Defining the repertoire of interactions that a particular protein can undergo is crucial for understanding its function and regulation. Characterizing where and when these interactions occur is a major goal of cell biology. Although biochemical approaches (e.g., immunoprecipitation, pull-down assays, and cross-linking) are indispensable for identifying protein-protein interactions, they do not provide spatial and temporal information in the context of an intact cell. Conversely, immunofluorescence localization or genetically encoded fluorescent tags such as green fluorescent proteins (GFP) can provide spatial information regarding an individual protein, but little insight into its interacting partners. While co-localization of two proteins (e.g., in the same organelle) is a prerequisite for their interaction, an interaction cannot be concluded just because two proteins are co-localized by fluorescence microscopy. Clearly, a combination of fluorescence-based visualization of proteins (in their cellular context) that simultaneously provides subnanometer resolution of their proximities (i.e., whether they can physically interact) is highly desirable in nearly all areas of cell biology. For this reason, numerous approaches have been developed to meet these demands.
Because a protein's localization is one of its most basic features, there are an enormous number of reagents to visualize individual proteins by fluorescence microscopy. These include an ever-growing collection of fluorescent protein–tagged constructs as well as high-affinity mono-specific antibodies suitable for immunofluorescence. Given the wide range of color variants of both fluorescent proteins and fluorescent dyes, visualizing two or more proteins simultaneously is now routine. To convert this basic methodology to additionally report on close (subnanometer) proximities of the fluorescently marked proteins, one needs to employ fluorescence resonance energy transfer (FRET). In essence, measurement of FRET between two appropriately labeled proteins containing fluorophores with suitable properties can be used to infer the spatial and temporal characteristics of protein interactions in their native cellular environment.
How does this work? FRET refers to the nonradiative transfer of energy from one fluorescent molecule (the donor) to another fluorescent molecule (the acceptor; UNITS 4.14 & 17.1). Hence, energy that is captured by the donor upon its excitation is transferred to the acceptor. This results in the donor failing to emit a photon, while the acceptor emits a photon at its characteristic wavelength (despite the fact it was not directly excited). Although a wide variety of parameters influences the probability of FRET (see Matyus, 1992; Clegg, 1995; Wouters et al., 2001 for detailed discussions), the most important are the distance separating the donor and acceptor, and their respective fluorescence spectra. Multiple experimental methods and instruments exist for measuring FRET (Jares-Erijman and Jovin, 2003). Selecting the appropriate method and instrumentation can be daunting, even for experienced fluorescence microscopists. Each of the techniques has particular advantages and disadvantages, and the appropriateness of a technique depends on the nature of the hypothesis being tested.
The method that can be most widely and simply implemented, quantified, and interpreted is the acceptor-photobleaching FRET technique (also see UNIT 17.1). In this method, the presence of FRET between a donor and acceptor is revealed upon destruction (by photobleaching) of the acceptor. If the donor fluorescence now gets brighter, one can infer that it had been in sufficiently close proximity to the acceptor to undergo FRET. The extent of increase is a quantitative and direct measure of FRET efficiency. As described below, acceptor photobleaching represents a robust technique that can be exploited to detect changes in the composition and organization of subunit proteins within a multiprotein complex and even to gain insight into relative stoichiometries of proteins within the complex.
To obtain high-quality FRET data, care must be taken to select appropriate controls, maximize the signal-to-noise ratio, and perform sufficient numbers of measurements for the intended questions. In addition, subtleties of FRET theory have implications for accurate interpretation of experimental results. In this unit, strategies, tools, and background for designing and interpreting acceptor-photobleaching FRET experiments in cells are described. As mentioned above, the proteins of interest can be labeled in many ways: fluorescent antibodies, genetically encoded fluorescent protein tags, direct conjugation to dyes, or even fluorescent ligands. Different combinations of all of these methods have been exploited in various FRET methods. For this unit discussion will be limited to two proteins both labeled with fluorescent antibodies. However, the principles, particularly those related to the planning and interpretation stages of the experiment, can be applied easily to other methods of labeling. The basic protocols for cell fixation, labeling, dye-labeling of antibodies, and acceptor photobleaching are provided in UNIT 17.1 and will be referred to as appropriate.
BACKGROUND INFORMATION
Fluorescence resonance energy transfer (FRET) refers to the nonradiative transfer of energy from an excited donor fluorescent molecule to an acceptor molecule. Multiple parameters influence the probability of FRET (see Matyus, 1992; Clegg, 1995; Wouters et al., 2001; and UNITS 4.14 & 17.1 for detailed discussions). The most important parameters are the distance separating the donor and acceptor, and their respective fluorescence spectra. Because FRET efficiency is inversely dependent on the sixth power of the distance separating the donor and acceptor, it is a highly sensitive measure of even small (subnanometer) changes in the relative proximities of the dyes. For a single donor and acceptor fluorophore, the probability of FRET upon excitation of the donor is 1/[1 + (r/R0)6], where r is the distance separating the fluorophores, and R0 is the distance at which a 50% probability of FRET is observed (the so-called Föorster distance; Föorster, 1948).
The applications of FRET are numerous. For the cell biologist, FRET has been used to create biosensors of ions (i.e., the calcium-sensing cameleon fluorescent indicators; Miyawaki et al., 1997) or the active state of a protein, measure protein proximities, and measure changes in organization or composition of a protein complex. FRET biosensor assays have been well characterized and typically are measured using sensitized emission (see the Commentary in UNIT 17.1; Miyawaki and Tsien, 2000; Van Rheenen et al., 2004) or fluorescence lifetime imaging microscopy (FLIM; UNIT 4.14). These FRET assays benefit from the presence of both FRET dyes on the same molecule, which obviates the need to separately express donor and acceptor molecules at comparable levels. In contrast, studies of complexes containing multiple proteins are more theoretically and technically complex. The appropriate design and interpretation of such FRET experiments depend upon a careful consideration of the theoretical expectations.
A critical yet often overlooked concept in understanding FRET measurements is that FRET is a stochastic, all-or-nothing phenomenon. In other words, for any given donor molecule and acceptor molecule, FRET either happens or it doesn't happen; there is no such thing as partial transfer of energy. If FRET is an all-or-nothing phenomenon, why aren't reported FRET values either 0% or 100%, but something in between? The short answer is that FRET measurements in cells and solutions reflect the averaged probability of energy transfer between a very large number of donor and acceptor molecules in the sample. This means that a FRET value is the mean detected energy transfer efficiency for multiple FRET events. Furthermore, each measurement also reflects whether FRET occurs for all of the fluorophore molecules in each pixel of an image. A fluorescence image is a collection of fluorescence photon intensity values for each pixel (Michalet et al., 2003). A single pixel can contain multiple fluorophores. The intensity value of a pixel also reflects the time for collecting photons at that point, either the dwell time of a scanning laser in a confocal microscope or the detection time for a charge-coupled device (CCD) on a widefield microscope. Therefore, a typical FRET measurement for each pixel in a cell is an ensemble measurement that averages numerous FRET events. For this reason, FRET measurements are often described as per cent energy transfer efficiency. Thus, a measurement reflects how frequently FRET events occur for a population of fluorophores under the given conditions.
Often, investigators focus on the Föorster distance of a donor/acceptor pair in FRET studies, the rapid drop in energy transfer efficiency with distance, and the power of FRET measurements as a “spectroscopic ruler” (Stryer and Haugland, 1967). In the case of single-molecule studies or well-defined and homogeneous biochemical samples, FRET can indeed be used to measure absolute distances between fluorophores. However, interpretation of FRET measurements between pairs of proteins expressed in cells is complicated by the number of proteins being assayed and by how the donor and acceptor proteins are labeled.
