Abstract
Introduction
Dermatomyositis (DM) is an autoimmune disease involving muscle and skin. Perifascicular atrophy (PFA) of myofibers is a specific and characteristic DM pathological lesion. Interferon-stimulated gene 15 (ISG15) is a ubiquitin-like modifier with a poorly understood immunological role.
Methods
We generated microarray data measuring the expression of approximately 18,000 genes in each of 113 human muscle biopsy specimens. Biopsy specimens and cultured skeletal muscle were further studied using immunohistochemistry, immunoblotting, proteomic profiling by liquid chromatography/mass spectrometry, real-time quantitative PCR, and laser capture microdissection.
Results
Transcripts encoding ISG15-conjugation pathway proteins were upregulated in DM with PFA (DM-PFA) muscle, with marked elevation of ISG15 (339-fold), HERC5 (62-fold), and USP18 (68-fold) present in all DM-PFA patients but none of 99 non-DM samples. Combined analysis with publicly available microarray datasets further showed marked ISG15 and USP18 transcript elevation had 100% sensitivity and specificity for 28 biopsies from adult DM-PFA and juvenile DM compared to 199 other muscle samples from a wide range of muscle diseases. Free ISG15 and ISG15-conjugated proteins were found by immunoblot only in DM-PFA muscle. Cultured human skeletal muscle exposed to type 1 interferons produced similar transcripts and both ISG15 protein and ISG15 conjugates. Laser capture microdissection followed by proteomic analysis showed deficiency of titin in DM perifascicular atrophic myofibers.
Conclusion
A large-scale microarray study of muscle samples from a diverse collection of muscle diseases revealed that the autoimmune disease dermatomyositis was uniquely associated with overactivation of the ISG15 conjugation pathway. Exposure of human skeletal muscle cell culture to type 1 interferons produces a molecular picture highly similar to that of human DM muscle biopsy specimens. Perifascicular atrophic myofibers in DM are deficient in a number of skeletal muscle proteins, most markedly titin.
Dermatomyositis (DM) is an autoimmune disease of unknown cause primarily affecting skeletal muscle and skin. The characteristic muscle pathology of DM is notable for capillary abnormalities and small, abnormal appearing muscle fibers around the periphery of some muscle fascicles bordering the perimysial connective tissue, a lesion called perifascicular atrophy (PFA). Recent microarray and pathological studies have pointed towards a mechanism of tissue injury in DM associated with the overexpression of type 1 interferon-inducible genes. Over 85% of the highest expressed transcripts in muscle from DM compared to normal and other inflammatory myopathies are from genes known to be induced by type 1 interferons.1 Plasmacytoid dendritic cells (pDCs), cells that produce high levels of type 1 interferons, have infiltrated DM muscle1 and skin.2 Expression of the type 1 interferon-inducible protein MxA occurs in the characteristic sites of DM pathology, the perifascicular myofibers and capillaries. These findings suggest that injury to capillaries and myofibers may result directly from the intracellular overproduction of one or more type 1 interferon-inducible transcripts or proteins.
Interferon-stimulated gene 15 (ISG15), a type 1 interferon-inducible protein, is an ubiquitin-like modifier with poorly understood function. The enzymatic pathway for ISG15 conjugation to a target protein involves three conjugating enzymes, the E1 Ube1L,3 the E2 Ube2L6,4 and the E3 HERC5,5, 6 and a deconjugating enzyme USP18.7 Immunoprecipitates of ISG15 from interferon-β stimulated HeLa cells contain many putative ISG15-conjugated proteins.8 The conjugation of ISG15 to proteins in a human tissue sample has only been reported in 3 endometrial and 2 colon cancer biopsy specimens;9 the identities of any such proteins are unknown.
We previously reported that ISG15 is the single most overexpressed gene in DM muscle compared to both normal muscle and muscle from patients with other inflammatory myopathies.1 It is also highly expressed in circulating mononuclear blood cells of patients with DM.10 In this study, we have examined the ISG15 pathway in DM and other inflammatory myopathy muscle biopsy specimens and in cultured human skeletal muscle exposed to type 1 interferons.
Methods
Patients and Tissue Samples
We studied 113 patients with a variety of muscle diseases, of which 76 had immune-mediated inflammatory myopathies, 26 had non-inflammatory myopathies, and 11 had no evidence of a neuromuscular disease. Of the 76 patients with immune-mediated inflammatory myopathies, 14 had dermatomyositis, 24 inclusion body myositis (IBM), and 38 had polymyositis (PM), non-specific myositis, or necrotizing myopathy. Diagnostic criteria were used as previously described.11 Muscle tissue was obtained at the time of a diagnostic biopsy and immediately frozen for RNA, protein, and immunohistochemical studies. Institutional review boards approved these studies.
Cell culture
Human skeletal muscle cells (ScienCell Research Laboratories cat #3500) were expanded and studied at 2nd or 3rd passage. Cells were plated in 96-well microtiter plates and proliferated for 3 days until approximately 75% confluent, at which time media was changed to 2% FBS. Cells then differentiated into multinucleated myotubes. Cells were treated after initial plating with human IFN-α or IFN-β 1U/ml or 10U/ml (PBL Biomedical, Inc.) every other day with media changes for 13 days.
Transcript measurements: human muscle biopsy samples
RNA extraction and microarray experiments were performed on 113 human muscle biopsy samples as previously described using Affymetrix HU-133A arrays representing approximately 18,000 genes.1 Gene expression levels were calculated using GC-Content Robust Multichip Analysis (GCRMA).12 Combined analyses with publicly available Affymetrix HU-133A datasets were performed after normalizing all raw data CEL files together, again using GC-RMA.
Transcript measurements: human skeletal muscle cell culture
Human skeletal muscle cultured cells underwent total RNA extraction using the Zymo Research Mini RNA Isolation I kit (Orange, CA). RNA purity and concentration were determined spectrophotometrically (260/280>1.9). RNA quality was assessed on an Agilent 2100 Bioanalyzer using the RNA 6000 Nano LabChip. The generation of single-stranded cDNA from 15 ng of total RNA was accomplished with the Invitrogen SuperScript First-Strand Synthesis SuperMix kit (Carlsbad, CA).
Microarray studies were performed in duplicate with Affymetrix U2.0 whole genome arrays and gene expression levels were calculated using GC-Content Robust Multichip Analysis (GCRMA).12 The Fluidigm Biomark system was used for high-throughput real-time PCR. A mixture of 48 TaqMan Gene Expression assays (Applied Biosystems), including ISG15, HERC5, and USP18, was prepared for use with the TaqMan PreAmp Master Mix Kit (Applied Biosystems). All samples were pre-amplified for 10-cycles of PCR according to the manufacturer’s directions.
For real-time PCR, all samples were run in triplicate against a set of 48 TaqMan Gene Expression Assays in BioMark 48.48 Dynamic Array chips (Fluidigm Corp). The untreated (0-hr.) controls were included on every array. Dynamic arrays were loaded using a NanoFlex 4-IFC Controller (Fluidigm Corp), and real-time reactions were performed and analyzed using BioMark Real-Time PCR System and Analysis software (Fluidigm Corp), respectively. Cts above 30 were excluded from the calculation. Delta-delta Cts (ΔΔCt) were calculated using the mean of the 2 reference genes (ACTB and UBC) and a calibrator sample and were converted to fold expression change by the following formula: 2−ΔΔCt. ABI Taqman assays, used in these experiments, have been optimized to 100% +/− 10% efficiencies and the ΔΔCt method correlates best with fold changes according to the manufacturer (Applied Biosystems document cms_040377.pdf).
Immunohistochemistry and immunofluorescence microscopy
Immunohistochemical studies were performed on 20 biopsy specimens (5 each with DM, IBM, PM, and normal). Primary antibodies were directed against: MxA (mouse monoclonal clone M143, isotype IgG1, 1:1000 dilution, 12.5 hours incubation at room temperature; provided by Dr. Otto Haller, Department of Virology, University of Freiburg, Freiburg, Germany); interferon-gamma inducible protein IFI-16 (mouse monoclonal antibody clone 1G7, isotype IgG1, 1:50 dilution, 4 µg/ml, 80 minutes incubation; sc8023 from Santa Cruz Biotechnology, Inc., Santa Cruz, CA); ISG15 (rabbit polyclonal antibody l:50 dilution, 10 µg/ml or 1:250 dilution, 2 µg/ml, 80 minutes incubation; ab14374 from Abcam, Inc., Cambridge, MA); and CD31 (monoclonal clone JC/70A, isotype IgG1, 1:25 dilution, 70 minutes incubation, Dako, Carpinteria, CA). A Zeiss Axioimager with an Apotome optical sectioning device and Axiovision software (Carl Zeiss Inc., Oberkochen, Germany) was used for microscopy and imaging. Tissue processing, secondary antibodies, immunofluorescence studies, and controls are further described in Supplementary Methods.
Human muscle biopsy and cell culture ISG15 and MxA immunoblots
Immunoblots for ISG15 were performed on 23 biopsy samples (7 with DM-PFA; 4 each with DM-NO-PFA, IBM, PM, and normal) and immunoblots of MxA were performed on 9 biopsy samples (6 with DM-PFA; 3 normal). Whole muscle lysates (WML) and cultured skeletal muscle P1S1 fractions were prepared and immunoblots performed as described in Supplementary Methods. For ISG15 positive control, 0.25 µg of recombinant human ISG15 (Cat# UL-601, BostonBiochem, Cambridge, MA), diluted in 1 × LDS loading buffer and 10 mM DTT, was run in one of the lanes on the gel along with the muscle samples. In separate experiments, cross-reactivity of the anti-ISG15 antibody against ubiquitin was excluded through blots containing ISG15 and ubiquitin (4 × the molar amount of ISG15) in separate lanes (220 ng and 110 ng respectively). Staining with anti-ISG15 antibody following the same procedure and dilutions described in Supplementary Methods resulted in strong reactivity against ISG15 and no staining against ubiquitin.
Ex vivo stimulation of whole blood (WB) with cytokines
Ex vivo stimulation was conducted on WB collected from either 3 or 4 (depending on the specific cytokine experiment) healthy donors (MedImmune, LLC.) as previously described.13 WB samples were exposed to vehicle (1× PBS) control, or to IFN-α (350 IU/ml), IFN-β (600 IU/ml), IFN-γ (3 ng/ml), GM-CSF (240 pg/ml), IL-10 (6 ng/ml), IL-13 (3 ng/ml), IL-1β (30 pg/ml), or TNF-α (100 pg/ml), all at concentrations of 3×EC50. The cytokines were purchased from R&D Systems (Minneapolis, MN). Following dosing, the WB was incubated at 37°C, 5% CO2 for 4 hours, transferred to PAXgene RNA tubes, inverted 8 to 10 times, incubated at room temperature for 2 hours, and then frozen until processed.
Proteomic profiling of muscle biopsy samples
Large-scale quantification of proteins present in 4 DM muscle homogenates was performed using liquid chromatography and tandem mass spectrometry (LC/MS) as previously described.14
Laser capture microdissection for isolation of dermatomyositis and normal muscle perifascicular and central-fascicular myofibers
Muscle was cut into 7-µm sections using a Leica microtome and placed on a non-plus glass slide. HistoGene kit (Arcturus # KIT0401, Mountain View, CA) was used for staining as per manufacturer’s protocol. A Veritas microdissection system (Arcturus) and CapSure HS LCM caps (Arcturus # LCM0214) were used for separate laser capture microdissection of perifascicular and central-fascicular regions of muscle. 30 µl of extraction buffer (100 mM NH4HCO3; 1% SDS, 8 M urea, 10 mM DTT) was added to each cap and the resulting solution incubated for 2 hours at 37°C. The extracted proteins were removed by centrifugation at 5000 × g for 5 minutes. BCA assay (Pierce # 23225, Rockford IL) was used to determine protein concentration and the samples were stored at −80°C. LC/MS shotgun proteomics and data analysis were performed on laser captured muscle protein preparations as previously described.14
Results
Marked increased gene expression of the ISG15 conjugation pathway is specific to dermatomyositis muscle among all muscle diseases
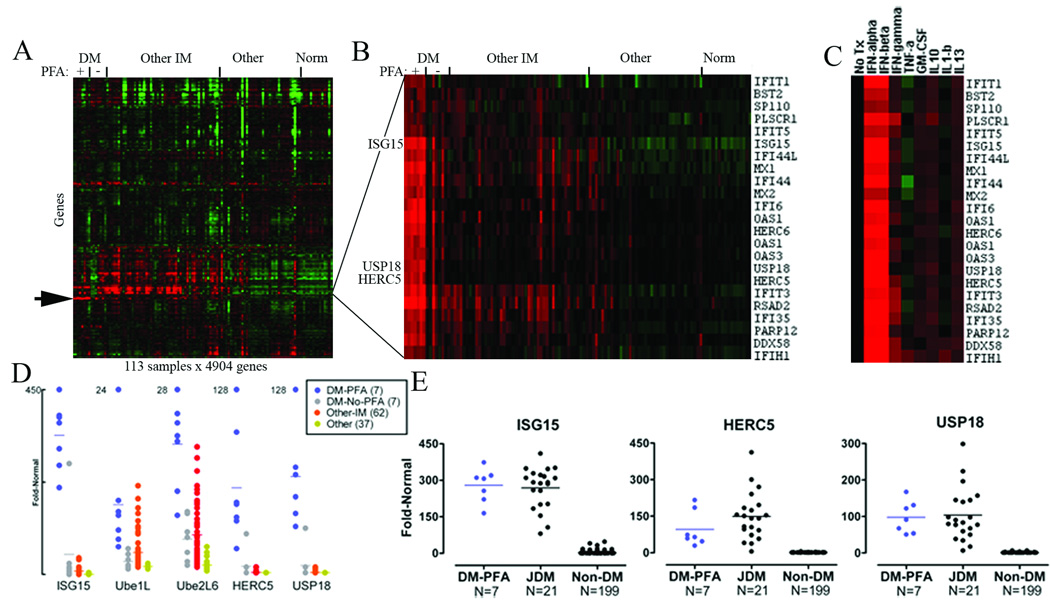
Analysis of microarray data for 22,283 transcript probesets for each of 113 muscle samples disclosed a cluster of highly upregulated type 1 interferon-inducible transcripts specifically in DM muscle that were not seen in other subtypes of inflammatory myopathy, including inclusion body myositis and polymyositis (Figure 1A,B). Within this cluster, 3 members of the ISG15 conjugation pathway were present. ISG15 was the second single highest differentially expressed gene in DM muscle compared to normal, increased by 194-fold compared to an 8-fold increase in other inflammatory myopathies. HERC5 and USP18 expression were elevated 34-fold and 37-fold in DM compared to 1.2-fold and 1.3-fold in other inflammatory myopathies.
Figure 1.
A marked increase in ISG15 transcript levels in DM was highly correlated to the presence of perifascicular atrophy (PFA) on hematoxylin and eosin stained sections. ISG15 transcript levels were significantly different for DM patients with (N=7) and without (N=7) perifascicular atrophy (p=0.0001). We compared transcript abundance for the major known members of the ISG15 conjugation pathway (ISG15; the conjugating enzymes Ube1L, Ube2L6, HERC5; and the deconjugating enzyme USP18) among muscle biopsy samples (Table). Of 14 DM patient biopsy samples, all 7 with PFA and 1 without PFA had ISG15, HERC5, and USP18 transcript levels exceeding those found in all 105 other muscle samples examined (Figure 1D). For patients with PFA, the mean fold-ratios compared to normal were 339-fold for ISG15, 62-fold for HERC5, and 68-fold for USP18. Transcript levels for Ube1L and Ube2L6 were less specifically elevated in DM-PFA.
Table 1.
Fold-ratio compared to normal for microarray measured ISG15 pathway transcripts in dermatomyositis muscle with and without PFA (DM-PFA N=7 and DM-NO-PFA N=7) compared to other inflammatory myopathies (OtherIM; N=62), both compared to normal muscle (N=12). ISG15 pathway transcript elevation for ISG15, HERC5, and USP18 are relatively specific to DM and highly specific to patients with DM and the presence of perifascicular atrophy (PFA) present on H&E stained muscle sections.
| Transcript | Ubiquitin-like (Ubl) role |
DM-PFA Fold |
DM-NO- PFA |
OtherIM- Fold |
p-value All DM vs Other IM |
p-value DM-PFA vs DM-NO-PFA |
|---|---|---|---|---|---|---|
| ISG15 | Ub-like modifier | 339 | 49/13* | 8 | 0.001 | 0.0001 |
| Ube1L | E1 enzyme | 9 | 2/2* | 3 | 0.160 | 0.0317 |
| Ube2L6 | E2 enzyme | 21 | 6/6* | 6 | 0.018 | 0.0007 |
| HERC5 | E3 enzyme | 62 | 6/1.7* | 1.2 | 0.010 | 0.0093 |
| USP18 | De-Ubl enzyme | 68 | 6/1.7* | 1.3 | 0.005 | 0.0013 |
1 of 7 DM-NO-PFA samples had very high ISG15, HERC5, and USP18 transcript levels as shown in Figure 1D; the mean fold-ratios for the remaining 6 DM-NO-PFA samples is indicated by *.
We compared our findings with all publicly available microarray datasets in The National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database. Two datasets from other investigators contained DM and normal muscle data (GEO accession numbers GDS1956 and GDS2153). Analysis of these data with respect to the ISG15 conjugation pathway has not been published.15, 16 We found that ISG15 was the 2nd most abundantly differentially expressed transcript among 22,283 probesets in both datasets, one with 21 juvenile DM (JDM) samples compared to 100 other biopsy samples from 13 different disease categories representing a wide range of neuromuscular disorders and the other with 5 adult DM samples compared to 4 normal muscle samples. All 21 JDM samples had higher ISG15 and USP18 transcript levels than present in any of the 100 other samples, while 20 of the 21 JDM samples had higher HERC5 transcript levels than any of the 100 other samples. Combining our data with these data, every ISG15, HERC5, and USP18 transcript level in all 7 adult DM-PFA samples was higher than any other ISG15, HERC5, or USP18 level among 199 non-DM biopsy samples (Figure 1E).
Type 1 interferons but not other cytokines examined cause this pattern of transcript expression in blood cells
Many of the transcripts highly elevated in DM muscle are well characterized as type 1 interferon inducible. We further addressed potential causes of their induction by exposing human blood mononuclear cells to a range of cytokines including IFN-α, IFN-β, IFN-γ, TNF-α, GM-CSF, IL-10, IL-1β, and IL-13. Only the type 1 interferons (IFN-α and IFN-β) resulted in marked upregulation of the same cluster of transcripts that we found to be highly elevated in DM biopsy samples (Figure 1C).
ISG15 protein and ISG15-conjugated proteins are present in DM-PFA muscle
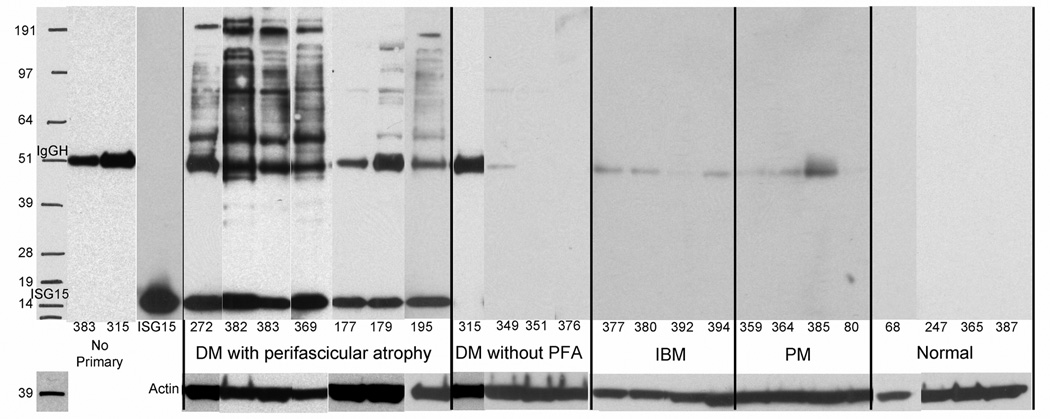
Whole muscle lysates were fractionated by gel electrophoresis, transferred to nitrocellulose membranes, and probed with anti-ISG15 antibody, or in control experiments in which primary antibody was omitted or replaced with anti-actin antibody, for 23 patient samples (DM-PFA N=7; DM-NO-PFA N=4; IBM N=4; PM N=4, Normal N=4). All 7 patient DM-PFA samples, but none of the other 16 samples, had prominent 17 kDa free ISG15 bands. In all 7 DM-PFA samples, there were multiple other bands suggesting that ISG15 was covalently conjugated to other proteins (Figure 2). Two DM-NO-PFA samples showed a single faint band at ~85 kDa; none of the other samples showed any ISG15 or putative ISG15 conjugated proteins. The patterns of ISG15 immunoreactive bands were distinct from the pattern of total protein detected by Coomasie staining of parallel whole muscle lysate gels. ISG15-conjugated proteins were seen faintly at lower molecular weights in several DM-PFA samples, but remarkably were largely restricted to protein-conjugate complexes of molecular weight greater than approximately 51 kDa.
Figure 2.
Molecular similarity of cultured human skeletal muscle treated with interferon-alpha or interferon-beta to human dermatomyositis muscle biopsy samples
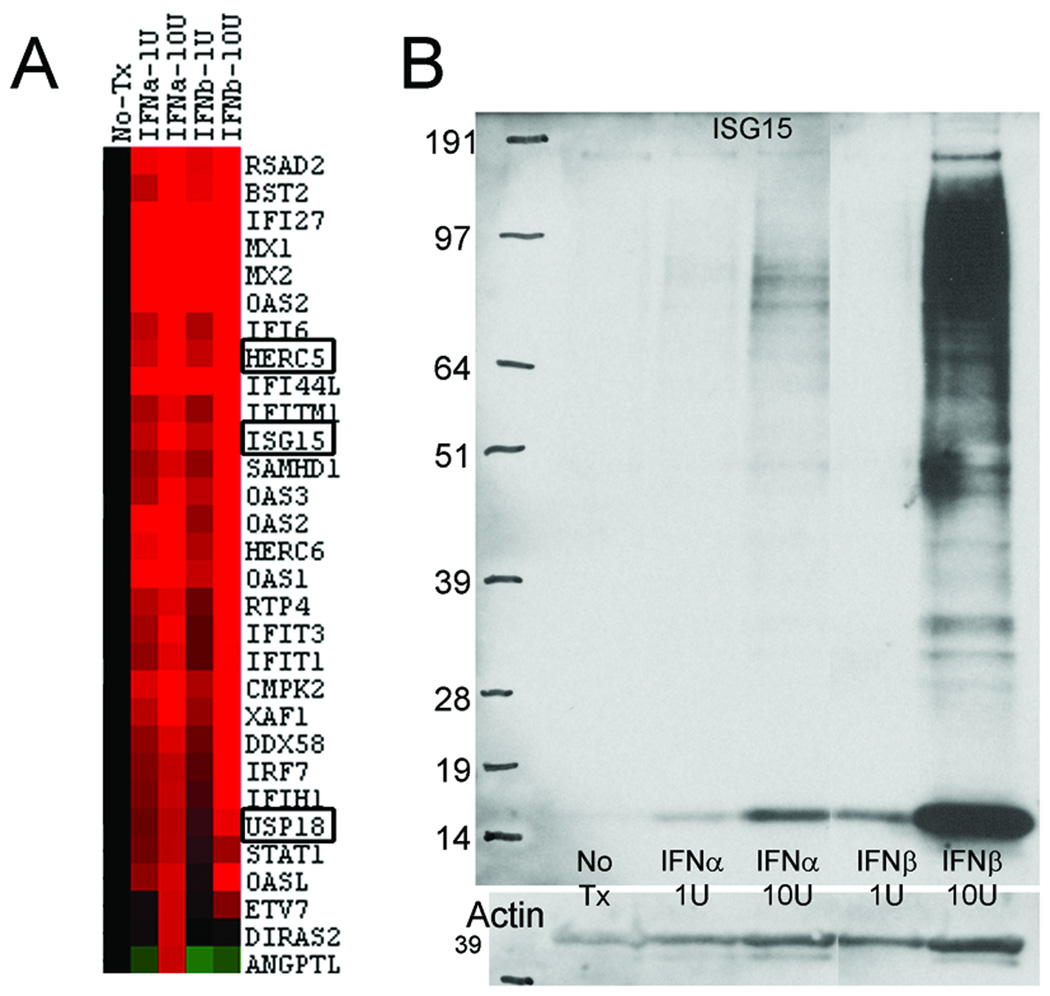
Stimulation of cultured human muscle skeletal cells with type 1 interferons and measurement of transcripts through whole genome microarrays with 55,000 probesets demonstrated a molecular profile highly similar to human DM-PFA muscle biopsy samples. Of the 23 transcripts present within the unique cluster in DM-PFA samples (Figure 1B), 16 were among the highest 30 produced transcripts in the cell culture experiments (Figure 3A). High-throughput quantitative RT-PCR experiments similarly showed a marked upregulation of transcripts in cell culture similar to that present in human DM-PFA samples (Supplementary Table).
Figure 3.
Cultured muscle cells produced free ISG15 protein which was functionally active, conjugating to other proteins in patterns similar to those present in human DM samples (Figure 3B), in a dose-dependent fashion, and more observable with IFN-β than IFN-α.
Localization of ISG15 protein to sites of DM muscle pathology: perifascicular myofibers and capillaries
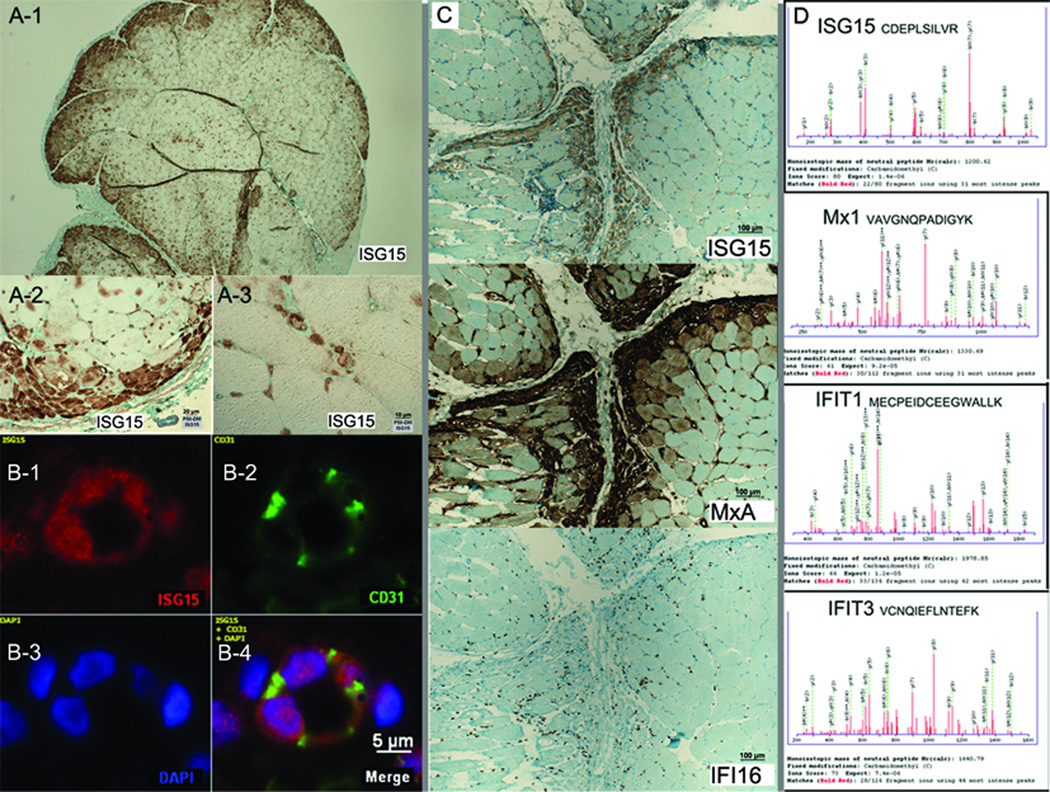
The correlation of ISG15 transcript with the presence of perifascicular atrophy in DM suggested that ISG15 protein might be present in perifascicular atrophic myofibers. In 2 of the 5 DM samples (P58 and P195), but in none of 15 non-DM controls (5 each with PM, IBM, and normal), intense ISG15 staining was present in myofibers in the regions of perifascicular atrophy (Figure 4A). This was confirmed in both immunohistochemical peroxidase based and immunofluorescent studies. A gradient of ISG15 intensity was evident, with most intense staining in the perifascicular fibers bordering the perimysial tissue and less intense staining with progressively deeper myofibers; staining was absent in central-fascicular fibers. In 3 other DM samples (P179, P382, and P383), ISG15 staining resulted in diffuse immunoreactivity of most myofibers, without accentuation of the perifascicular fibers. This pattern suggests either diffuse myofiber expression or no myofiber expression, with non-specific immunoreactivity, so that the localization of ISG15 to myofibers in these samples is uncertain.
Figure 4.
In addition to myofiber staining, ISG15 expression was evident in many capillaries (not quantitated) in all 5 DM but none of the 15 non-DM controls. Peroxidase based immunohistochemistry showed expression in structures in the typical location of capillaries (Figure 4A-3). Studies with multiple color immunofluorescence optical sectioning confirmed that ISG15 expression was high in capillaries (identified by CD31 expression), both within the endothelial cell cytoplasm and nucleus, identified by DAPI (Figure 4B).
Transcript and protein studies for MxA, an ISG15-conjugation candidate
Because MxA may be an ISG15 conjugated protein,8 we examined its location in relationship to ISG15 in serial sections by immunohistochemistry. In all 5 DM samples, MxA was expressed in perifascicular myofibers and capillaries; in the 2 DM cases that had intense ISG15 myofiber expression, both MxA and ISG15 were present in the same myofibers and capillaries on adjacent sections (Figure 4C). MxA myofiber expression was more extensive, and in many myofibers not immunoreactive to ISG15. In contrast, the interferon-gamma-induced protein 16 (IFI16) was not detected by immunostaining in these abnormal fibers. MxA was not seen in any myofibers or capillaries in the 15 control samples (5 each PM, IBM, and normal muscle).
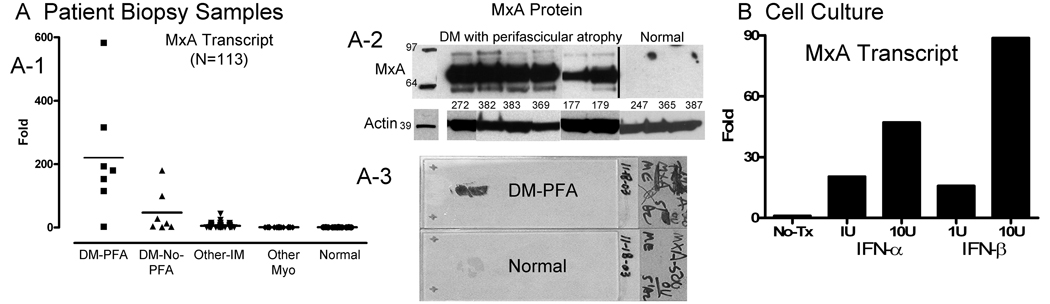
We furthermore examined both MxA transcript and protein by immunoblot in human DM samples. Across 113 muscle biopsy samples, MxA transcript levels were 220-fold increased compared to normal in DM-PFA, 47-fold in DM-NO-PFA, and 5-fold in other inflammatory myopathies (Figure 5A-1). Immunoblots from 6 DM-PFA and 3 normal samples similarly showed large bands for MxA protein and none in normal muscle (Figure 5A-2). Immunohistochemically prepared whole muscle sections on glass slides sometimes had MxA immunoreactivity so pronounced it was visible to the naked eye (Figure 5A-3).
Figure 5.
Cultured human skeletal muscle cells treated with interferon-α or interferon-β produced MxA transcript, in a dose-dependent fashion. In response to IFN-β 10U/ml, MxA transcript was increased 89-fold by microarray and 38-fold by quantitative RT-PCR (Figure 5B; and Supplementary Table).
Other ISG15-conjugation candidates are identified in DM muscle through LC/MS profiling
From liquid chromatography mass spectrometry (LC/MS) experiments that generated proteomic profiles of each of 4 DM patient muscle samples, we detected in 1 DM-PFA sample ISG15, MxA, IFIT1, and IFIT3 (Figure 4D), other proteins putatively conjugated to ISG15.8 IFIT1 and IFIT3 transcripts were also specifically elevated in DM-PFA samples (Figure 1B), 18- and 184-fold increased in DM-PFA compared with 1.4- and 7.7-fold in other inflammatory myopathies.
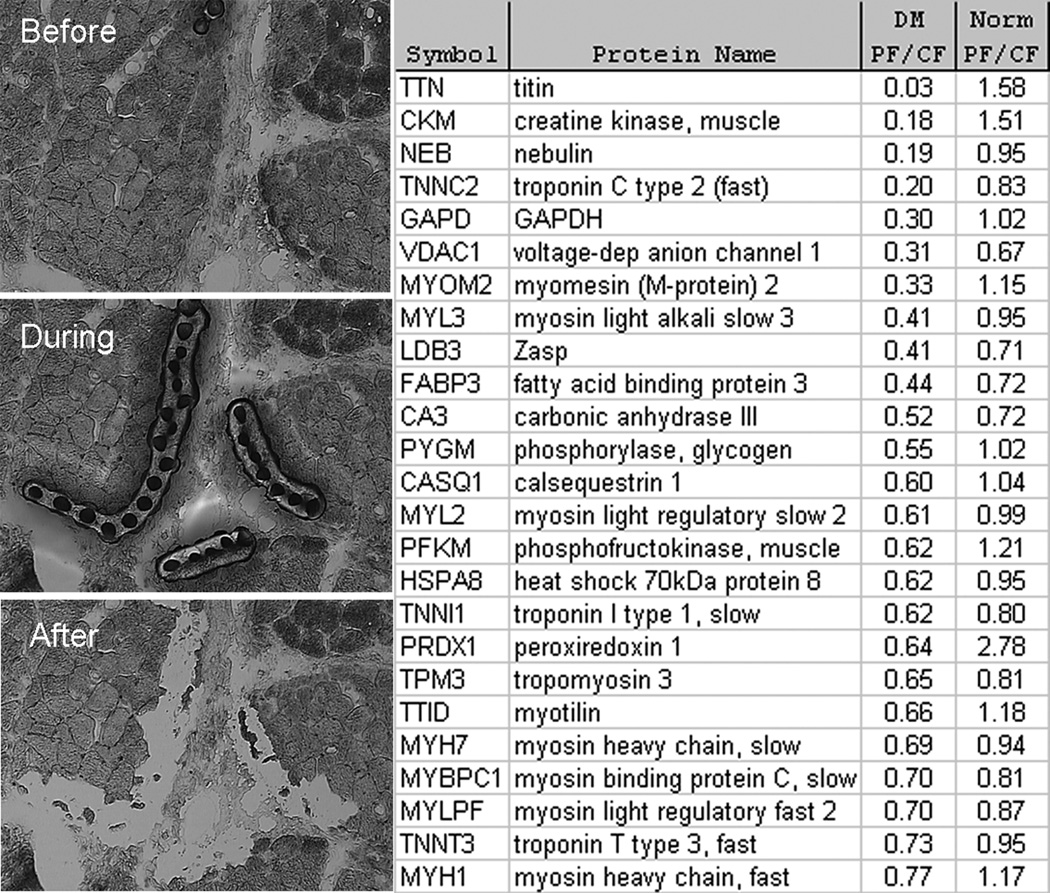
Titin deficiency in DM perifascicular atrophic myofibers
To better understand the nature of perifascicular myofibers, we selectively isolated DM and normal perifascicular and central-fascicular myofibers by laser capture microdissection and studied the proteins that were present in these minute specimens using shotgun proteomics.14 Initially, a total of 8 separate myofiber preparations, each containing approximately 100 7-µm thick myofiber cross-sections, were prepared (perifascicular × 2; central-fascicular × 2 from 1 patient with DM and 1 normal); each of these 8 samples underwent LC/MS based proteomic studies for protein abundances. These studies found a substantial reduction in the abundance of titin in DM perifascicular atrophic myofibers (Figure 6), present at 3% the amount seen in DM central-fascicular myofibers and 0.7% the amount present in normal muscle perifascicular myofibers. In the 2 normal muscle perifascicular myofiber preparations, a mean of 135 titin peptide identifications were made (111 and 158), second in abundance only to myosin heavy chain 7 (MYH7; mean of 145 peptide identifications). In the 2 DM perifascicular myofiber preparations, a mean of 0.5 (0 and 1) titin peptide identifications were made, with a mean of 99.5 (100 and 99) MYH7 peptides identified. The perifascicular myofiber titin reduction to 3% compared with the MYH7 reduction to 69% the central-fascicular amount indicates that the extreme reduction in titin was unlikely to be due to non-specific loss of abundant myofibrillar proteins. A second laser-capture/proteomics experiment with a different DM and normal sample retrieved smaller amounts of protein but again showed the complete absence of titin peptide identifications in only the DM perifascicular sample with more abundant amounts in other samples (DM central-fascicular – 5 peptide identifications; normal perifascicular – 9 peptides; and normal central-fascicular – 7 peptides).
Figure 6.
Discussion
Relevance of ISG15 Conjugation to Dermatomyositis
Dermatomyositis is an autoimmune disease with poorly understood mechanisms of myofiber, skin, and other tissue injury. Distinct from other inflammatory myopathies, DM muscle has two unique pathological features, capillary injury that includes tubuloreticular inclusions (TRIs; also called lupus inclusions)17–19 and perifascicular atrophy (PFA). Understanding these unique pathologies offers potential insight into the unique mechanisms present in DM. Although studies performed over 20 years ago firmly linked the formation of TRIs in vivo and in cultured endothelial and other cells to interferon-α and interferon-β,20–25 these data were not linked to DM until recently.1
We performed a large microarray study measuring the expression of approximately 18,000 genes in each of 113 muscle samples and found a small unique cluster of transcripts specific to dermatomyositis muscle samples with perifascicular atrophy. These transcripts included ISG15 and the ISG15 conjugation pathway members HERC5 and USP18. Marked ISG15, HERC5, and USP18 transcript elevation and presence of ISG15 protein and ISG15 protein conjugates were 100% specific to DM-PFA samples. Studies of ISG15 protein found it spatially associated with the main sites of pathology, capillaries in all 5 patients examined and perifascicular myofibers in 2 of them, and conjugated to other proteins of unknown identity.
Additionally, we analyzed publicly available microarray data that had not previously been studied with respect to ISG15. We found similar unreported findings in these existing datasets, further highlighting the remarkable specificity for ISG15 conjugation in DM-PFA among muscle diseases. Every measured ISG15, HERC5, and USP18 transcript level in all 7 adult DM-PFA samples exceeded those of all 199 other non-DM samples. Furthermore, it is likely that ISG15 conjugation in muscle occurs in children with DM given that marked ISG15 and USP18 transcript elevation was greater in all 21 juvenile DM muscle samples (and HERC5 greater in 20 of the 21 juvenile DM samples) than all 199 other non-DM samples. Whether these juvenile DM samples had PFA is not known.
ISG15 is believed to function both as a ubiquitin-like modifier conjugating other proteins and as a cytokine.26, 27 ISG15-conjugates have been demonstrated from cells in culture,6, 8, 26, 28–30 animal models,31–33 normal human circulating blood cells26 and thymus tissue,32 and 5 human cancer biopsy specimens.9 We provide here the first demonstration that ISG15 conjugates are present in autoimmune human diseased tissue samples. ISG15-conjugates in DM muscle were almost exclusively high-molecular weight protein-conjugate complexes, greater than approximately 51 kDa. This restriction to relatively higher molecular weights is similar to the findings shown in published immunoblot images of ISG15-conjugates in multiple other studies.9, 26, 28, 30, 32, 33 Identities of ISG15-conjugated proteins are not determined by these studies, nor has the identity of any ISG15 conjugated protein in a human tissue sample yet been reported. Three proteins we have identified in DM muscle (MxA by immunohistochemistry, immunoblot, and mass spectrometry; IFIT1 and IFIT3 by mass spectrometry) were previously identified as ISG15-conjugated in interferon-β stimulated HeLa cells, based on mass spectrometric identification in immunoprecipitated material.8 These proteins may be conjugated in DM muscle as well.
The factors driving ISG15 production and conjugation in DM-PFA muscle are not identified in this study. ISG15 production by cells has been well characterized as following from type 1 interferon stimulation. We are not aware of any other molecules other than type 1 interferons that have been reported capable of inducing ISG15 transcription and protein production. Nevertheless, what drives this production in DM is uncertain. Although it seems likely that a type 1 interferon is responsible, we did not detect in DM-PFA substantial increases in interferon-α or interferon-β transcript by microarray studies or interferon-α protein by immunohistochemistry in previous studies reporting on interferon-α detection in muscle-infiltrating pDCs.1 Those studies did sometimes suggest a gradient of interferon-α extending from a perimysial location into neighboring perifascicular myofibers (Supplementary Figure 1), but technical concerns limit emphasis of this finding. Certainly it might be the case that some other type 1 interferon, or an as yet undiscovered molecule that is not a type 1 interferon, might drive ISG15 conjugation.
A human cell culture model of DM
The development of a human cell culture model that accurately represents the molecular processes leading to myofiber injury in people with DM would be an important advance. The marked upregulation of ISG15 transcript and ISG15 conjugated proteins appears unique to DM among the inflammatory myopathies and reproduction of these abnormalities by exposure of human skeletal muscle cultured cells to interferon-α and interferon-β suggests a bona fide culture model of DM myofiber injury. Whether ISG15 production or the production of some other type 1 interferon-inducible transcript or protein mediates myofiber injury was not addressed in this study. This potential model can be used to study cellular events that might lead to myofiber injury.
Towards Understanding the Nature of Perifascicular Atrophy
As perifascicular atrophy, perhaps better called perimysial perifascicular atrophy,34 is a highly specific feature of DM, understanding its nature may provide insight into the events leading to DM myofiber injury. Although traditionally attributed to ischemia,35 we know of no evidence that this lesion develops from ischemia. No transcripts or proteins induced by ischemia in model systems have been reported to be significantly upregulated in DM whole muscle or in perifascicular myofibers. Histochemical and immunohistochemical methods used in this and previous studies have identified proteins present within perifascicular atrophic myofibers (MHCn, NCAM, desmin, alpha B crystallin, STAT1, MHC1, MxA, and ISG15; reviewed in 36), but have not identified proteins underexpressed or absent from these myofibers. The approach taken here, of removing such fibers by laser capture microdissection and performing unbiased exploratory analyses of their protein content,37 may allow for deeper understanding of the nature of the myofiber injury in DM. Although technically challenging, these studies do suggest that a major skeletal muscle protein that provides the scaffold for the sarcomere,38 titin, is severely deficient in these fibers. This finding is consistent with previous electron microscopic studies that found a loss of visible sarcomeric structure within DM perifascicular atrophic myofibers.17 Indeed, titin gene M-line exon 1 and 2 knockout mice develop marked myofiber atrophy and loss of myofiber striation,39 the latter finding present in DM perifascicular atrophic myofibers as well (Supplementary Figure 2).
The relationship between DM titin and other protein reductions (Figure 6) to ISG15 conjugation is uncertain, but one speculation is that DM myofiber injury results from inappropriate myofiber intracellular ISG15 production and conjugation to other proteins necessary for the maintenance of mature skeletal muscle myofibers. The perimysial perifascicular location of myofiber injury in DM, similar in topology to the keratinocyte injury in DM skin,34 may relate to greater concentrations of type 1 interferons in the perimysial regions. The determination of the identities of ISG15 conjugated proteins in specific human tissues and their precise locations in DM muscle might clarify the mechanism of this disease and the potential for therapeutic targeting of this pathway or upstream inducers of ISG15. In addition to DM, ISG15 conjugation in injured tissue might turn out to be present in systemic lupus erythematosus (SLE), given that ISG15 transcript has been reported highly upregulated in SLE kidney,40 blood,41,42 and skin.13
Supplementary Material
Acknowledgements
We thank Alan H. Beggs, PhD for advice and Peter Kiener, PhD for review of the manuscript.
Supported by grants to S.A.G. from the National Institutes of Health (R01NS43471 and R21NS057225), the Muscular Dystrophy Association MDA3878, and MedImmune, LLC. Human muscle microarray data were generated by the Children’s Hospital Gene Expression Core (NS40828).
References
- 1.Greenberg SA, Pinkus JL, Pinkus GS, et al. Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol. 2005;57:664–678. doi: 10.1002/ana.20464. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel J, Schmidt R, Proelss J, et al. Type I interferon-associated skin recruitment of CXCR3+ lymphocytes in dermatomyositis. Clin Exp Dermatol. 2006;31:576–582. doi: 10.1111/j.1365-2230.2006.02150.x. [DOI] [PubMed] [Google Scholar]
- 3.Yuan W, Krug RM. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. Embo J. 2001;20:362–371. doi: 10.1093/emboj/20.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao C, Beaudenon SL, Kelley ML, et al. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. PNAS. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dastur A, Beaudenon S, Kelley M, et al. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J Biol Chem. 2006;281:4334–4338. doi: 10.1074/jbc.M512830200. [DOI] [PubMed] [Google Scholar]
- 6.Wong JJ, Pung YF, Sze NS, Chin KC. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. PNAS. 2006;103:10735–10740. doi: 10.1073/pnas.0600397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malakhov MP, Malakhova OA, Kim KI, et al. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem. 2002;277:9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 8.Zhao C, Denison C, Huibregtse JM, et al. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. PNAS. 2005;102:10200–10205. doi: 10.1073/pnas.0504754102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai SD, Haas AL, Wood LM, et al. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006;66:921–928. doi: 10.1158/0008-5472.CAN-05-1123. [DOI] [PubMed] [Google Scholar]
- 10.Walsh RJ, Kong SW, Yao Y, et al. Type I interferon-inducible gene expression in blood is present and reflects disease activity in dermatomyositis and polymyositis. Arthritis Rheum. 2007;56:3784–3792. doi: 10.1002/art.22928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenberg SA, Sanoudou D, Haslett JN, et al. Molecular profiles of inflammatory myopathies. Neurology. 2002;59:1170–1182. doi: 10.1212/wnl.59.8.1170. [DOI] [PubMed] [Google Scholar]
- 12.Wu Z, Irizarry RA. Stochastic models inspired by hybridization theory for short oligonucleotide arrays. J Comput Biol. 2005;12:882–893. doi: 10.1089/cmb.2005.12.882. [DOI] [PubMed] [Google Scholar]
- 13.Yao Y, Richman L, Higgs BW, et al. Neutralization of IFN-alpha/beta-inducible genes and downstream effect in a phase I trial of an anti-IFN-alpha monoclonal antibody in SLE. Arthritis Rheum. 2009 doi: 10.1002/art.24557. In Press. [DOI] [PubMed] [Google Scholar]
- 14.Parker KC, Kong SW, Walsh RJ, et al. Fast-twitch sarcomeric and glycolytic enzyme protein loss in inclusion body myositis. Muscle Nerve. 2009;39:739–753. doi: 10.1002/mus.21230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakay M, Wang Z, Melcon G, et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- 16.Chen YW, Shi R, Geraci N, et al. Duration of chronic inflammation alters gene expression in muscle from untreated girls with juvenile dermatomyositis. BMC Immunol. 2008;9:43. doi: 10.1186/1471-2172-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banker BQ. Dermatomyostis of childhood, ultrastructural alteratious of muscle and intramuscular blood vessels. J Neuropathol Exp Neurol. 1975;34:46–75. doi: 10.1097/00005072-197501000-00005. [DOI] [PubMed] [Google Scholar]
- 18.Carpenter S, Karpati G, Rothman S, Watters G. The childhood type of dermatomyositis. Neurology. 1976;26:952–962. doi: 10.1212/wnl.26.10.952. [DOI] [PubMed] [Google Scholar]
- 19.De Visser M, Emslie-Smith AM, Engel AG. Early ultrastructural alterations in adult dermatomyositis. Capillary abnormalities precede other structural changes in muscle. J Neurol Sci. 1989;94:181–192. doi: 10.1016/0022-510x(89)90228-1. [DOI] [PubMed] [Google Scholar]
- 20.Grimley PM, Davis GL, Kang YH, et al. Tubuloreticular inclusions in peripheral blood mononuclear cells related to systemic therapy with alpha-interferon. Lab Invest. 1985;52:638–649. [PubMed] [Google Scholar]
- 21.Grimley PM, Rutherford MN, Kang YH, et al. Formation of tubuloreticular inclusions in human lymphoma cells compared to the induction of 2'-5'-oligoadenylate synthetase by leucocyte interferon in dose-effect and kinetic studies. Cancer Res. 1984;44:3480–3488. [PubMed] [Google Scholar]
- 22.Kuyama J, Kanayama Y, Mizutani H, et al. Formation of tubuloreticular inclusions in mitogen-stimulated human lymphocyte cultures by endogenous or exogenous alpha-interferon. Ultrastruct Pathol. 1986;10:77–85. doi: 10.3109/01913128609015565. [DOI] [PubMed] [Google Scholar]
- 23.Norton WL, Velayos E, Robison L. Endothelial inclusions in dermatomyositis. Ann Rheum Dis. 1970;29:67–72. doi: 10.1136/ard.29.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rich SA. Human lupus inclusions and interferon. Science. 1981;213:772–775. doi: 10.1126/science.6166984. [DOI] [PubMed] [Google Scholar]
- 25.Rich SA, Owens TR, Bartholomew LE, Gutterman JU. Immune interferon does not stimulate formation of alpha and beta interferon induced human lupus-type inclusions. Lancet. 1983;1:127–128. doi: 10.1016/s0140-6736(83)91771-3. [DOI] [PubMed] [Google Scholar]
- 26.D'Cunha J, Knight E, Jr, Haas AL, et al. Immunoregulatory properties of ISG15, an interferon-induced cytokine. Proc Natl Acad Sci U S A. 1996;93:211–215. doi: 10.1073/pnas.93.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Recht M, Borden EC, Knight E., Jr A human 15-kDa IFN-induced protein induces the secretion of IFN-gamma. J Immunol. 1991;147:2617–2623. [PubMed] [Google Scholar]
- 28.Loeb KR, Haas AL. The interferon-inducible 15-kDa ubiquitin homolog conjugates to intracellular proteins. J. Biol. Chem. 1992;267:7806–7813. [PubMed] [Google Scholar]
- 29.Giannakopoulos NV, Luo JK, Papov V, et al. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem Biophys Res Commun. 2005;336:496–506. doi: 10.1016/j.bbrc.2005.08.132. [DOI] [PubMed] [Google Scholar]
- 30.Pitha-Rowe I, Hassel BA, Dmitrovsky E. Involvement of UBE1L in ISG15 Conjugation during Retinoid-induced Differentiation of Acute Promyelocytic Leukemia. J. Biol. Chem. 2004;279:18178–18187. doi: 10.1074/jbc.M309259200. [DOI] [PubMed] [Google Scholar]
- 31.Hamerman JA, Hayashi F, Schroeder LA, et al. Serpin 2a Is Induced in Activated Macrophages and Conjugates to a Ubiquitin Homolog. J Immunol. 2002;168:2415–2423. doi: 10.4049/jimmunol.168.5.2415. [DOI] [PubMed] [Google Scholar]
- 32.Malakhov MP, Kim KI, Malakhova OA, et al. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J Biol Chem. 2003;278:16608–16613. doi: 10.1074/jbc.M208435200. [DOI] [PubMed] [Google Scholar]
- 33.Ritchie KJ, Malakhov MP, Hetherington CJ, et al. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 2002;16:2207–2212. doi: 10.1101/gad.1010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Greenberg SA, Fiorentino D. Similar topology of injury to keratinocytes and myofibers in dermatomyositis skin and muscle. Br J Derm. 2009;160:464–465. doi: 10.1111/j.1365-2133.2008.08967.x. [DOI] [PubMed] [Google Scholar]
- 35.Dalakas MC, Hohlfeld R. Polymyositis and dermatomyositis. Lancet. 2003;362:971–982. doi: 10.1016/S0140-6736(03)14368-1. [DOI] [PubMed] [Google Scholar]
- 36.Greenberg SA. Proposed immunologic models of the inflammatory myopathies and potential therapeutic implications. Neurology. 2007;69:2008–2019. doi: 10.1212/01.WNL.0000291619.17160.b8. [DOI] [PubMed] [Google Scholar]
- 37.Parker KC, Walsh RJ, Salajegheh M, et al. Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J Proteome Res. 2009 doi: 10.1021/pr800873q. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 38.Gregorio CC, Granzier H, Sorimachi H, Labeit S. Muscle assembly: a titanic achievement? Curr Opin Cell Biol. 1999;11:18–25. doi: 10.1016/s0955-0674(99)80003-9. [DOI] [PubMed] [Google Scholar]
- 39.Peng J, Raddatz K, Labeit S, et al. Muscle atrophy in titin M-line deficient mice. J Muscle Res Cell Motil. 2005;26:381–388. doi: 10.1007/s10974-005-9020-y. [DOI] [PubMed] [Google Scholar]
- 40.Peterson KS, Huang JF, Zhu J, et al. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest. 2004;113:1722–1733. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bennett L, Palucka AK, Arce E, et al. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao Y, Higgs BW, Morehouse C, et al. Development of Potential Pharmacodynamic and Diagnostic Markers for Anti-IFN-alpha Monoclonal Antibody Trials in Systemic Lupus Erythematosus. Human Genomics and Proteomics. 2009 doi: 10.4061/2009/374312. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.