential effects on mitochondrial dynamics to include fission/fusion, trafficking and autophagy, acting as a sensor or switch to either stabilize or dismantle the mitochondrial network depending on whether or not the damage can be repaired. Ultimately, obtaining a better understanding of the processing, post-translational modification and role of endogenous PINK1 under normal and stressed conditions may be just as important as identifying substrates in understanding PINK1-related pathophysiology.
FUNDING
C.T.C. is supported, in part, by funding from the National Institutes of Health (AG026389, AG026389-03S109, NS059806-01A26235), and is recipient of the AFAR Julie Martin Mid-Career Award in Aging Research sponsored by the Ellison Medical Foundation.
ACKNOWLEDGEMENTS
Ruben Dagda performed the electron microscopy and data quantification for Figure 2.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Dickson D.W., Braak H., Duda J.E., Duyckaerts C., Gasser T., Halliday G.M., Hardy J., Leverenz J.B., Del Tredici K., Wszolek Z.K., et al. Neuropathological assessment of Parkinson's disease: refining the diagnostic criteria. Lancet Neurol. 2009;8:1150–1157. doi: 10.1016/S1474-4422(09)70238-8. doi:10.1016/S1474-4422(09)70238-8. [DOI] [PubMed] [Google Scholar]
- 2.Gao H.M., Kotzbauer P.T., Uryu K., Leight S., Trojanowski J.Q., Lee V.M. Neuroinflammation and oxidation/nitration of alpha-synuclein linked to dopaminergic neurodegeneration. J. Neurosci. 2008;28:7687–7698. doi: 10.1523/JNEUROSCI.0143-07.2008. doi:10.1523/JNEUROSCI.0143-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dagda R.K., Zhu J., Chu C.T. Mitochondrial kinases in Parkinson's disease: converging insights from neurotoxin and genetic models. Mitochondrion. 2009;9:289–298. doi: 10.1016/j.mito.2009.06.001. doi:10.1016/j.mito.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee R., Starkov A.A., Beal M.F., Thomas B. Mitochondrial dysfunction in the limelight of Parkinson's disease pathogenesis. Biochim. Biophys. Acta. 2009;1792:651–663. doi: 10.1016/j.bbadis.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winklhofer K.F., Haass C. Mitochondrial dysfunction in Parkinson's disease. Biochim. Biophys. Acta. 1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Zimprich A., Biskup S., Leitner P., Lichtner P., Farrer M., Lincoln S., Kachergus J., Hulihan M., Uitti R.J., Calne D.B., et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. doi:10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Paisan-Ruiz C., Jain S., Evans E.W., Gilks W.P., Simon J., van der Brug M., Lopez de Munain A., Aparicio S., Gil A.M., Khan N., et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. doi:10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 8.Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. doi:10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 9.Chartier-Harlin M.C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364:1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 10.Kitada T., Asakawa S., Matsumine H., Hattori N., Minoshima S., Shimizu N., Mizuno Y. Positional cloning of the autosomal recessive juvenile parkinsonism (AR-JP) gene and its diversity in deletion mutations. Parkinsonism Relat. Disord. 1999;5:163–168. doi: 10.1016/s1353-8020(99)00032-2. doi:10.1016/S1353-8020(99)00032-2. [DOI] [PubMed] [Google Scholar]
- 11.Bonifati V., Rizzu P., Squitieri F., Krieger E., Vanacore N., van Swieten J.C., Brice A., van Duijn C.M., Oostra B., Meco G., et al. DJ-1 (PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol. Sci. 2003;24:159–160. doi: 10.1007/s10072-003-0108-0. doi:10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- 12.Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304:1158–1160. doi: 10.1126/science.1096284. doi:10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 13.Ramirez A., Heimbach A., Grundemann J., Stiller B., Hampshire D., Cid L.P., Goebel I., Mubaidin A.F., Wriekat A.L., Roeper J., et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006;38:1184–1191. doi: 10.1038/ng1884. doi:10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 14.Mizuno Y., Ikebe S., Hattori N., Nakagawa-Hattori Y., Mochizuki H., Tanaka M., Ozawa T. Role of mitochondria in the etiology and pathogenesis of Parkinson's disease. Biochim. Biophys. Acta. 1995;1271:265–274. doi: 10.1016/0925-4439(95)00038-6. [DOI] [PubMed] [Google Scholar]
- 15.Przedborski S., Jackson-Lewis V. Mechanisms of MPTP toxicity. Mov. Disord. 1998;13(Suppl. 1):35–38. [PubMed] [Google Scholar]
- 16.Betarbet R., Sherer T.B., Di Monte D.A., Greenamyre J.T. Mechanistic approaches to Parkinson's disease pathogenesis. Brain Pathol. 2002;12:499–510. doi: 10.1111/j.1750-3639.2002.tb00468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Callio J., Oury T.D., Chu C.T. Manganese superoxide dismutase protects against 6-hydroxydopamine injury in mouse brains. J. Biol. Chem. 2005;280:18536–18542. doi: 10.1074/jbc.M413224200. doi:10.1074/jbc.M413224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulich S., Horbinski C., Patel M., Chu C. 6-Hydroxydopamine induces mitochondrial ERK activation. Free Radic. Biol. Med. 2007;43:372–383. doi: 10.1016/j.freeradbiomed.2007.04.028. doi:10.1016/j.freeradbiomed.2007.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T.M., Thomas B., Ko H.S., Sasaki M., Ischiropoulos H., Przedborski S., et al. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl Acad. Sci. USA. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. doi:10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu C.T. Tickled PINK1: mitochondrial homeostasis and autophagy in recessive parkinsonism. Biochim. Biophys. Acta. 2010;1802:20–28. doi: 10.1016/j.bbadis.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitworth A.J., Pallanck L.J. The PINK1/Parkin pathway: a mitochondrial quality control system? J. Bioenerg. Biomembr. 2009;41:499–503. doi: 10.1007/s10863-009-9253-3. doi:10.1007/s10863-009-9253-3. [DOI] [PubMed] [Google Scholar]
- 22.Narendra D., Tanaka A., Suen D.F., Youle R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. doi:10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shavali S., Brown-Borg H.M., Ebadi M., Porter J. Mitochondrial localization of alpha-synuclein protein in alpha-synuclein overexpressing cells. Neurosci. Lett. 2008;439:125–128. doi: 10.1016/j.neulet.2008.05.005. doi:10.1016/j.neulet.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono T., Isobe K., Nakada K., Hayashi J.I. Human cells are protected from mitochondrial dysfunction by complementation of DNA products in fused mitochondria. Nat. Genet. 2001;28:272–275. doi: 10.1038/90116. doi:10.1038/90116. [DOI] [PubMed] [Google Scholar]
- 25.Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. doi:10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dagda R.K., Cherra S.J., 3rd, Kulich S.M., Tandon A., Park D., Chu C.T. Loss of pink1 function promotes mitophagy through effects on oxidative stress and mitochondrial fission. J. Biol. Chem. 2009;284:13843–13855. doi: 10.1074/jbc.M808515200. doi:10.1074/jbc.M808515200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Voos W. Mitochondrial protein homeostasis: the cooperative roles of chaperones and proteases. Res. Microbiol. 2009;160:718–725. doi: 10.1016/j.resmic.2009.08.003. doi:10.1016/j.resmic.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Martinelli P., Rugarli E.I. Emerging roles of mitochondrial proteases in neurodegeneration. Biochim. Biophys. Acta. 1797:1–10. doi: 10.1016/j.bbabio.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Santel A., Frank S. Shaping mitochondria: the complex posttranslational regulation of the mitochondrial fission protein DRP1. IUBMB Life. 2008;60:448–455. doi: 10.1002/iub.71. doi:10.1002/iub.71. [DOI] [PubMed] [Google Scholar]
- 30.Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Lohr F., Popovic D., Occhipinti A., et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. doi: 10.1038/embor.2009.256. doi:10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang J., Ney P.A. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. doi:10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehrpour M., Esclatine A., Beau I., Codogno P. Autophagy in health and disease: 1. Regulation and significance of autophagy—an overview. Am. J. Physiol. Cell Physiol. 2010;298:C776–C785. doi: 10.1152/ajpcell.00507.2009. [DOI] [PubMed] [Google Scholar]
- 33.Dagda R.K., Zhu J., Kulich S.M., Chu C.T. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress. Autophagy. 2008;4:770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorin S., Borges A., Ribeiro Dos Santos L., Souquere S., Pierron G., Ryan K.M., Codogno P., Djavaheri-Mergny M. c-Jun NH2-terminal kinase activation is essential for DRAM-dependent induction of autophagy and apoptosis in 2-methoxyestradiol-treated Ewing sarcoma cells. Cancer Res. 2009;69:6924–6931. doi: 10.1158/0008-5472.CAN-09-1270. doi:10.1158/0008-5472.CAN-09-1270. [DOI] [PubMed] [Google Scholar]
- 35.Chu C.T., Plowey E.D., Dagda R.K., Hickey R.W., Cherra S.J.C., III, Clark R.S. Autophagy in neurite injury and neurodegeneration: in vitro and in vivo models. Methods Enzymol. 2009;453:217–249. doi: 10.1016/S0076-6879(08)04011-1. doi:10.1016/S0076-6879(08)04011-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lamark T., Kirkin V., Dikic I., Johansen T. NBR1 and p62 as cargo receptors for selective autophagy of ubiquitinated targets. Cell Cycle. 2009;8:1986–1990. doi: 10.4161/cc.8.13.8892. [DOI] [PubMed] [Google Scholar]
- 37.Yue Z., Friedman L., Komatsu M., Tanaka K. The cellular pathways of neuronal autophagy and their implication in neurodegenerative diseases. Biochim. Biophys. Acta. 2009;1793:1496–1507. doi: 10.1016/j.bbamcr.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherra S.J., III, Chu C.T. Autophagy in neuroprotection and neurodegeneration: a question of balance. Future Neurol. 2008;3:309–323. doi: 10.2217/14796708.3.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J.Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y.S., Pandey U.B., Kaushik S., Tresse E., Lu J., et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., Shimizu S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–659. doi: 10.1038/nature08455. doi:10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J.H., Horbinski C., Guo F., Watkins S., Uchiyama Y., Chu C.T. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am. J. Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. doi:10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scarlatti F., Maffei R., Beau I., Codogno P., Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008;15:1318–1329. doi: 10.1038/cdd.2008.51. doi:10.1038/cdd.2008.51. [DOI] [PubMed] [Google Scholar]
- 43.Yee K.S., Wilkinson S., James J., Ryan K.M., Vousden K.H. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–1145. doi: 10.1038/cdd.2009.28. doi:10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plowey E.D., Cherra S.J., III, Liu Y.J., Chu C.T. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J. Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. doi:10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He C., Levine B. The Beclin 1 interactome. Curr. Opin. Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kissova I., Deffieu M., Manon S., Camougrand N. Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 2004;279:39068–39074. doi: 10.1074/jbc.M406960200. doi:10.1074/jbc.M406960200. [DOI] [PubMed] [Google Scholar]
- 47.Kanki T., Wang K., Cao Y., Baba M., Klionsky D.J. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 2009;17:98–109. doi: 10.1016/j.devcel.2009.06.014. doi:10.1016/j.devcel.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12:119–131. doi: 10.1038/ncb2012. doi:10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 49.Hoepken H.H., Gispert S., Morales B., Wingerter O., Del Turco D., Mulsch A., Nussbaum R.L., Muller K., Drose S., Brandt U., et al. Mitochondrial dysfunction, peroxidation damage and changes in glutathione metabolism in PARK6. Neurobiol. Dis. 2007;25:401–411. doi: 10.1016/j.nbd.2006.10.007. doi:10.1016/j.nbd.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 50.Morais V., Verstreken P., Roethig A., Smet J., Snellinx A., Vanbrabant M., Haddad D., Frezza C., Mandermakers W., Vogt-Weisenhorn D., et al. Parkinson's disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol. Med. 2009;1:99–111. doi: 10.1002/emmm.200900006. doi:10.1002/emmm.200900006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gautier C.A., Kitada T., Shen J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl Acad. Sci. USA. 2008;105:11364–11369. doi: 10.1073/pnas.0802076105. doi:10.1073/pnas.0802076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gegg M.E., Cooper J.M., Schapira A.H., Taanman J.W. Silencing of PINK1 expression affects mitochondrial DNA and oxidative phosphorylation in dopaminergic cells. PLoS ONE. 2009;4:e4756. doi: 10.1371/journal.pone.0004756. doi:10.1371/journal.pone.0004756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gandhi S., Wood-Kaczmar A., Yao Z., Plun-Favreau H., Deas E., Klupsch K., Downward J., Latchman D.S., Tabrizi S.J., Wood N.W., et al. PINK1-associated Parkinson's disease is caused by neuronal vulnerability to calcium-induced cell death. Mol. Cell. 2009;33:627–638. doi: 10.1016/j.molcel.2009.02.013. doi:10.1016/j.molcel.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weihofen A., Thomas K.J., Ostaszewski B.L., Cookson M.R., Selkoe D.J. Pink1 forms a multiprotein complex with Miro and Milton, linking Pink1 function to mitochondrial trafficking (dagger) Biochemistry. 2009;48:2045–2052. doi: 10.1021/bi8019178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu W., Vives-Bauza C., Acin-Perez R., Yamamoto A., Tan Y., Li Y., Magrane J., Stavarache M.A., Shaffer S., Chang S., et al. PINK1 defect causes mitochondrial dysfunction, proteasomal deficit and alpha-synuclein aggregation in cell culture models of Parkinson's disease. PLoS ONE. 2009;4:e4597. doi: 10.1371/journal.pone.0004597. doi:10.1371/journal.pone.0004597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pridgeon J.W., Olzmann J.A., Chin L.S., Li L. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 2007;5:e172. doi: 10.1371/journal.pbio.0050172. doi:10.1371/journal.pbio.0050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang D., Qian L., Xiong H., Liu J., Neckameyer W.S., Oldham S., Xia K., Wang J., Bodmer R., Zhang Z. Antioxidants protect PINK1-dependent dopaminergic neurons in Drosophila. Proc. Natl Acad. Sci. USA. 2006;103:13520–13525. doi: 10.1073/pnas.0604661103. doi:10.1073/pnas.0604661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood-Kaczmar A., Gandhi S., Yao Z., Abramov A.S., Miljan E.A., Keen G., Stanyer L., Hargreaves I., Klupsch K., Deas E., et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE. 2008;3:e2455. doi: 10.1371/journal.pone.0002455. doi:10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mei Y., Zhang Y., Yamamoto K., Xie W., Mak T.W., You H. FOXO3a-dependent regulation of Pink1 (Park6) mediates survival signaling in response to cytokine deprivation. Proc. Natl Acad. Sci. USA. 2009;106:5153–5158. doi: 10.1073/pnas.0901104106. doi:10.1073/pnas.0901104106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y., Lu B. Mitochondrial morphogenesis, distribution, and Parkinson disease: insights from PINK1. J. Neuropathol. Exp. Neurol. 2009;68:953–963. doi: 10.1097/NEN.0b013e3181b2048c. doi:10.1097/NEN.0b013e3181b2048c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandebring A., Thomas K.J., Beilina A., van der Brug M., Cleland M.M., Ahmad R., Miller D.W., Zambrano I., Cowburn R.F., Behbahani H., et al. Mitochondrial alterations in PINK1 deficient cells are influenced by calcineurin-dependent dephosphorylation of dynamin-related protein 1. PLoS ONE. 2009;4:e5701. doi: 10.1371/journal.pone.0005701. doi:10.1371/journal.pone.0005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lutz A.K., Exner N., Fett M.E., Schlehe J.S., Kloos K., Lammermann K., Brunner B., Kurz-Drexler A., Vogel F., Reichert A.S., et al. Loss of Parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J. Biol. Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. doi:10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gispert S., Ricciardi F., Kurz A., Azizov M., Hoepken H.H., Becker D., Voos W., Leuner K., Muller W.E., Kudin A.P., et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS ONE. 2009;4:e5777. doi: 10.1371/journal.pone.0005777. doi:10.1371/journal.pone.0005777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haque M.E., Thomas K.J., D'Souza C., Callaghan S., Kitada T., Slack R.S., Fraser P., Cookson M.R., Tandon A., Park D.S. Cytoplasmic Pink1 activity protects neurons from dopaminergic neurotoxin MPTP. Proc. Natl Acad. Sci. USA. 2008;105:1716–1721. doi: 10.1073/pnas.0705363105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weihofen A., Ostaszewski B., Minami Y., Selkoe D.J. Pink1 Parkinson mutations, the Cdc37/Hsp90 chaperones and Parkin all influence the maturation or subcellular distribution of Pink1. Hum. Mol. Genet. 2008;17:602–616. doi: 10.1093/hmg/ddm334. [DOI] [PubMed] [Google Scholar]
- 66.Lin W., Kang U.J. Characterization of PINK1 processing, stability, and subcellular localization. J. Neurochem. 2008;106:464–474. doi: 10.1111/j.1471-4159.2008.05398.x. doi:10.1111/j.1471-4159.2008.05398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Silvestri L., Caputo V., Bellacchio E., Atorino L., Dallapiccola B., Valente E.M., Casari G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005;14:3477–3492. doi: 10.1093/hmg/ddi377. doi:10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 68.Gandhi S., Muqit M.M., Stanyer L., Healy D.G., Abou-Sleiman P.M., Hargreaves I., Heales S., Ganguly M., Parsons L., Lees A.J., et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129:1720–1731. doi: 10.1093/brain/awl114. doi:10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 69.Zhou C., Huang Y., Shao Y., May J., Prou D., Perier C., Dauer W., Schon E.A., Przedborski S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl Acad. Sci. USA. 2008;105:12022–12027. doi: 10.1073/pnas.0802814105. doi:10.1073/pnas.0802814105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiong H., Wang D., Chen L., Choo Y.S., Ma H., Tang C., Xia K., Jiang W., Ronai Z., Zhuang X., et al. Parkin, PINK1 and DJ-1 form a ubiquitin E3 ligase complex promoting unfolded protein degradation. J. Clin. Invest. 2009;119:650–660. doi: 10.1172/JCI37617. doi:10.1172/JCI37617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shiba K., Arai T., Sato S., Kubo S.I., Ohba Y., Mizuno Y., Hattori N. Parkin stabilizes PINK1 through direct interaction. Biochem. Biophys. Res. Commun. 2009;383:331–335. doi: 10.1016/j.bbrc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 72.Um J.W., Stichel-Gunkel C., Lubbert H., Lee G., Chung K.C. Molecular interaction between parkin and PINK1 in mammalian neuronal cells. Mol. Cell Neurosci. 2009;40:421–432. doi: 10.1016/j.mcn.2008.12.010. doi:10.1016/j.mcn.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 73.Vives-Bauza C., Zhou C., Huang Y., Cui M., de Vries R.L., Kim J., May J., Tocilescu M.A., Liu W., Ko H.S., et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc. Natl Acad. Sci. USA. 107:378–383. doi: 10.1073/pnas.0911187107. doi:10.1073/pnas.0911187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pfanner N., Meijer M. Protein sorting. Pulling in the proteins. Curr. Biol. 1995;5:132–135. doi: 10.1016/s0960-9822(95)00033-9. [DOI] [PubMed] [Google Scholar]
- 75.Narendra D.P., Jin S.M., Tanaka A., Suen D.F., Gautier C.A., Shen J., Cookson M.R., Youle R.J. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8:e1000298. doi: 10.1371/journal.pbio.1000298. doi:10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Marongiu R., Spencer B., Crews L., Adame A., Patrick C., Trejo M., Dallapiccola B., Valente E.M., Masliah E. Mutant Pink1 induces mitochondrial dysfunction in a neuronal cell model of Parkinson's disease by disturbing calcium flux. J. Neurochem. 2009;108:1561–1574. doi: 10.1111/j.1471-4159.2009.05932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cherra S.J., III, Dagda R., Chu C.T. Autophagy and neurodegeneration: survival at a cost? Neuropathol. Appl. Neurol. 2010;36:125–132. doi: 10.1111/j.1365-2990.2010.01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dagda R.K., Chu C.T. Mitochondrial quality control: insights on how Parkinson's disease related genes PINK1, parkin, and Omi/HtrA2 interact to maintain mitochondrial homeostasis. J. Bioenerg. Biomembr. 2009;41:473–479. doi: 10.1007/s10863-009-9255-1. doi:10.1007/s10863-009-9255-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007;26:1749–1760. doi: 10.1038/sj.emboj.7601623. doi:10.1038/sj.emboj.7601623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Michiorri S., Gelmetti V., Giarda E., Lombardi F., Romano F., Marongiu R., Nerini-Molteni S., Sale P., Vago R., Arena G., et al. The Parkinson-associated protein PINK1 interacts with Beclin1 and promotes autophagy. Cell Death Differ. 2010 doi: 10.1038/cdd.2009.200. in press. [DOI] [PubMed] [Google Scholar]
- 81.Kim Y., Park J., Kim S., Song S., Kwon S.K., Lee S.H., Kitada T., Kim J.M., Chung J. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem. Biophys. Res. Commun. 2008;377:975–980. doi: 10.1016/j.bbrc.2008.10.104. doi:10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 82.Xue L., Fletcher G.C., Tolkovsky A.M. Mitochondria are selectively eliminated from eukaryotic cells after blockade of caspases during apoptosis. Curr. Biol. 2001;11:361–365. doi: 10.1016/s0960-9822(01)00100-2. doi:10.1016/S0960-9822(01)00100-2. [DOI] [PubMed] [Google Scholar]
- 83.Chakrabarti L., Eng J., Ivanov N., Garden G.A., La Spada A.R. Autophagy activation and enhanced mitophagy characterize the Purkinje cells of pcd mice prior to neuronal death. Mol. Brain. 2009;2:24. doi: 10.1186/1756-6606-2-24. doi:10.1186/1756-6606-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wright G., Terada K., Yano M., Sergeev I., Mori M. Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp. Cell Res. 2001;263:107–117. doi: 10.1006/excr.2000.5096. doi:10.1006/excr.2000.5096. [DOI] [PubMed] [Google Scholar]
- 85.Keeney P.M., Quigley C.K., Dunham L.D., Papageorge C.M., Iyer S., Thomas R.R., Schwarz K.M., Trimmer P.A., Khan S.M., Portell F.R., et al. Mitochondrial gene therapy augments mitochondrial physiology in a Parkinson's disease cell model. Hum. Gene Ther. 2009;20:897–907. doi: 10.1089/hum.2009.023. doi:10.1089/hum.2009.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rintoul G.L., Reynolds I.J. Mitochondrial trafficking and morphology in neuronal injury. Biochim. Biophys. Acta. 1802:143–150. doi: 10.1016/j.bbadis.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 87.Plun-Favreau H., Klupsch K., Moisoi N., Gandhi S., Kjaer S., Frith D., Harvey K., Deas E., Harvey R.J., McDonald N., et al. The mitochondrial protease HtrA2 is regulated by Parkinson's disease-associated kinase PINK1. Nat. Cell Biol. 2007;9:1243–1252. doi: 10.1038/ncb1644. doi:10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 88.Yang Y., Gehrke S., Imai Y., Huang Z., Ouyang Y., Wang J.W., Yang L., Beal M.F., Vogel H., Lu B. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl Acad. Sci. USA. 2006;103:10793–10798. doi: 10.1073/pnas.0602493103. doi:10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whitworth A.J., Lee J.R., Ho V.M., Flick R., Chowdhury R., McQuibban G.A. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson's disease factors Pink1 and Parkin. Dis. Model. Mech. 2008;1:168–174. doi: 10.1242/dmm.000109. (discussion 173) doi:10.1242/dmm.000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yun J., Cao J.H., Dodson M.W., Clark I.E., Kapahi P., Chowdhury R.B., Guo M. Loss-of-function analysis suggests that Omi/HtrA2 is not an essential component of the PINK1/PARKIN pathway in vivo. J. Neurosci. 2008;28:14500–14510. doi: 10.1523/JNEUROSCI.5141-08.2008. doi:10.1523/JNEUROSCI.5141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kuroda Y., Mitsui T., Kunishige M., Shono M., Akaike M., Azuma H., Matsumoto T. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum. Mol. Genet. 2006;15:883–895. doi: 10.1093/hmg/ddl006. doi:10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 92.Samaranch L., Lorenzo-Betancor O., Arbelo J.M., Ferrer I., Lorenzo E., Irigoyen J., Pastor M.A., Marrero C., Isla C., Herrera-Henriquez J., et al. PINK1-linked parkinsonism is associated with Lewy body pathology. Brain. 2010;133:1128–1142. doi: 10.1093/brain/awq051. doi:10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]