For this unit, it is assumed that the investigator will label the proteins of interest with at least one antibody and either a variant fluorescent protein (i.e., GFP), a small dye (FlAsh and ReAsh), or another antibody. The dimensions of the antibody probes (~ 14 nm) are substantially larger than the distances over which FRET occurs, and the number and distribution of dyes on the antibody surface are random. Furthermore, the antibodies, as well as the dyes conjugated to them, are flexible enough to substantially influence their absolute positions. These and other variables complicate the relationship between the observed FRET and the distance separating the antigens to which donor and acceptor antibodies are bound (Dewey and Hammes, 1980; Haas and Steinberg, 1984). The consequences of these properties are that FRET cannot be used as a “spectroscopic ruler” for measuring absolute distances of native cellular proteins. This is not to imply that FRET can't be used to detect discrete changes in protein proximity, but rather to emphasize the difference between measuring absolute versus comparative distances. For most cell biologists, the absolute FRET values are far less important to the interpretation of the results than the relative differences obtained for direct comparisons. For example, the absolute FRET values are generally not used to calculate or draw conclusions regarding distances between components; rather, it is the changes in FRET that are used to infer changes in complex organization or structure.
OPTIMIZATION OF ACCEPTOR-PHOTOBLEACHING FRET
Acceptor-Photobleaching FRET in Cells
There are several ways to measure FRET (Jares-Erijman and Jovin, 2003) in cells including sensitized emission (Miyawaki and Tsien, 2000), FLIM (Deniz et al., 2001; Haj et al., 2002; UNIT 4.14), fluorescence anisotropy (Krishnan et al., 2001; Rizzo and Piston, 2005), and acceptor photobleaching (Kenworthy and Edidin, 1998; Haj et al., 2002; Snapp et al., 2004; UNIT 17.1). Acceptor-photobleaching FRET has several advantages that make it suitable for studying protein interactions in cells. First, FLIM and anisotropy measurements require access to instruments that often must be custom designed and built by the user. In contrast, suitable laser scanning confocal microscopes that can be used for acceptor-photobleaching experiments are both commercially available and accessible at most institutions. In addition, even a standard fluorescence microscope with a mercury lamp can be used to perform acceptor photobleaching (Kenworthy and Edidin, 1998). Second, sensitized emission depends on acceptor emission and often suffers from signal bleed-through from the donor, requires multiple correction factors, and provides no information about the relative populations of associated proteins (see Commentary in UNIT 17.1). In contrast, acceptor photobleaching can be quantitated with a simple arithmetic equation, is unaffected by bleed-through, and can be used to gain insights into associated and unassociated populations of proteins. A noteworthy disadvantage of acceptor-photobleaching FRET is that it requires destruction of the probe, which prevents more than one measurement in a region of a cell. In addition, photobleaching to background levels of fluorescence intensity is often slow and either requires cells to be fixed or the proteins of interest to be relatively immobile.
An important consideration for experimental design is whether the goal of the study is to distinguish between two distinct readout states or a continuum of states. It is often not feasible to synchronize multiple protein complexes in their dynamic changes. In contrast, measuring the change between a treated and untreated sample is more experimentally tractable for samples in cells. The measurement of discrete states permits cells to be fixed. While the aesthetic and intellectual appeal of live cell data is undeniable, the actual requirement for using live cells is worth considering. If the experimental readout is treated versus untreated cells, then cells in the two states can be fixed and assayed. Fixation of cells for 15 min in 3.7% formaldehyde retains substantial cell structure and does not significantly promote nonspecific protein interactions (Jackson, 1999; Metz et al., 2004; Snapp et al., 2004). Formaldehyde is a remarkably specific cross-linker and has been used for years in chromatin immunoprecipitation (ChIP; UNIT 17.7) assays to identify proteins that bind to unique DNA sequences in the nucleus (Jackson, 1999). It is still possible to perform live-cell acceptor-photobleaching FRET studies, though the investigator must now contend with the issue of diffusion. Many proteins are not immobile (Lippincott-Schwartz et al., 2001). Because complete photobleaching of a fluorophore can take several seconds to minutes, acceptor photobleaching benefits substantially from immobilizing proteins with fixation.
Based on the background information above, acceptor photobleaching of FRET of native proteins in cells is most appropriate for the following types of questions. Does a pair of proteins interact to a significant degree in a particular region of a cell? Does a protein complex undergo changes in composition? Do protein subunits undergo changes in their proximity or organization in a protein complex? Are two different protein subunits present at equivalent amounts in a complex? In contrast, acceptor photobleaching is poor at detecting small subpopulations of interacting proteins (e.g., only 10% of a protein is in a complex, and the complexes are homogenously distributed throughout the cell), quantitating absolute distances between protein subunits, and detecting small continuous changes in a mixed population of protein complexes.
Before investing the time and resources into performing FRET experiments, the investigator will benefit from critical analyses of the questions to be addressed, the available reagents, and anticipated outcomes under idealized conditions. For example, a failure to consider expression levels when choosing which of two proteins to label as the donor, or attempting to measure absolute distances between proteins with antibody-based FRET will result in low probabilities of success. To enhance the probability of detecting the highest possible FRET signal for a pair of proteins, several conditions need to be empirically optimized.
Determine Which Protein Will Be the Donor and the Acceptor
In acceptor-photobleaching experiments, this choice can significantly impact the observed FRET value. While the consequences of being the donor or acceptor are discussed in detail in the Commentary??, there are two key points to consider. First, unpaired donors (i.e., those not in complex with the acceptor) dilute the detectable FRET signal. Second, unpaired acceptors have little effect on acceptor-photobleaching FRET values, and thus, are effectively invisible in this assay format. Thus, the donor in most cases should be the protein of lower stoichiometry to minimize the percent of unpaired molecules. Note that, because of this asymmetry, a discrepancy in FRET values upon exchanging donor and acceptor status can provide insights into the stoichiometry of complex components.
Select an Appropriate Labeling Scheme
The method of labeling the proteins of interest will directly affect the ability of FRET to detect changes in protein proximities. For this unit, it is assumed that proteins of interest will be proteins expressed in the cell. These proteins can be labeled either with fluorescent dye-labeled antibodies, variant fluorescent proteins (i.e., GFP) or newer fluorescent tags including FlAsh and ReAsh (Adams et al., 2002). After designating which protein will be the donor and which will be the acceptor, the investigator must choose a suitable labeling scheme. If a donor protein is present at low levels and overexpression changes a cellular phenotype, then antibody labeling is likely to be better than using a fluorescent fusion protein. Not only will the antibody boost the fluorescent signal due to multiple dyes on the antibody, but the multiple dyes can also enhance the probability of detecting FRET (see Fig. 17.9.1). If the donor and acceptor are abundant proteins and the donor remains functional when fused to a fluorescent protein, such as GFP, then addition of a fluorescent protein tag or epitope tag may provide flexibility in experimental design. It is not recommended to have an antibody donor and a fluorescent protein acceptor. In this case, there would be multiple dyes on the antibody that might fail to exhibit FRET with a single fluorophore acceptor. Again, the key principle is to avoid situations with excess donor molecules or limiting acceptor molecules.
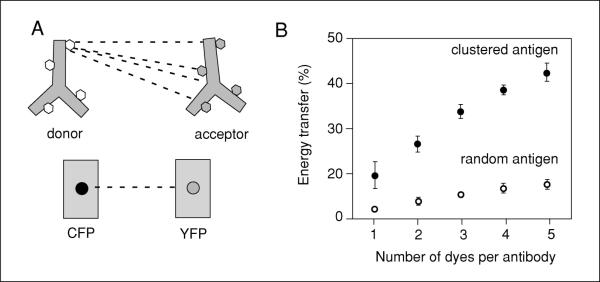
Figure 17.9.1.
Illustration of the advantage of using antibody probes labeled with multiple dyes. (A) Cartoon illustrates how each donor dye on an antibody has the potential to transfer energy to four different acceptor dyes on an acceptor antibody. In contrast, a cyan fluorescent protein (CFP) molecule can only potentially transfer energy to one yellow fluorescent protein (YFP) acceptor. The consequence is that multiple dye-labeled acceptor antibodies enhance the probability of detecting FRET. (B) The relationship of dye number on each donor and acceptor antibody as plotted with simulated data for antibodies bound to a multiprotein complex. See Snapp et al. (2004) for details concerning the simulation parameters. Note that the value calculated for a single dye will not be equivalent to the CFP-YFP pair, because the simulated dye placement on the antibodies was random and could include distances up to three times greater than for the fluorescent fusion proteins.
If using antibodies to label the protein(s) of interest, either monoclonal or mono-specific polyclonal antibodies (raised against short peptide sequences of ~8 to 20 amino acids) are preferable to broad specificity antibodies. A single protein epitope is likely to ensure that only a single antibody will bind a single protein. This is an especially useful quality when an investigator is trying to determine whether a complex contains more than one copy of a protein. A single epitope will provide more specific information concerning protein organization and can aid in experimental design. For example, an epitope against the cytoplasmic domain of a membrane protein has a higher probability of undergoing FRET with the cytoplasmic epitope of a partner protein.
Select the Donor and Acceptor Fluorophores
The donor should be a fluorophore that is excited at a lower wavelength than the acceptor, and with an emission spectrum that overlaps significantly with the excitation spectrum of the acceptor. The degree of spectral overlap is a key determinant of the efficiency of FRET. A useful measure of the suitability of a donor-acceptor pair for FRET is known as the Förster value: the distance at which the probability of FRET between the donor and acceptor is 50%. Optimal FRET pairs are constantly being updated. A popular and well-characterized donor-acceptor pair is the dyes Cy3 and Cy5, with a Förster distance of ~5 nm (Bastiaens et al., 1996). Another useful FRET pair in cells is GFP and Cy3 with a Förster distance of 6 nm (Haj et al. 2002). A second parameter important in FRET efficiency is the brightness and stability of the fluorophores. Photostability is a desirable quality in the donor and less desirable for the acceptor. For example, Alexa fluorophores photobleach poorly and will thus be poor choices as acceptor, but they are reasonable choices for donors.
Optimize Antibody Dye Labeling Ratios
See UNIT 17.1 for the Cy3/Cy5 dye labeling protocol. Addition of dyes to antibodies requires balancing of three properties: maximal FRET signal, antibody intensity quenching, and antibody binding inhibition. For Cy3 and Cy5 dyes, four dye molecules per antibody generally give excellent results (Snapp et al., 2004). This number of dye molecules tends to maximize the fluorescent signal and FRET efficiency while minimizing the disruption of binding activity. It goes without saying that it should be confirmed that the directly conjugated antibody recognizes antigen with an affinity similar to the unconjugated starting antibody.
Identify Relevant Positive and Negative Controls
The importance of the choice of controls cannot be overstated. Quite simply, controls define the limits and scale for interpretation of experimental FRET values. Regardless of the predicted Förster distance for a FRET pair, actual FRET data is very much dependent on the properties of the system and must take into account the geometry of the protein complex, cellular autofluorescence, the method of FRET measurement, the size of the fluorescent label, and effects of the cellular environment on fluorophore properties. Therefore, the investigator should select controls that: (1) use the same FRET pair of fluorophores; (2) mimic the spatial environment of the proteins of interest (i.e., if membrane proteins are being investigated, the controls should also be membrane proteins); (3) label cells with similar fluorescence intensities relative to the experimental labeling; and (4) display co-localization in immunofluorescence images for both positive and negative controls. This last point is obvious for a positive control because proteins that do not co-localize at the level of light microscopy will not exhibit FRET. However, using co-localized proteins for the negative control is also important because the goal of the experiment is to get spatial resolution that is higher than light microscopy can deliver. Thus, it is important to demonstrate that noninteracting proteins close enough to co-localize in the cell do not give significant FRET. For acceptor-photobleaching FRET using antibodies, a simple but nice positive control is an acceptor-labeled primary antibody bound by a donor-labeled secondary antibody. In this instance, all donor-labeled antibodies are necessarily adjacent to an acceptor-labeled antibody, and should necessarily yield highly efficient FRET.
Empirically Determine Antibody Labeling Conditions
It is assumed that the investigator is familiar with the basic operation of a confocal microscope. The investigator should understand both the concept and the operation of scan speed, zoom, detector gain, laser power, photobleaching, and collection of a time series on a laser scanning confocal microscope. A series of simple immunofluorescence experiments should be performed to determine the several imaging and labeling conditions (see below). Cells can be assayed with a fluorescence microscope fitted with a charge-coupled device (CCD) or a confocal microscope with a photomultiplier tube (PMT). It is important to collect the data and quantitate intensities using software provided by the microscope maker or a program such as NIH Image, ImageJ, or Metamorph. The human eye is remarkably poor at quantifying subtle differences in intensity. Also, carefully consider the method used for monitoring intensities. If the proteins of interest localize to a discrete structure or only a few dispersed structures, it will be more informative to assay a region of interest that includes only the structure of interest. Large regions of interest that include large unlabeled regions will average out differences in the fluorescence intensities of structures.
Initially, fix, permeabilize, and label cells separately with donor and acceptor antibodies to separately optimize each to maximize signal intensity and reduce nonspecific labeling. Typically, the donor antibody is used at ~0.2to1.0 μg/ml and the acceptor is used at ~2to4 μg/ml (this refers to the concentration of the specific antibody; hence, crude IgG from a polyclonal serum would be used at ~10-fold higher concentrations since only ~10% is specific antibody). A donor/acceptor ratio at or below 1:4 (usually ~1:8) will help maximize FRET efficiency without making the donor fluorescence intensity too dim to easily visualize. Incubation times will vary depending on antibody affinity and will need to be determined for each antibody. In general, 60 to 120 min is sufficient for optimal antibody labeling, as assessed by maximal fluorescence intensity and minimal nonspecific staining. Each of these modifications increases the efficiency of FRET as would be expected if the occupancy of antigens were improved. The goal is to maximize occupancy of the antigens bound by the acceptor antibody and have a sufficiently bright but highly specific labeling of the donor. Recall that excess acceptor is effectively invisible to FRET, as measured by acceptor photobleaching, so some nonspecific binding by the acceptor is acceptable. In contrast, optimization of conditions that give highly specific donor labeling is critical.
Another important control is to label cells separately with donor followed by acceptor, acceptor followed by donor, and donor and acceptor simultaneously and then measure mean fluorescence intensities of the whole cell or relevant structures. This control will reveal whether the antibodies sterically affect accessibility of protein epitopes and potentially affect labeling. A similar control experiment that tests the same parameter is to systematically change the concentration of one antibody and determine whether the efficiency of labeling by another antibody in cells is affected.
Identify Optimal Photobleaching Conditions for the Acceptor
Photobleaching the acceptor by high-intensity laser illumination is influenced by laser power, magnification, dwell time of the laser, number of bleach iterations, and sample preparation. The goal should be to optimize conditions where more than 90% of acceptor fluorescence can be bleached. Dyes such as Cy5 may require hundreds of iterations of laser photobleaching to deplete acceptor fluorescence to background levels. If a large area is to be photobleached or the viewing field is at a low magnification, then the time required to achieve sufficient photobleaching will become burdensome. So choose the highest magnification that allows the regions of interest to be visualized completely. Also, perform the optimization at different magnifications (optimum conditions will differ) so that in the future, a complete set of bleach conditions for whatever application might arise has been obtained. Once optimized, be sure that the selected imaging conditions do not result in photobleaching of the cell outside of the photobleach region of interest.
Make and Quantify FRET Measurements
Once the above parameters are optimized, the samples (along with the predetermined positive and negative controls) can be prepared in which both donor and acceptor are labeled. To make measurements, find some appropriate cells or regions of interest. This is best done by visualization of the cells using the absolute lowest illumination as possible to prevent premature partial bleaching of the sample. One can use phase-contrast microscopy to find suitable cells, or if fluorescence microscopy is used, illuminate only the donor while searching. Once the area is focused and imaging/bleaching conditions are set, the procedure is to take pre-bleach images of the donor and acceptor fluorescence, photobleach the acceptor in a region of interest, and take a second set of post-bleach images of the donor and acceptor. It is critical to avoid sample movement, change in focal plane, or any change in imaging conditions between the pre- and post-bleach images. Once completed, there should be four images for the measurement: pre- and post-bleach donor images, and pre- and post-bleach acceptor images. Although only the donor images are absolutely required for the calculation of FRET efficiency, capture and save the acceptor images because they contain additional information that aids in the interpretation.
