Summary
In Escherichia coli, the TolC–AcrAB complex forms a major antibiotic efflux system with broad substrate specificity. During the complex assembly, the periplasmic helices and bottom turns of TolC are thought to interact with a hairpin helix of AcrA and hairpin loops of AcrB respectively. In the present study we show that a four-residue substitution in TolC’s turn 1, which connects outer helices 3 and 4 proximal to TolC’s periplasmic aperture, confers antibiotic hypersensitivity without affecting TolC-mediated phage or colicin infection. However, despite the null-like drug sensitivity phenotype, chemical cross-linking analysis revealed no apparent defects in the ability of the mutant TolC protein to physically interact with AcrA and AcrB. A role for TolC turn 1 residues in the functional assembly of the tripartite efflux pump complex was uncovered through isolating suppressor mutations of the mutant TolC protein that mapped within acrA and by utilizing a labile AcrA protein. The data showed that AcrA-mediated suppression of antibiotic sensitivity was achieved by dilating the TolC aperture/channel in an AcrB-dependent manner. The results underscore the importance of the periplasmic turn 1 of TolC in the functional assembly of the tripartite efflux complex and AcrA in transitioning TolC from its closed to open state.
Introduction
The outer membrane protein TolC, together with the periplasmic adapter protein AcrA and the inner membrane pump protein AcrB, forms the major antibiotic efflux system in Escherichia coli (Fralick, 1996). High resolution structures of all three proteins have been solved (Koronakis et al., 2000; Murakami et al., 2002; Yu et al., 2003; Mikolosko et al., 2006; Murakami et al., 2006; Seeger et al., 2006; Das et al., 2007; Sennhauser et al., 2007; Bavro et al., 2008; Drew et al., 2008), and several models have been proposed that envisage the mechanism by which drugs are expelled by the TolC–AcrAB complex (Koronakis et al., 2000; Andersen et al., 2002; Koronakis, 2003; Eswaran et al., 2004; Fernández-recio et al., 2004; Tamura et al., 2005; Lobedanz et al., 2007; Bavro et al., 2008; Misra and Bavro, 2009; Pos, 2009). These studies have suggested that the transitioning of TolC’s periplasmic aperture from its resting closed state to a transport-active open state is a key event during drug extrusion. For this to occur, TolC’s periplasmic helices guarding the tunnel entrance are proposed to undergo considerable realignments in order to open the aperture in an iris-like fashion (Koronakis et al., 2000). Experiments have identified crucial hydrogen bonds and salt bridges in the proximity of TolC’s aperture that are important for keeping the aperture in a closed state, presumably by constraining the periplasmic helices (Andersen et al., 2002). Independent verification of the involvement of one of the key residues, R367, in aperture closing came from the isolation of a TolC mutant in which a R367H substitution kept TolC’s aperture in a constitutively open state and caused a hypersensitivity phenotype (Augustus et al., 2004). Recent structural resolution of a TolC mutant in its partial open-state confirmed in vivo data and earlier predictions and showed realignment of TolC’s inner helices relative to outer helices in transition to an open aperture (Bavro et al., 2008). While structural and mutational data have been helpful in identifying key TolC residues involved in controlling aperture diameter, precisely how TolC’s outer and inner helices undergo realignments to facilitate aperture widening and what triggers this transition remain poorly understood.
Physical interactions between any two components of the TolC–AcrAB complex in the absence of the third component have been reported (Husain et al., 2004; Tikhonova and Zgurskaya, 2004; Touze et al., 2004; Tamura et al., 2005). However, it is reasonable to expect that TolC’s aperture opening under normal circumstances is accomplished only in the presence of the two other components and when their interacting surfaces are properly juxtaposed (Bavro et al., 2008; Misra and Bavro, 2009). Consistent with this notion that proper interfacing between the three components of the efflux complex is required, expression of efflux components from heterologous bacterial sources does not often lead to the functional assembly of an efflux-competent complex (Bokma et al., 2006; Stegmeier et al., 2006; Vediyappan et al., 2006; Krishnamoorthy et al., 2008) despite the fact that the heterologous components can physically interact, as shown by chemical cross-linking analysis (Bokma et al., 2006; Stegmeier et al., 2006; Vediyappan et al., 2006). However, gain-of-function compensatory mutations can be obtained that make heterologous complexes partially functional, presumably by establishing quasi-normal interfacing between heterologous partners (Bokma et al., 2006; Vediyappan et al., 2006; Krishnamoorthy et al., 2008). On the other hand, functional interactions among native partners can be disrupted by loss-of-function missense mutations in one component and restored by compensatory alterations in the other interacting component (Gerken and Misra, 2004). These compensatory alterations often identify residues or regions of proteins that directly or indirectly influence interfacing between the interacting partners.
Since proper interfacing between TolC, AcrA and AcrB is required for the assembly of an active efflux complex, it is important to identify residues critical for the complex assembly. Based on systematic cysteine-mediated cross-linking analysis, it has been proposed that the exposed residues along TolC’s α-helices H3 and H7 located below the equatorial domain most likely interact with residues of the α-helical hairpin domain of AcrA (Lobedanz et al., 2007). Further support for this notion came from mapping of the gain-of-function suppressor alteration of either an efflux-defective TolC mutant (Gerken and Misra, 2004; this study) or a non-functional heterologous TolC-AcrA-MexB pump (Krishnamoorthy et al., 2008) in the α-helical region of AcrA. Conversely, suppressor alterations of a non-functional heterologous VceC-AcrAB complex were found in the α-helical domain of VceC (Vediyappan et al., 2006). Similarly, modelling and experimental data have indicated that the tip of AcrB’s hairpin domain docks with the bottom of TolC’s α-helical domain (Fernández-recio et al., 2004; Tamura et al., 2005; Bavro et al., 2008). More specifically, based on analyses of in vivo cross-linking between non-native cysteine residues placed at the two proposed interacting regions of AcrB and TolC, it has been concluded that residues of the AcrB hairpin loop are in close proximity to residues of the TolC turn regions between the inner helices H7 and H8 and the outer helices H3 and H4 (Tamura et al., 2005).
In this study, we investigated the role of four residues located in the turn region between the outer helices H3 and H4 of TolC in efflux function. Simultaneous alterations of four turn residues from 147GLVA150 to AGSG obliterated efflux activity without affecting cross-linkable interactions between TolC and AcrA or TolC and AcrB. Characterization of antibiotic resistant revertants revealed compensatory changes in AcrA, which induced widening of the TolC aperture. This indicated that the inability of the mutant TolC protein to properly dock with AcrB prevented it from undergoing an AcrAB-mediated transition from the closed to open state. However, compensatory alterations in AcrA could overcome this block by inducing opening of the TolC aperture in an AcrB-dependent manner.
Results
Mutational analysis of the periplasmic turn 1 residues of TolC
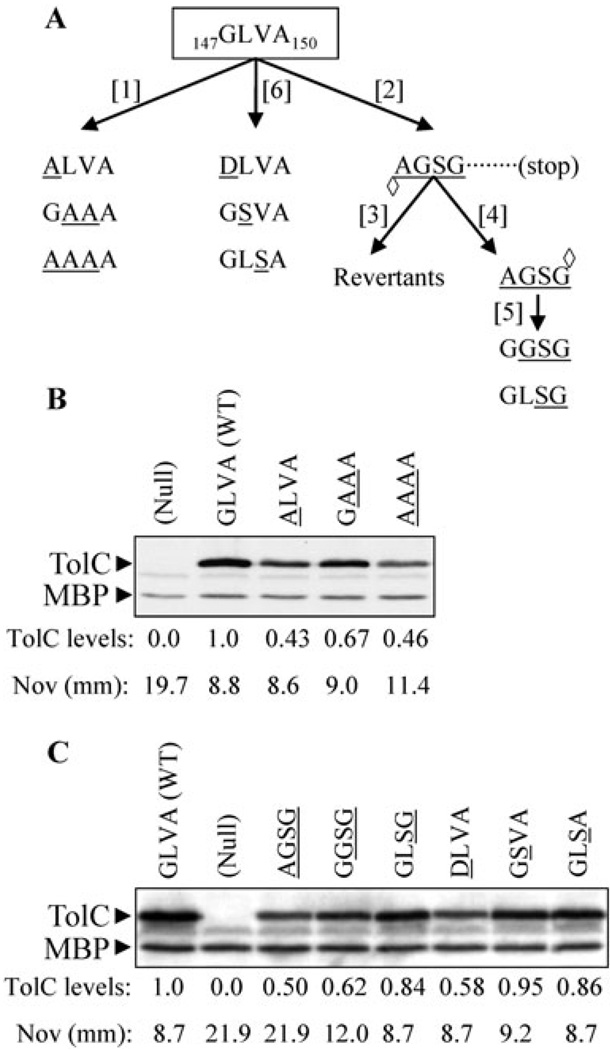
Based on the AcrB structure, Murakami et al. (2002) proposed that hairpins protruding from the top end of AcrB intermesh with the α-helix-turn-α-helix structure extending from the bottom of TolC to form a contiguous drug expulsion passage crossing the two membranes. Experimental evidence for this proposal came from demonstrating that certain cysteine-modified residues of the TolC turns could form disulphide bonds with the corresponding cysteine-modified residues of the AcrB loops in vivo (Tamura et al., 2005). In this study we investigated what roles in pump assembly and drug efflux the TolC turn 1 residues 147GLVA150, present between the static H3 and H4 helices play, through two different genetic approaches involving alanine scanning and localized frameshift mutagenesis (Fig. 1A).
Fig. 1. Mutagenesis of the TolC turn 1 residues and characterization of various TolC turn 1 mutants.
A. The wild type TolC turn 1 residues 147G LVA150 were substituted with alanine [1] or subjected to −2 frameshift [2] mutagenesis. The TolC frame was subsequently restored either through reversion analysis [3] or by +2 frameshift site-directed mutagenesis [4]. Open diamonds point to the sites of frameshift mutations. The two residues of the resulting frameshift mutant [4] were further altered by site-directed mutagenesis [5]. Individual alterations of the wild type turn 1 residues were made by site-directed mutagenesis [6]. All mutant residues are underlined. Western blot analysis to determine levels of various TolC turn 1 alanine (B) and frameshift-derived (C) mutants (amino acid substitutions in mutants are underlined). Protein extracts from approximately 5 × 107 cells grown overnight at 37°C were analysed by SDS-PAGE and electro-transferred onto PVDF membranes. Membranes were blotted with primary antibodies against TolC-MBP (maltose binding protein). MBP was used as a gel loading control. Protein levels were quantified with Quantity One software (Bio-Rad). Wild type TolC level was taken as 1 and other values were adjusted relative to wild type TolC. Zones of inhibition around a pre-soaked novobiocin disk (30 µg) are shown in millimeters (mm). Average inhibition zones recorded from three independent assays are shown, with zones varying no greater than 10%.
Of three alanine mutants, 147ALVA150, 147GAAA150 and 147AAAA150, only the 147AAAA150 variant displayed a modest drug sensitivity phenotype (Fig. 1A and B). Alanine mutagenesis also revealed that G147 is important for TolC folding/stability, but not for antibiotic efflux (Fig. 1B). This is consistent with our previous observation (Vakharia et al., 2001), showing that a G147D substitution lowers TolC levels without affecting its efflux-associated activity (Fig. 1C). The modest drug sensitivity of the 147AAAA150 variant (Fig. 1B) is unlikely due to a disruption of the side-chain-mediated interactions, as the wild type TolC turn 1 residues either lack a side-chain (G) or have neutral side-chains (LVA). Interestingly, the 147GAAA150 variant, which had 20% higher TolC levels than the 147AAAA150 variant, displayed negligible drug sensitivity (Fig. 1B). On the other hand, the 147ALVA150 and 147AAAA150 have identical protein levels, yet the former displays no novobiocin sensitivity, while the latter is moderately sensitive (Fig. 1B). Thus the significance of G147A in efflux function becomes apparent only when the 148LV149 residues are simultaneously substituted with alanine. Together, these data suggest that a reduced overall hydrophobicity of the side-chains coupled with lower TolC levels causes the modest hypersensitivity phenotype of the 147AAAA150 variant.
In the second mutagenesis approach, we introduced a −2 frameshift mutation by deleting the last two nucleotides of the G147 codon, thereby disrupting the TolC reading frame downstream of the V146 codon (Fig. 1A). This mutant displayed a null phenotype owing to the premature appearance of a stop codon 25 codons downstream from the site of the −2 frameshift mutation. We then either restored the reading frame past the codon 150 to create a 147AGSG150 variant or sought spontaneous revertants of the −2 frameshift mutant to gauge the ‘structural flexibility’ of the turn 1 residues and those present in the vicinity of turn 1, in the drug efflux function. However, repeated efforts yielded revertants in which the original reading frame was restored, indicating the extremely narrow sequence flexibility of the affected region to synthesize an efflux-proficient TolC protein.
We further investigated the 147AGSG150 variant, which had half the TolC level relative to wild type but displayed a null-like hypersensitivity drug phenotype (Fig. 1C). Because the importance of G147 in TolC’s folding/stability has already been demonstrated (see above), we therefore changed A147 of 147AGSG150 back to the wild type G residue to see if elevated TolC levels can reduce or abolish the drug sensitivity phenotype of this mutant. Although the TolC levels in the resulting 147GGSG150 variant rose nominally, this was accompanied by a significant reduction in the drug sensitivity phenotype (Fig. 1C). A second change of G148 back to the wild type L residue (147GLSG150) restored almost wild type TolC levels and the mutant no longer displayed drug sensitivity (Fig. 1C). It is worth noting that the 147DLVA150 variant, which we originally isolated among TolC mutants defective in hemolysin secretion (Vakharia et al., 2001), produces almost same TolC levels as the 147AGSG150 mutant, and yet the 147DLVA150 variant displays no drug sensitivity while the 147AGSG150 mutant is hypersensitive. Thus reduced TolC levels alone do not contribute to the drug hypersensitivity phenotype of the 147AGSG150 mutant. The introduction of a small polar serine residue at position 148 or 149 in the wild type turn 1 sequence neither affected the protein level nor produced a drug sensitivity phenotype (Fig. 1C). Together, these results indicated that the 147GL148 residues of turn 1 are the most critical for protein’s folding/stability and its efflux-associated activity.
Detailed characterization of the TolC turn 1 mutant
We focused on the 147AGSG150 variant because its strong, null-like hypersensitivity phenotype is best suited for dissecting the cause of its defects and shed some light on the role of the TolC turn 1 region in efflux function. We examined various TolC activities through employing seven different agents that require TolC for either entry into (TLS phage and colicin E1) or exit from (novobiocin, erythromycin, CCCP and HlyA) the cell. We also used vancomycin, which we have found to be a useful in vivo probe in ascertaining the open or closed state of TolC’s periplasmic aperture (Augustus et al., 2004).
With regard to novobiocin, erythromycin and CCCP, the TolC turn 1 147AGSG150 mutant behaved just like a TolC null mutant (Table 1). Similarly, the mutant TolC protein failed to secrete active hemolysin, an observation consistent with our previous report where we showed that a TolC mutant with a G147D alteration in the turn 1 region was impaired in hemolysin secretion (Vakharia et al., 2001). In contrast to these defects, the TolC 147AGSG150 mutant behaved just like the wild type strain with respect to TLS phage and colicin E1 infections, reflecting that its cell surface properties and hence its insertion into the outer membrane were not affected. Therefore, reduced levels of the TolC turn 1 147AGSG150 mutant is likely due to defects in earlier steps of its assembly, leading to the degradation of roughly half of the newly synthesized nascent polypeptide chains. Finally, the TolC turn 1 147AGSG150 mutant did not display any sensitivity towards vancomycin (Table 1), showing that the hypersensitivity towards novobiocin, erythromycin and CCCP is due to the loss of efflux activity and not to a gross breach in the outer membrane permeability barrier caused by mis-insertion of the mutant protein, or a constitutively open TolC aperture. The TolC147AGSG150 mutant is therefore a first of its kind, being completely impaired in its export activities but fully functional in its import activities.
Table 1.
Sensitivity of wild type, null and mutant TolC proteins to various inhibitors.
| Sensitivity to inhibitorsa |
|||
|---|---|---|---|
| Inhibitors | TolCWT | TolC− | TolCQb |
| Novobiocin | 15.80 | 1.02 | 1.01 |
| Erythromycin | 61.60 | 1.90 | 1.82 |
| CCCP | 11.20 | 1.13 | 0.97 |
| Vancomycin | (8.38) | (6.60) | (6.60) |
| HlyA | + | − | − |
| TLS | 1 | >10−6 | 1 |
| E1 | 1 | 2−12 | 1 |
Numbers for novobiocin, erythromycin and CCCP represent minimum inhibitory concentrations. For vancomycin, zones of inhibition in mm are shown in parenthesis. A 10 µl of solution containing 75 µg of vancomycin was spotted on paper disks of 6.5 mm diameter. Average inhibition zones recorded from three independent assays are shown, with diameter varying no greater than 10%. Plus and minus signs indicate presence (+) or absence (−) of hemolytic zones on blood agar medium. Sensitivity to TLS phage is measured as efficiency of plaguing. Colicin E1 sensitivity data report inhibition zones after spotting 10 µl of twofold serial dilutions of colicin E1 stock on an agar plate overlayed with bacterial cultures.
TolCQ denotes TolC turn 1 147AGSG150 quadruple mutant.
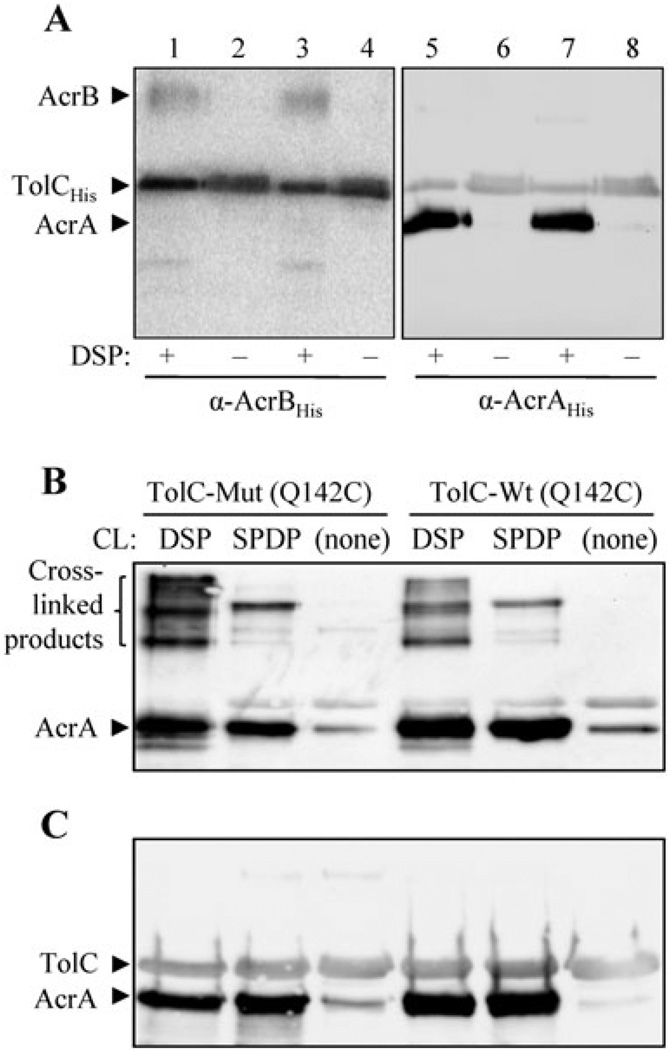
To assess whether the drug hypersensitivity phenotype was due to a loss of interaction with other members of the TolC–AcrAB complex, in vivo cross-linking was carried out using DSP as described in Fig. 2A. The analysis failed to uncover any obvious defects in the mutant TolC protein’s ability to simultaneously pull down AcrA and AcrB in a DSP-dependent manner (Fig. 2A). Because TolC is thought to share a greater surface area with AcrA than with AcrB, it is possible that TolC–AcrA interactions are perturbed in the TolC turn 1 147AGSG150 mutant. To test this possibility, we used SPDP, which facilitates amine-to-sulfhydryl cross-links. As TolC does not contain any cysteine residues, we introduced a Q142C substitution that has proven to be useful in probing TolC–AcrA interactions (Lobedanz et al., 2007). As with a 6xHis tag, the presence of the Q142C substitution did not alter the phenotype of wild type or the TolC turn 1 147AGSG150 mutant protein (data not shown). Both wild type and the TolC turn 1 mutant efficiently pulled down AcrA regardless of whether DSP or SPDP was used as a cross-linker (Fig. 2B and C), thus showing no obvious defects in the ability of the mutant TolC protein to ‘physically’ interact with AcrA.
Fig. 2. In vivo cross-linking analysis to probe TolC–AcrAB and TolC–AcrA interactions.
A. To analyse TolC–AcrAB interactions, freshly grown bacterial cultures were incubated with or without DSP. TolCHis was purified through a Ni2+ affinity column, and AcrB and AcrA from two identical sets of eluates were probed by Western analysis using AcrBHis or AcrAHis antibodies. These antibodies also recognize TolC due to the presence of a C-terminal 6xHis tag in TolC. Note that the wild type (lanes 1, 2, 5 and 6) and mutant TolC147AGSG150 (lanes 3, 4, 7 and 8) proteins can pull down AcrB and AcrA only in the presence of DSP cross-linker. All protein samples were boiled in sample buffer containing β-mercaptoethanol prior to gel electrophoresis. B and C. To analyse TolC–AcrA interaction, a Q142C substitution was introduced into both wild type (TolC-Wt) and the TolC turn 1 147AGSG150 mutant (TolC-Mut) to facilitate SPDP-mediated cross-linking. Both wild type and mutant TolC proteins contain a 6xHis tag at the C-terminal end of the protein for affinity purification. TolC from cultures incubated with DSP, SPDP, or no cross-linker (CL) was affinity purified through Ni2+ affinity columns, AcrA and TolC from two identical sets of eluates were detected through Western blots using AcrAHis antibodies, which also recognize TolCHis. Prior to gel electrophoresis, protein samples were boiled in sample buffer either without (B) or with (C) β-mercaptoethanol.
Suppressors of the TolC turn 1 147AGSG150 mutant
Because cross-linking analysis failed to reveal any gross defects in the TolC turn 1 147AGSG150 mutant’s ability to interact with AcrA or AcrB, we resorted to reversion analysis in an attempt to identify suppressor mutations that could provide insight into the reason for the mutant TolC’s defect. Antibiotic resistant revertants were isolated by simultaneously utilizing two antibiotics, novobiocin and erythromycin, in the selection medium to avoid mutations that alter the cellular targets of these antibiotics. Approximately 5 × 108 cells from 24 independent cultures were plated onto selection medium and incubated at 37°C for 20 h. Nineteen cultures produced a total of 55 spontaneous antibiotic resistant mutants at a frequency of about 10−8, indicating the prevalence of missense mutations. To test whether the mutations in these isolates mapped within the plasmid DNA from which the mutant tolC gene was expressed, the plasmids were extracted and transformed into a ΔtolC strain and transformants were tested for their ability to grow on medium supplemented with novobiocin and erythromycin. In no cases did the mutations conferring antibiotic resistance move with the plasmid, showing that they all resided on the chromosome. We then carried out P1 transductional mapping using the selectable marker Tn10-Tcr, which is linked to the acrAB genes (40% co-transducible by P1 phage). For all 55 revertants, mutations conferring antibiotic resistance moved with the acrAB-linked Tn10 marker at the expected frequency, indicating that the mutations mapped in or near the acrAB genes.
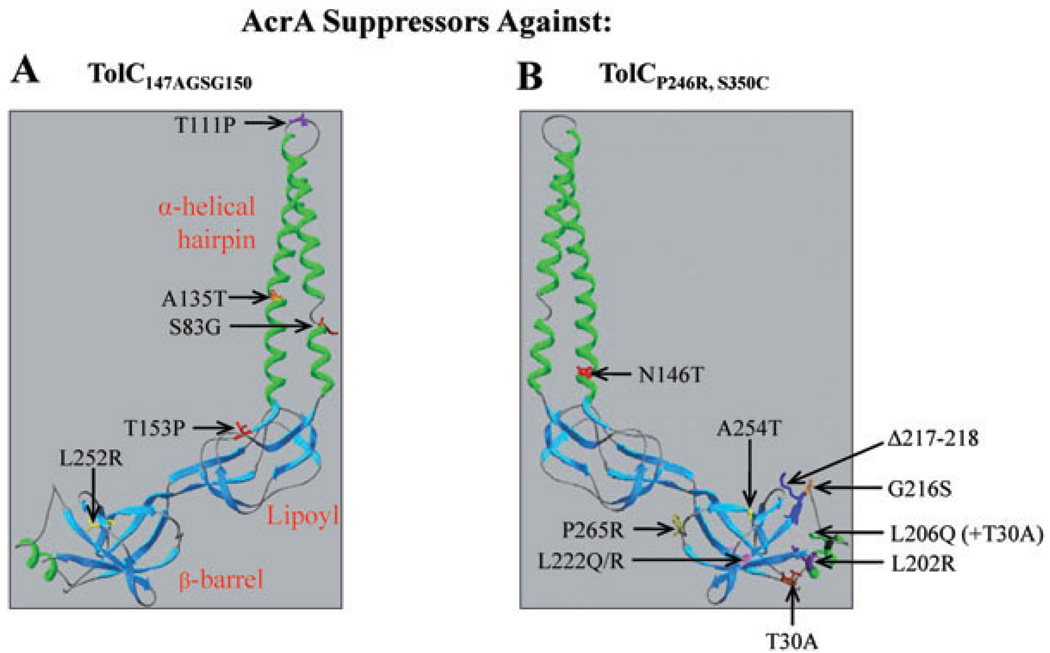
Twenty-four of the 55 revertants, representing multiple independent isolates of each distinct phenotype, were analysed. Nucleotide sequence analysis of PCR-amplified acrAB DNA templates obtained from these 24 mutants revealed five different missense mutations within the acrA gene, resulting in a single amino acid substitution in the mature AcrA sequence: an A135T substitution was obtained once, while S83G, T111P, T153P and L252R substitutions were found in seven, three, eight and five isolates, respectively. These alterations were localized in the α-helical hairpin (S83G, T111P and A135T), lipoyl (T153P) or β-barrel (L252R) domain of AcrA (Fig. 3A). Interestingly, despite affecting some of the same domains of AcrA, the five AcrA suppressor alterations isolated in this study affected different AcrA residues than 10 previous AcrA suppressors isolated against an assembly-defective TolC protein (Fig. 3B).
Fig. 3.
A cartoon showing X-ray structures of AcrA (2F1M). Locations of five AcrA substitutions obtained in this study and those obtained previously (Gerken and Misra, 2004) are shown in A and B respectively. AcrA residue numbering corresponds to that of the mature protein.
As expected, all five AcrA suppressors reduced the hypersensitivity phenotype of the TolC turn 1 147AGSG150 mutant (Table 2). Note that an AcrA suppressor-mediated reduction in CCCP sensitivity is consistent with our data (Table S2) and that of Colmer et al. (1998) showing that AcrAB confers resistance against CCCP independent of EmrAB. Accordingly, AcrAL252R, the strongest of the five AcrA suppressors, reduces the CCCP sensitivity of TolC147AGSG150 independent of EmrAB (data not shown). Western blot analysis data showed that suppression was achieved without significantly elevating the mutant TolC protein level (Table S3). We also examined levels of AcrA mutants and found that only the mutant carrying a T111P substitution had somewhat reduced AcrA levels, regardless of whether it was expressed in a wild type or TolC turn 1 147AGSG150 mutant background (Table S3). Thus the reduced AcrAT111P level is not caused by an aberrant interaction between AcrAT111P and the mutant TolC protein. Rather, it appears that the T111P substitution affects AcrA’s conformation and stability independent of TolC. The isolation of TolC turn 1 147AGSG150 suppressor mutations in acrA suggested either a defective functional interaction between the TolC mutant protein and wild type AcrA or that the AcrA suppressors actually mend a possible aberrant interaction between the mutant TolC protein and AcrB.
Table 2.
Sensitivity of wild type, mutant TolC and mutant TolC containing AcrA suppressors to various inhibitors.
| Sensitivity to inhibitorsb | ||||
|---|---|---|---|---|
| TolC and AcrA proteinsa |
Novobiocin | Erythromycin | CCCP | HlyA |
| TolCWT AcrAWT | 15.80 | 61.60 | 11.20 | + |
| TolCQ AcrAWT | 1.00 | 1.82 | 0.97 | − |
| TolCQ AcrAS83G | 2.00 | 7.15 | 1.05 | − |
| TolCQ AcrAT 111P | 3.60 | 24.60 | 2.50 | − |
| TolCQ AcrAA135T | 3.80 | 13.50 | 1.20 | − |
| TolCQ AcrAT153P | 7.70 | 13.70 | 2.70 | − |
| TolCQ AcrAL252R | 14.50 | 31.10 | 4.20 | − |
TolCQ denotes TolC turn 1 147AGSG150 quadruple mutant.
Numbers for novobiocin, erythromycin and CCCP represent minimum inhibitory concentrations. Plus and minus signs indicate presence (+) or absence (−) of hemolytic zones on blood agar medium. For hemolysin sensitivity tests, strains shown in the table were transformed with a plasmid expressing the entire hemolysin operon.
Mechanism of suppression
It has been proposed that AcrA plays an active role in transducing conformational energy from drug/proton-bound AcrB to TolC to help open TolC’s channel (Fernández-recio et al., 2004; Murakami et al., 2006; Seeger et al., 2006; Bavro et al., 2008; Misra and Bavro, 2009). Accordingly, once AcrB is proton/drug bound, con-formational signals received by the AcrB-proximal β-barrel and lipoyl domains of AcrA are relayed, via AcrA’s α-helical hairpin domain, to the TolC helices guarding the aperture. This would allow for a stable engagement between the AcrA hairpin helices and intra-protomer grooves of the TolC helices, leading to full dilation of the TolC aperture and successful drug extrusion. We considered the possibility that the AcrA suppressors isolated in this study represent constitutively ‘activated’ AcrA forms, which the wild type protein normally assumes only transiently upon receiving conformational signals from AcrB. If the AcrA suppressors are indeed constitutively activated, do they then also transform TolC into a constitutively open state? To test this possibility, we resorted to the vancomycin sensitivity assay which we have shown to be an effective way to monitor the open or closed state of the TolC aperture/channel in vivo (Augustus et al., 2004; Bavro et al., 2008).
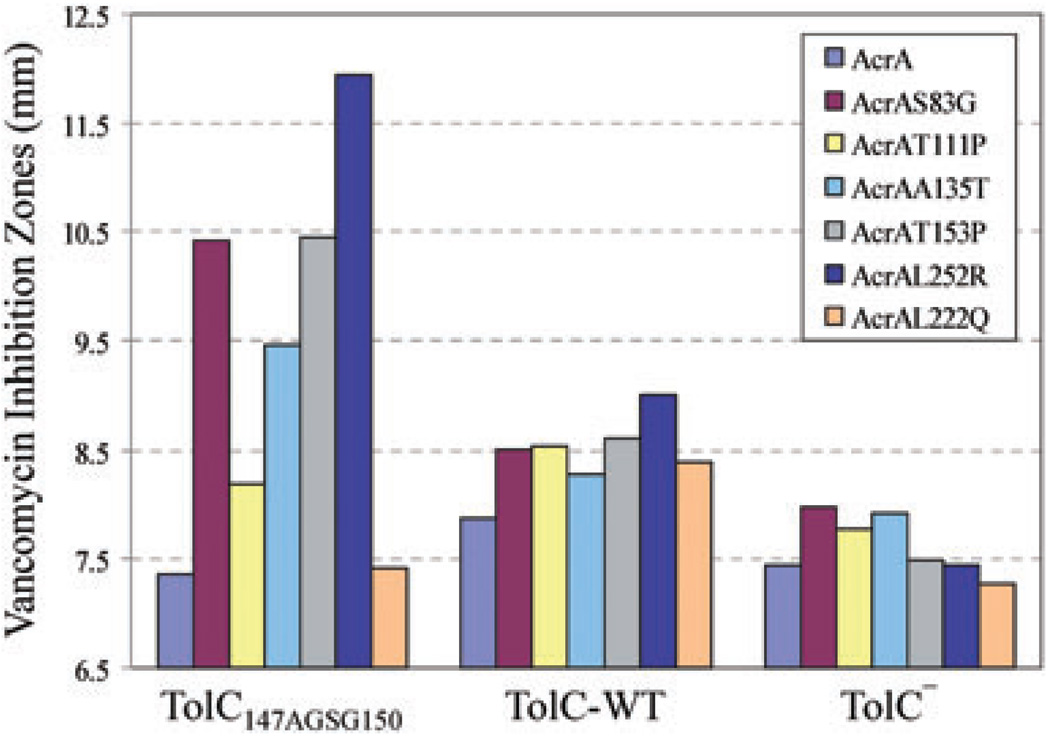
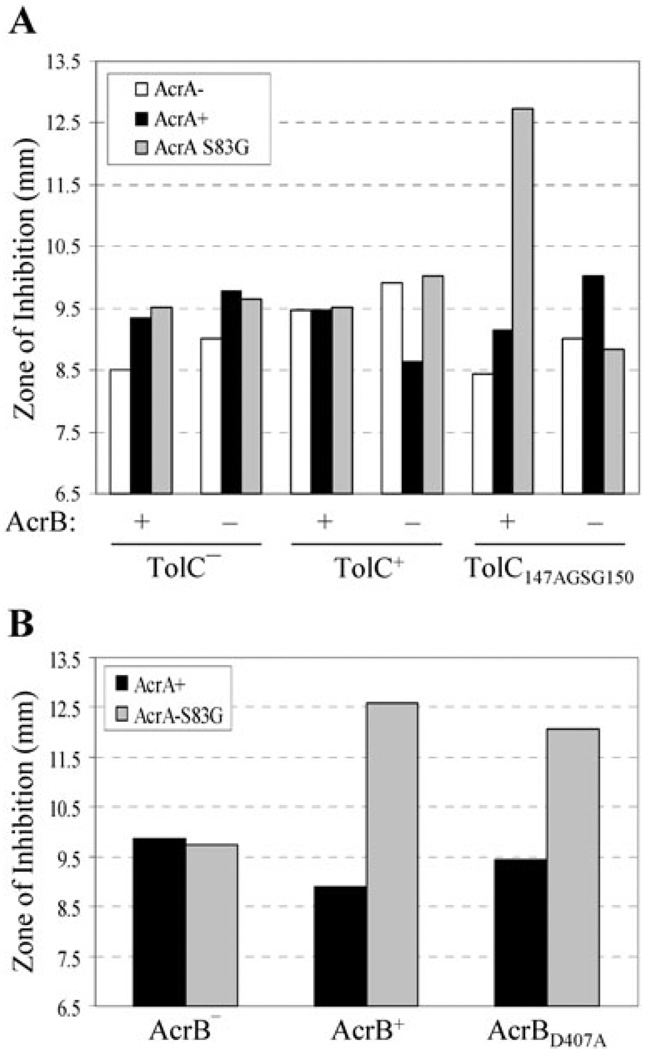
Remarkably, four of the five AcrA suppressors isolated against TolC147AGSG150 significantly increased sensitivity to vancomycin in a TolC147AGSG150-specific manner (Figs 4 and 6), indicating that their mode of suppression involves constitutively dilating the mutant TolC aperture/tunnel. The remaining suppressor, with a T111P substitution, produced a modest (11%) increase in vancomycin sensitivity despite efficiently reducing novobiocin sensitivity. It is worth noting that AcrAT111P is present at approximately 60% that of wild type AcrA level, so a modest increase in vancomycin sensitivity in the case of AcrAT111P could be in part due to its reduced level compared with other AcrA suppressors (Table S3). To see whether suppressor AcrA-mediated vancomycin sensitivity is dependent on AcrB, we expressed one of the suppressor AcrA mutants, AcrAS83G, from a plasmid in ΔacrAacrB+ and ΔacrAB backgrounds. Vancomycin sensitivity was observed only in an ΔacrAacrB+ background expressing TolC147AGSG150 (Fig. 5A), indicating that the presumed activated state of AcrA induces opening of the mutant TolC aperture/channel in an AcrB-dependent manner. Finally, we asked whether the presence of an efflux-active AcrB protein is required for the observed vancomycin sensitivity. For this we introduced a D407A substitution in AcrB that abolishes AcrB’s pump activity by disrupting the proton translocation pathway of AcrB (Guan and Nakae, 2001; Murakami et al., 2002). Interestingly, AcrAS83G-mediated vancomycin sensitivity was observed even in a nonfunctional AcrBD407A background (Fig. 5B). This supports the notion that the AcrA suppressor proteins have adopted a constitutively activated state, thus requiring AcrB only as a scaffold to induce opening of TolC147AGSG150 aperture.
Fig. 4.
AcrA suppressor alterations induce opening of the mutant TolC aperture. Vancomycin sensitivity, which reflects an open state of the TolC aperture, was tested in wild type and suppressor AcrA strains expressing TolC147AGSG150, wild type TolC (TolC-WT), or no TolC (TolC−). A strain expressing AcrAL222Q, which does not suppress TolC147AGSG150, was included as a control. Zones of inhibition around disks soaked with vancomycin (75 µg) were measured after incubating plates for 16 h at 37°C. Average inhibition zones recorded from three independent assays are shown, with zones varying no greater than 10%.
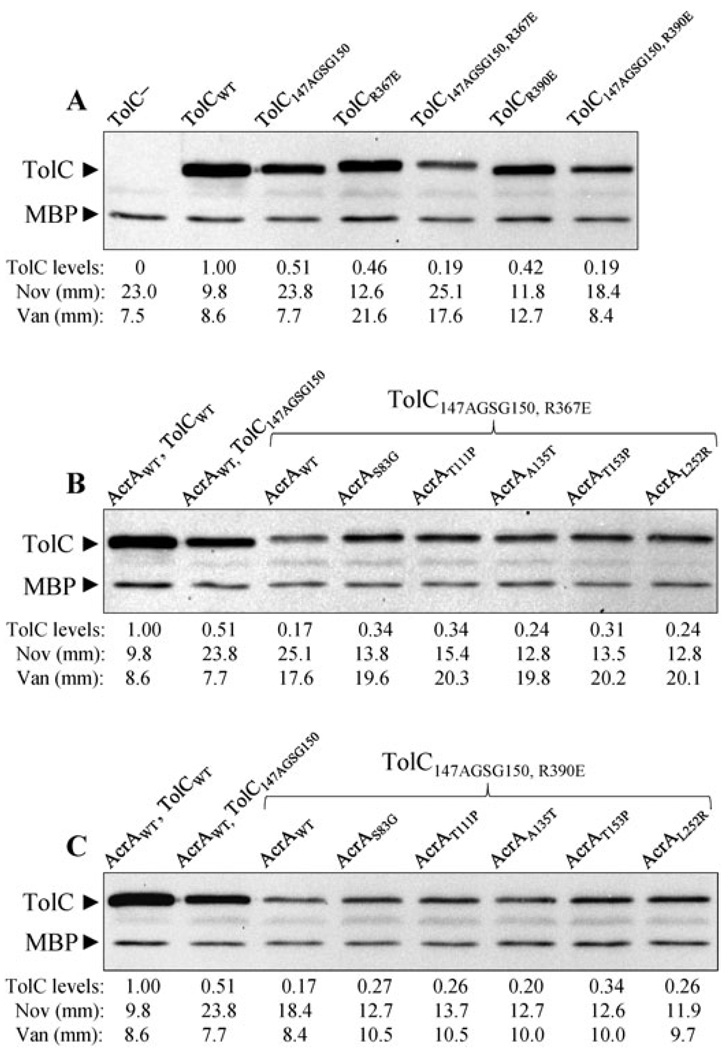
Fig. 6. Combined effects of TolC alterations, which influence the TolC aperture/channel, and various AcrA suppressors on TolC147AGSG150.
A. Effects of R367E and R390E that increase the TolC aperture/channel opening on TolC levels and vancomycin and novobiocin sensitivities.
B and C. Effects of various AcrA suppressors on TolC147AGSG150, R367E (B) and TolC147AGSG150, R390E (C) levels and novobiocin and vancomycin sensitivities. Proteins were detected by Western blots as described in Fig. 1 legend. Antibiotic disk sensitivity assays were carried out as described in Figs 1 and 4 legends. Average inhibition zone diameters were plotted from two independent experiments, with zones varying no greater than 10%.
Fig. 5. AcrB is required for AcrA suppressor-induced opening of the mutant TolC aperture.
A. Vancomycin sensitivity was determined, as described in Fig. 4 legend, from 18 different genetic backgrounds shown in the graph.
B. An active AcrB protein is not required for the AcrAS83G-mediated vancomycin sensitivity. Average inhibition zone diameters were plotted from three independent experiments, with zones varying no greater than 10%.
Do alterations that constitutively open the TolC aperture/channel reverse the drug sensitivity phenotype of TolC147AGSG150?
The AcrA suppressor data suggest that the efflux function is partially restored in part by inducing opening of the mutant TolC aperture/channel. This prompted us to ask whether an alteration in TolC that constitutively induces TolC aperture/channel opening will also reduce the drug sensitivity phenotype of TolC147AGSG150. To test this, we used two different TolC alterations, R367E and R390E, which constitutively transform the TolC aperture/channel into an open state. R367E destabilizes the network of ionic interactions at the entrance of TolC channel (Andersen et al., 2002), while R390E is thought to affect the aperture/channel by influencing the supercoiling of the coiled coils (Bavro et al., 2008). Alterations affecting R367 and R390 were originally obtained among drug sensitive (leaky) TolC mutants (Augustus et al., 2004).
The presence of R367E or R390E in an otherwise wild type TolC backbone elevated sensitivity to novobiocin and vancomycin, confirming the leaky nature of these mutant TolC channels, with R367E conferring a leakier phenotype than R390E (Fig. 6A). When these two alterations were independently introduced into a TolC147AGSG150 mutant, R367E failed to reduce the novobiocin sensitivity of TolC147AGSG150, while R390E did (Fig. 6A). Interestingly, in both cases a significant reduction in vancomycin sensitivity was observed. Western blot analysis revealed that the presence of R367E or R390E in the TolC147AGSG150 backbone significantly lowered the mutant TolC levels (Fig. 6A), which would at least in part explain the reduced vancomycin sensitivity. However, despite diminished protein levels, the reduced novobiocin sensitivity of the TolC147AGSG150, R390E double mutant compared with the TolC147AGSG150 single mutant indicated that forced opening of the TolC aperture/channel can indeed improve the efflux function of TolC147AGSG150. The inability of R367E to reduce the novobiocin sensitivity of TolC147AGSG150 is likely due to a negative effect on the mutant protein’s stability and the acute leaky nature of the TolC aperture/channel (Fig. 6A).
Combined effects of AcrA suppressors and leaky alterations on TolC147AGSG150
Vancomycin sensitivity data in Fig. 4 indicated that AcrA suppressors reduce the novobiocin sensitivity of the TolC147AGSG150 mutant in part by inducing TolC aperture/channel opening. This hypothesis was further corroborated by data showing that the presence of R390E, which only moderately opens the TolC aperture/channel, also reduces the novobiocin sensitivity phenotype of TolC147AGSG150. However, manifestation of a synthetic phenotype, i.e. reduced protein level, when 147AGSG150 was combined with R390E (Fig. 6A) made it difficult to determine whether the AcrA suppressors and R390E acted via the same mechanism, i.e. by opening the TolC aperture/channel, to reduce the novobiocin sensitivity of TolC147AGSG150. Nevertheless, to see whether the two mutations would act synergistically or antagonistically when combined, we introduced the AcrA suppressor alterations in a TolC147AGSG150, R390E background. We also analysed the effect of AcrA suppressors on TolC147AGSG150, R367E, which display a null-like hypersensitivity phenotype towards novobiocin (Fig. 6A).
The presence of the five AcrA suppressor alterations in TolC147AGSG150, R367E (Fig. 6B) and TolC147AGSG150, R390E (Fig. 6C) backgrounds significantly increased the mutant TolC levels, which rose from 18 to 200%, indicating stabilization of the complex. However, unlike the TolC147AGSG150 background where the five AcrA suppressors increased the vancomycin sensitivity to varying degrees (Fig. 4) without significantly altering the mutant TolC levels (Table S3), in the TolC147AGSG150, R367E and TolC147AGSG150, R390E backgrounds the AcrA suppressor-mediated increases in the vancomycin sensitivity were very similar (Fig. 6B and C). This indicated that the increased vancomycin sensitivity seen in the TolC double mutant backgrounds is primarily due to the AcrA suppressor-mediated stabilization of the mutant TolC proteins and not due to the unique effects of individual AcrA suppressors on the TolC aperture/channel. In other words, this increase in vancomycin sensitivity is not due to the additive effects of the TolC open state alterations and the action of the AcrA suppressors on the TolC aperture/channel. As observed in the TolC147AGSG150 background, all AcrA suppressors reduced the novobiocin sensitivity of the two TolC double mutants (Fig. 6B and C).
Suppression specificity
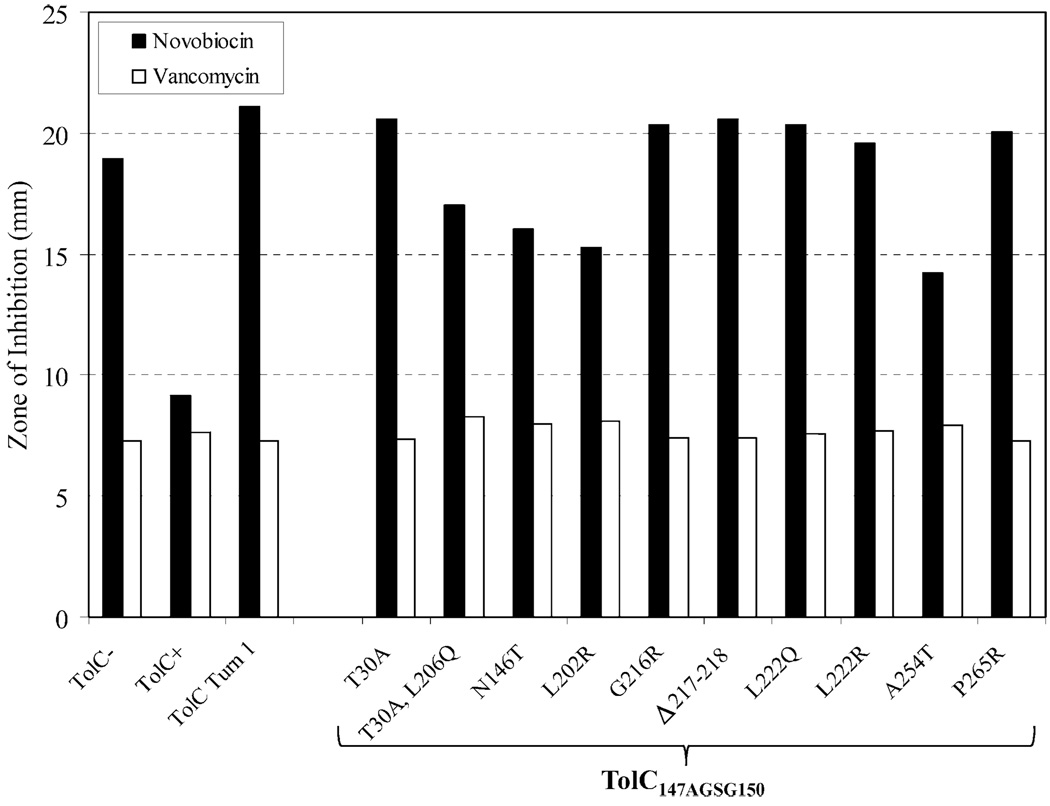
We have previously described an assembly-defective TolC protein (TolCP246R, S350C) and its antibiotic resistant AcrA suppressors (Gerken and Misra, 2004). As mentioned above, these suppressors affected different AcrA residues than those isolated here against TolC147AGSG150 (Fig. 3). So we asked whether these 10 different AcrA suppressors isolated against TolCP246R, S350C could reverse the antibiotic sensitivity defect of the TolC147AGSG150 mutant, and if they do, whether their mode of suppression also involves constitutive dilation of the TolC aperture/channel, resulting in vancomycin sensitivity. Six of the 10 AcrA suppressors isolated against TolCP246R, S350C failed to reverse the novobiocin sensitivity of TolC147AGSG150, thus showing suppression specificity (Fig. 7). The remaining four AcrA suppressors modestly reduced the novobiocin sensitivity of TolC147AGSG150 but conferred only a very small increase in vancomycin sensitivity (less than 8% of wild type AcrA; Fig. 7). In contrast, the majority of AcrA suppressors (four out of five) isolated against TolC147AGSG150 elevated vancomycin sensitivity from 28 to 62% over the wild type AcrA background (Fig. 4). Thus in four cases where cross-suppression was observed, the mechanism of suppression appears to be complex stabilization rather than constitutive dilation of the TolC aperture/channel.
Fig. 7.
Effects of different AcrA suppressors on efflux function and TolC aperture were assessed by measuring novobiocin and vancomycin sensitivities, respectively, in a background expressing TolC turn 1 mutant (TolC147AGSG150). Inhibition zones were also measured in AcrA+ control strains expressing no TolC (TolC−), wild type TolC (TolC-WT) and TolC turn 1 mutant (TolC147AGSG150). Zones of inhibition around disks soaked with novobiocin (30 µg) and vancomycin (75 µg) were measured after incubating plates for 16 h at 37°C. Average inhibition zone diameters from three independent experiments were plotted, with zones varying no greater than 10%.
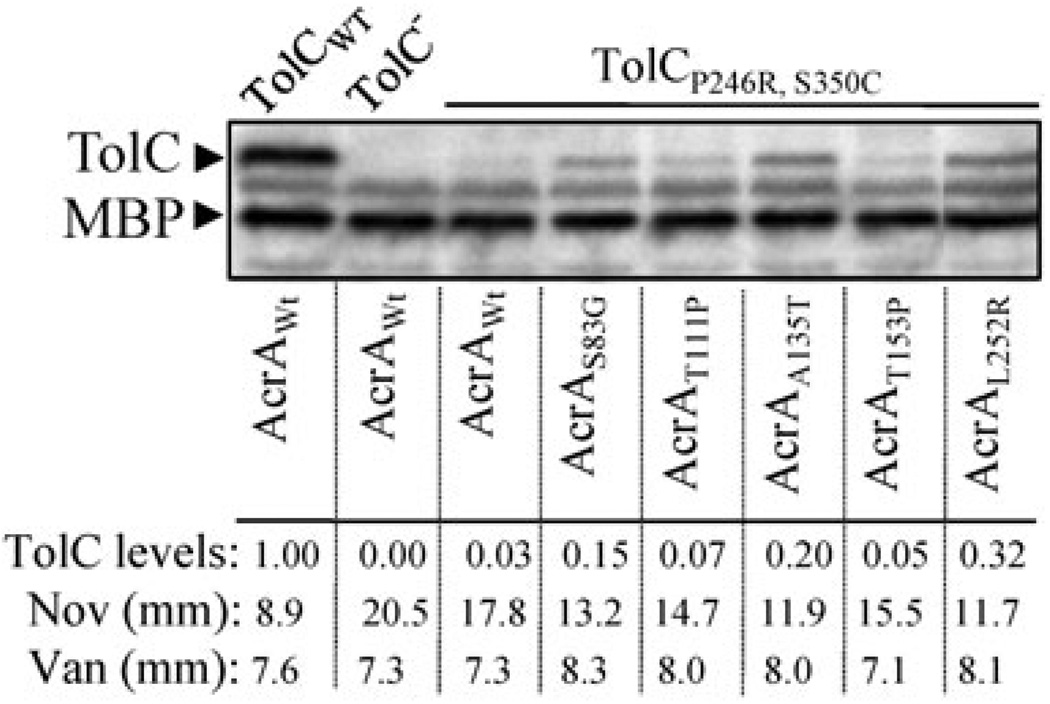
Next we carried out a reciprocal analysis and tested whether the AcrA suppressors isolated against TolC147AGSG150 are specific to this TolC protein or if they also suppress the assembly-defective TolCP246R, S350C protein (Gerken and Misra, 2004). The hypersensitivity phenotype of this TolC mutant was partially reversed by the AcrA suppressors obtained in this study (Fig. 8). Curiously, all the AcrA suppressors also elevated TolCP246R, S350C levels from around two- to greater than 10-fold, reflecting stabilized interactions with the mutant TolC protein (Fig. 8). Moreover, there appeared to be good correlation between the AcrA suppressors’ ability to stabilize the mutant TolC protein and reduce antibiotic sensitivity (Fig. 8). Interestingly, however, none of the five AcrA suppressors significantly increased vancomycin sensitivity (Fig. 8), indicating that their mode of suppression in the case of TolCP246R, S350C involved complex stabilization rather than constitutively dilating the TolC aperture/channel.
Fig. 8.
Effects of wild type and suppressor AcrA variants on TolCP246R, S350C. Protein extracts, obtained from cultures grown overnight at 37°C, were used to detect TolC and MBP, a gel loading control, by Western blots using TolC-MBP antibodies. TolC was expressed from a plasmid replicon under the control of an IPTG-inducible pTrc promoter; acrA alleles were expressed from their chromosomal locations. Wild type TolC levels were taken as 1 and levels of TolCP246R, S350C in various AcrA backgrounds were calculated relative the wild type TolC levels. Zones of inhibition around pre-soaked novobiocin (30 µg) and vancomycin (75 µg) disks are also shown in millimeters (mm).
Together, these data showed that while there were some clear cases of suppression specificity, when cross-suppression was observed the mechanism of suppression appears to be complex stabilization and not TolC aperture/channel opening, which was unique to only AcrA suppressors isolated against TolC147AGSG150.
Re-probing of TolC–AcrA interactions using a labile AcrA protein
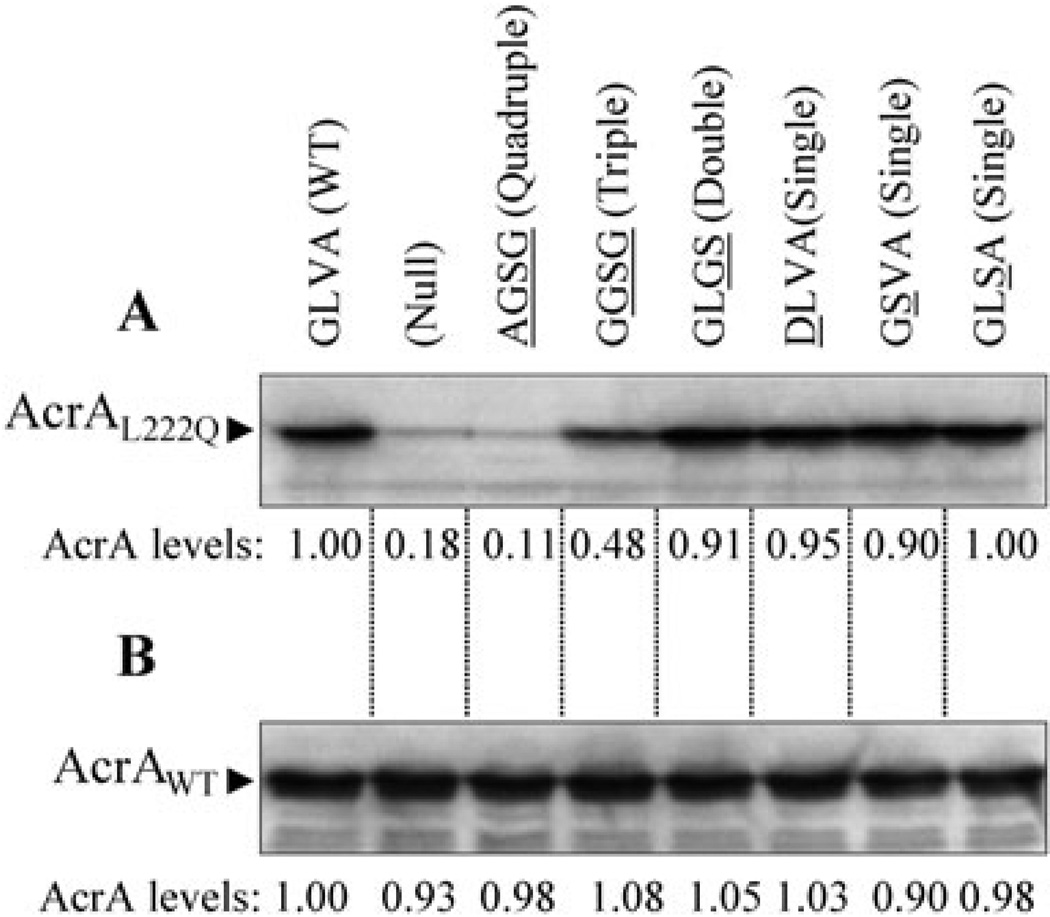
The isolation of suppressor mutations of the TolC turn 1 147AGSG150 mutant in acrA suggested that the mutant TolC protein is unable to make productive interactions with AcrA. Alternatively, such defects may lie between the mutant TolC protein and AcrB but changes in AcrA can overcome them. Because we could not ascertain any obvious defects in the mutant TolC protein’s ability to interact with AcrA or AcrB from chemical cross-linking analyses (Fig. 2), we resorted to a second and perhaps more sensitive approach that indirectly probes in vivo interactions between TolC, AcrA and AcrB. We have previously shown that the in vivo stability of a mutant AcrA protein (AcrAL222Q) is highly dependent on its ability to properly interact with TolC; without TolC, AcrAL222Q is rapidly degraded by a periplasmic protease DegP (Gerken and Misra, 2004). We also noted that the stability of AcrAL222Q is dependent on AcrB, although to a lesser extent than it is on TolC (Fig. S1). TolC–AcrAB interactions were probed through utilizing a genetic background expressing AcrAL222Q from the chromosome and various TolC turn 1 mutants from a plasmid replicon. As expected, in the absence of TolC, AcrAL222Q was rapidly degraded (Fig. 9A). Expressions of TolC turn 1 mutants carrying single and double substitutions stabilized AcrAL222Q just as effectively as the wild type TolC protein, indicating normal interactions. In contrast, TolC turn 1 mutants with triple (147GGSG150) and quadruple (147AGSG150) substitutions showed intermediate or no protection, respectively, of AcrAL222Q (Fig. 9A). Coincidently, only these two TolC mutants displayed a drug-sensitive phenotype in the presence of wild type AcrA (Fig. 1C), thus further corroborating the notion of an impaired interaction with AcrA or aberrant TolC–AcrAB complex assembly. Figure 9B shows data from a control experiment demonstrating that wild type AcrA is an intrinsically stable protein regardless of the status of the TolC protein. Thus AcrAL222Q stability data presented here more closely mirror the efflux phenotype and functional tripartite complex assembly status than the in vivo chemical cross-linking data.
Fig. 9.
Effects of TolC on AcrAL222Q and wild type AcrA levels. AcrA levels from approximately 5 × 107 cells grown overnight at 37°C were determined by Western blot analysis using antibodies against AcrA6His. AcrAL222Q (A) and wild type AcrA (B) levels from the wild type TolC background were taken as 1 and other values were adjusted relative to it.
Discussion
The outward movement of the inner helices H7 and H8 is thought to be crucial for the opening of TolC’s periplasmic aperture. This movement is most likely initiated when TolC is properly interfaced with AcrA and AcrB. Recent genetic and biochemical data have begun to shed light on the potential interacting regions of TolC, AcrA and AcrB (Elkins and Nikaido, 2003; Gerken and Misra, 2004; Touze et al., 2004; Bokma et al., 2006; Stegmeier et al., 2006; Vediyappan et al., 2006; Lobedanz et al., 2007; Krishnamoorthy et al., 2008; Symmons et al., 2009). From these studies, it has become apparent that the grooves between the inner and outer helices H7/H8/H3 of TolC juxtapose the N-terminal α-helix 1 of AcrA.
The available data suggest the involvement of TolC’s periplasmic turns 1 and 2, located between the bottom of outer helices H3 and H4 and inner helices H7 and H8, respectively, in docking with AcrB (Tamura et al., 2005). In this study, we examined the role of TolC periplasmic turn 1 residues in antibiotic efflux through site-directed mutagenesis and isolating extragenic suppressors of a turn 1 TolC mutant. As the residues of turn 1 are thought to be in close proximity to AcrB hairpin loop residues (Tamura et al., 2005), alterations in the turn region encompassing these residues are expected to produce a drug sensitivity phenotype by altering TolC–AcrB interactions. Indeed it was found that simultaneous replacement of the four turn 1 residues 147GLVA150 with the small and neutral residues, AGSG, obliterated TolC’s efflux function. The effect was less pronounced when 147GLVA150 were replaced by 147AAAA150, while single or double alterations affecting the four wild type residues produced no efflux defects. These results, coupled with the fact that the four wild type turn residues either lack a side-chain (G147) or have inert side-chains (148LVA150), suggest that backbone interactions and/or the overall hydrophobicity/packing of the turn 1 region are probably critical for TolC’s interaction with other pump components.
Despite a null-like drug hypersensitivity phenotype, the TolC turn 1 147AGSG150 mutant could be efficiently cross-linked to AcrA and AcrB in vivo, indicating that the physical interactions between the three proteins are not grossly defective. Therefore, the defect must stem from either weaker or non-functional interactions between them. Genetic analysis involving in vivo stabilization of a labile AcrA protein, AcrAL222Q, provided important clues concerning the roles of TolC’s turn 1 region and AcrA in pump assembly. Stability of AcrAL222Q is strongly dependent on TolC and weakly on AcrB (Gerken and Misra, 2004; this study). Just as we found in a strain lacking TolC, the TolC turn 1 147AGSG150 mutant failed to stabilize AcrAL222Q, indicating its inability to maintain stable interactions with AcrAL222Q. Importantly, there was remarkable correlation between the various TolC mutants’ abilities to stabilize AcrAL222Q and their efflux function. The single and double alterations in TolC’s turn 1 neither affected TolC’s efflux function nor the protein’s ability to stabilize AcrAL222Q. On the other hand, the triple (147GGSG150) and quadruple (147AGSG150) alterations in TolC’s turn 1 led to either partial (triple) or complete (quadruple) loss of efflux function and the proteins’ ability to fully stabilize AcrAL222Q. These results indicate that the TolC turn 1 residues play a role in the functional complex assembly. It is important to emphasize that chemical cross-linking does not distinguish weak or non-functional interactions from those that are normal and functional (Bokma et al., 2006; Stegmeier et al., 2006; Vediyappan et al., 2006; this study). Consistent with this, Touze et al. (2004) showed efficient cross-linking between TolC and AcrB in the absence of AcrA, but found no stable interactions between TolC and AcrB by isothermal calorimetry. In contrast to cross-linking analyses, the labile AcrAL222Q and TolCP246R, S350C proteins used here provide a sensitive means of monitoring the existence of functional TolC–AcrAB tripartite complexes in vivo. It is interesting to note that the observed instability of AcrAL222Q in the absence of TolC is similar to that occurring naturally for wild type HlyD, AcrA’s counterpart of the TolC-HlyBD hemolysin secretion complex (Pimenta et al., 1999).
The drug hypersensitivity phenotype of the TolC turn 1 147AGSG150 mutant provided the opportunity to isolate antibiotic resistant revertants whose characterization was expected to help understand the basis of the mutant TolC’s defect. Characterization of these revertants revealed five different compensatory alterations in the AcrA protein. This was somewhat unexpected because TolC’s turn 1 is projected to be in close vicinity to one of AcrB’s hairpin loops, which was later confirmed by spontaneous disulfide bond formation between cysteine residues introduced in these regions of TolC and AcrB (Tamura et al., 2005). Instead, the isolation of compensatory alterations in AcrA initially suggested a possible defect in the mutant TolC protein’s ability to properly interface with AcrA, thus precluding TolC’s tunnel aperture from undergoing the transition from its resting closed state to an efflux-competent open state.
If TolC’s periplasmic turn 1 region is in close proximity to AcrB, then why were extragenic suppressors of the TolC turn 1 mutant repeatedly isolated in acrA and not in acrB? Also, it was surprising that we failed to obtain any intragenic tolC mutations. Our inability to isolate suppressor mutations in tolC could be due to the fact that restoration of the antibiotic resistant phenotype may require multiple substitutions within the turn 1 or adjacent region – an extremely rare event. Similarly, we cannot rule out the possibility that the primary defect of the TolC mutant stems from its aberrant interaction with AcrB, but alterations in AcrA can partially restore these interactions. However, as TolC and AcrB are expected to share a much smaller overlapping surface area between them than with AcrA (Symmons et al., 2009), a much smaller number of potential sites in AcrB are expected to yield a suppressor phenotype, compared with those in AcrA.
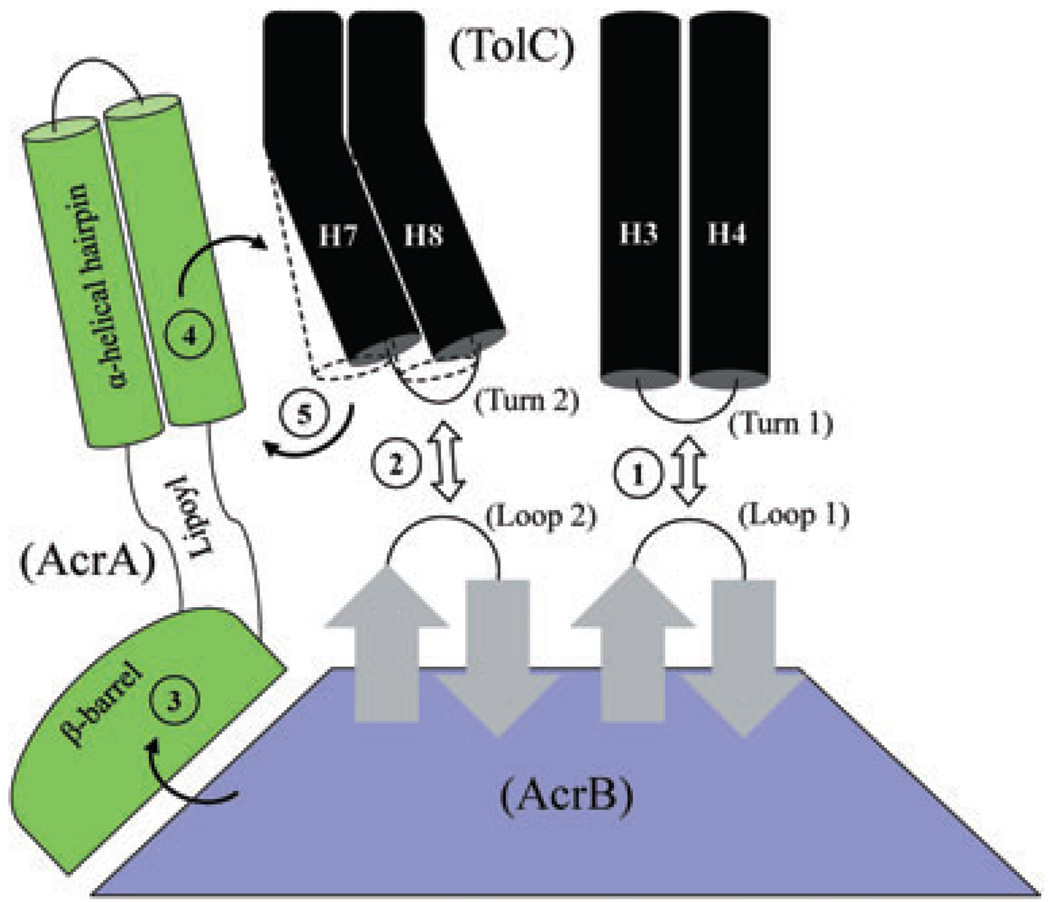
The genetic data pointing to the importance of TolC’s turn 1 region and AcrA in pump assembly are consistent with a recently proposed multi-step pump assembly model (Bavro et al., 2008). According to this model, AcrB docking of TolC results in a partial opening of TolC’s aperture, causing a deepening of the intra-protomer grooves of the TolC helices (Fig. 10; steps 1 and 2). AcrA engages with TolC through these surface grooves, further extending the TolC helices and achieving full dilation and stabilization of the TolC aperture/channel (Fig. 10; steps 3–5). We suspect that the TolC turn 1 147AGSG150 mutant is unable to properly dock with AcrB’s periplasmic crown (Fig. 10; step 1), thus not allowing the proper engagement of AcrA’s hairpin helices with the TolC helices to achieve full dilation of the TolC aperture/channel. The compensatory changes in AcrA must allow the mutant TolC protein to re-engage with the complex in a manner which partially restores efflux activity.
Fig. 10.
A cartoon depicting possible stepwise interactions between TolC, AcrA and AcrB, leading to full dilation of the TolC aperture/channel. Static (H3/H4) and mobile (H7/H8) paired helices guarding the TolC aperture are shown as black cylinders. In the resting stage, the TolC aperture is kept closed by intra- and inter-subunit ionic bridges. The three domains of AcrA and only the top half of AcrB, including its TolC-docking domain, are shown. It is envisaged that initial interactions between AcrB hairpin loops and TolC turns (steps 1 and 2) can trigger partial opening of the TolC aperture; however, these bilateral interactions are not sufficient to fully dilate the TolC aperture. Binding of substrates and protons induce conformational changes in AcrB that are then transduced via AcrA’s β-barrel and lipoyl domains (step 3) to its TolC-proximal α-helical hairpin domain. Direct interactions between the conformationally induced α-helical hairpin domain of AcrA and intra-protomer grooves formed between H3/H4 and H7/H8 pairs (step 4) allow for outward extension of the TolC helices (outlined by dashed lines), leading to full dilation of the TolC aperture (step 5). The model is based on proposals by Fernández-Recio et al. (2004), Murakami et al. (2006), Seeger et al. (2006) and Bavro et al. (2008). The AcrA suppressors obtained in this study dilate the TolC aperture in an AcrB-dependent manner but without receiving its conformational energy generated by the drug/proton translocation cycle.
The vancomycin sensitivity data provided mechanistic clues. Normally, E. coli strains are insensitive to vancomycin except for mutants with defective outer membrane biogenesis (Vuong et al., 2008) or expressing TolC variants with a constitutively open aperture/channel (Augustus et al., 2004; Bavro et al., 2008). This TolC mutant-mediated vancomycin sensitivity is independent of AcrA and AcrB (Bavro et al., 2008). The five AcrA suppressors isolated in this study elevated vancomycin sensitivity in a mutant TolC147AGSG150-specific manner. Using one of the AcrA suppressors isolated in this study, AcrAS83G, we showed that the mutant AcrA-mediated increase in vancomycin sensitivity is dependent on AcrB. Thus, AcrA alterations achieve suppression by inducing opening of the TolC aperture/channel only in the context of the tripartite pump complex. We believe that the AcrA suppressors isolated here transform AcrA into an ‘activated’ state, which the wild type AcrA protein transiently adopts upon receiving conformational signals from AcrB during the normal course of pump assembly and drug efflux (Fig. 10; steps 3–5). Consistent with the notion that the AcrA suppressors have adopted an ‘activated’ state, we found that AcrAS83G, one of suppressors we tested, increases the vancomycin sensitivity even in the presence of AcrBD407A, which is completely disabled in efflux function due to a disruption in its ability to translocate protons (Guan and Nakae, 2001; Murakami et al., 2002). Therefore, AcrA suppressors appear to employ AcrB only as a scaffold to interact with TolC and induce its aperture/channel opening. An independent verification of our assertion that the mechanism of suppression of the hypersensitivity phenotype of TolC147AGSG150 involves its aperture/channel opening comes from the finding that the introduction of an alteration, R390E, which moderately widens the TolC aperture/channel, also reduces the novobiocin sensitivity of TolC147AGSG150.
The five suppressor alterations isolated in this study affect residues located in three separate domains of the AcrA protein (Mikolosko et al., 2006). The S83, T111 and A135 residues are located in the α-helical hairpin domain; L252 in the β-barrel domain; and T153 in the lipoyl domain sandwiched between the α-helical hairpin and β-barrel domains (Fig. 2). Recent cysteine-mediated cross-linking analysis showed that exposed residues of the N-terminal α-helix 1 of AcrA make contacts with TolC (Lobedanz et al., 2007). Therefore, substitutions at S83 and T111 are likely to directly influence AcrA’s conformation in a manner which facilitates interactions with the mutant TolC protein without adversely affecting interactions with wild type TolC. As A135 is present in AcrA’s C-terminal α-helix 2, which is thought to be positioned away from TolC’s helices, changes at this site may influence the TolC–AcrA interface indirectly.
The remaining two suppressor alterations, T153P and L252R, affect AcrA residues present in the lipoly and β-barrel domains, respectively, which are positioned distal from TolC’s periplasmic helices (Fig. 3). The drastic nature of the T153P and L252R changes suggests that they may broadly impact the folding of their respective domains and thus produce long-range conformational changes that influence the TolC-proximal α-helical domain of AcrA. New asymmetric AcrB structures revealed no significant conformational changes in AcrB at the TolC–AcrB interface during the drug capture and expulsion cycle (Murakami et al., 2006; Seeger et al., 2006). In contrast, significant conformational changes are observed in AcrB’s external clefts at the predicted AcrB–AcrA interface. Based on this, it has been speculated that conformational changes in AcrB, triggered by proton/drug binding, are transduced to TolC via long-range conformational changes in AcrA to cause the opening of the TolC channel (Murakami et al., 2006; Seeger et al., 2006). Normally, such AcrB-induced conformational changes in wild type AcrA would be sufficient to trigger the opening of the wild type TolC aperture/channel. However, in the case of the TolC turn 1 mutant, a further conformational nudge, generated by suppressor alterations in AcrA, is needed to achieve mutant TolC aperture/channel opening.
Even though the five AcrA suppressors isolated here continue to function normally with wild type TolC and are even able to suppress a different TolC mutant (TolCP246R, S350C), vancomycin sensitivity data indicate that the mechanism of suppression may be different between the two TolC mutants. In the case of TolCP246R, S350C, where the protein’s stability is a major concern, suppression appears to be achieved through protein/complex stabilization. On the other hand, in the case of TolC147AGSG150, where the tripartite complex appears to be stable but non-functional, presumably due to defective TolC–AcrB interface, the suppression mechanism employed involves stimulating TolC aperture/channel opening. The vancomycin data shown here provide experimental validation to the hypothesis of AcrB-assisted, AcrA-mediated dilation of the TolC aperture/channel (Fernández-recio et al., 2004; Murakami et al., 2006; Seeger et al., 2006; Bavro et al., 2008).
Experimental procedures
Strains, culture conditions and chemicals
All the strains and plasmids used in this study are listed in Table 3. Luria broth (LB) and LB agar (LBA) media were prepared as described by Silhavy et al. (1984). When required, ampicillin (50 µg ml−1), chloramphenicol (12.5 µg ml−1), kanamycin (25 µg ml−1), tetracycline (10 µg ml−1), isopropyl-β-D-thiogalactopyranoside (IPTG; 0.4 mM), L-arabinose (0.2%) was added to bacterial cultures. ECF substrate was purchased from Amersham Pharmacia Biotechnologies. All other chemicals were of analytical grade.
Table 3.
Bacterial strains and plasmids used in this study.
| Strain/plasmid | Characteristics | Reference or source |
|---|---|---|
| Strains | ||
| MC4100 | FaraD139 Δ(argF-lac)U139 rpsL 150 flbB5301 ptsF25 deoC1 thi-1 rbsR relA | Casadaban, 1976 |
| RAM1129 | MC4100 ΔtolC∷Kmr | Augustus et al., 2004 |
| RAM1130 | MC4100 ΔtolC∷Cmr | Augustus et al., 2004 |
| RAM1292 | MC4100 Δara714 | Werner and Misra, 2005 |
| RAM1330 | RAM1292 ΔtolC∷Kmr | Masi et al., 2007 |
| RAM1181 | RAM1292 ΔacrA-scar | Gerken and Misra, 2004 |
| RAM1182 | RAM1181 ΔtolC∷Kmr | Gerken and Misra, 2004 |
| RAM1197 | RAM1292 ΔacrAB-scar | Gerken and Misra, 2004 |
| RAM1198 | RAM1197 ΔtolC∷Kmr | Gerken and Misra, 2004 |
| RAM1335 | MC4100 Δara714 ΔtolC-scar | Husain et al., 2004 |
| RAM1350 | RAM1130 ΔtolC∷Cmr acrA (L222Q) zba∷Tn10–10.5 | Gerken and Misra, 2004 |
| RAM1351 | RAM1130 ΔtolC∷Cmr acrA (L222Q) ΔacrB∷Kmr zba∷Tn10–10.5 | This study |
| RAM1418 | RAM1335 ΔacrA∷Kmr | This study |
| RAM1419 | RAM1335 ΔacrAB∷Kmr | This study |
| Plasmids | ||
| pTrc99A | Apr; expression vector | Pharmacia |
| pTrc-tolC (NcoI clone) | Apr; expresses wild type TolC | Vakharia et al., 2001 |
| pTrc-tolC (NcoI clone) | Apr; expresses TolCP246R, S350C | Gerken and Misra, 2004 |
| pTrc-tolC (BspHI clone) | Apr; expresses wild type TolC | Augustus et al., 2004 |
| pTrc-tolC (BspHI clone) | Apr; expresses TolC147AGSG150 | This study |
| pTrc-tolC (6His) | Apr; expresses TolC6His | Husain et al., 2004 |
| pTrc-tolC (6His) | Apr; expresses TolC147AGSG150, 6His | This study |
| pTrc-tolC (6His) | Apr; expresses TolCQ142C, 6His | This study |
| pTrc-tolC (6His) | Apr; expresses TolCQ142C, 147AGSG150, 6His | This study |
| pCP20 | Apr, Cmr ts replicon, thermal induction of FLP synthesis | Datsenko and Wanner, 2000 |
| pKD46 | Apr; Lambda-red recombinase | Datsenko and Wanner, 2000 |
| pKD4 | Kmr | Datsenko and Wanner, 2000 |
| pACYC184 | Tcr, Cmr; cloning vector | Chang and Cohen, 1978 |
| pSF4000-hlyCABD + | Cmr, expresses Haemolysin proteins | Welch et al., 1981 |
| pACYC184-acrA | Cmr; expresses wild type AcrA | This study |
| pACYC184-acrAB | Cmr; expresses wild type AcrAB | Husain et al., 2004 |
| pBAD33 | Cmr; expression vector | Guzman et al., 1995 |
| pBAD33-acrA | Cmr; expresses AcrA | This study |
DNA manipulations
The acrA gene was cloned into pACYC184 (Chang and Cohen, 1978) and pBAD33 (Guzman et al., 1995) plasmid vectors. acrA sequence from the chromosome was amplified using a forward primer: 5′-AGATCTCATGA ACAATCC GACTTGTC-3′ and a reverse primer, 5′-GTCTTAACGGATCCTGTTTAAGTTAAG-3′ (underlined sequence indicate the BspHI and BamHI cut sites, respectively) and ligated into appropriately digested pACYC184. acrA from this clone was expressed from its native promoter. To clone acrA into pBAD33, acrA sequence was amplified using a forward primer: 5′-GCAGGTACCGGACACTCGAGGTTTACATATG-3′ and a reverse primer: 5′-GCTCTAGAAGCTTAGTGAT GGTGATGGTGATGAGACTTGGACTGTTCAGGCTGACG-3′ (underlined sequence indicates KpnI and HindIII restriction sites respectively). Expression of acrA from this clone was controlled by an arabinose-inducible pBAD promoter. Deletion of the chromosomal tolC, acrA and acrB genes was described previously (Augustus et al., 2004) and was carried out the method described by Datsenko and Wanner (2000). The acrB locus from an acrAL222Q acrB + strain was replaced by a kanamycin resistance (Kmr) gene amplified from the pKD4 plasmid. Gene deletion was confirmed by PCR and DNA sequence analyses. Plasmid transformation and P1 transduction were performed according to the standard laboratory protocols.
In order to modify the TolC turn 1 sequence, two-step site-directed mutagenesis was carried out to first delete the last two bases of the 147th codon creating a frameshift, followed by the insertion of GC after the last base of the 150th codon, thus restoring the original frame, but altering 147GLVA150 residues to AGSG. Site-directed mutagenesis was carried out by using the Quick Change Site Directed mutagenesis kit (Stratagene) according to manufacturer’s instructions. Primers used in site directed mutagenesis are listed in Table S1.
Western blots
Whole-cell extracts were analysed by mini sodium dodecyl sulfate (SDS)-polyacrylamide (11%) gel electrophoresis (PAGE) and transferred onto Immobilon-P polyvinylidenedifluoride membranes (Millipore). Membrane blots were blocked overnight in 5% (wt/vol) non-dairy cream. After blocking, membranes were incubated for 1.5 h in primary antibodies raised against TolC-MBP and/or AcrA6His, followed for two 15 min washes and 1 h incubation in secondary antibody (goat anti-rabbit alkaline phosphatase or horseradish peroxidase-conjugated IgG). Detection of hybridized proteins bands was carried out using ECF (when using alkaline phosphatase) or immunostar HRP substrate (when using horseradish peroxidase). Proteins bands were visualized with Molecular Dynamics Storm Imager or Bio-Rad Molecular Imager ChemiDoc XRS System.
Antibiotic sensitivity assays
Sensitivity to antibiotics was analysed by placing pre-soaked antibiotic disks (Becton Dickinson) on bacterial lawn grow on LBA. Typically, zones of inhibition were measured after 8 h of incubation at 37°C, except in case of vancomycin where zones were measured after 16 h of incubation. All antibiotic sensitivity assays were carried in triplicates.
Minimum inhibitory concentrations (MICs) were determined by measuring growth of bacterial cultures on media containing different concentrations of various inhibitors. Approximately 1 × 106 cells, mixed with twofold serial dilutions of inhibitors in microtiter plates, were incubated at 37°C for 18 h. OD600 was measured by a microtitre plate reader (Molecular Devices VERSAmax), and values were plotted against inhibitor concentrations. MIC values were extrapolated from linear regressions obtained from OD600/concentration plots. Growth was measured from two independent cultures and in duplicates.
In vivo cross-linking
Amine-reactive dithiobis (succinimidylpropionate) (DSP) and hetero-bi functional, amine- and sulfyhydryl-reactive (N-succinimidyl 3-[2-pyridyldithio]-propionate) (SPDP) were used to carry out in vivo cross-linking of proteins. His-tagged variants of wild type TolC and TolC turn 1 quadruple mutant were expressed from a pTrc99A plasmid vector and were used as baits to pull down AcrA and AcrB proteins expressed from a pACYC clone. Cross-linking was carried out essentially as described previously (Thanabalu et al., 1998; Husain et al., 2004; Vuong et al., 2008).
Supplementary Material
Acknowledgements
This work was supported by a grant from the National Institutes of Health (R01-GM066988). We are thankful to Muriel Masi and Phu Vuong for critically reading the earlier versions of this manuscript and Leanne Misra for general comments. We would also like to thank Robin Truer for her initial involvement in suppressor isolation and Cindy Castellanos for assistance with AcrB cross-linking experiments.
Footnotes
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Andersen C, Koronakis E, Bokma E, Eswaran J, Humphreys D, Hughes C, Koronakis V. Transition to the open state of the TolC periplasmic tunnel entrance. Proc Natl Acad Sci USA. 2002;99:11103–11108. doi: 10.1073/pnas.162039399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustus AM, Celaya T, Husain F, Humbard M, Misra R. Antibiotic-sensitive TolC mutants and their suppressors. J Bacteriol. 2004;186:1851–1860. doi: 10.1128/JB.186.6.1851-1860.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bavro VN, Pietras Z, Furnham N, Pérez-Cano L, Fernández-Recio J, Pei XY, et al. Assembly and channel opening in a bacterial drug efflux machine. Mol Cell. 2008;30:114–121. doi: 10.1016/j.molcel.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokma E, Koronakis E, Lobedanz S, Hughes C, Koronakis V. Directed evolution of a bacterial efflux pump: Adaptation of the E. coli TolC exit duct to the Pseudomonas MexAB translocase. FEBS Lett. 2006;58:5339–5343. doi: 10.1016/j.febslet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to select promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976;141:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chang ACY, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colmer JA, Fralick JA, Hamood AN. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholera. Mol Microbiol. 1998;27:63–72. doi: 10.1046/j.1365-2958.1998.00657.x. [DOI] [PubMed] [Google Scholar]
- Das D, Xu QS, Lee JY, Ankoudinova I, Huang C, Lou Y, et al. Crystal structure of the multidrug efflux transporter AcrB at 3.1 Å resolution reveals the N-terminal region with conserved amino acids. Struct Biol. 2007;158:494–502. doi: 10.1016/j.jsb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew D, Klepsch MM, Newstead S, Flaig R, de Gier JW, Iwata S, Beis K. The structure of the efflux pump AcrB in complex with bile acid. Mol Membr Biol. 2008;25:677–682. doi: 10.1080/09687680802552257. [DOI] [PubMed] [Google Scholar]
- Elkins CA, Nikaido H. Chimeric analysis of AcrA function reveals the importance of its C-terminal domain in its interaction with the AcrB multidrug efflux pump. J Bacteriol. 2003;185:5349–5356. doi: 10.1128/JB.185.18.5349-5356.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaran J, Koronakis E, Higgins MK, Hughes C, Koronakis V. Three’s company: component structures bring a closer view of tripartite drug efflux pumps. Curr Opin Struct Biol. 2004;14:741–747. doi: 10.1016/j.sbi.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Fernández-Recio J, Walas F, Federici L, Venkatesh Pratap J, Bavro VN, Miguel RN, et al. A model of a transmembrane drug-efflux pump from Gram-negative bacteria. FEBS Lett. 2004;578:5–9. doi: 10.1016/j.febslet.2004.10.097. [DOI] [PubMed] [Google Scholar]
- Fralick JA. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J Bacteriol. 1996;178:5803–5805. doi: 10.1128/jb.178.19.5803-5805.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerken H, Misra R. Genetic evidence for functional interactions between TolC and AcrA proteins of a major antibiotic efflux pump of Escherichia coli. Mol Microbiol. 2004;54:620–631. doi: 10.1111/j.1365-2958.2004.04301.x. [DOI] [PubMed] [Google Scholar]
- Guan L, Nakae T. Identification of essential charged residues in transmembrane segments of the multidrug transporter MexB of Pseudomonas aeruginosa. J Bacteriol. 2001;183:1734–1739. doi: 10.1128/JB.183.5.1734-1739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain F, Humbard M, Misra R. Interaction between the TolC and AcrA proteins of a multidrug efflux system of Escherichia coli. J Bacteriol. 2004;186:8533–8536. doi: 10.1128/JB.186.24.8533-8536.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V. TolC-the exit duct for proteins and drugs. FEBS Lett. 2003;555:66–71. doi: 10.1016/s0014-5793(03)01125-6. [DOI] [PubMed] [Google Scholar]
- Koronakis V, Sharff A, Koronakis E, Luisi B, Hughes C. Crystal structure of the bacterial membrane protein TolC central to multidrug efflux and protein export. Nature. 2000;405:914–919. doi: 10.1038/35016007. [DOI] [PubMed] [Google Scholar]
- Krishnamoorthy G, Tikhonova EB, Zgurskaya HI. Fitting periplasmic membrane fusion proteins to inner membrane transporters: mutations that enable Escherichia coli AcrA to function with Pseudomonas aeruginosa MexB. J Bacteriol. 2008;190:691–698. doi: 10.1128/JB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobedanz S, Bokma E, Symmons MF, Koronakis E, Hughes C, Koronakis V. A periplasmic coiled-coil interface underlying TolC recruitment and the assembly of bacterial drug efflux pumps. Proc Natl Acad Sci USA. 2007;104:4612–4617. doi: 10.1073/pnas.0610160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masi M, Vuong P, Humbard M, Malone K, Misra R. Initial steps of colicin E1 import across the outer membrane of Escherichia coli. J Bacteriol. 2007;189:2667–2676. doi: 10.1128/JB.01448-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikolosko JK, Bobyk K, Zgurskaya HI, Ghosh P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure. 2006;14:577–587. doi: 10.1016/j.str.2005.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra R, Bavro VN. Assembly and transport mechanism of tripartite drug efflux systems. Biochim Biophys Acta. 2009;1794:817–825. doi: 10.1016/j.bbapap.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419:587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- Pimenta AL, Young J, Holland IB, Blight MA. Antibody analysis of the localization, expression and stability of HlyD, the MFP component of the E. coli haemolysin translocator. Mol Gen Genet. 1999;261:122–132. doi: 10.1007/s004380050949. [DOI] [PubMed] [Google Scholar]
- Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794:782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Klaas M, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- Sennhauser G, Amstutz P, Christophe B, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:106–113. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Berman M, Enquist L. Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- Stegmeier JF, Polleichtner G, Brandes N, Hotz C, Andersen C. Importance of the adaptor (membrane fusion) protein hairpin domain for the functionality of multidrug efflux pumps. Biochemistry. 2006;45:10303–10312. doi: 10.1021/bi060320g. [DOI] [PubMed] [Google Scholar]
- Symmons MF, Bokma E, Koronakis E, Hughes C, Koronakis V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc Natl Acad Sci USA. 2009;106:7173–7178. doi: 10.1073/pnas.0900693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura N, Murakami S, Oyama Y, Ishiguro M, Yamaguchi A. Direct interaction of multidrug efflux transporter AcrB and outer membrane channel TolC detected via site-directed disulfide cross-linking. Biochemistry. 2005;44:11115–11121. doi: 10.1021/bi050452u. [DOI] [PubMed] [Google Scholar]
- Thanabalu T, Koronakis E, Hughes C, Koronakis V. Substrate-induced assembly of a contiguous channel for protein export from E. coli: reversible bridging of an inner-membrane translocase to an outer membrane exit pore. EMBO J. 1998;17:6487–6496. doi: 10.1093/emboj/17.22.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova EB, Zgurskaya HI. AcrA, AcrB, and TolC of Escherichia coli form a stable intermembrane multidrug efflux complex. J Biol Chem. 2004;279:32116–32124. doi: 10.1074/jbc.M402230200. [DOI] [PubMed] [Google Scholar]
- Touze T, Eswaran J, Bokma E, Koronakis E, Hughes C, Koronakis V. Interactions underlying assembly of the Escherichia coli AcrAB-TolC multidrug efflux system. Mol Microbiol. 2004;53:697–706. doi: 10.1111/j.1365-2958.2004.04158.x. [DOI] [PubMed] [Google Scholar]
- Vakharia H, German GJ, Misra R. Isolation and characterization of Escherichia coli tolC mutants defective in secreting enzymatically active a-hemolysin. J Bacteriol. 2001;183:6908–6916. doi: 10.1128/JB.183.23.6908-6916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vediyappan G, Borisova T, Fralick JA. Isolation and characterization of VceC gain-of-function mutants that can function with the AcrAB multiple-drug-resistant efflux pump of Escherichia coli. J Bacteriol. 2006;188:3757–3762. doi: 10.1128/JB.00038-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RA, Dellinger EP, Minshew B, Falkow S. Hemolysin contributes to virulence of extra-intestinal Escherichia coli infections. Nature. 1981;294:665–667. doi: 10.1038/294665a0. [DOI] [PubMed] [Google Scholar]
- Werner J, Misra R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol Microbiol. 2005;57:1450–1459. doi: 10.1111/j.1365-2958.2005.04775.x. [DOI] [PubMed] [Google Scholar]
- Yu EW, McDermott G, Zgurskaya HI, Nikaido H, Koshland DE., Jr Structural basis of multiple drug-binding capacity of the AcrB multidrug efflux pump. Science. 2003;300:976–980. doi: 10.1126/science.1083137. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.