Abstract
Inclusion body myopathy associated with Paget's disease of the bone and fronto-temporal dementia (IBMPFD) is a progressive autosomal dominant disorder caused by mutations in p97/VCP (valosin-containing protein). p97/VCP is a member of the AAA+ (ATPase associated with a variety of activities) protein family and participates in multiple cellular processes. One particularly important role for p97/VCP is facilitating intracellular protein degradation. p97/VCP has traditionally been thought to mediate the ubiquitin-proteasome degradation of proteins; however, recent studies challenge this dogma. p97/VCP clearly participates in the degradation of aggregate-prone proteins, a process principally mediated by autophagy. In addition, IBMPFD mutations in p97/VCP lead to accumulation of autophagic structures in patient and transgenic animal tissue. This is likely due to a defect in p97/VCP-mediated autophagosome maturation. The following review will discuss the evidence for p97/VCP in autophagy and how a disruption in this process contributes to IBMPFD pathogenesis.
INTRODUCTION
Inclusion body myopathy (IBM) associated with Paget's disease of the bone (PDB) and fronto-temporal dementia (FTD), or IBMPFD (OMIM 167320), is a multisystem degenerative disorder caused by mutations in p97/VCP (valosin-containing protein) on chromosome 9p12-13 (1). IBMPFD is autosomal dominantly inherited and primarily affects muscle, brain and bone tissue. Of IBMPFD's phenotypic features, a myopathy is the most common and it is present in 90% of affected individuals. It is characterized by adult-onset (∼44 years), proximal and distal muscle weakness with associated atrophy. Affected skeletal muscle contains ‘rimmed vacuoles’ (RVs) and both myonuclear and sarcoplasmic inclusions (2). These inclusions are in some instances congophilic and are immunoreactive for ubiquitin and TARDNA-binding protein-43 (TDP-43) (3). The penetrance of FTD in IBMPFD patients is ∼30% and its onset is at a later age (∼54 years) than myopathy. IBMPFD CNS pathology is tau-negative and ubiquitin-positive consistent with a fronto-temporal lobar degeneration with ubiquitinated inclusions (FTLD-U) (4). IBMPFD patient CNS tissue has prominent intranuclear ubiquitinated and TDP-43-positive inclusions in affected brain regions distinguishing it from other FTLD-U subtypes and placing it in its own category FTLD-U subtype 4 (4,5). PDB manifests in ∼50% of IBMPFD patients at a similar age to the myopathy. Pagetoid osteoclasts have ubiquitinated nuclear and cytosolic inclusions as well (6). Cellular degeneration and ubiquitinated protein inclusions unify the pathologies of these three disparate tissues in IBMPFD.
p97/VALOSIN-CONTAINING PROTEIN
p97/VCP (also termed cdc48 and ter94) is a ubiquitously and highly expressed member of the type II AAA+ (ATPase associated with various activities) ATPase family (7). It is an essential and evolutionarily highly conserved cellular protein (p97/VCP knockout is lethal in yeast, C. elegans and mice) (8–10). p97/VCP has a tripartite structure consisting of an N-terminal domain and two central D1 and D2 AAA+ domains (Fig. 1) (7). The N-terminal domain is necessary for substrate and cofactor association, whereas the D1 and D2 domains are needed for ATP binding and hydrolysis (11). A p97/VCP monomer assembles into a functioning stable homo-hexamer with a central cylinder formed by the D1/2 domains surrounded by the N domains. The D1 domain in association with ATP is largely responsible for p97/VCP hexamerization; whereas the D2 domain performs the majority of ATP hydrolysis (12). Hence, the measures of the basal ATPase activity come predominantly from the D2 domain. During D2-mediated ATP hydrolysis, the largest conformational change occurs between the N and D1 domains (13,14). This structural change enables p97/VCP to function as a chaperone or ‘molecular motor’ that interacts with a diverse group of adaptors (15). These adaptors dictate VCP's promiscuous involvement in many cellular functions (16).
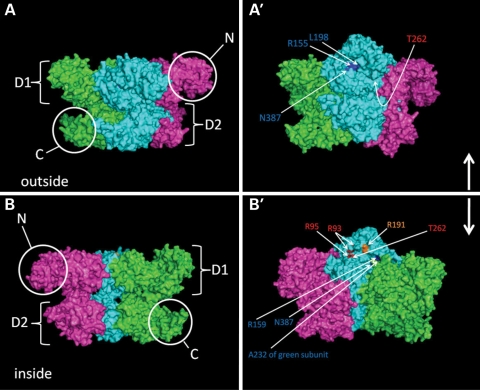
Figure 1.
IBMPFD mutations in the p97/VCP cluster at the interface between the N and D1 domains. (A) Exterior view of a p97/VCP hexamer. Three subunits are shown each individually colored. (A′) The view in (A) is tilted upward by 30° to allow visualization of mutant residues labeled and colored in blue or red. (B) Interior view of a p97/VCP hexamer. Again three subunits are shown each individually colored. (B′) The view in (B) is tilted downward by 30° to allow visualization of mutant residues labeled and colored in blue, red or orange.
As implied by its protein genre name, AAA+ protein, p97/VCP mediates a wide variety of essential cellular processes (7). These include post-mitotic nuclear envelope reformation and Golgi reassembly, cell cycle progression, DNA damage repair and protein degradation via the ubiquitin-proteasome system (UPS) and endoplasmic reticulum-associated protein degradation (ERAD) pathways. Loss of p97/VCP or expression of an ATP hydrolysis-deficient p97/VCP mutant leads to the accumulation of undegraded ubiquitinated proteins in cells (17,18). This initially implicated p97/VCP as a key player in the UPS. Subsequently, p97/VCP was found to associate with ubiquitinated substrates and many components of the UPS including E3/E4 ligases and deubiquitylating enzymes (16). p97/VCP is also instrumental in ERAD (19). Substrate-trapped hydrolysis-deficient p97/VCP localizes to the cytosolic face of endoplasmic reticulum (ER) (18). In addition, p97/VCP interacts with the putative retrotranslocon, Derlin-1, and is itself essential for the retrotranslocation of ERAD substrates (20). Specifically, p97/VCP in a multiprotein complex with UFD1 and Npl4 serves as the ‘motor’ needed to extract ERAD substrate proteins from the ER lumen or membrane into the cytoplasm for subsequent degradation by the 26S proteasome (20). p97/VCP may be necessary for the degradation of all ERAD and UPS substrates or only a specific set. For example, studies have shown that p97/VCP mediates the turnover of several specific cytosolic UPS substrates including IκB (inhibitor of kappa B), Unc-45b (skeletal muscle myosin chaperone) and Hif-1α (hypoxia inducible factor 1α) (21–23). Another putative degradation substrate for p97/VCP is the protein aggregate (24). p97/VCP localizes to pathogenic protein inclusions in multiple neurodegenerative disorders such as Huntington's disease, Parkinson's disease and amyotrophic lateral sclerosis (25). p97/VCP also binds polyglutamine-containing proteins in vitro (26). p97/VCP binds to the polyglutamine-containing protein ataxin-3. Ataxin-3 is associated with spinocerebellar ataxia type 3 (26). Interestingly, p97/VCP preferentially binds ataxin-3 when it contains as a pathologically expanded polyglutamine repeat.
IBMPFD MUTANT p97/VCP
IBMPFD-associated mutations are located in three different domains, I27V, R93C, R95C/G, P137L, R155H/P/C/S/L, G157R and R159H/C are located within the N domain, R191Q and L198W are within the linker connecting N and D1 domains and A232E, T262A, N387H and A439S are within the D1 domain (2,27,28). This totals 19 missense mutations at 13 different residues. Mutations of the arginine at residue 155 comprise the most frequent mutations in IBMPFD patients (1). Initial studies found that the R155H mutation in VCP did not affect its ability to hexamerize or hydrolyze ATP (29). Subsequent in vitro studies found that the IBMPFD mutants A232E and R155P have an increase in basal ATPase activity (30). We have evaluated the basal ATPase activities of eight additional IBMPFD mutants in addition to R155H, R155P and A232E and find that they all have increased ATPase activity when compared with wild-type VCP (VCP-WT; unpublished observations). How in vitro activity reflects in vivo function is unclear. These in vitro studies do not accurately reflect the 1:1 stoichiometry of endogenous VCP-WT to IBMPFD mutant VCP containing hexamers. Moreover, it is not known if mutant monomers even mix with WT monomers in patients.
Molecular modeling of a VCP hexamer places all IBMPFD mutant residues in a similar location at the N–D1 domain interface (Fig. 1). In fact, mutant N domain residues R95 and R155 may interact with mutant D1 domain residues A232 and N387, respectively (11). The largest movement throughout the ATPase cycle occurs between the N and D1 domains (13). This region is also important for substrate and cofactor selectivity (7). Preliminary studies have failed to show a defect in the association of IBMPFD mutants with three cofactors Ufd1, Npl4 and ataxin-3 (31). Whether IBMPFD mutations affect protein conformation during the ATPase cycle or substrate and cofactor binding is not known, but these defects could explain how a mutant VCP could have an increase in ATPase activity with impaired function.
Expression of IBMPFD mutations in cell culture and transgenic mice leads to the accumulation of ubiquitinated proteins similar to that seen with siRNA knockdown of VCP or proteasome inhibition (24,29,32,33). However, studies aimed at defining a clear pathogenic role for impairment in the UPS or ERAD pathway have yielded conflicting results. One study found that the steady-state levels of the ERAD substrate ΔF508-CFTR were elevated and accumulated as ER-associated ubiquitinated inclusions in cultured myoblasts expressing IBMPFD mutant R155H or R95G similar to that found with the expression of a hydrolysis-deficient mutant VCP (29). In contrast, another study found that IBMPFD mutant R155H and A232E expression did not impair the ERAD of CD3δ-YFP (34). The same study failed to see a defect in the UPS-mediated degradation of the ubiquitin fusion domain protein Ub-G76V-GFP when IBMPFD mutant R155H or A232E were co-expressed (34). Conflicting results were seen once again when the UPS-mediated degradation of the specific VCP substrate, UNC-45B instead of Ub-G76V-GFP, was evaluated. In cultured cells expressing IBMPFD mutant VCP and in IBMPFD transgenic mouse skeletal muscle, UNC-45B levels were elevated consistent with its failure to be degraded via the UPS (22,32). These data suggest that IBMPFD mutations in p97/VCP do not affect the ERAD or UPS degradation of all substrates but instead only a select group.
IBMPFD MUTANT p97/VCP DISRUPTS PROTEIN INCLUSION FORMATION AND CLEARANCE
p97/VCP interacts with aggregated proteins (25,26). This suggests that p97/VCP may be involved in protein inclusion formation or aggregate clearance from the cell. p97/VCP binds to histone deacetylase 6 (HDAC6) (35). HDAC6 is a cytosolically localized deacetylase (36). One of its functions is to bind ubiquitinated proteins and facilitate their delivery to the aggresome in times of proteotoxic stress (36). An aggresome is an actively generated, microtubule-dependent, perinuclear cellular structure (37). It contains ubiquitinated and aggregated proteins as well as the machinery needed to degrade them (e.g. proteasomal subunits and autophagic components). p97/VCP is also needed for aggresome formation (17). The loss of p97/VCP activity via RNA interference or following expression of an ATP hydrolysis-deficient mutant which can serve as a potent dominant-negative inhibitor of p97/VCP results in dispersed aggregates of ubiquitinated proteins and prevents aggresome formation following proteasome inhibition (17,24,38).
IBMPFD mutant p97/VCP expression has a similar effect on aggresome formation (24). In cells expressing IBMPFD mutant VCPs R155H, A232E or R95G, proteasome inhibition failed to induce a single perinuclear aggresome as was seen in control or p97/VCP-WT expressing cells (24). Similarly, in IBMPFD mutant cell lines, expression of an expanded polyQ failed to generate a larger perinuclear aggresome (24). Instead, smaller ubiquitin-positive polyQ aggregates were found throughout the cytoplasm as opposed to them consolidating in an aggresome. These smaller cytosolic inclusions did not co-localize with autophagic machinery such as LC3, p62 and HDAC6 (24). This resulted in the accumulation of insoluble polyQ protein, a decrease in the clearance of polyQ inclusions and cell death in IBMPFD mutant expressing cells (24). The decrease in polyQ aggregate clearance was also found in IBMPFD mutant expressing transgenic mouse muscle (24,39).
Although autophagic proteins did not co-localize to the inclusion in IBMPFD mutant expressing cells, p97/VCP did localize to polyQ aggregates both at the aggresome in control and p97/VCP-WT cells and at the smaller non-aggresomal polyQ inclusions in IBMPFD mutant expressing cells (24). This raises the question; what is the role of p97/VCP on protein aggregates and how do IBMPFD mutants on these aggregates affect degradation. One possibility is that p97/VCP is ‘triaging’ an aggregated protein to the aggresome and autophagic pathways. Or, p97/VCP may be actively delivering aggregated proteins to the autophagosome. Alternatively, it is conceivable that p97/VCP is trapped within the polyQ inclusions in a futile attempt to disaggregate or deliver them to the UPS. To address this question and distinguish between the function of VCP-WT and the dysfunction of IBMPFD mutant VCP, fluorescently tagged p97/VCPs were co-expressed in polyQ expressing cells and visualized via live imaging (24). p97/VCP-WT protein was found to rim polyQ aggresomes (24). In addition, the aggregate-associated p97/VCP-WT protein was freely diffusible with cytosolic p97/VCP. This was in contrast to IBMPFD mutant p97/VCPs which associated with a polyQ inclusion but remained stuck on the aggregate (24). An analogous finding was seen via co-immunoprecipitation in which IBMPFD mutant VCPs bound more polyQ protein than VCP-WT. This was similar to what was seen when a hydrolysis-deficient p97/VCP was evaluated on a polyQ aggregate (24).
p97/VCP requires ATP binding at the D2 domain to engage with substrates and cofactors. ATP hydrolysis then provides the energy necessary for substrate disengagement (15). Therefore, point mutations in the Walker B motif of the D2 domain can generate a p97/VCP that binds substrate but fails to release; effectively serving as ‘substrate-trap’ mutant (18). These data suggest that IBMPFD mutants behave similarly to a hydrolysis-deficient mutant and bind substrate (e.g. protein aggregates) but fail to release or deliver them to the UPS, autophagy or aggresomal pathways. However, in vitro ATPase activity assays show that IBMPFD mutants are actually ‘hyperactive’ and have higher basal activity rates (30). This would be inconsistent with a ‘substrate-trap’ mutant that is unable to hydrolyze ATP. There are several plausible explanations for this discrepancy. For example, in vitro basal ATPase activity does not reflect the ‘stimulated’ ATPase activity that likely occurs with substrate engagement. Many AAA+ proteins have low basal ATPase activity that increases several fold with the addition of substrate and/or cofactors. A potentially suitable in vitro substrate for VCP has been recently identified and is a fragment of synaptotagmin-1 (40). This peptide stimulates the ATPase activity of VCP by 4-fold (40). Whether IBMPFD mutants have a similar stimulation remains to be determined. Another possibility is that IBMPFD mutations lead to a structural uncoupling of the D2 and N domains during the ATP hydrolysis cycle. This scenario could in fact result in a VCP protein with increased ATPase activity since the steric hindrance of the N domain would be removed. One could imagine that IBMPFD disease mutations within the linker region between the N and D1 domains or at the N–D1 domain interface would lead to a poorly moving N domain that could bind substrate but fail to release.
IBMPFD MUTANT p97/VCP IMPAIRS AUTOPHAGOSOME MATURATION
A defect in protein aggregate degradation raised the possibility that IBMPFD mutations in p97/VCP-disrupted autophagy. It has been postulated that protein aggregates such as expanded polyglutamine-containing proteins are resistant to the proteasome and are instead degraded via autophagy (41). Defects in autophagy are associated with several other vacuolar myopathies such as Pompe's and Danon's diseases (42,43). Mutations in the endosomal sorting complex required for transport (ESCRT) pathway protein CHMP2B cause an FTD similar to IBMPFD with prominent ubiquitinated inclusions (44). ESCRT family members including CHMP2B are necessary for autophagy (45,46). Finally, mutations in p62, a key autophagic adaptor protein, cause familial and sporadic PDB (47).
Consistent with a disruption in autophagy, IBMPFD patients and VCP mutant R155H expressing transgenic mouse muscle accumulate the autophagic substrates p62 and LC3II (39). The degree of p62 and LC3II accumulation is similar to that seen when autophagy is inhibited via chronic administration of the lysosomal inhibitor chloroquine. Using a cell culture model of IBMPFD mutant expression, it was found that this was due to the accumulation of immature autophagosomes (39). Subsequent studies found that VCP itself is necessary for autophagosome maturation into autolysosomes. Expression of a dominant-negative VCP mutant or siRNA knockdown of VCP also led to the accumulation of p62, LC3II and immature autophagosomes (39). These autophagosomes failed to co-localize with acidic- and LAMP1-positive vesicles (39). These data suggest that VCP is needed for autophagosome–lysosme fusion.
Recently, it has become clear that basal autophagy or ‘quality-control’ autophagy is particularly important for the degradation of ubiquitinated inclusions (48,49). This type of autophagy contrasts ‘induced’ autophagy which occurs in the setting of nutrient deprivation or other signaling event. The loss of basal autophagy in the CNS leads to the accumulation of ubiquitinated inclusions and neurodegeneration (48,49). The autophagy mediated by VCP may be important for basal autophagy. In IBMPFD mutant R155H and A232E expressing cells, immature autophagic structures accumulated under basal conditions and contained ubiquitin (34). This contrasted blocking autophagosome maturation with bafilomycin A in which few ubiquitin-positive autophagosomes were present (34). This finding is very intriguing and suggests that VCP may be particularly important in the selective autophagic degradation of ubiquitinated proteins.
Further supporting a role for VCP in the maturation of ubiquitin-containing autophagosomes comes from a recent study implicating HDAC6 in a similar process. The loss of HDAC6 in cultured cells resulted in the accumulation of immature autophagosomes under basal conditions (50). However, when autophagy was induced via starvation, this effect was not apparent and measures of protein degradation were unaffected in HDAC6 knockout cells (50). These data suggested that HDAC6 was dispensable for the induced autophagy seen with nutrient deprivation. This is similar to when IBMPFD mutant expressing cells were similarly treated with nutrient deprivation (34). These data highlight an emerging type of autophagy—quality-control autophagy. This model suggests that the autophagosomes that accumulate in IBMPFD cells and HDAC6 KO cells are specialized. They are specifically involved in the clearance of ubiquitinated proteins.
DEFECTS IN AUTOPHAGOSOME MATURATION EXPLAIN IBMPFD PATHOLOGY
An impairment in autophagosome maturation may explain the pathology seen in IBMPFD including TDP-43 accumulation. In IBMPFD skeletal muscle, the most striking pathology is the ‘RV’ (2). RVs are accumulations of discontinuous membranous and proteinaceous debris. In some cases, RVs contain amyloid, ubiquitin and TDP-43 immunoreactive debris (2). In IBMPFD patient and transgenic mouse skeletal muscle, many RVs are decorated by LC3 and p62, suggesting that they are autophagic in origin (Fig. 2A–C) (39). Interestingly, in contrast to other autophagic vacuolar myopathies such as acid maltase deficiency, Danon's disease or even other hereditary IBMs, the RVs in IBMPFD do not label with hydrolytic enzyme stains such as acid phosphatase and non-specific esterase, suggesting that they are not lysosomal in origin (2,51). Instead, an RV in IBMPFD transgenic mice and patients contains autophagosomal proteins such as p62 and LC3 that do not co-localize with lysosomal proteins such as LAMP1/2 (39). Other pathologies in IBMPFD muscle include the accumulation of p62, LC3II and high molecular weight ubiquitinated proteins; all of which accumulate under conditions of impaired autophagy (33,39).
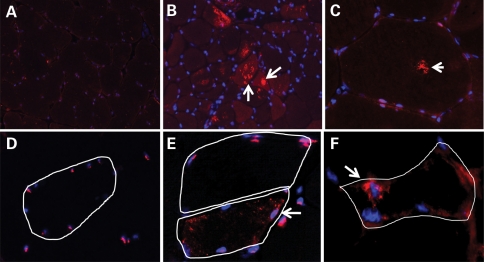
Figure 2.
IBMPFD muscle contains LC3-positive RVs and TDP-43 inclusions. (A) Normal patient muscle immunostained with an antibody against LC3 (red). Myonuclei are labeled blue. (B and C) Muscle from a patient with IBMPFD (R155H mutation in p97/VCP) immunostained with anti-LC3 (red) and myonuclei in blue. Note large LC3-positive structures in many muscle fibers and a large LC3-positive RV in a normal appearing myofiber. (D) Normal mouse muscle immunostained with an antibody to TDP-43. TDP-43 localizes to myonuclei in normal muscle. (E) Age-matched IBMPFD transgenic mouse muscle immunostained with anti-TDP-43. Note sarcoplasmic TDP-43 and loss from the nuclei. A normal fiber is adjacent which retains nuclear TDP-43 staining. (F) IBMPFD transgenic mouse muscle immunostained with an antibody to phosphorylated TDP-43. Phospho-TDP-43 is localized around muscle nuclei in an angular fiber.
Markers of autophagy have not been systematically evaluated in IBMPFD patient or transgenic mouse CNS tissue. However, it is clear that both ubiquitin- and TDP-43-positive inclusions accumulate in affected brain tissue (4,32,52). Several studies suggest that TDP-43 itself is an autophagic substrate that may accumulate under conditions of dysfunctioning autophagy. The evidence for this comes from cell culture experiments that have evaluated the degradation and/or accumulation of TDP-43 after pharmacologic manipulation of autophagy or the proteasome (53,54). An alternative hypothesis is that TDP-43 is leaving the nucleus and accumulating in the cytosol of affected neurons and muscle in response to impaired autophagy or changes in proteostasis. TDP-43 was found to redistribute to the cytosol in IBMPFD mutant expressing cells and transgenic mouse muscle (Fig. 2D–F) (39,55). This was similar to that seen when autophagosome maturation was impaired with bafilomycin A or chloroquine in cells and chronically treated mice (39). Although these studies do not confirm whether TDP-43 is an autophagic substrate, they do suggest that the TDP-43 pathology seen in IBMPFD can be explained by impaired autophagosome maturation.
Although current cellular and transgenic models of IBMFPD recapitulate most aspects of the disease, one hallmark feature seen in IBMPFD-associated FTLD-U has not been demonstrated. Specifically, intranuclear ubiquitinated and TDP-43-positive inclusions have not been seen in IBMPFD cell and animal models. These are the pathologic feature of IBMPFD-associated FTLD-U that resulted in its categorization as a unique subtype (4,52). It should be noted that intranuclear inclusions of TDP-43 are not present in IBMPFD patient skeletal muscle (3). Instead, TDP-43 is cleared from myonuclei in inclusion bearing muscle fibers. Moreover, vacuolation is not seen in IBMPFD patient neurons (4). This suggests that although the pathogenesis of IBMPFD dementia and muscle weakness may be similar, there are still distinct differences. It may be that a disruption in quality-control or basal autophagy is the principal defect in the CNS of IBMPFD patients, whereas in skeletal muscle the large accumulation of membranous and proteinaceous debris is pathogenic. Support for this comes from tissue-specific knockout of key autophagic proteins in the brain or skeletal muscle. The loss of ATG5 or ATG7 in the CNS leads to profound neurodegeneration after several weeks (48,49). In contrast, the loss of ATG5 or ATG7 in the skeletal muscle results in preserved muscle function for several months with evidence of type II muscle atrophy after 7 months (56,57). This discrepancy suggests that neurons and skeletal muscle have different requirements for basal autophagy, and the pathogenesis of IBMPFD may be different in these two tissues.
p97/VCP IS A MEDIATOR OF PROTEIN HOMEOSTASIS
The recent studies identifying a role for p97/VCP in autophagy and the UPS, place it at a unique position within these protein degradation pathways of the cell. Both UPS and autophagy are clearly important for intracellular protein homeostasis. The concerted or counterbalanced action of these two pathways has traditionally been thought to behave as parallel processes degrading distinct subsets of proteins within the cell. However, recent studies suggest that the UPS and autophagy pathways communicate and may degrade similar substrates. For example, proteasome inhibition or an excess of ubiquitinated misfolded proteins induces autophagy (58,59). Conversely, impaired autophagy results in the accumulation of ubiquitinated proteins and proteasome-specific substrates (48,60). Within an organism, mice bearing a skeletal muscle-specific conditional knockout allele of Atg5 or Atg7 show the loss of muscle mass and elevated levels of muscle-specific ubiquitin ligases (both atrogin-1 and MuRF1) (56,57). Similarly, CNS-specific knockout of Atg5 or Atg7 results in the accumulation of ubiquitinated proteins (48,49). This strongly suggests that the UPS and autophagy are functionally interrelated, but the compensatory or cross-talk mechanisms between the two pathways are largely unknown.
Several proteins with ubiquitin-association domains, HDAC6, p62, NBR1 and the FYVE domain containing protein Alfy, serve as adapters that traffic ubiquitinated proteins to autophagic machinery (61). p62 also has a role in the UPS (62). It can deliver ubiquitinated proteins to the proteasome and the autophagosome (62,63). In settings of impaired autophagy, the accumulation of p62 results in a sequestration of UPS substrates (60). This limits their proteasomal degradation. This proteasomal impairment can be overcome by the expression of p97/VCP (60). How p97/VCP does this is not clear. p97/VCP does not clearly bind to ubiquitinated proteins alone but instead indirectly associates with ubiquitinated substrates through cofactors such as Ufd1/Npl4 and p47 that contain ubiquitin-association domains. Whether these factors are necessary to overcome the secondary impairment in the UPS during autophagic inhibition is not known.
We suggest that p97/VCP is well-suited to mediate the interplay between these two proteolytic systems (Fig. 3). p97/VCP itself may be able to recognize a proteasomal (i.e. misfolded) versus autophagic (i.e. aggregated) substrate. p97/VCP would effectively triage these substrates to their respective degradation pathways. Alternatively, it may be that the availability of p97/VCP-specific cofactors in the cell dictates whether a substrate is degraded via the proteasome or autophagy. In this model, p97/VCP would serve as a facilitator and the cell would decide how a protein is degraded. For example, an increase in UFD1/Npl4's association with p97/VCP may favor proteasomal degradation, whereas higher levels of HDAC6 may favor degradation of substrates by autophagy. Finally, p97/VCP may be regulated and directed to associate with specific cofactors. p97/VCP is phosphorylated and acetylated at multiple residues (64). c-Src kinase, p34cdc2 kinase and Akt are examples of kinases that phosphorylate p97/VCP (65–67). In the case of Akt, it has been shown that the phosphorylation of specific serine residues on p97/VCP mediates its binding to ubiquitinated proteins (65). Depending upon the proteostatic state of the cell, p97/VCP may be post-translationally modified to favor association with different degradation pathway components.
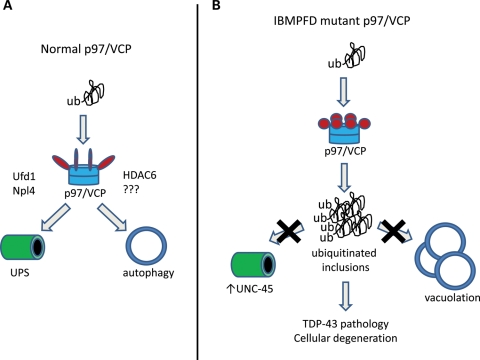
Figure 3.
p97/VCP triages ubiquitinated proteins to the UPS or autophagy pathways. (A) Normal p97/VCP associates with ubiquitinated proteins via interactions with cofactors such as UFD1 and Npl4 which favor proteasomal degradation. Alternatively, p97/VCP in association with HDAC6 facilitates the autophagic degradation of ubiquitinated proteins. (B) IBMPFD mutations in p97/VCP disrupt the degradation of ubiquitinated proteins via both the UPS and autophagy leading to inclusion formation. In the case of UPS-mediated protein degradation, specific p97/VCP substrates such as UNC-45b accumulate. Whereas disruptions in p97/VCP-mediated autophagy results in the accumulation of non-degradative autophagosomes and vacuolation. The impairment in cellular proteostasis results in TDP-43 redistribution from the nucleus to the cytoplasm and subsequent cellular degeneration.
CONCLUSION
p97/VCP is a pivotal regulatory molecule necessary for multiple key biological processes. This review highlighted an additional function for p97/VCP in the intracellular catabolic pathways, besides its well-established role in protein degradation via the UPS. The mechanism by which p97/VCP mediates the autophagic degradation of ubiquitinated substrates is currently unknown. It is conceivable that one may be able to manipulate p97/VCP and shift the degradation of substrates from the UPS to autophagy or vice versa. Thus, p97/VCP may serve to balance the proteostatic state of the cell.
Conflict of Interest statement. None declared.
FUNDING
C.C.W. is funded by the Muscular Dystrophy Association and the National Institutes of Health grants R01AG031867 and K08AG026271.
REFERENCES
- 1.Watts G.D., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., Kimonis V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. doi:10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 2.Weihl C.C., Pestronk A., Kimonis V.E. Valosin-containing protein disease: inclusion body myopathy with Paget's disease of the bone and fronto-temporal dementia. Neuromuscul. Disord. 2009;19:308–315. doi: 10.1016/j.nmd.2009.01.009. doi:10.1016/j.nmd.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weihl C.C., Temiz P., Miller S.E., Watts G., Smith C., Forman M., Hanson P.I., Kimonis V., Pestronk A. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry. 2008;79:1186–1189. doi: 10.1136/jnnp.2007.131334. doi:10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forman M.S., Mackenzie I.R., Cairns N.J., Swanson E., Boyer P.J., Drachman D.A., Jhaveri B.S., Karlawish J.H., Pestronk A., Smith T.W., et al. Novel ubiquitin neuropathology in frontotemporal dementia with valosin-containing protein gene mutations. J. Neuropathol. Exp. Neurol. 2006;65:571–581. doi: 10.1097/00005072-200606000-00005. doi:10.1097/00005072-200606000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Neumann M., Kwong L.K., Truax A.C., Vanmassenhove B., Kretzschmar H.A., Van Deerlin V.M., Clark C.M., Grossman M., Miller B.L., Trojanowski J.Q., et al. TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J. Neuropathol. Exp. Neurol. 2007;66:177–183. doi: 10.1097/01.jnen.0000248554.45456.58. doi:10.1097/01.jnen.0000248554.45456.58. [DOI] [PubMed] [Google Scholar]
- 6.Kimonis V.E., Fulchiero E., Vesa J., Watts G. VCP disease associated with myopathy, Paget disease of bone and frontotemporal dementia: review of a unique disorder. Biochim. Biophys. Acta. 2008;1782:744–748. doi: 10.1016/j.bbadis.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Halawani D., Latterich M. p97: the cell's molecular purgatory? Mol. Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. doi:10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Muller J.M., Deinhardt K., Rosewell I., Warren G., Shima D.T. Targeted deletion of p97 (VCP/CDC48) in mouse results in early embryonic lethality. Biochem. Biophys. Res. Commun. 2007;354:459–465. doi: 10.1016/j.bbrc.2006.12.206. doi:10.1016/j.bbrc.2006.12.206. [DOI] [PubMed] [Google Scholar]
- 9.Yamanaka K., Okubo Y., Suzaki T., Ogura T. Analysis of the two p97/VCP/Cdc48p proteins of Caenorhabditis elegans and their suppression of polyglutamine-induced protein aggregation. J. Struct. Biol. 2004;146:242–250. doi: 10.1016/j.jsb.2003.11.017. doi:10.1016/j.jsb.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 10.Madeo F., Schlauer J., Frohlich K.U. Identification of the regions of porcine VCP preventing its function in Saccharomyces cerevisiae. Gene. 1997;204:145–151. doi: 10.1016/s0378-1119(97)00535-0. doi:10.1016/S0378-1119(97)00535-0. [DOI] [PubMed] [Google Scholar]
- 11.Pye V.E., Beuron F., Keetch C.A., McKeown C., Robinson C.V., Meyer H.H., Zhang X., Freemont P.S. Structural insights into the p97-Ufd1-Npl4 complex. Proc. Natl Acad. Sci. USA. 2007;104:467–472. doi: 10.1073/pnas.0603408104. doi:10.1073/pnas.0603408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q., Song C., Li C.C. Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem. Biophys. Res. Commun. 2003;300:253–260. doi: 10.1016/s0006-291x(02)02840-1. doi:10.1016/S0006-291X(02)02840-1. [DOI] [PubMed] [Google Scholar]
- 13.Pye V.E., Dreveny I., Briggs L.C., Sands C., Beuron F., Zhang X., Freemont P.S. Going through the motions: the ATPase cycle of p97. J. Struct. Biol. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Wang Q., Song C., Yang X., Li C.C. D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J. Biol. Chem. 2003;278:32784–32793. doi: 10.1074/jbc.M303869200. doi:10.1074/jbc.M303869200. [DOI] [PubMed] [Google Scholar]
- 15.Dalal S., Hanson P.I. Membrane traffic: what drives the AAA motor? Cell. 2001;104:5–8. doi: 10.1016/s0092-8674(01)00186-6. doi:10.1016/S0092-8674(01)00186-6. [DOI] [PubMed] [Google Scholar]
- 16.Dreveny I., Pye V.E., Beuron F., Briggs L.C., Isaacson R.L., Matthews S.J., McKeown C., Yuan X., Zhang X., Freemont P.S. p97 and close encounters of every kind: a brief review. Biochem. Soc. Trans. 2004;32:715–720. doi: 10.1042/BST0320715. [DOI] [PubMed] [Google Scholar]
- 17.Wojcik C., Yano M., DeMartino G.N. RNA interference of valosin-containing protein (VCP/p97) reveals multiple cellular roles linked to ubiquitin/proteasome-dependent proteolysis. J. Cell. Sci. 2004;117:281–292. doi: 10.1242/jcs.00841. doi:10.1242/jcs.00841. [DOI] [PubMed] [Google Scholar]
- 18.Dalal S., Rosser M.F., Cyr D.M., Hanson P.I. Distinct roles for the AAA ATPases NSF and p97 in the secretory pathway. Mol. Biol. Cell. 2004;15:637–648. doi: 10.1091/mbc.E03-02-0097. doi:10.1091/mbc.E03-02-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rabinovich E., Kerem A., Frohlich K.U., Diamant N., Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. doi:10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Y., Meyer H.H., Rapoport T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. doi:10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 21.Alexandru G., Graumann J., Smith G.T., Kolawa N.J., Fang R., Deshaies R.J. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. doi:10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janiesch P.C., Kim J., Mouysset J., Barikbin R., Lochmuller H., Cassata G., Krause S., Hoppe T. The ubiquitin-selective chaperone CDC-48/p97 links myosin assembly to human myopathy. Nat. Cell. Biol. 2007;9:379–390. doi: 10.1038/ncb1554. doi:10.1038/ncb1554. [DOI] [PubMed] [Google Scholar]
- 23.Dai R.M., Chen E., Longo D.L., Gorbea C.M., Li C.C. Involvement of valosin-containing protein, an ATPase Co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J. Biol. Chem. 1998;273:3562–3573. doi: 10.1074/jbc.273.6.3562. doi:10.1074/jbc.273.6.3562. [DOI] [PubMed] [Google Scholar]
- 24.Ju J.S., Miller S.E., Hanson P.I., Weihl C.C. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP associated disease. J. Biol. Chem. 2008;283:30289–30299. doi: 10.1074/jbc.M805517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuno Y., Hori S., Kakizuka A., Okamoto K. Vacuole-creating protein in neurodegenerative diseases in humans. Neurosci. Lett. 2003;343:77–80. doi: 10.1016/s0304-3940(03)00280-5. doi:10.1016/S0304-3940(03)00280-5. [DOI] [PubMed] [Google Scholar]
- 26.Hirabayashi M., Inoue K., Tanaka K., Nakadate K., Ohsawa Y., Kamei Y., Popiel A.H., Sinohara A., Iwamatsu A., Kimura Y., et al. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ. 2001;8:977–984. doi: 10.1038/sj.cdd.4400907. doi:10.1038/sj.cdd.4400907. [DOI] [PubMed] [Google Scholar]
- 27.Kumar K.R., Needham M., Mina K., Davis M., Brewer J., Staples C., Ng K., Sue C.M., Mastaglia F.L. Two Australian families with inclusion-body myopathy, Paget's disease of bone and frontotemporal dementia: novel clinical and genetic findings. Neuromuscul. Disord. 2010;20:330–334. doi: 10.1016/j.nmd.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Stojkovic T., Hammouda el H., Richard P., Lopez de Munain A., Ruiz-Martinez J., Gonzalez P.C., Laforet P., Penisson-Besnier I., Ferrer X., Lacour A., et al. Clinical outcome in 19 French and Spanish patients with valosin-containing protein myopathy associated with Paget's disease of bone and frontotemporal dementia. Neuromuscul. Disord. 2009;19:316–323. doi: 10.1016/j.nmd.2009.02.012. doi:10.1016/j.nmd.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 29.Weihl C.C., Dalal S., Pestronk A., Hanson P.I. Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Hum. Mol. Genet. 2006;15:189–199. doi: 10.1093/hmg/ddi426. doi:10.1093/hmg/ddi426. [DOI] [PubMed] [Google Scholar]
- 30.Halawani D., LeBlanc A.C., Rouiller I., Michnick S.W., Servant M.J., Latterich M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Mol. Cell. Biol. 2009;29:4484–4494. doi: 10.1128/MCB.00252-09. doi:10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hubbers C.U., Clemen C.S., Kesper K., Boddrich A., Hofmann A., Kamarainen O., Tolksdorf K., Stumpf M., Reichelt J., Roth U., et al. Pathological consequences of VCP mutations on human striated muscle. Brain. 2007;130:381–393. doi: 10.1093/brain/awl238. doi:10.1093/brain/awl238. [DOI] [PubMed] [Google Scholar]
- 32.Custer S.K., Neumann M., Lu H., Wright A.C., Taylor J.P. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum. Mol. Genet. 2010;19:1741–1755. doi: 10.1093/hmg/ddq050. [DOI] [PubMed] [Google Scholar]
- 33.Weihl C.C., Miller S.E., Hanson P.I., Pestronk A. Transgenic expression of inclusion body myopathy associated mutant p97/VCP causes weakness and ubiquitinated protein inclusions in mice. Hum. Mol. Genet. 2007;16:919–928. doi: 10.1093/hmg/ddm037. doi:10.1093/hmg/ddm037. [DOI] [PubMed] [Google Scholar]
- 34.Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.P., Dantuma N.P., Taylor J.P. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6:1–11. doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyault C., Gilquin B., Zhang Y., Rybin V., Garman E., Meyer-Klaucke W., Matthias P., Muller C.W., Khochbin S. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. doi:10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. doi:10.1016/S0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 37.Johnston J.A., Ward C.L., Kopito R.R. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1998;143:1883–1898. doi: 10.1083/jcb.143.7.1883. doi:10.1083/jcb.143.7.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kobayashi T., Manno A., Kakizuka A. Involvement of valosin-containing protein (VCP)/p97 in the formation and clearance of abnormal protein aggregates. Genes Cells. 2007;12:889–901. doi: 10.1111/j.1365-2443.2007.01099.x. doi:10.1111/j.1365-2443.2007.01099.x. [DOI] [PubMed] [Google Scholar]
- 39.Ju J.S., Fuentealba R.A., Miller S.E., Jackson E., Piwnica-Worms D., Baloh R.H., Weihl C.C. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. doi:10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLaBarre B., Christianson J.C., Kopito R.R., Brunger A.T. Central pore residues mediate the p97/VCP activity required for ERAD. Mol. Cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. doi:10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Rubinsztein D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. doi:10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- 42.Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J.E., Oh S.J., Koga Y., et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. doi:10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- 43.Nishino I. Autophagic vacuolar myopathies. Curr. Neurol. Neurosci. Rep. 2003;3:64–69. doi: 10.1007/s11910-003-0040-y. doi:10.1007/s11910-003-0040-y. [DOI] [PubMed] [Google Scholar]
- 44.Skibinski G., Parkinson N.J., Brown J.M., Chakrabarti L., Lloyd S.L., Hummerich H., Nielsen J.E., Hodges J.R., Spillantini M.G., Thusgaard T., et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat. Genet. 2005;37:806–808. doi: 10.1038/ng1609. doi:10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 45.Filimonenko M., Stuffers S., Raiborg C., Yamamoto A., Malerod L., Fisher E.M., Isaacs A., Brech A., Stenmark H., Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. doi:10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J.A., Beigneux A., Ahmad S.T., Young S.G., Gao F.B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. doi:10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 47.Morissette J., Laurin N., Brown J.P. Sequestosome 1: mutation frequencies, haplotypes, and phenotypes in familial Paget's disease of bone. J. Bone Miner. Res. 2006;21(Suppl. 2):P38–P44. doi: 10.1359/jbmr.06s207. [DOI] [PubMed] [Google Scholar]
- 48.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. doi:10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 49.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 50.Lee J.Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y.S., Pandey U.B., Kaushik S., Tresse E., Lu J., et al. HDAC6 controls autophagosome maturation essential forubiquitin-selective quality-control autophagy. EMBO J. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malicdan M.C., Noguchi S., Nonaka I., Saftig P., Nishino I. Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscul. Disord. 2008;18:521–529. doi: 10.1016/j.nmd.2008.04.010. doi:10.1016/j.nmd.2008.04.010. [DOI] [PubMed] [Google Scholar]
- 52.Neumann M., Mackenzie I.R., Cairns N.J., Boyer P.J., Markesbery W.R., Smith C.D., Taylor J.P., Kretzschmar H.A., Kimonis V.E., Forman M.S. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J. Neuropathol. Exp. Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. doi:10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 53.Wang X., Fan H., Ying Z., Li B., Wang H., Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci. Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. doi:10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 54.Caccamo A., Majumder S., Deng J.J., Bai Y., Thornton F.B., Oddo S. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J. Biol. Chem. 2009;284:27416–27424. doi: 10.1074/jbc.M109.031278. doi:10.1074/jbc.M109.031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gitcho M.A., Strider J., Carter D., Taylor-Reinwald L., Forman M.S., Goate A.M., Cairns N.J. VCP mutations causing frontotemporal lobar degeneration disrupt localization of TDP-43 and induce cell death. J. Biol. Chem. 2009;284:12384–12398. doi: 10.1074/jbc.M900992200. doi:10.1074/jbc.M900992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Masiero E., Agatea L., Mammucari C., Blaauw B., Loro E., Komatsu M., Metzger D., Reggiani C., Schiaffino S., Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. doi:10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 57.Raben N., Hill V., Shea L., Takikita S., Baum R., Mizushima N., Ralston E., Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum. Mol. Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. doi:10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwata A., Riley B.E., Johnston J.A., Kopito R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. doi:10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 59.Tannous P., Zhu H., Johnstone J.L., Shelton J.M., Rajasekaran N.S., Benjamin I.J., Nguyen L., Gerard R.D., Levine B., Rothermel B.A., et al. Autophagy is an adaptive response in desmin-related cardiomyopathy. Proc. Natl Acad. Sci. USA. 2008;105:9745–9750. doi: 10.1073/pnas.0706802105. doi:10.1073/pnas.0706802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Korolchuk V.I., Mansilla A., Menzies F.M., Rubinsztein D.C. Autophagy inhibition compromises degradation of ubiquitin-proteasome pathway substrates. Mol. Cell. 2009;33:517–527. doi: 10.1016/j.molcel.2009.01.021. doi:10.1016/j.molcel.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirkin V., McEwan D.G., Novak I., Dikic I. A role for ubiquitin in selective autophagy. Mol. Cell. 2009;34:259–269. doi: 10.1016/j.molcel.2009.04.026. doi:10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 62.Babu J.R., Geetha T., Wooten M.W. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J. Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. doi:10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 63.Ichimura Y., Kumanomidou T., Sou Y.S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. doi:10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 64.Mori-Konya C., Kato N., Maeda R., Yasuda K., Higashimae N., Noguchi M., Koike M., Kimura Y., Ohizumi H., Hori S., et al. p97/valosin-containing protein (VCP) is highly modulated by phosphorylation and acetylation. Genes Cells. 2009;14:483–497. doi: 10.1111/j.1365-2443.2009.01286.x. doi:10.1111/j.1365-2443.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 65.Klein J.B., Barati M.T., Wu R., Gozal D., Sachleben L.R., Jr, Kausar H., Trent J.O., Gozal E., Rane M.J. Akt-mediated valosin-containing protein 97 phosphorylation regulates its association with ubiquitinated proteins. J. Biol. Chem. 2005;280:31870–31881. doi: 10.1074/jbc.M501802200. doi:10.1074/jbc.M501802200. [DOI] [PubMed] [Google Scholar]
- 66.Marti F., King P.D. The p95-100 kDa ligand of the T cell-specific adaptor (TSAd) protein Src-homology-2 (SH2) domain implicated in TSAd nuclear import is p97 Valosin-containing protein (VCP) Immunol. Lett. 2005;97:235–243. doi: 10.1016/j.imlet.2004.10.021. doi:10.1016/j.imlet.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 67.Mayr P.S., Allan V.J., Woodman P.G. Phosphorylation of p97(VCP) and p47 in vitro by p34cdc2 kinase. Eur. J. Cell Biol. 1999;78:224–232. doi: 10.1016/S0171-9335(99)80055-7. [DOI] [PubMed] [Google Scholar]