The calculation of FRET efficiency from these images can be done manually, or automated using relatively straightforward custom macros for a program like NIH image (see section on automated image analysis). To manually calculate FRET, measure the donor intensity in both the pre- and post-bleach images within the region that was bleached (referred to here as Dpre and Dpost). Subtract background intensity (i.e., the value obtained in an area where there are no cells) from both intensity values. Once these values are obtained, the % FRET (also referred to in various publication as % energy transfer, %E,orjust E) can be calculated as:
All that has been done is to calculate the percent of total donor fluorescence that had been quenched in the presence of the acceptor. It is important to confirm that in an area of the image that was NOT subject to photobleaching, the donor intensity does not change significantly between the pre- and post-bleach images. If it does, it means that the sample may have moved or changed focus between the two images, or that perhaps the acceptor was partially bleached unintentionally between capturing the two donor images. If conditions have been optimized as outlined above, this should not happen (except for occasional sample movement or change in focus).
An increase in donor intensity selectively in the region of acceptor photobleaching is indicative of FRET (with numerous caveats outlined in the next section). Instead of doing the manual calculation on the entire bleached region as a whole, one can calculate FRET for subregions of the sample to gain insight into the spatial distribution of FRET (which could for example reflect differences in protein-protein interaction in different regions of the cell). Although this can be done manually, it is cumbersome. The macro the authors developed for NIH image does this automatically on individual 8 × 8–pixel regions throughout the image and displays the results in a color-coded map of FRET. This can be extremely useful for visualizing spatial differences in FRET throughout the cell.
INTERPRETATION OF RESULTS
Of equal importance to the actual collection of FRET data is a rational interpretation of the results (also see UNIT 17.1). This is critical to ensure that any observed FRET is actually due to the proximities of the proteins of interest. Sometimes it is easy to forget that the FRET values are indirect measurements based on a series of assumptions (such as specificity of the antibodies) that may or may not have been thoroughly validated. Furthermore, other potential artifacts must be taken into account and excluded to maximize the likelihood of correct interpretation. Some of these issues are listed below.
Restrict Quantitative Comparisons to Experiments Performed at the Same Time
Experimental FRET values can be affected by batch-to-batch differences including variations in cell density, cell cycle, alignment of the photobleaching laser (which will affect completeness of the photobleach), and antibody degradation. While the overall trends and relationships in the data should be robustly repeatable, the absolute values in the data will tend to vary by as much as 20% between experiments. This observation also emphasizes the importance of including a positive control with every experiment to assist in detecting batch-to-batch variability.
Be Aware of Artifactual Reasons for Changes in Donor Fluorescence
It was recently reported that upon photobleaching, the Cy5 fluorophore is photoconverted into a fluorescein-like fluorophore that has the potential to confound FRET analyses using the acceptor-photobleaching method (Nichols, 2003; Snapp et al. 2004). This conversion is particularly relevant if the Cy3 fluorescence is extremely dim relative to Cy5 and/or if the Cy3 excitation light intensity or Cy3 detector gains are set at very high levels. Because of the potential to substantially influence the apparent observed FRET, it is important and worthwhile to carefully consider this phenomenon in interpreting the results. One approach to the problem is to correct for this photoconverted product by using a standard curve, which can indicate that the FRET signal is significantly over-represented due to photoconversion (e.g., see Nichols, 2003). However, data correction is often not necessary. In many cases, photoconversion has contributed to less than ~5% of the value of observed FRET signals. Changing the position of the donor antibody while keeping the Cy5 acceptor constant changes the FRET signal without a resulting change in the intensity of either the donor or acceptor argues against a substantial contribution from photoconversion. This phenomenon can be monitored both by comparing per cent energy transfer efficiency maps with fluorescence intensities in images and by plotting %E (or % FRET) as a function of acceptor intensity for different regions of an image (Fig. 17.9.2). An alternative solution to the photoconversion problem is to use different donor/acceptor fluorophore pairs. This can be difficult if one has already invested substantial time and effort in one FRET pair, but it may be feasible early in the experimental design or project.
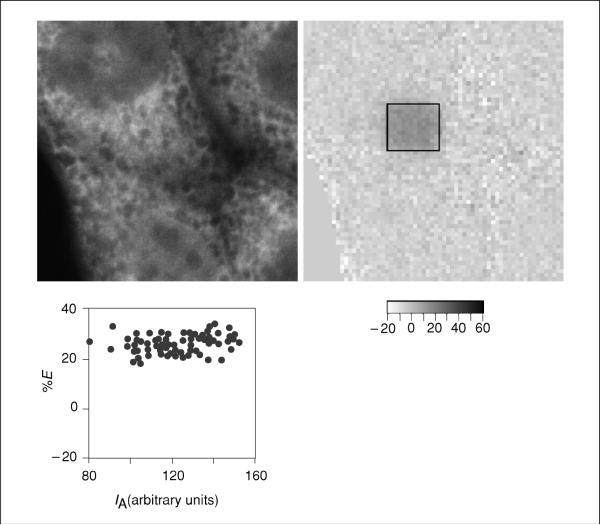
Figure 17.9.2.
Control for acceptor density dependence. The left panel is an image of the pre-bleach acceptor-labeled cell. The middle panel shows the transfer efficiency map, and the plot in the right panel is of acceptor fluorophore intensity (IA) on the x-axis and transfer efficiency (%E)on the y-axis. Note that similar %E values are observed over a broad range of acceptor intensities, demonstrating that FRET is not acceptor-density dependent for this experiment.
The artifactual FRET signal due to photoconversion shows a direct dependence on Cy5 intensity (e.g., Nichols, 2003; Snapp et al. 2004). A useful control is to measure the effect of decreasing the donor concentration (with constant acceptor concentration) on FRET efficiency. All other things being equal, FRET efficiency should be independent of donor intensity. If photoconversion is contributing significantly to the Cy3 fluorescence measurements, halving the donor concentration will substantially increase the apparent FRET signal. This is because the photoconverted product continues to contribute the same amount of fluorescence to the Cy3 measurements. However, the starting donor intensity is decreased. Thus, the photoconverted product will cause the Cy3 measurements to increase by a much higher percent upon Cy5 photobleaching, resulting in erroneously high FRET values. In contrast, genuine FRET without interference from photoconversion should be largely independent of donor intensity and occupancy. Indeed, a useful analysis of one's FRET data is to plot %E vs. absolute donor intensity. The relationship should be horizontal line. If FRET efficiency increases with donor intensity, then the investigator should suspect nonspecific clustering effects or photoconversion of the acceptor fluorophore to a donor-like spectral emission.
Ensure That the Antibodies Do Not Interact Directly with Each Other
It is assumed that antibodies do not directly interact with each other to generate FRET, but instead simply mark the positions of the antigens against which they are directed. Thus, a FRET signal between antibodies is taken to reflect the proximities of the antigens to which the antibodies are bound, and not to nonspecific interactions among the antibodies themselves. It is therefore critical to the interpretation of the results that the antibodies not interact with each other. FRET between directly interacting antibodies is largely insensitive to changes in concentration of the acceptor antibody (Fig. 17.9.3A). This is because the only donor labeling that occurs is via binding to an acceptor. Thus, although reduced labeling occurs due to the reduced acceptor concentration, all donors are still adjacent to an acceptor and therefore generate a high FRET signal (see Fig. 17.9.3A).
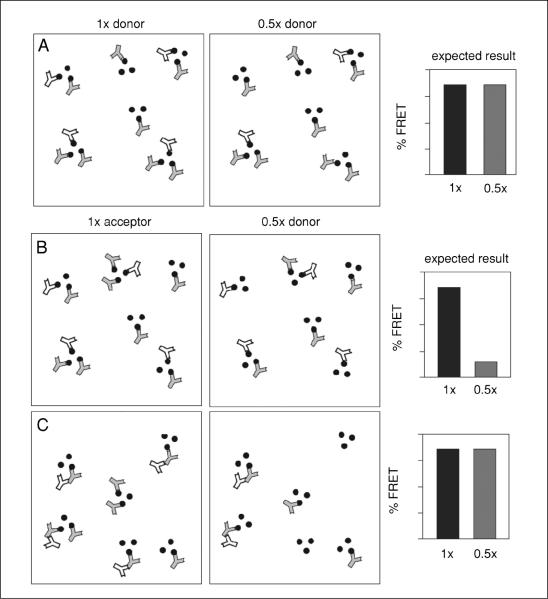
Figure 17.9.3.
Relationship of donor and acceptor concentrations to FRET. (A) Schematic diagram illustrating the effect of donor concentration on FRET. Oligomers of antigens (black dots) are shown randomly labeled with donor (white) and acceptor (gray) antibodies. The right diagram indicates one half the amount of donor antibody than that in the left panel. Although the donor fluorescence is expected to be lower for the right panel, the proximity of each donor to acceptor antibodies predicts that FRET efficiency should stay the same. The situation is very different for acceptor antibody concentration. (B and C) Antibody distributions are illustrated for donor (white) and acceptor (gray) antibodies on a hypothetical clustered three-antigen oligomer (black dots). Panel (B) shows the situation where the donor and acceptor antibodies bind their antigens, but do not interact with each other. Panel (C) shows the situation where the acceptor binds its antigen, but the donor antibody interacts with the acceptor antibody. In both cases, the right panel shows the consequence of reducing the acceptor concentration by one half, and the predicted effect on FRET.
In marked contrast, FRET between the antibodies as a consequence of the fact that their antigens are in close proximity is highly sensitive to acceptor concentration (Fig. 17.9.3B). Here, the donor antibodies still bind to their antigens, but some of the acceptor antigens will now be unoccupied by acceptor antibodies (Fig. 17.9.3C). The presence of donor antibodies unaccompanied by nearby acceptors reduces the FRET signal and helps rule out inappropriate interactions between antibodies.
For an additional control, measure FRET between the donor and acceptor antibodies with and without a peptide competitor corresponding to the antigen for the acceptor antibody. Inclusion of the peptide during the antibody incubation should result in both decreased labeling of cells by the acceptor antibody (but not the donor antibody), and a loss of the FRET signal. Taken together, these controls help confirm that FRET between dye-labeled antibodies is due to the proximities of the antigens to which the antibodies bind, and not due to interactions between the antibodies themselves.
Optimize Sample Size
Part of the power of FRET methods is the ability to quantitate small changes in protein proximities. However, achieving this quantitation requires sufficient statistical sampling. The amount of sampling dramatically increases as the investigator attempts to monitor single nanometer changes. At a minimum, perform multiple, at least n≥10, measurements to permit statistical analysis to identify significant changes in FRET efficiency values.
Expression levels of the proteins of interest will affect both the cell and the sensitivity of the planned measurements. Proteins, such as kinases or transcription factors, tend to be expressed at nanomolar concentrations (Huang and Ferrell., 1996). This translates to a few hundred copies of a protein or less per cell, in some cases. Consider that a homogeneously distributed fluorescent protein must be present at 200 nM to be visualized over background cellular fluorescence (Niswender et al., 1995). Thus, experimental questions involving cellular expression of a fluorescently tagged protein may require unnaturally high expression levels of a protein that may affect a cell phenotype. In contrast, dye-labeled antibodies rarely require overexpression of a protein of interest and also avoid problems related to whether a fluorescent fusion protein is functional (UNIT 21.4). Measurement sensitivity will be dependent on the available number of proteins to be assayed in a region of interest. Too few proteins will result in a low signal-to-noise ratio and low sampling sizes.
For purposes of experimental design, measuring absolute states of associated and nonassociated proteins should be readily discernable even for low protein sampling sizes. This is illustrated in Figure 17.9.4. The first graph of Figure 17.9.4B (n = 1) shows a scatter plot of the range of FRET values obtained for any single antibody pair separated by various distances. At a separation distance of 8 nm, the FRET efficiencies ranged broadly from less than 5% to nearly 80% (Fig. 17.9.4B; n = 1). This tremendous variability reflects the stochastic distributions of the donor and acceptor dyes over a large volume, combined with the extreme sensitivity of FRET to small changes in the distances separating them. Indeed, it has been shown previously that if the number of sampled molecules is small, such dramatic fluctuations can be anticipated (Haas and Steinberg, 1984).
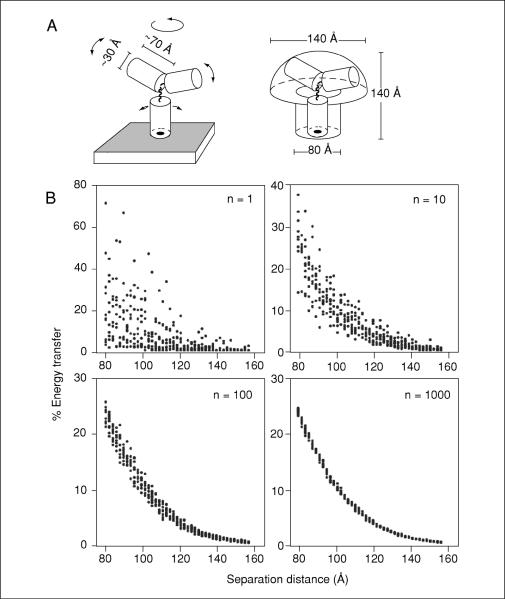
Figure 17.9.4.
Sampling size and the resolving power of antibody-mediated FRET. (A) Diagram of a model IgG molecule bound to an antigen on the membrane surface (left). The Fc and each Fab domain are modeled as a cylinder of diameter 3 nm, height 7 nm, connected by flexible hinges. Arrows indicate directions of allowed rotational flexibility. The range of potential positions that can be occupied by dyes conjugated to the antibody surface is indicated on the right. Dyes are allowed to be on the surface of the stalk of the mushroom-shaped space, and anywhere in the volume of the head. (B) A simulated donor and acceptor labeled antibody (4 dyes/IgG, randomly distributed as described in panel A) were bound to antigens separated by distances of between 8 and 16 nm, and the % energy transfer between the dyes calculated and plotted. Each datum represents the average of between 1 and 1000 such simulations as indicated (n) on each graph. Custom macros (available upon request from the authors) were written for NIH Image 1.62 to perform the antibody-mediated FRET simulations. In essence, an algorithm was designed to simulate the stochastic binding of a mixture of donor- and acceptor-labeled antibodies to a set of antigens on a membrane surface, followed by a calculation of the FRET between the randomly distributed dyes on all of the bound antibodies. The algorithm encompassed the following steps: (1) the x-y positions of the appropriate number of antigens were distributed on a hypothetical surface of defined area (usually 0.5 × 0.5 μm) at the indicated density and configuration (either randomly distributed, or in clusters of three). Clusters were not allowed to overlap, and the minimal distance separating adjacent antigens was limited to 8 nm, as determined by the steric hindrance of bound IgG molecules. (2) Each antigen was randomly assigned to either be unoccupied, bound by a donor antibody, or bound by an acceptor antibody. The relative probabilities of each assignment were determined by the desired occupancy and donor/acceptor ratio. (3) The x-y-z positions of dyes were randomly chosen relative to each antigen by the criteria outlined in the text. (4) Once the x-y-z positions for all of the donor and acceptor dyes were set, the summed FRET efficiency that would be expected for this distribution of dyes was calculated according to previously established equations (Förster, 1948; Dewey and Hammes, 1980).
Although a trend is observed in which increased separation distance between the antibodies results in lower FRET, a single interaction cannot be used to discriminate different antigen positions (except to say that antigens are either within ~15 nm of each other, or further away). However, the resolving power increases substantially as more antibody pair interactions are sampled and averaged (Fig. 17.9.4B; n = 10 through n = 1000). At a sampling size of 1000, differences in separation distance of between 0.2 and 0.4 nm can be resolved with confidence. Thus, subnanometer changes in antigen separation can be resolved using FRET between dye-conjugated antibodies despite the highly flexible and large nature of the probes, the stochastic distribution of the dyes bound to them, and the complex relationships for FRET between ensembles of donor and acceptor fluorophores.
Create a Model for the FRET Experiment
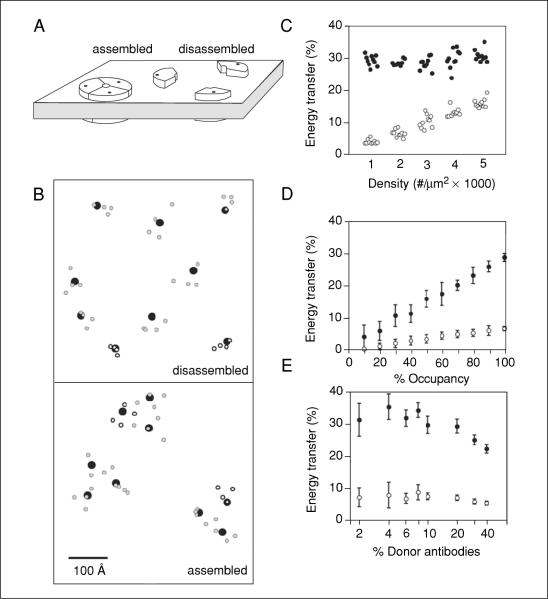
If sufficient information about the proteins of interest is available, it is worthwhile to model and simulate FRET experiments to assist with experimental design and expectations for results. Simulated results serve to illustrate the capabilities, sensitivity, and specificity of an approach and provide boundaries for the type of questions that can be addressed. In particular, two issues are of direct relevance to most studies of protein complex assembly: the discrimination of assembled from disassembled multiprotein (or oligomeric) complexes and the discrimination of small changes in the structure of a complex that remains assembled in the same general configuration.
Monte Carlo simulations, rather than theoretical calculations using simplifying assumptions, can provide insight into not only the expected FRET efficiency, but also the degree of variability that can be anticipated from measurement to measurement due to the stochastic nature of many of the variables involved (e.g., see Haas and Steinberg, 1984). For example, FRET between subunits of a membrane protein complex containing three subunits was modeled (Fig. 17.9.5A). Donor and acceptor antibodies labeled with four dyes at random positions were simulated in binding to the proteins with varying ratios. A visual representation of the relative proximities of subunit proteins and fluorescent dyes in the disassembled and assembled states for a small section of membrane is shown in Figure 17.9.5B as a two-dimensional illustration. Then the expected FRET efficiency for each ensemble of fluorophores was calculated according to previously established equations (Dewey and Hammes, 1980).
Figure 17.9.5.
Simulations of FRET for assembled versus unassembled oligomers. (A) Idealized configurations of an oligomeric channel in the membrane in an assembled and disassembled configuration. The positions of a putative antigen present as three copies in the channel are indicatedwithablack dot.(B) Surface view of a 70 × 70–nm section of membrane containing antigens (black dots) at 2000 copies per mm2 in assembled and disassembled configurations as in (A). The two dimensional projection of the positions of donor (green) and acceptor (red) dyes on antibodies bound to these antigens is also indicated. An antibody was assigned a 20% probability of containing donor dyes. (C) Monte Carlo simulations were used to calculate the FRET efficiencies in a 0.25 μm2 section of membrane containing donor and acceptor labeled antibodies (at a donor/acceptor ratio of 1:4, each containing 4 dye molecules per IgG) bound to antigens (with 100% occupancy) at densities of 1000 to 5000 copies per μm2. The antigens were either randomly distributed or assembled into clusters of three (around a 10-nm circle) that were randomly distributed in the membrane surface (closed circles). The simulation was repeated ten times for each condition, with each point representing the FRET from a single simulation. Note that FRET in the nonclustered configuration displays a clear density-dependence that is not seen in the clustered configuration. (C-E) FRET as a function of the degree of occupancy of the antigen by antibody, and proportion of antibodies that carry donor dyes were simulated as in (C). Randomly distributed antigens (open circles) were compared to clustered antigens (closed circles). The mean ± SD of ten simulations is plotted for each condition. When not being specifically varied, antibodies contained four dyes each, 20% of antibodies were donors, antigen occupancy was 100%, and antigen density was 2000 copies per μm2.
The putative assembled versus disassembled states of a protein complex represent the extremes of configurations, thereby making them easily distinguishable by antibody-mediated FRET. Because the active versus inactive configurations of the complex may instead represent a more subtle change in arrangement of its components, the authors used the simulations to ask whether such small changes could also be distinguished. As seen in Figure 17.9.4, even such small changes can be distinguished given sufficient sampling. Under these conditions, twenty areas of 0.25 μm2, each containing assemblies at a density of ~667 per μm2 (Fig. 17.9.6), was sufficient to distinguish a 0.4-nm difference. This modest sampling size (e.g., a total of ~5 μm2) represents the lower limit for distinguishing such small changes using idealized conditions. Thus, changes to a multiprotein structure, whether it is simple or complex, are likely to be detectable with antibody-mediated FRET. While different geometries of a protein complex can produce different absolute FRET values compared to an idealized complex, changes within the confines of a different structure are still detectable with antibody-mediated FRET (Snapp et al., 2004). These simulations were in fact very useful in concluding that antibody-mediated FRET is a feasible method to monitor both gross and subtle changes in complex structure in cells.
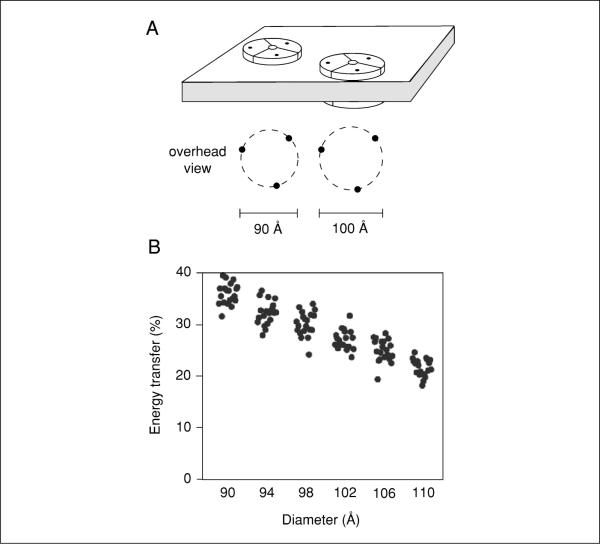
Figure 17.9.6.
Simulations of FRET for multiprotein complexes undergoing a conformational change. (A) Scale diagram of a complex of 90- versus 100-Å diameter. The modeled complex is of three antigens around a circle of the indicated diameter. (B) Clusters of three antigens in circles of diameters ranging from 9 to 11-nm were randomly distributed in 0.25 μm2 areas, and the FRET between the bound antibodies calculated by Monte Carlo simulations as in Figure 17.9.5. Each set of 20 measurements was statistically significant from an adjacent set.
The details of the simulation model system benefit from data on the size, structure, and abundance of the complex to be studied. Parameters to test include: simulated FRET between fluorophores with varying parameters such density of antigens (Fig. 17.9.5C), percent occupancy of antigens by antibodies (Fig. 17.9.5D), relative ratios of donor to acceptor antibodies (Fig. 17.9.5E), proximities of protein subunits, and configuration of antigens (e.g., randomly distributed versus assembled into clusters of defined size). It is beyond the scope of this unit to provide a detailed guide for model design and simulation. For more background on designing molecular simulations, the investigator is referred to Frenkel and Smit (1996). Examples of simulation design are described in the supplemental data sections for Snapp et al. (2004) and Sharma et al. (2004) and the appendix in Kenworthy and Edidin (1998).
RECIPROCAL FRET
When a protein complex contains more than two different proteins, determining the relative stoichiometries of the protein subunits can be valuable. The choice of acceptor and donor will significantly affect the FRET values if one protein is present at lower levels of expression (Fig. 17.9.7). The inherently asymmetric relationship between donor and acceptor molecules during acceptor-photobleaching FRET measurements makes it possible to assess whether protein subunits are present at comparable stoichiometries. While acceptors adjacent to a donor are essential for FRET to occur, the presence or absence of additional acceptor molecules that are unaccompanied by an adjacent donor does not change the overall FRET efficiency. In contrast, the presence of donor molecules that are unaccompanied by an adjacent acceptor causes an overall reduction of FRET efficiency. This is because the excess donors increase the overall donor fluorescence intensity without changing the absolute amount of energy that is transferred to the acceptor, thereby resulting in a net decrease in the percent of total energy transferred. That is, excess acceptors are effectively invisible and do not affect FRET values, while excess donors decrease FRET values.
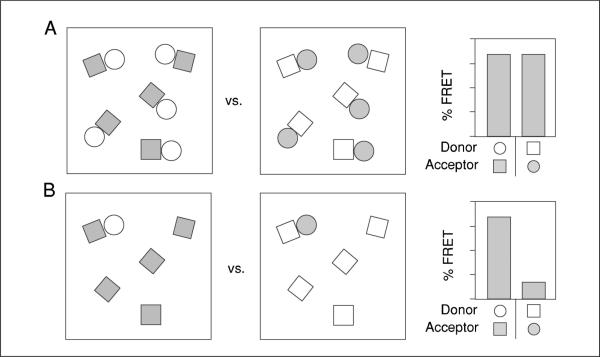
Figure 17.9.7.
FRET-based analysis of multiprotein complex component interactions and stoichiometry. (A and B) Cartoon of reciprocal FRET measurements between a hypothetical interacting pair of molecules (squares and circles) at different stoichiometries. The two fields in each panel represent situations in which each shape is labeled as donor (open) or acceptor (gray). The histograms to the right represent relative FRET efficiencies for each situation. Equal numbers of paired squares and circles (A) yields equivalent FRET values regardless of which protein is the donor or acceptor. In contrast, a substoichiometric number of circles, with excess unpaired squares, results in a discordance in FRET between the reciprocal measurements (B).
A schematic illustration of this principle is shown in Figure 17.9.7. In panel 17.9.4A, all copies of two interacting components (represented by squares and circles) are paired with each other, with neither containing excess copies. The FRET efficiency in this scenario should be equivalent regardless of which component serves as the donor or acceptor. In Figure 17.9.7B, however, one of two interacting components is present in excess (i.e., the squares). In this case, the FRET efficiency will be substantially lower if the squares serve as the donor than if the circles serve as the donor. Thus, a greater than 2-fold discordance in the FRET efficiencies upon switching the donor and acceptor molecules can be used to infer that at least some copies of one of the components exists in the absence of the other component. Reciprocal FRET will not necessarily reveal the precise stoichiometry, as the FRET relationships are not inherently linear. Nonetheless, the method should enable the investigator to distinguish between proteins present as single or multiple copies in a complex, and whether this changes in response to some treatment or manipulation. In general, it is not appropriate to perform reciprocal FRET when one of the proteins is labeled with a fluorescent fusion protein. Expressing a fluorescent fusion protein may significantly increase the levels of the protein of interest and result in protein that isn't incorporated into complexes or overexpression-related changes in cell behavior (UNIT 21.4).
To perform Reciprocal FRET, use the optimized acceptor-photobleaching protocol established in the acceptor-photobleaching FRET strategy with the following modifications:
When using antibodies to label donor and acceptor proteins, the experimental protocol is reversed, though the labeling protocol for the antibodies needs to be optimized. Determine labeling conditions (see Empirically Determine Antibody Labeling Conditions). Specifically, identify appropriate antibody labeling ratios for each antibody, maintain a ratio of ~1:4 to 1:8 donor/acceptor antibodies, and optimize amounts of antibodies to achieve maximal labeling intensity without causing nonspecific labeling or a decrease in FRET due to excess donor. In addition, confirm that order of addition of antibodies does not affect labeling efficiency and that FRET values are independent of donor intensities.
Using a fluorescent microscope, perform acceptor-photobleaching FRET for each combination of donor and acceptor, and then switch the labeling of each protein to make the initial donor now the acceptor and the initial acceptor now the donor. If one protein is present at significantly lower concentration than the other or if one protein is always bound to a partner, while the other protein is only sometimes bound, a significant difference in FRET under the two conditions will be observed. Because the absolute FRET efficiencies can vary somewhat depending on the number of dyes per antibody and labeling conditions, it is suggested that only discordance differences more than 2-fold be considered meaningful.
AUTOMATED IMAGE ANALYSIS
To process multiple data sets, automation of FRET analysis can be readily performed and one such scheme and macro are described. The authors have created an acceptor-photobleaching FRET analysis macro for NIH Image 1.62 (freely available from the authors). The macro quantitates FRET within the photobleached area and generates an energy transfer map. Briefly, the macro performs the following operations in sequence: (1) The pre-bleach and post-bleach images are registered to optimal alignment. (2) The area of photobleaching is identified. (3) The percent change in donor intensity (after background subtraction) is calculated in each 8 × 8–pixel region (0.56 × 0.56 μm) of the image and drawn as a pseudo-colored map. (4) The average change within the entire photobleached and non-bleached regions is calculated. The user is free to either write her own or modify the authors' macro as necessary for specific applications. To use the macro, the user must perform the following steps:
Name image files as 101000.tif, 101001.tif, 102000.tif, 102001.tif. for consecutive files. Note that the nomenclature is as follows: the first digit is the sample number (or the chamber number to indicate which chamber of an 8-well chambered coverslip is being used); the next two digits are the trial number within this sample; and the last three digits are necessarily “000” to indicate a pre-bleach image, and “001” to indicate a post-bleach image. Each image must be an 8-bit 512 × 512–pixel RGB file in which the red channel contains the acceptor image and the green channel contains the donor image. The photobleached region must be a 75 × 75–pixel region; the portion of the image that contains the bleach box does not matter (the macro automatically finds this).
Place all images in a single folder. Within that folder, the user must create a new folder entitled “processed images.”
Load the macro into NIH image and run. Before the macro starts, the user is asked a series of questions as follows. The user will be asked how many chambers. This is usually between 1 and 8. The user is then asked how many experiments are in a chamber (e.g., how many trials for each sample). Enter the number of experiments from 1 to 99. The user is next asked for a threshold (background value). The default is 10, but the user can decrease or increase this value as necessary based on the imaging conditions used. The user is asked the size of the sampling square. The default value is 8. This means that the FRET calculations are performed on each 8 × 8–pixel square within the image and the average FRET value within this area is plotted in the map. The averaging helps reduce image noise. It also affects the speed of the program. A smaller sampling square will slow the analysis because more pixels must be individually calculated.
When the user is asked to open the first image of the set, select OK. Then NIH image will open a browsing window to find the folder containing the images. Open this folder, select the first image, and select Open. This points the program to the place where the data set of interest is stored. The program will now run until all of the images in this folder have been processed.
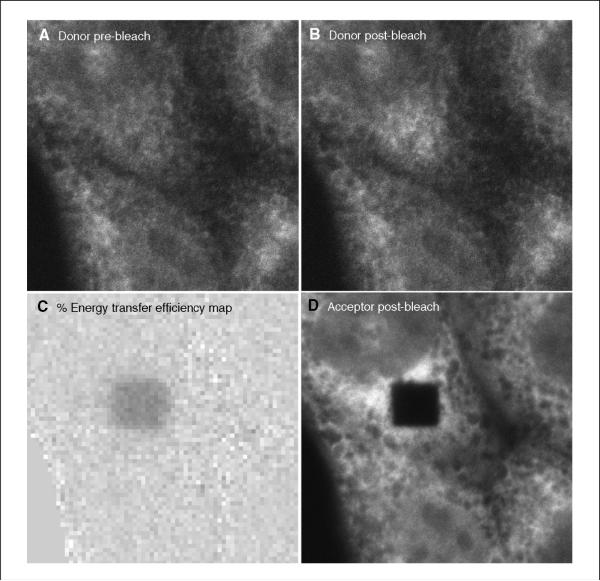
At the end of the program, the processed data file will include data images and a quantitation text file. The data images will contain the pre- and post-bleach donor images, an energy map, and the post-bleach acceptor image, in that order (Fig. 17.9.8).
Figure 17.9.8.
Output from the macro developed by the authors to assist in data interpretation (see Automated Image Analysis section). (A) Donor pre-bleach: ?? (B) Donor post-bleach: ?? (C) Energy transfer efficiency map: ?? (D) Acceptor post-bleach: ??
The quantitation file can be opened in a spreadsheet such as Microsoft Excel. The first column is the chamber number. The second column is the experiment number. The third column is the background subtracted post-bleach intensity of the donor and the fourth column is the background-subtracted pre-bleach donor intensity for the bleach region of interest. The final column is the calculated percent energy transfer. This information can be used to make graphs, do statistical analysis, etc.
CONCLUSION
The strategies described in this unit and UNIT 17.1 should permit the investigator to design, carry out, and interpret an acceptor-photobleaching FRET experiment to study protein interactions in cells. The investigator can use the same methodology to gain insight into comparative stoichiometry of proteins in a complex. Finally, the modeling methods described allow the investigator to simulate potential results to optimize the experimental setup and interpretation. Table 17.9.1 discusses methods to troubleshoot acceptor-photobleaching FRET.
Table 17.9.1.
Troubleshooting Guide for Acceptor-Photobleaching FRET
| Problem | Possible cause | Solution |
|---|---|---|
| FRET efficiency low, even for positive control | Acceptor fluorophore photobleached before performing FRET measurement | Do not image labeled cells with a fluorescent lamp, which will cause significant photobleaching; instead use low intensity laser power, using the donor fluorescence to find cells and focus them. Keep imaging time to a minimum before making the FRET measurement. Any loss of acceptor fluorescence will reduce the difference between the quenched and unquenched donor fluorophore. |
| Acceptor fluorophore incompletely photobleached | Be sure to identify photobleaching conditions to reduce fluorescence of the acceptor to background levels; otherwise, the donor will not be completely dequenched. | |
| High and low FRET observed along edges of fluorescent structures | Pre- and post-bleach images not registered, causing a slight shift of positions of areas with low fluorescence in a pre-bleach image to areas with significant fluorescence and resulting in an apparent increase in the donor channel | Align pre- and post-bleach images in an image manipulation program (e.g., Photoshop, ImageJ or NIH Image; then perform FRET analysis with the modified images. Note that the authors' macro includes a registration step. |
| FRET detected when using fluorescent fusion proteins as negative controls. (if the proteins used authentically do not interact) | Fused fluorescent proteins dimerizing (may occur at high concentrations; see Zacharias et al., 2002 and Snapp et al., 2003) | Be sure to make fluorescent fusion proteins with fluorescent proteins that contain monomerizing mutations. |
| FRET observed between negative control proteins in a fluorescence intensity-dependent manner | FRET due to density-dependent clustering of proteins most likely caused by (1) nonspecificity of an antibody or (2) labeled proteins are present at such high concentrations that their proximities are sufficiently close for FRET to occur | Confirm specificity by preincubating antibody with peptide that antibody was raised against; restrict analysis to regions of cells in which FRET occurs in an intensity independent manner (problem may be unavoidable in cases where the protein is concentrated within an organelle); use smaller fluorescent probes instead of antibodies; or consider other FRET methods including FLIM and fluorescence anisotropy (see supplementary data and appendix in Kenworthy and Edidin, 1998 and Sharma et al., 2004). |
LITERATURE CITED
- Adams SR, Campbell RE, Gross LA, Martin BR, Walkup GK, Yao Y, Tsien RY. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: Synthesis and biological applications. J. Am. Chem. Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- Bastiaens PI, Majoul IV, Verveer PJ, Soling HD, Jovin TM. Imaging the intra-cellular trafficking and state of the AB5 quaternary structure of cholera toxin. EMBO J. 1996;15:4246–4253. [PMC free article] [PubMed] [Google Scholar]
- Clegg RM. Fluorescence resonance energy transfer. Curr. Opin. Biotechnol. 1995;6:103–110. doi: 10.1016/0958-1669(95)80016-6. [DOI] [PubMed] [Google Scholar]
- Deniz AA, Laurence TA, Dahan M, Chemla DS, Schultz PG, Weiss S. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 2001;52:233–253. doi: 10.1146/annurev.physchem.52.1.233. [DOI] [PubMed] [Google Scholar]
- Dewey TG, Hammes GG. Calculation on fluorescence resonance energy transfer on surfaces. Biophys. J. 1980;32:1023–1035. doi: 10.1016/S0006-3495(80)85033-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster T. Zwischenmolekulare energiewanderung und fluoreszenz. Ann. Phys. 1948;2:57–75. [Google Scholar]
- Frenkel D, Smit B. Understanding Molecular Simulations. Academic Press; San Diego: 2002. [Google Scholar]
- Haas E, Steinberg IZ. Intramolecular dynamics of chain molecules monitored by fluctuations in efficiency of excitation energy transfer. A theoretical study. Biophys. J. 1984;46:429–437. doi: 10.1016/S0006-3495(84)84040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haj FG, Verveer PJ, Squire A, Neel BG, Bastiaens PI. Imaging sites of receptor dephosphorylation by PTP1B on the surface of the endoplasmic reticulum. Science. 2002;295:1708–1711. doi: 10.1126/science.1067566. [DOI] [PubMed] [Google Scholar]
- Huang CY, Ferrell JE. Ultrasensitivity in the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10078–10083. doi: 10.1073/pnas.93.19.10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson V. Formaldehyde cross-linking for studying nucleosomal dynamics. Methods. 1999;17:125–139. doi: 10.1006/meth.1998.0724. [DOI] [PubMed] [Google Scholar]
- Jares-Erijman E, Jovin TM. FRET imaging. Nat. Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK. Imaging protein-protein interactions using fluorescence resonance energy transfer microscopy. Methods. 2001;24:289–296. doi: 10.1006/meth.2001.1189. [DOI] [PubMed] [Google Scholar]
- Kenworthy AK, Edidin M. Distribution of a glycosylphosphatidylinositol-anchored protein at the apical surface of MDCK cells examined at a resolution of <100 Å using imaging fluorescence resonance energy transfer. J. Cell Biol. 1998;142:69–84. doi: 10.1083/jcb.142.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan RV, Varma R, Mayor S. Fluorescence methods to probe nanometer-scale organization of molecules in living cell membranes. J. Fluor. 2001;11:211–226. [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Matyus L. Fluorescence resonance energy transfer measurements on cell surfaces. A spectroscopic tool for determining protein interactions. J. Photochem. Photobiol. B. 1992;12:323–337. doi: 10.1016/1011-1344(92)85039-w. [DOI] [PubMed] [Google Scholar]
- Metz B, Kersten GFA, Hoogerhout P, Brugghe HF, Timmermans HAM, de Jong A, Meiring H, ten Hove J, Hennink WE, Crommelin DJA, Jiskoot W. Identification of formaldehyde-induced modifications in proteins—reactions with model peptides. J. Biol. Chem. 2004;279:6235–6243. doi: 10.1074/jbc.M310752200. [DOI] [PubMed] [Google Scholar]
- Michalet X, Kapanidis AN, Laurence T, Pinaud F, Doose S, Pflughoefft M, Weiss S. The power and prospects of fluorescence microscopies and spectroscopies. Annu. Rev. Biophys. Biomol. Struct. 2003;32:161–182. doi: 10.1146/annurev.biophys.32.110601.142525. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent protein and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- Miyawaki A, Tsien RY. Monitoring protein conformations and interactions by fluorescence resonance energy transfer between mutants of green fluorescent protein. Methods Enzymol. 2000;327:472–500. doi: 10.1016/s0076-6879(00)27297-2. [DOI] [PubMed] [Google Scholar]
- Nichols BJ. GM1-containing lipid rafts are depleted within clathrin-coated pits. Curr. Biol. 2003;13:686–690. doi: 10.1016/s0960-9822(03)00209-4. [DOI] [PubMed] [Google Scholar]
- Niswender KD, Blackman SM, Rohde L, Magnuson MA, Piston DW. Quantitative imaging of green fluorescent protein in cultured cells: Comparison of microscopic techniques, use in fusion proteins and detection limits. J. Microsc. 1995;180:109–116. doi: 10.1111/j.1365-2818.1995.tb03665.x. [DOI] [PubMed] [Google Scholar]
- Rizzo MA, Piston DW. High-contrast imaging of fluorescent protein FRET by fluorescence polarization microscopy. Biophys. J. 2005;88:L14–L16. doi: 10.1529/biophysj.104.055442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P, Varma R, Sarasij RC, Ira, Gousset K, Krishnamoorthy G, Rao M, Mayor S. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–589. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- Snapp E, Hegde R, Francolini M, Lombardo F, Colombo S, Pedrazzini E, Borgese N, Lippincott-Schwartz J. