Abstract
Activated macrophages acquire a proinflammatory (classic) or antiinflammatory (alternative) phenotype that influences atherosclerosis. The present study investigated whether sphingosine-1-phosphate (S1P), with its known antiinflammatory effects, could regulate the inflammatory phenotype of lipopolysaccharide (LPS)-stimulated mouse macrophages. Activation of macrophages by LPS significantly increases proinflammatory cytokine secretion. Pretreatment of macrophages with 500 nmol/L S1P markedly reduced LPS-mediated secretion of tumor necrosis factor-α, monocyte chemoattractant protein-1, and interleukin-12. Such antiinflammatory actions were also evident in LPS-stimulated macrophages treated with the S1P1 receptor–specific agonist SEW2871. Pharmacological antagonism of the S1P1 receptor on macrophages using the S1P1-specific antagonist VPC44116 also blocked proinflammatory cytokine secretion in response to LPS. Studies using bone marrow–derived macrophages from S1P2-deficient mice revealed that the S1P2 receptor did not play a pivotal role in this process. Thus, activation of the S1P1 receptor in mouse macrophages limits the expression of proinflammatory cytokines. Furthermore, we demonstrated that S1P increased arginase I activity and inhibited LPS-induced inducible NO synthase activity in LPS-treated macrophages, again through S1P1 receptor activation on macrophages. Analysis of a 1.7-kb region of the murine inducible NO synthase promoter revealed the presence of putative nuclear factor κB, activator protein-1, and STAT-1 response elements. Using inducible NO synthase promoter-reporter constructs, we found that S1P significantly reduced the nuclear factor κB–mediated induction of inducible NO synthase. These findings demonstrate an important role for S1P in the regulation of macrophage phenotypic switching. Therefore, we conclude that S1P promotes the production of an alternative antiinflammatory macrophage phenotype through activation of the macrophage S1P1 receptor.
Keywords: macrophage, sphingosine-1-phosphate, arginase I, iNOS, NFκB, inflammation
Atherosclerosis is a chronic inflammatory disease.1 Monocytes/macrophages play key roles in the initiation and progression of atherosclerosis, and can alter their phenotype in response to changes in the local cytokine environment.2 Macrophages can be distinctly activated to either a classically activated, or M1, phenotype by proinflammatory molecules such as interferon-γ and lipopolysaccharide (LPS) or to an “alternatively activated,” or M2, phenotype by Th-2 antiinflammatory cytokines such as interleukin (IL)-4.3 Macrophages can also display a M2b activated phenotype when stimulated by immune complexes and LPS.4 The M1 and M2 phenotypes are specifically distinguished by the cytokines produced and by the way in which arginine is processed, whereas the M2b phenotype is mainly distinguished by high IL-10 and low IL-12 levels. In classically activated macrophages, Th-1 cytokines trigger the induction of inducible NO synthase (iNOS), causing the production of NO. However, in alternatively activated macrophages, Th-2 cytokines induce arginase (Arg) I, which converts arginine to ornithine and urea.5,6 Several studies have shown that on macrophage activation by Th-2 cytokines, the upregulation of Arg activity results in the depletion of arginine and subsequent inhibition of nitric oxide production.7,8 However, the exact functional properties of classically and alternatively activated macrophages in vivo remain unclear. Further understanding of the mechanisms contributing to the generation of these macrophage phenotypes and their functions in vivo will provide important information regarding the development of novel targeted therapies for treatment of inflammatory diseases, including atherosclerosis.
Sphingosine-1-phosphate (S1P), a biologically active sphingolipid, plays important roles in the regulation of a variety of cellular processes, including cell survival and vascular maturation, by binding to a family of G protein–coupled receptors (termed S1P1–5).9 FTY720, a synthetic analogue of sphingosine and a potent agonist of 4 of 5 S1P receptors (S1P1, S1P3, S1P4, and S1P5), has been shown to modulate lymphocyte trafficking in mice through activation of the S1P1 receptor.10 Ogawa and colleagues reported that the novel S1P1 receptor agonist KRP-203 reduced experimental autoimmune myocarditis in rats.11 Klingenberg et al recently reported that FTY720 modulated lymphocyte distribution in apolipoprotein E–deficient mice.12 Major unresolved questions in the field of atherosclerosis are whether S1P promotes antiinflammatory responses in macrophages by interacting with specific macrophage S1P receptors and, thus, whether specific S1P receptors significantly influence atherosclerosis development. Recent studies show an important role for S1P receptors in macrophage function in atherogenesis. Recently, Nofer et al showed that FTY720 can modulate macrophage activation and reduce atherosclerosis development in low-density lipoprotein receptor–deficient mice.13 Keul et al similarly reported reduced atherosclerosis in apolipoprotein E–deficient mice treated with FTY720 through modulation of monocyte chemotaxis.14 Additionally, Brune and colleagues have demonstrated that the production of S1P by sphingosine kinase protects macrophages from apoptosis and triggers macrophage production of the antiinflammatory cytokine transforming growth factor-β.15 Thus, S1P can regulate survival pathways as well as promote antiinflammatory signals in macrophages, which are important for atherogenesis.
In the present study, we report that during acute inflammation, S1P switches the phenotype of macrophages from a proinflammatory to an antiinflammatory phenotype. We also provide evidence that this switch to an antiinflammatory phenotype is regulated by the S1P1 receptor. Furthermore, we identify that S1P regulates Arg I and iNOS expression and that regulation of macrophage iNOS expression by S1P is mediated primarily through inhibition of nuclear factor (NF)κB.
Materials and Methods
An expanded Materials and Methods section is available in the online data supplement at http://circres.ahajournals.org.
Mice
Ten-week-old C57BL/6 mice were purchased from The Jackson Laboratory (Bar Harbor, Me). S1P2 receptor–deficient mice on a C57BL/6 background were generated by the laboratory or R.L.P. (NIH).16 These S1P2-deficient mice have been backcrossed for 7 generations onto C57BL/6J. Wild-type littermates from the colony were used as controls for all studies using S1P2-deficient mice. All experiments followed University of Virginia Animal Care and Use Committee guidelines, and approval for use of rodents was obtained from the University of Virginia.
Isolation of Mouse Peritoneal Macrophages
Eight- to 10-week-old C57BL/6 mice were injected with 2 mL of 3% thioglycollate medium. On day 5 postinjection, mice were anesthetized with isoflurane and injected IP with 5 mL cold PBS with 10 mmol/L EDTA. The PBS was then removed with a syringe, and the process was repeated 3 times. Macrophages from each mouse were plated separately in RPMI/10% FBS overnight. The next day, nonadherent cells were removed by aspiration, and the remaining macrophages were washed thoroughly with PBS and used for experiments.
Isolation of Bone Marrow–Derived Macrophages
Cells were obtained from the tibia and femur bone marrow of wild-type littermates and S1P2 receptor–deficient mice (age, 8 to 10 weeks) and were cultured in the presence of L-929 conditioned medium as described in detail previously.17
Cell Culture Studies
Macrophages were maintained in RPMI medium 1640/10% FBS as described above. For studies, LPS was added at 10 ng/mL in the absence or presence of 500 nmol/L S1P, 1 μmol/L SEW2871, 10 μmol/L VPC44116, 100 ng/mL pertussis toxin, or BAY11-7085 (5 to 10 μmol/L) as described in each figure legend.
Promoter Cloning and Site-Directed Mutagenesis
The 1749-bp fragment of the 5′-flanking region of the murine iNOS gene was subcloned into the Kpn1 site of pGL3-Basic. The presence of the iNOS promoter was confirmed by PCR using oligonucleotides specific for regions within the promoter. Mutations in the distal NFκB site were generated in the piNOS-luc construct by using the Quik-change site-directed mutagenesis kit (Stratagene, La Jolla, Calif) using the following oligonucleotides: 5′-TAA CTT GCA CAC CCA ACT AAA AAA AAA AAC TTT GGG AAC A-3′ and 3′-ATT GAA CGT GTG GGT TGA TTT TTT TTT TTG AAA CCC TTG T-5′.
Results
S1P Receptor Expression in Mouse Macrophages
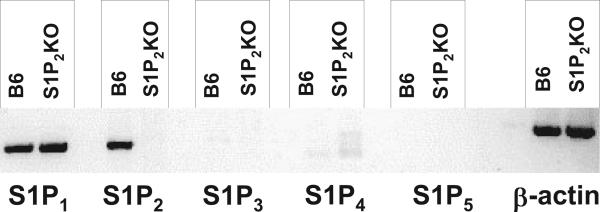
We isolated bone marrow derived macrophages from C57Bl/6J (B6) mice and measured the relative mRNA expression of each S1P receptor using RT-PCR. S1P1 and S1P2 mRNA were expressed in bone marrow–derived macrophages; however, we found no expression of S1P3, S1P4, and S1P5 mRNA (Figure 1) in either B6 or S1P2 knockout (KO) macrophages. We have confirmed similar S1P receptor expression in peritoneal macrophages (data not shown). Furthermore, we did not observe any compensatory upregulation of S1P1 receptor expression in the S1P2 KO macrophages (Figure 1).
Figure 1.
Macrophages express S1P1 and S1P2 receptors. Expression of S1P receptor mRNA in C57BL/6J and S1P2-deficient bone marrow–derived macrophages were analyzed by conventional RT-PCR. There was no detectable expression of S1P3, S1P4, or S1P5 mRNAs. There was no compensatory upregulation of S1P1 in the S1P2-deficient macrophages. β-Actin was used as a control for sample loading. Data represent pooled RNA samples from 3 mice per group.
S1P Regulation of LPS-Stimulated, Proinflammatory Cytokine Production by Primary Murine Macrophages
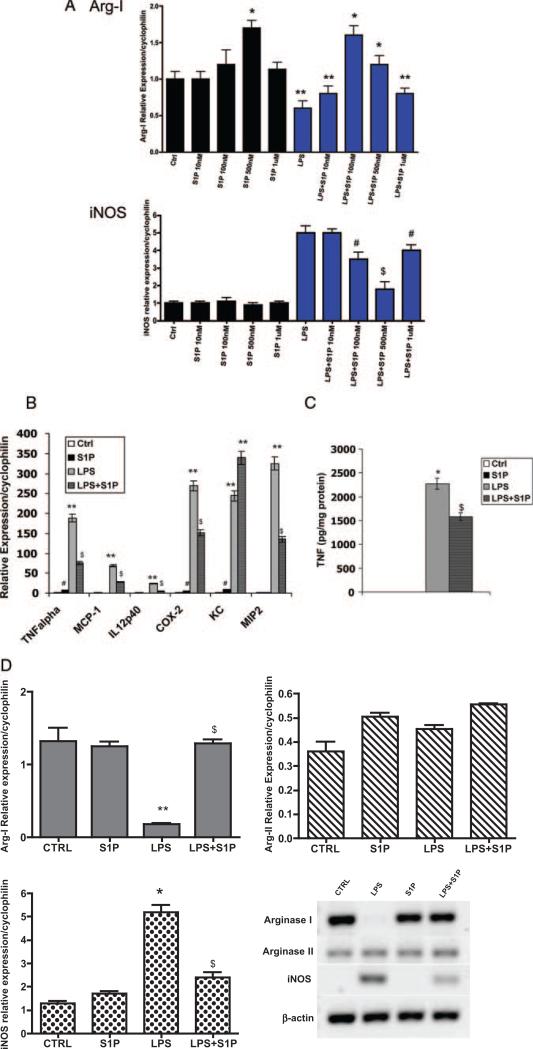
LPS has been shown to downregulate Arg I and upregulate iNOS expression.18,19 Increased Arg I activity is associated with downregulation of the inflammatory response, whereas iNOS activity is associated with a proinflammatory macrophage response.20 In the present study, we examined whether S1P could modulate the effects of LPS on macrophage activation. S1P is present in the range of 300 nmol/L to 1 μmol/L in plasma; therefore, we first performed a dose curve of S1P on mRNA expression of Arg I and iNOS in B6 macrophages.21 B6 peritoneal macrophages were incubated for 4 hours with 10 ng/mL LPS in the absence or presence of S1P. As shown in Figure 2A, S1P effectively increased Arg I expression and reduced iNOS expression at 100 and 500 nmol/L concentrations of S1P. Because we observed maximal effects using 500 nmol/L S1P, we chose the 500 nmol/L concentration for our experiments. We then examined levels of cytokine mRNA in response to LPS and 500 nmol/L S1P. In the absence of LPS stimulation, no cytokine expression could be detected in macrophages (Figure 2B). Treatment of macrophages with S1P suppressed LPS-induced tumor necrosis factor (TNF)α, monocyte chemoattractant protein-1, IL-12, cyclooxygenase-2, and macrophage inflammatory protein-2 mRNA expression by 3-fold, 2-fold, 6-fold, 5-fold, and 7-fold, respectively (P<0.001; Figure 2B). However, S1P had no effect on the LPS-mediated induction of KC mRNA (Figure 2B). We also measured TNFα secretion by macrophages using ELISA (Figure 2C). Naïve, unstimulated macrophages secreted very little TNFα; however, incubation of macrophages with 10 ng/mL LPS induced TNFα secretion by 3-fold. S1P reduced TNFα secretion by 30%. Surprisingly, we observed a small, yet significant, increase in TNFα mRNA expression in the presence of S1P alone (Figure 2B), but this did not translate into detectable TNFα protein secretion (Figure 2C). Thus, within the 4-hour time frame of our study, S1P significantly reduced LPS-mediated TNFα secretion. We also observed that S1P reduced interferon-γ–mediated increases in TNFα, as well as oxidized low-density lipoprotein–mediated increases in TNFα and CD36 (data not shown), indicating that the S1P-mediated phenotypic switching of macrophages is not restricted to LPS.
Figure 2.
S1P reduces LPS-induced inflammatory gene expression. A, S1P dose response. Total RNA was isolated from C57BL/6J peritoneal macrophages treated with S1P at concentrations of 10 nmol/L, 100 nmol/L, 500 nmol/L, and 1 μmol/L in the absence (black bars) or presence (blue bars) of 10 ng/mL LPS for 4 hours. Quantitative real-time PCR for murine Arg I and iNOS were performed. Cyclophilin was measured as a housekeeping gene for normalization purposes. Data are expressed using the relative expression method using B6 control as set to 1. *P<0.001 vs control (Ctrl), **P<0.0001 vs Ctrl, #P<0.01 vs LPS, $P<0.001 vs LPS by ANOVA. Data are from 3 independent experiments using 4 mice per group. B, Inflammatory gene expression. Cytokine mRNA expression was determined using quantitative real-time PCR. C57BL/6J peritoneal macrophages were treated with media alone (Ctrl) or with 10 ng/mL LPS in the absence or presence of 500 nmol/L S1P for 4 hours. **P<0.0001 vs Ctrl, $P<0.005 vs LPS, #P<0.05 vs Ctrl by ANOVA. C, TNFα secretion. TNFα secretion by C57BL/6J peritoneal macrophages was measured using ELISA. Values were normalized to total cellular protein. *P<0.001 vs Ctrl, $P<0.005 vs LPS by ANOVA. D, Arg I and iNOS expression. Levels of mRNA expression in C57BL/6J peritoneal macrophages were determined using quantitative real-time PCR as described in Materials and Methods and are graphically shown. **P<0.0001 vs Ctrl, $P<0.009 vs LPS, *P<0.001 vs Ctrl by ANOVA. Data represent 5 mice per group performed in triplicate. Conventional RT-PCR products of Arg I, Arg-2, iNOS, and β-actin are also shown in a representative gel.
Sphingosine-1-Phosphate Regulates Arg I and iNOS Expression in Macrophages
Given that iNOS and Arg I use the common substrate arginine, for which they compete, and because we observed changes in enzyme expression in response to S1P (Figure 2A), we investigated in more detail the effect of S1P on Arg I and iNOS in B6 macrophages stimulated with LPS. First, we examined the potential regulation of different Arg isoforms by S1P. The addition of LPS to B6 macrophages inhibited Arg I mRNA expression (Figure 2D). Incubation of macrophages with S1P restored Arg I mRNA expression to control levels (Figure 2D). In contrast, neither LPS nor S1P affected Arg-2 mRNA expression. Furthermore, we found that S1P significantly reduced LPS-mediated iNOS mRNA expression in primary macrophages (Figure 2D).
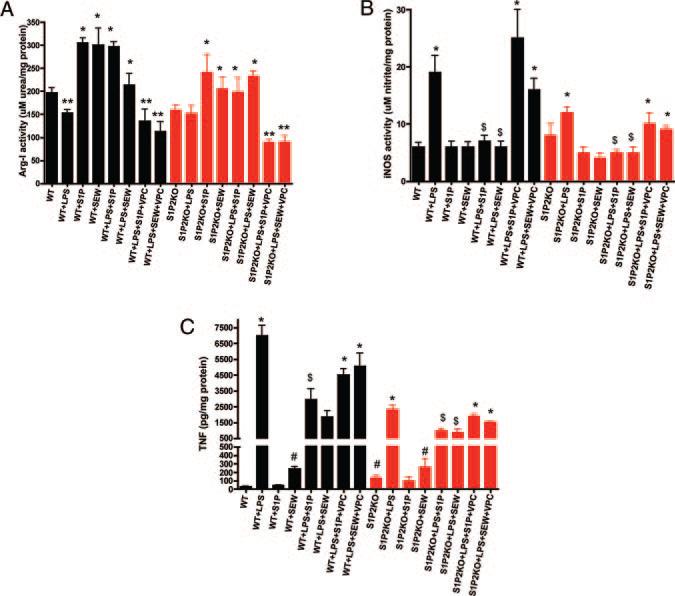
We next measured Arg I and iNOS enzyme activities. In wild-type macrophages, LPS suppressed Arg I activity, and both S1P and SEW stimulated Arg I activity, as measured by macrophage urea production (see black bars in Figure 3A). These results indicate that Arg I is indeed responsive to the S1P pathway in macrophages. Furthermore, in agreement with our iNOS mRNA results, we observed a significant decrease in LPS-induced iNOS enzymatic activity in macrophages on stimulation with S1P (see black bars in Figure 3B). Collectively, these data strongly suggest that S1P shifts nitrogen use away from the iNOS pathway toward the Arg I pathway. Thus, we hypothesize that S1P acts in macrophages, at least in part, by a dual mechanism of both suppressing the induction of iNOS and enhancing Arg I expression.
Figure 3.
The S1P1 receptor is antiinflammatory in macrophages. Bone marrow–derived macrophages from either wild-type littermate controls (WT) (black bars) or S1P2-deficient (S1P2KO; red bars) mice (n=5 mice per group) were treated with LPS (10 ng/mL) for 4 hours. In some cases, macrophages were treated with 500 nmol/L S1P, 1 μmol/L SEW2871 (+SEW), or with 10 μmol/L VPC44116 (+VPC) for 4 hours. A, Arg activity. Arg activity was quantified in cell lysates by measuring the conversion of l-arginine to urea. *P<0.001 vs WT, **P<0.01 vs WT by ANOVA. B, iNOS activity. Total nitrate and nitrite levels were measured with the Griess reagent as described in Materials and Methods. *P<0.001 vs WT, $P<0.01 vs LPS by ANOVA. C, TNFα secretion. TNFα levels were measured using ELISA. *P<0.001 vs WT, #P<0.05 vs WT, $P<0.01 vs LPS by ANOVA.
Identification of S1P1 As an Antiinflammatory Receptor in Macrophages
We next wished to identify the specific receptor target that mediates the antiinflammatory S1P response in macrophages. As shown in Figure 1, macrophages express S1P1 and S1P2 receptors. Mice that are deficient in the S1P1 receptor show embryonic lethality attributable to the critical role that S1P1 plays in vascular maturation.22 Thus, we used pharmacological reagents that are selective for S1P1. SEW2871 [5-(4-Phenyl-5-trifluoromethylthiophen-2-yl)-3-(3-(3-trifluoromethyphenyl)-1,2,4-oxadiazole] is a selective S1P1 receptor agonist that is 30-fold less potent than S1P at S1P1, with no agonist activity at S1P2 at concentrations up to 10 micromolar.23 VPC44116, an N-arylamide phosphonate, is a selective S1P1 receptor antagonist that exhibits a Ki of 30 nmol/L for S1P1.24 VPC44116 has recently been shown to prevent the protective effects of FTY720 on acute renal injury in mice.25 We found a significant reduction in iNOS activity (see black bars in Figure 3B) and TNFα secretion (see black bars in Figure 3C) in wild-type macrophages treated with SEW2871 compared to LPS. We observed a significant increase in Arg I activity in SEW2871-treated macrophages (see black bars in Figure 3A). The activation of S1P1 by SEW2871 paralleled the S1P-mediated changes in macrophage activation in all cases, suggesting that the S1P1 receptor mediates the antiinflammatory effects of S1P in macrophages.
To confirm the role of the S1P1 receptor, we used bone marrow–derived macrophages from S1P2-deficient mice (see red bars in Figure 3).16 These mice are viable, but have been found recently to be deaf.16 We confirmed the absence of S1P2 expression in these macrophages; also, there was no compensatory upregulation of S1P1 in these cells (Figure 1). In S1P2-deficient macrophages, LPS caused a slightly blunted response in TNFα secretion (red bars in Figure 3C) and iNOS activity (red bars in Figure 3B), suggesting that the absence of the S1P2 receptor impacts the magnitude of the LPS response in macrophages. Arg I activity, however, appeared similar in both wild-type and S1P2-deficient macrophages (Figure 3A). Both S1P and SEW2871 again significantly reduced LPS-induced iNOS activity and TNFα secretion by these S1P2-deficient macrophages (red bars in Figure 3). S1P and SEW2871 also increased Arg I activity in both B6 and S1P2 KO macrophages to similar extents (Figure 3).
We also found that VPC44116, a selective S1P1 receptor antagonist, was a potent inhibitor of both S1P and SEW2871 action in wild-type and S1P2KO macrophages (see VPC bars in Figure 3). VPC44116 worked as effectively in S1P2KO macrophages as in wild-type macrophages to prevent S1P and SEW2871 action on TNFα secretion as well as iNOS and Arg I activities (see red bars in Figure 3).
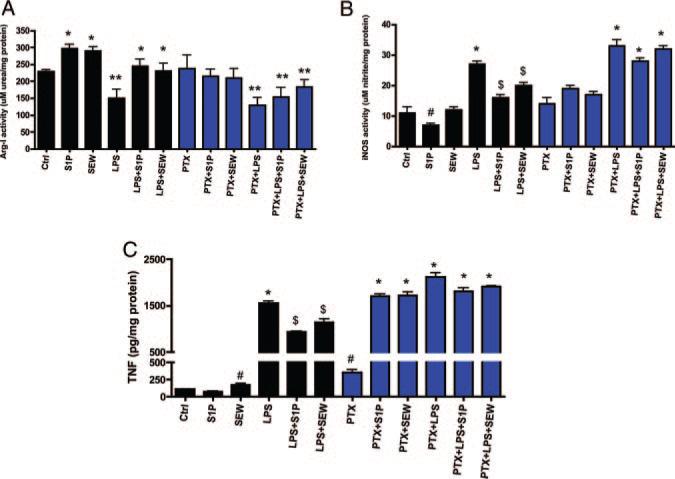
S1P1 is solely coupled to Gαi whereas S1P2 couples primarily to Gαq. To uncouple Gαi signaling in macrophages, we treated macrophages overnight with 100 ng/mL pertussis toxin as another means to target S1P1. As shown in Figure 4, treatment of macrophages with pertussis toxin reduced the action of both S1P and SEW2871 on Arg I and iNOS activities (blue bars in Figure 4A and 4B), as well as TNFα secretion (blue bars in Figure 4C).
Figure 4.
Uncoupling of Gαi signaling in macrophages blocks S1P action. Peritoneal macrophages from C57BL/6J mice (black bars) were incubated with media alone (Ctrl) or with 10 ng/mL LPS in the absence or presence of 500 nmol/L S1P for 4 hours. In some cases, cells were pretreated overnight with pertussis toxin (PTX) (blue bars) before LPS or S1P incubation to uncouple Gαi signaling. A, Arg activity. Arg activity was quantified in cell lysates by measuring the conversion of l-arginine to urea. *P<0.001 vs Ctrl, **P<0.01 vs Ctrl by ANOVA. B, iNOS activity. Total nitrate and nitrite levels were measured with the Griess reagent as described in Materials and Methods. #P<0.05 vs Ctrl, *P<0.001 vs Ctrl, $P<0.01 vs LPS by ANOVA. C, TNFα secretion. TNFα levels were measured using ELISA. #P<0.05 vs Ctrl, *P<0.001 vs Ctrl, $P<0.01 vs LPS by ANOVA.
As another approach to target S1P1, we used small interfering RNA to reduce expression of S1P1 in macrophages. We achieved a 40% knockdown of S1P1 in RAW macrophages. This resulted in an ≈40% reduction in the ability of either S1P or SEW2871 to either induce Arg I or suppress iNOS or TNFα mRNA expression (see Figure I in the online data supplement). The S1P1 small interfering RNA reduced S1P action in S1P2KO macrophages (supplemental Figure I), again supporting the notion that the S1P1 receptor is mediating many of the antiinflammatory effects of S1P on macrophage function.
iNOS Promoter Activity in RAW264.7 Macrophages
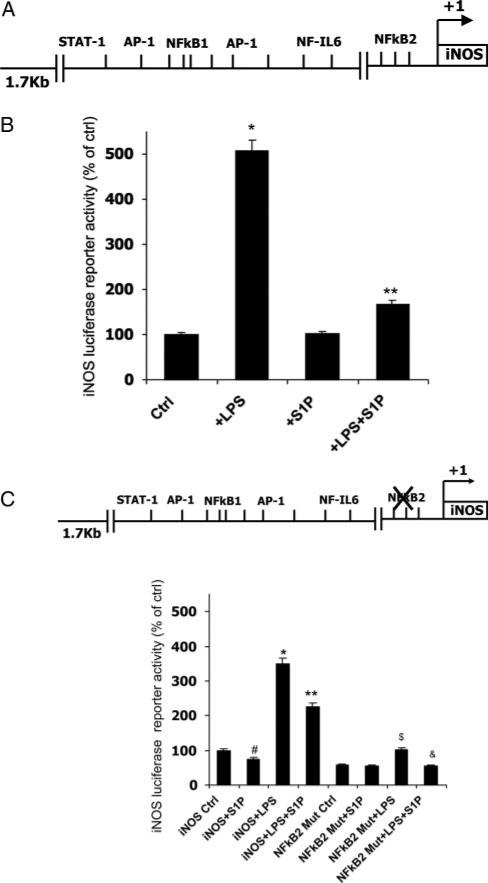
Because iNOS is such an important mediator of macrophage inflammatory responses, we wanted to further examine the regulation of iNOS promoter activity by S1P. We cloned a 1.7-kb region of 5′-flanking DNA located upstream of the transcriptional start site of the murine iNOS gene. This promoter region contains putative NFκB, activator protein (AP)-1, and STAT-1 response elements (Figure 5A). Xie et al have shown that the LPS-mediated induction of iNOS transcriptional activity is primarily attributable to 1 of 2 NFκB binding sites (termed NFκB2 in Figure 5A) located within this promoter region.25 We subcloned this 1.7-kb iNOS promoter region into pGL3-luciferase and transfected RAW macrophages. We found that LPS stimulated iNOS promoter activity by 400%, whereas S1P significantly reduced the LPS-mediated activation of the iNOS promoter (Figure 5B). Next, we mutated the distal NFκB site that confers LPS induction of iNOS (termed NFκB2 in Figure 5C). Mutation of this single NFκB site reduced the ability of LPS to induce iNOS promoter activity by 75% to 80% (Figure 5C). However, although the bulk of the S1P effect was lost, there remained slight residual S1P activity (≈25%) in the absence of this functional NFκB2 site (Figure 5C).
Figure 5.
S1P modulates mouse iNOS promoter activity through the transcription factor NFκB. A, Diagram of putative transcription factor binding sites in the 1.7-kb murine iNOS promoter region. B, iNOS promoter-reporter activity. RAW264.7 macrophages were transiently transfected with a luciferase reporter plasmid containing 1.7 kb upstream of the transcriptional start site of the murine iNOS promoter. At 24 hours after transfection, macrophages were incubated in media alone (Ctrl), 10 ng/mL LPS (+LPS), 500 nmol/L S1P (+S1P), or both (+LPS+S1P) for 24 hours. Cells were harvested, and luciferase activity was measured. Luciferase values were normalized to Renilla luciferase. *P<0.001 vs Ctrl, **P<0.001 vs LPS by ANOVA. Values are set to 100% for comparison between transfections; n=4 experiments performed in triplicate. C, Mutant iNOS promoter-reporter activity. RAW264.7 macrophages were transfected with a luciferase reporter plasmid containing the 1.7 kb upstream of the transcriptional start site of the murine iNOS promoter in which the NFκB site no. 2 was mutated (NFκB2 Mut). Luciferase values were normalized to Renilla luciferase values. Values are set to 100% for comparison between transfections; n=3 experiments performed in triplicate. *P<0.001 vs Ctrl, **P<0.005 vs LPS, #P<0.05 vs Ctrl, $P<0.01 vs NFκB Mut Ctrl, &P<0.02 vs NFκB Mut+LPS by ANOVA.
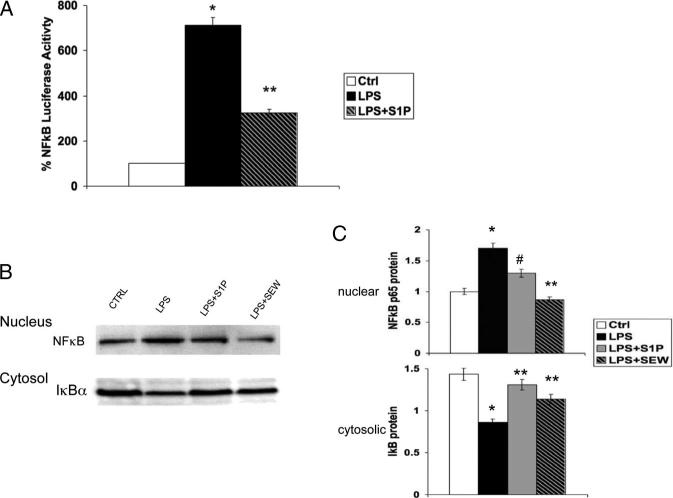
Because we previously reported that S1P inhibits NFκB translocation to the nucleus in aortic endothelial cells,26 we focused our efforts on NFκB-mediated regulation of iNOS and TNFα. Using a NFκB promoter-reporter construct that contained 3 tandem NFκB elements linked to the luciferase gene, we confirmed that LPS activated NFκB and that S1P significantly reduced NFκB activation (Figure 6A). NFκB resides in the cytosol in an inactive state, where it is sequestered by IκB. Degradation of IκB frees NFκB, allowing it to mobilize to the nucleus to initiate gene transcription. We found that S1P both reduced the amount of p65-NFκB that mobilized to the nucleus and also increased the amount of IκB remaining in the cytosol (Figure 6B and 6C). Thus, we anticipate that regulation of IκB expression is 1 mechanism by which S1P reduces NFκB activation to regulate expression of inflammatory genes, such as iNOS and TNFα, in macrophages.
Figure 6.
S1P modulates NFκB activation in macrophages. A, NFκB reporter activation. RAW264.7 macrophages were transfected with a NFκB-luciferase reporter plasmid. After 48 hours, cells were incubated in media alone (Ctrl), 10 ng/mL LPS (+LPS), or LPS +500 nmol/L S1P (+LPS+S1P) for 4 hours. Luciferase activity was then measured. *P<0.001 vs Ctrl, **P<0.001 vs LPS by ANOVA. B, Bone marrow–derived macrophages from C57BL/6J mice were incubated with 10 ng/mL LPS, 500 nmol/L S1P, or 1 μmol/L SEW2871 for 2 hours. Nuclear and cytosol extracts were collected and analyzed by SDS-PAGE for NFκB p65 and IκBα. A representative immunoblot is shown. Top, Nuclear extract probed with anti-NFκB p65 antibody. Bottom, Cytosolic extract probed with anti-IκBα antibody. C, Densitometry. Graphical representation of immunoblots in B normalized to histone H1 (nuclear) and tubulin (cytosol) from 3 independent experiments. *P<0.001 vs Ctrl, #P<0.003 vs LPS, **P<0.001 vs LPS by ANOVA.
Finally, we confirmed that S1P acts through the NFκB pathway to regulate TNFα secretion. As shown in supplemental Figure II, macrophages incubated with LPS show significant elevations in TNFα secretion. Treatment of macrophages with a well-characterized NFκB inhibitor caused a dose-dependent reduction in TNFα secretion by these macrophages. S1P was unable to further reduce TNFα secretion, again supporting the notion that S1P inhibits NFκB signaling in macrophages.
Discussion
Atherosclerosis development in the aortic wall is initiated by endothelial and monocyte/macrophage activation. The present study demonstrates a role for S1P in mediating antiinflammatory actions in macrophages by switching the macrophage phenotype from a proinflammatory classic phenotype to an antiinflammatory alternative phenotype primarily through action on the macrophage S1P1 receptor. LPS stimulates macrophage production of proinflammatory cytokines such as TNFα. In the present study, we show that S1P significantly reduces LPS-mediated expression of proinflammatory cytokines and the enzyme iNOS and stimulates Arg I expression in macrophages, indicating that S1P promotes an antiinflammatory macrophage phenotype (see Figure 2). This notion of phenotypic switching in macrophages by S1P was further supported by studies showing a direct effect of S1P on macrophage Arg and iNOS enzymatic activities (Figure 3). In addition to upregulation of Arg I, we also found increased production of the antiinflammatory cytokines transforming growth factor-β and IL-10 in S1P-treated macrophages (data not shown). Thus, S1P promotes a novel macrophage phenotype that is antiinflammatory in nature.
Furthermore, in the present study, we observed antiinflammatory effects of S1P at concentrations of 100 nmol/L to 1 μmol/L, although the maximal effect on induction of Arg I and reduction of iNOS was at 500 nmol/L S1P rather than 1 μmol/L S1P. There is an inverse relationship between the concentration of S1P and its effectiveness in modulating LPS responses in macrophages (Figure 2A). We have found this to be the case in studies of endothelial cells.27 The physiological concentrations of S1P in blood are in the nanomolar range; the affinity of S1P for the S1P class of receptors lies in the nanomolar range.9 Consistently, we have found that 1 to 10 μmol/L concentrations of S1P actually blunt the antiinflammatory effects of S1P in both macrophages and endothelial cells in a dose-dependent manner. One likely explanation for such findings is that the higher concentrations of S1P may activate receptors (other than S1P receptors) in a nonspecific manner that results in stimulation of proinflammatory signaling pathways.
Using a combination of KO mice and pharmacological reagents, we found that most of the phenotypic switching is driven by S1P action on the S1P1 receptor. S1P1-deficient mice display embryonic lethality, so we used SEW2871, a pharmacological agonist of the S1P1 receptor, and VPC44116, a pharmacological antagonist of the S1P1 receptor, to delineate the role of S1P1 in mediating these effects in macrophages. Because macrophages express only S1P1 and S1P2 receptors (Figure 1), we used S1P2-deficient mice to delineate the role of S1P2 in these processes. We found that S1P1 receptor signaling was responsible for the upregulation of Arg I and the downregulation of TNFα and iNOS (Figures 2 through 4 and supplemental Figure I). We confirmed that iNOS, TNFα, and Arg I regulation was mediated by the S1P1 receptor in that S1P2-deficient macrophages responded in a similar manner as wild-type macrophages to S1P and SEW2871 (Figure 3). We did observe that the response of S1P2-deficient macrophages to LPS, S1P, and SEW2871 was blunted compared with wild-type macrophages. It is possible that S1P could activate S1P2 to somehow oppose signaling through S1P1 and, thereby, reduce the antiinflammatory effect of the macrophage S1P-S1P1 axis. The role of S1P2 in macrophage phenotypic switching is unknown; we anticipate that direct S1P signaling through S1P2 may also have antiinflammatory effects. Indeed, we did find that S1P2 mediated the upregulation of IL-10 in macrophages (data not shown). S1P caused a significant 4-fold induction of IL-10 mRNA and protein in macrophages from C57BL/6J mice, but this induction was not observed in macrophages isolated from S1P2-deficient mice and treated with S1P (data not shown). Thus, the S1P2 receptor does appear to play a role in regulating the phenotype of macrophages, and this role may also have significance in atherosclerosis through upregulation of Th2-like responses via IL-10 signaling.
Previously, we demonstrated that S1P decreases NFκB nuclear translocation in endothelial cells.26 Based on this knowledge, we wondered whether the S1P-mediated reduction of iNOS expression was through inhibition of NFκB. As shown in Figure 5, we confirmed that NFκB is an important transcription factor that regulates iNOS activation by LPS. Mutation of the NFκB2 binding site in the murine iNOS promoter dramatically inhibited LPS-mediated activation of iNOS (Figure 5C). However, despite the mutation of the NFκB2 site, S1P was still able to downregulate iNOS promoter activity by approximately 25%. In preliminary studies, we have found that S1P reduces AP-1 and STAT-1 activation (data not shown), so the additional action(s) of S1P on iNOS regulation could be either through inhibition of the remaining NFκB1 site or through inhibition of AP-1 and/or STAT-1 activation. All 3 of these transcription factors have been shown to be important for iNOS activation by LPS, although NFκB is believed to be the primary regulatory transcription factor. It is unlikely that S1P would have a direct effect on transcription factors or bind directly to DNA. Most likely, S1P inhibits an upstream target of the transcription factor. For instance, we found that S1P activates mitogen-activated protein kinase phosphatases (data not shown), which inhibit mobilization of c-Jun and extracellular signal-regulated kinase 1/2 to the nucleus for initiation of gene transcription. Inhibition of c-Jun translocation to the nucleus would reduce AP-1 activation, for example. Furthermore, we found that S1P significantly reduces NFκB activation in macrophages, and this appears to be attributable, at least in part, to regulation of IκB expression (Figure 6). We have previously reported that S1P induces IκB synthesis within 30 minutes of incubation, which is very much within the time-frame of the present studies.26 Taken together, our data indicate that the primary regulation of iNOS by LPS is through NFκB and that S1P serves to reduce NFκB activation, at least in part, through regulation of IκB expression. Our data also suggest that regulating NFκB activation may serve as an important signaling component for macrophage phenotypic switching in the vascular wall.
In summary, we show that S1P promotes phenotypic switching of macrophages to a novel, alternative antiinflammatory phenotype. These macrophages show decreased NFκB activation and have reduced production of proinflammatory cytokines and nitric oxide. This novel antiinflammatory macrophage phenotype is mostly driven by the S1PS1P1 receptor axis. Therefore, S1P could provide a beneficial therapeutic effect for reducing macrophage-mediated inflammation in the vessel wall through specific activation of the S1P1 receptor.
Supplementary Material
Supplemental Materials and Methods:
Reagents: RAW264.7 macrophages and PGL3 was a gift from Dr. Michael Smith (University of Virginia). The plasmid containing the full-length murine iNOS cDNA was obtained from Dr. Lisa Palmer (University of Virginia). The Griess Reagent assay was obtained from Cayman Chemicals. The arginase activity assay was from BioAssay Systems. FBS was ordered from HyClone (Logan, UT). Thioglycollate medium was obtained from Sigma. S1P was obtained from BIOMOL Research Laboratories BAY11-7085, LPS and SEW2871 were obtained from Sigma. Cytokine ELISA kits were obtained from R&D Systems.
Conventional and Quantitative real-time PCR: Peritoneal macrophages were freshly isolated from B6 and cultured as described above for mRNA measurements. Total cellular RNA was obtained from peritoneal macrophages as previously described1. For measurement of S1P receptor mRNA abundance by conventional RT-PCR, 2ul cDNA from each experimental group was utilized. Conventional RT-PCR conditions for arginase-I, arginase-II, iNOS, S1P receptors, and β-actin were as follows: 95°C 3min, followed by 38 cycles of 95°C 45 sec, 59.5°C 60 sec, 72°C 60 sec, with a final extension time of 7 min at 72°C. Bands were analyzed on a 1.0% agarose gel in 1X TAE buffer. PCR for β-actin was performed as a control for normalization purposes. For quantitative real-time PCR analysis of mRNA expression (arginase-I, arginase-II, iNOS, COX-2, cytokines), cDNA was obtained as described above, and was diluted 1:8; 4uL of this dilution was used for each PCR reaction. Reagents from the BioRad real-time PCR kit containing Sybr Green were used for quantitative PCR reactions. The PCR conditions were: 95 °C 3 min, followed by 40 cycles of 95 °C 10 secs, 60 °C 30 secs, and 72 °C 45 secs. Data were analyzed as previously described1;2. Primer sequences for all PCR reactions are shown in Online Table I and Online Table II.
Arginase enzymatic assay: Macrophages were plated in RPMI/10% FBS at 37 °C for 24h. Cells were then washed and treated in RPMI/1% FBS with 500nM S1P 30min prior to the stimulation with LPS for another 4h. Arginase activity was measured in cell lysates as described previously3.
Griess reagent assay: Supernatants from macrophages were collected and assayed by the Griess assay for inducible NO production according to manufacturer's instructions (Cayman Chemicals). Briefly, 80ul aliquots of supernatant or sodium nitrate standards were combined with equal volumes of fresh Griess reagent (1% sulfanilamide, 0.1% naphthylethylenediamine dihydrochloride, 2.5% H3PO4). Samples were incubated at room temperature for 10min, and the absorbance was measured at 540 nm. The concentration of nitrite (NO2-) and nitrate (NO2-) was determined using a sodium nitrate standard curve.
Cytokine measurements: Macrophages were cultured using RPMI containing 1% heat-inactivated FBS with 10ng/ml LPS ± 500nM S1P. Cell culture supernatant was quantitatively measured for TNFα protein levels using R&D Systems ELISA kit according to manufacturer's instructions.
Immunoblotting for NFκB and IκB: Macrophages were incubated in RPMI + 1% FBS with LPS (25ng/ml), S1P (500nM) or SEW2871 (1uM) for 1 hour. Cytosol and nuclear extracts were collected from macrophages using the NE-PER kit (Pierce) according to the manufacturer's instructions. 50 μg of protein was analyzed by SDS-PAGE on 4-12% gels (Invitrogen). Cytosol was probed for IκBα 1:2000 (Cell Signaling) and nuclear extract for NFκB p65 1:2000 (Santa Cruz). Cytosol and nuclear extracts were normalized to tubulin 1:5000 (Sigma) or histone H1 1:2000 (Santa Cruz) respectively.
Inhibition of S1P1 gene expression by SiRNA: SiRNA for murine S1P1 was expressed in pSuper (OligoEngine, Seattle, WA). The target nucleotide sequence for S1P1 is 5’-CTATGATATCATAGTCCGG-3’ Bone marrow derived macrophages from S1P2-deficient and littermate control mice were transfected with 2 μg S1P1SiRNA plasmid using a mouse macrophage nucleofector kit (Amaxa Biosystems). After 48hrs, macrophages were incubated with LPS, S1P and SEW2871 as described in the figure legends.
Transfections: RAW264.7 macrophages were transiently transfected using the Amaxa Nucleofector II and Cell Line Nucleofector Kit V according to manufacturers protocol. The D-032 setting was determined to provide the highest transfection efficiency (>70%) with the least amount of cell death (<10%). At 24h post-transfection, cells were treated for 4h with 10ng/ml LPS ± 500nM S1P. Luciferase activities in cell extracts were measured using the Dual-Luciferase® assay system (Promega) with a Berthold Syrius Luminometer (Fisher Scientific). Luciferase activity was corrected for transfection efficiency against Renilla luciferase activity and expressed relative to untreated controls.
Statistical Analyses: Data for all experiments were analyzed using the StatView 6.0 software program. Comparisons between groups were performed using oneway analysis of variance (ANOVA) methods. Data are graphically represented as mean + SE, in which each mean consists of 4 experiments performed in triplicate (unless noted otherwise in the figure legends) using 6-8 mice per group. Comparisons between groups and tests of interactions were made assuming a two-factor analysis with the interaction term testing each main effect with the residual error testing the interaction. All comparisons were made using Fisher's LSD procedure, so that multiple comparisons were made at the 0.05 level only if the overall F- test from the ANOVA was significant at p<0.05.
References
1. Mauldin JP, Srinivasan S, Mulya A, Gebre A, Parks JS, Daugherty A, Hedrick CC. Reduction in ABCG1 in Type 2 diabetic mice increases macrophage foam cell formation. J Biol Chem. 2006;281:21216-21224.
2. Bolick DT, Whetzel AM, Skaflen M, Deem TL, Lee J, Hedrick CC. Absence of the G protein-coupled receptor G2A in mice promotes monocyte/endothelial interactions in aorta. Circ Res. 2007;100:572-580.
3. Pesce J, Kaviratne M, Ramalingam TR, Thompson RW, Urban JF, Jr., Cheever AW, Young DA, Collins M, Grusby MJ, Wynn TA. The IL-21 receptor augments Th2 effector function and alternative macrophage activation. J Clin Invest. 2006;116:2044-2055.
Online Figure I. S1P1 siRNA reduces anti-inflammatory action of S1P in macrophages. Bone marrow-derived macrophages from either wild-type littermate controls (WT; black bars) or S1P2-deficient (S1P2KO; red bars) mice (n=5 mice per group) were transfected with S1PsiRNA as described in Materials and Methods. At 48h after transfection, macrophages were treated with LPS (10 ng/ml) in the absence or presence of 500nM S1P, or 1μM SEW2871 (+SEW) for 4h. Quantitative real-time PCR for murine arginase-I (Arg-I), iNOS and TNFα were performed. Cyclophilin was measured as a housekeeping gene for normalization purposes. Data are expressed using the relative expression method using B6 control as set to 1. Panel A, Arginase-I expression. *p<0.0001 versus WT/S1P2KO. Panel B iNOS expression. #p<0.005 versus WT/S1P2KO; **p<0.009 versus S1P2KO by ANOVA. Panel C, TNFα expression. TNFα levels were measured using ELISA. #p<0.005 versus WT/S1P2KO; $p<0.05 versus WT/S1P2KO, p<0.05; &p<0.01 versus S1P2KO+S1P1SiRNA+LPS by ANOVA.
Online Figure II. NFκB regulates LPS-mediated induction of TNFα in macrophages. Peritoneal macrophages from C57BL/6J mice were incubated for 4h with 10ng/ml LPS in the absence (+LPS) or presence of a 1h-pretreatment with either 5μM or 10μM BAY11-7085 (+Bay). In some cases, cells were also incubated with 500nM S1P (+ S1P). TNFα secretion into media was measured using ELISA and normalized to total cell protein. *p<0.0001 versus Ctrl; #p<0.001 versus LPS; **p<0.0001 versus LPS by ANOVA.
Online Table I
Quantitative Real-time PCR primers used in this study:
Online Table II
Conventional PCR primers used in this study:
Acknowledgments
We thank Dr Norbert Leitinger (University of Virginia) for helpful discussions and for reagents for Arg I, Dr Lisa Palmer (University of Virginia) for the murine iNOS promoter plasmid, David T. Bolick (University of Virginia) for assistance with NFκB signaling studies, and Jeremy P. Mauldin (University of Virginia) for assistance with NFκB mutation studies.
Sources of Funding
This research was supported by grants from the Juvenile Diabetes Research Foundation (to C.C.H. and K.R.L.), NIH grant HL079621 (to C.C.H.), and the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases, NIH (to R.L.P.).
Footnotes
Disclosures
None.
References
- 1.Ross R. Atherosclerosis–an inflammatory disease. N Engl J Med. 1999;340:115–126. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 2.Stout RD, Jiang C, Matta B, Tietzel I, Watkins SK, Suttles J. Macrophages sequentially change their functional phenotype in response to changes in microenvironmental influences. J Immunol. 2005;175:342–349. doi: 10.4049/jimmunol.175.1.342. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 4.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–212. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 5.Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol. 1998;160:5347–5354. [PubMed] [Google Scholar]
- 6.Hesse M, Modolell M, La Flamme AC, Schito M, Fuentes JM, Cheever AW, Pearce EJ, Wynn TA. Differential regulation of nitric oxide synthase-2 and arginase I by type 1/type 2 cytokines in vivo: granulomatous pathology is shaped by the pattern of L-arginine metabolism. J Immunol. 2001;167:6533–6544. doi: 10.4049/jimmunol.167.11.6533. [DOI] [PubMed] [Google Scholar]
- 7.Rutschman R, Lang R, Hesse M, Ihle JN, Wynn TA, Murray PJ. Cutting edge: Stat6-dependent substrate depletion regulates nitric oxide production. J Immunol. 2001;166:2173–2177. doi: 10.4049/jimmunol.166.4.2173. [DOI] [PubMed] [Google Scholar]
- 8.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, Hamid Q, Rothenberg ME. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–1874. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spiegel S, Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat Rev Mol Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- 10.Brinkmann V. Sphingosine 1-phosphate receptors in health and disease: mechanistic insights from gene deletion studies and reverse pharmacology. Pharmacol Ther. 2007;115:84–105. doi: 10.1016/j.pharmthera.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa R, Takahashi M, Hirose S, Morimoto H, Ise H, Murakami T, Yasue T, Kuriyama K, Hongo M, Kobayashi E, Ikeda U. A novel sphingosine-1-phosphate receptor agonist KRP-203 attenuates rat autoimmune myocarditis. Biochem Biophys Res Commun. 2007;361:621–628. doi: 10.1016/j.bbrc.2007.07.061. [DOI] [PubMed] [Google Scholar]
- 12.Klingenberg R, Nofer JR, Rudling M, Bea F, Blessing E, Preusch M, Grone HJ, Katus HA, Hansson GK, Dengler TJ. Sphingosine-1-phosphate analogue FTY720 causes lymphocyte redistribution and hypercholesterolemia in ApoE-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:2392–2399. doi: 10.1161/ATVBAHA.107.149476. [DOI] [PubMed] [Google Scholar]
- 13.Nofer JR, Bot M, Brodde M, Taylor PJ, Salm P, Brinkmann V, van Berkel T, Assmann G, Biessen EA. FTY720, a synthetic sphingosine 1 phosphate analogue, inhibits development of atherosclerosis in low–density lipoprotein receptor-deficient mice. Circulation. 2007;115:501–508. doi: 10.1161/CIRCULATIONAHA.106.641407. [DOI] [PubMed] [Google Scholar]
- 14.Keul P, Tolle M, Lucke S, von Wnuck LK, Heusch G, Schuchardt M, van der GM, Levkau B. The sphingosine-1-phosphate analogue FTY720 reduces atherosclerosis in apolipoprotein E-deficient mice. Arterioscler Thromb Vasc Biol. 2007;27:607–613. doi: 10.1161/01.ATV.0000254679.42583.88. [DOI] [PubMed] [Google Scholar]
- 15.Weigert A, Johann AM, von Knethen A, Schmidt H, Geisslinger G, Brune B. Apoptotic cells promote macrophage survival by releasing the antiapoptotic mediator sphingosine-1-phosphate. Blood. 2006;108:1635–1642. doi: 10.1182/blood-2006-04-014852. [DOI] [PubMed] [Google Scholar]
- 16.Kono M, Mi Y, Liu Y, Sasaki T, Allende ML, Wu YP, Yamashita T, Proia RL. The sphingosine-1-phosphate receptors S1P1, S1P2, and S1P3 function coordinately during embryonic angiogenesis. J Biol Chem. 2004;279:29367–29373. doi: 10.1074/jbc.M403937200. [DOI] [PubMed] [Google Scholar]
- 17.de Bruijn MF, Slieker WA, van der Loo JC, Voerman JS, van Ewijk W, Leenen PJ. Distinct mouse bone marrow macrophage precursors identified by differential expression of ER-MP12 and ER-MP20 antigens. Eur J Immunol. 1994;24:2279–2284. doi: 10.1002/eji.1830241003. [DOI] [PubMed] [Google Scholar]
- 18.Itoh N, Shibayama H, Kanekiyo M, Namphung D, Nakanishi T, Matsuyama A, Odani T, Tanaka K. Reduced bactericidal activity and nitric oxide production in metallothionein-deficient macrophages in response to lipopolysaccharide stimulation. Toxicology. 2005;216:188–196. doi: 10.1016/j.tox.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 19.Nelin LD, Wang X, Zhao Q, Chicoine LG, Young TL, Hatch DM, English BK, Liu Y. MKP-1 switches arginine metabolism from nitric oxide synthase to arginase following endotoxin challenge. Am J Physiol Cell Physiol. 2007;293:C632–C640. doi: 10.1152/ajpcell.00137.2006. [DOI] [PubMed] [Google Scholar]
- 20.Marathe C, Bradley MN, Hong C, Lopez F, Ruiz de Galarreta CM, Tontonoz P, Castrillo A. The arginase II gene is an anti-inflammatory target of liver X receptor in macrophages. J Biol Chem. 2006;281:32197–32206. doi: 10.1074/jbc.M605237200. [DOI] [PubMed] [Google Scholar]
- 21.Kimura T, Sato K, Kuwabara A, Tomura H, Ishiwara M, Kobayashi I, Ui M, Okajima F. Sphingosine 1-phosphate may be a major component of plasma lipoproteins responsible for the cytoprotective actions in human umbilical vein endothelial cells. J Biol Chem. 2001;276:31780–31785. doi: 10.1074/jbc.M104353200. [DOI] [PubMed] [Google Scholar]
- 22.Sanna MG, Liao J, Jo E, Alfonso C, Ahn MY, Peterson MS, Webb B, Lefebvre S, Chun J, Gray N, Rosen H. Sphingosine 1-phosphate (S1P) receptor subtypes S1P1 and S1P3, respectively, regulate lymphocyte recirculation and heart rate. J Biol Chem. 2004;279:13839–13848. doi: 10.1074/jbc.M311743200. [DOI] [PubMed] [Google Scholar]
- 23.Xie QW, Kashiwabara Y, Nathan C. Role of transcription factor NF-kappa B/Rel in induction of nitric oxide synthase. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 24.Foss FW, Jr, Snyder AH, Davis MD, Rouse M, Okusa MD, Lynch KR, Macdonald TL. Synthesis and biological evaluation of gamma-aminophosphonates as potent, subtype-selective sphingosine 1-phosphate receptor agonists and antagonists. Bioorg Med Chem. 2007;15:663–677. doi: 10.1016/j.bmc.2006.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awad AS, Ye H, Huang L, Li L, Foss FW, Jr, Macdonald TL, Lynch KR, Okusa MD. Selective sphingosine 1-phosphate 1 receptor activation reduces ischemia-reperfusion injury in mouse kidney. Am J Physiol Renal Physiol. 2006;290:F1516–F1524. doi: 10.1152/ajprenal.00311.2005. [DOI] [PubMed] [Google Scholar]
- 26.Whetzel AM, Bolick DT, Srinivasan S, Macdonald TL, Morris MA, Ley K, Hedrick CC. Sphingosine-1 phosphate prevents monocyte/endothelial interactions in type 1 diabetic NOD mice through activation of the S1P1 receptor. Circ Res. 2006;99:731–739. doi: 10.1161/01.RES.0000244088.33375.52. [DOI] [PubMed] [Google Scholar]
- 27.Bolick DT, Srinivasan S, Kim KW, Hatley ME, Clemens JJ, Whetzel A, Ferger N, Macdonald TL, Davis MD, Tsao PS, Lynch KR, Hedrick CC. Sphingosine-1-phosphate prevents tumor necrosis factor-{alpha}-mediated monocyte adhesion to aortic endothelium in mice. Arterioscler Thromb Vasc Biol. 2005;25:976–981. doi: 10.1161/01.ATV.0000162171.30089.f6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Materials and Methods:
Reagents: RAW264.7 macrophages and PGL3 was a gift from Dr. Michael Smith (University of Virginia). The plasmid containing the full-length murine iNOS cDNA was obtained from Dr. Lisa Palmer (University of Virginia). The Griess Reagent assay was obtained from Cayman Chemicals. The arginase activity assay was from BioAssay Systems. FBS was ordered from HyClone (Logan, UT). Thioglycollate medium was obtained from Sigma. S1P was obtained from BIOMOL Research Laboratories BAY11-7085, LPS and SEW2871 were obtained from Sigma. Cytokine ELISA kits were obtained from R&D Systems.
Conventional and Quantitative real-time PCR: Peritoneal macrophages were freshly isolated from B6 and cultured as described above for mRNA measurements. Total cellular RNA was obtained from peritoneal macrophages as previously described1. For measurement of S1P receptor mRNA abundance by conventional RT-PCR, 2ul cDNA from each experimental group was utilized. Conventional RT-PCR conditions for arginase-I, arginase-II, iNOS, S1P receptors, and β-actin were as follows: 95°C 3min, followed by 38 cycles of 95°C 45 sec, 59.5°C 60 sec, 72°C 60 sec, with a final extension time of 7 min at 72°C. Bands were analyzed on a 1.0% agarose gel in 1X TAE buffer. PCR for β-actin was performed as a control for normalization purposes. For quantitative real-time PCR analysis of mRNA expression (arginase-I, arginase-II, iNOS, COX-2, cytokines), cDNA was obtained as described above, and was diluted 1:8; 4uL of this dilution was used for each PCR reaction. Reagents from the BioRad real-time PCR kit containing Sybr Green were used for quantitative PCR reactions. The PCR conditions were: 95 °C 3 min, followed by 40 cycles of 95 °C 10 secs, 60 °C 30 secs, and 72 °C 45 secs. Data were analyzed as previously described1;2. Primer sequences for all PCR reactions are shown in Online Table I and Online Table II.
Arginase enzymatic assay: Macrophages were plated in RPMI/10% FBS at 37 °C for 24h. Cells were then washed and treated in RPMI/1% FBS with 500nM S1P 30min prior to the stimulation with LPS for another 4h. Arginase activity was measured in cell lysates as described previously3.
Griess reagent assay: Supernatants from macrophages were collected and assayed by the Griess assay for inducible NO production according to manufacturer's instructions (Cayman Chemicals). Briefly, 80ul aliquots of supernatant or sodium nitrate standards were combined with equal volumes of fresh Griess reagent (1% sulfanilamide, 0.1% naphthylethylenediamine dihydrochloride, 2.5% H3PO4). Samples were incubated at room temperature for 10min, and the absorbance was measured at 540 nm. The concentration of nitrite (NO2-) and nitrate (NO2-) was determined using a sodium nitrate standard curve.
Cytokine measurements: Macrophages were cultured using RPMI containing 1% heat-inactivated FBS with 10ng/ml LPS ± 500nM S1P. Cell culture supernatant was quantitatively measured for TNFα protein levels using R&D Systems ELISA kit according to manufacturer's instructions.
Immunoblotting for NFκB and IκB: Macrophages were incubated in RPMI + 1% FBS with LPS (25ng/ml), S1P (500nM) or SEW2871 (1uM) for 1 hour. Cytosol and nuclear extracts were collected from macrophages using the NE-PER kit (Pierce) according to the manufacturer's instructions. 50 μg of protein was analyzed by SDS-PAGE on 4-12% gels (Invitrogen). Cytosol was probed for IκBα 1:2000 (Cell Signaling) and nuclear extract for NFκB p65 1:2000 (Santa Cruz). Cytosol and nuclear extracts were normalized to tubulin 1:5000 (Sigma) or histone H1 1:2000 (Santa Cruz) respectively.
Inhibition of S1P1 gene expression by SiRNA: SiRNA for murine S1P1 was expressed in pSuper (OligoEngine, Seattle, WA). The target nucleotide sequence for S1P1 is 5’-CTATGATATCATAGTCCGG-3’ Bone marrow derived macrophages from S1P2-deficient and littermate control mice were transfected with 2 μg S1P1SiRNA plasmid using a mouse macrophage nucleofector kit (Amaxa Biosystems). After 48hrs, macrophages were incubated with LPS, S1P and SEW2871 as described in the figure legends.
Transfections: RAW264.7 macrophages were transiently transfected using the Amaxa Nucleofector II and Cell Line Nucleofector Kit V according to manufacturers protocol. The D-032 setting was determined to provide the highest transfection efficiency (>70%) with the least amount of cell death (<10%). At 24h post-transfection, cells were treated for 4h with 10ng/ml LPS ± 500nM S1P. Luciferase activities in cell extracts were measured using the Dual-Luciferase® assay system (Promega) with a Berthold Syrius Luminometer (Fisher Scientific). Luciferase activity was corrected for transfection efficiency against Renilla luciferase activity and expressed relative to untreated controls.
Statistical Analyses: Data for all experiments were analyzed using the StatView 6.0 software program. Comparisons between groups were performed using oneway analysis of variance (ANOVA) methods. Data are graphically represented as mean + SE, in which each mean consists of 4 experiments performed in triplicate (unless noted otherwise in the figure legends) using 6-8 mice per group. Comparisons between groups and tests of interactions were made assuming a two-factor analysis with the interaction term testing each main effect with the residual error testing the interaction. All comparisons were made using Fisher's LSD procedure, so that multiple comparisons were made at the 0.05 level only if the overall F- test from the ANOVA was significant at p<0.05.
References
1. Mauldin JP, Srinivasan S, Mulya A, Gebre A, Parks JS, Daugherty A, Hedrick CC. Reduction in ABCG1 in Type 2 diabetic mice increases macrophage foam cell formation. J Biol Chem. 2006;281:21216-21224.
2. Bolick DT, Whetzel AM, Skaflen M, Deem TL, Lee J, Hedrick CC. Absence of the G protein-coupled receptor G2A in mice promotes monocyte/endothelial interactions in aorta. Circ Res. 2007;100:572-580.
3. Pesce J, Kaviratne M, Ramalingam TR, Thompson RW, Urban JF, Jr., Cheever AW, Young DA, Collins M, Grusby MJ, Wynn TA. The IL-21 receptor augments Th2 effector function and alternative macrophage activation. J Clin Invest. 2006;116:2044-2055.
Online Figure I. S1P1 siRNA reduces anti-inflammatory action of S1P in macrophages. Bone marrow-derived macrophages from either wild-type littermate controls (WT; black bars) or S1P2-deficient (S1P2KO; red bars) mice (n=5 mice per group) were transfected with S1PsiRNA as described in Materials and Methods. At 48h after transfection, macrophages were treated with LPS (10 ng/ml) in the absence or presence of 500nM S1P, or 1μM SEW2871 (+SEW) for 4h. Quantitative real-time PCR for murine arginase-I (Arg-I), iNOS and TNFα were performed. Cyclophilin was measured as a housekeeping gene for normalization purposes. Data are expressed using the relative expression method using B6 control as set to 1. Panel A, Arginase-I expression. *p<0.0001 versus WT/S1P2KO. Panel B iNOS expression. #p<0.005 versus WT/S1P2KO; **p<0.009 versus S1P2KO by ANOVA. Panel C, TNFα expression. TNFα levels were measured using ELISA. #p<0.005 versus WT/S1P2KO; $p<0.05 versus WT/S1P2KO, p<0.05; &p<0.01 versus S1P2KO+S1P1SiRNA+LPS by ANOVA.
Online Figure II. NFκB regulates LPS-mediated induction of TNFα in macrophages. Peritoneal macrophages from C57BL/6J mice were incubated for 4h with 10ng/ml LPS in the absence (+LPS) or presence of a 1h-pretreatment with either 5μM or 10μM BAY11-7085 (+Bay). In some cases, cells were also incubated with 500nM S1P (+ S1P). TNFα secretion into media was measured using ELISA and normalized to total cell protein. *p<0.0001 versus Ctrl; #p<0.001 versus LPS; **p<0.0001 versus LPS by ANOVA.
Online Table I
Quantitative Real-time PCR primers used in this study:
Online Table II
Conventional PCR primers used in this study: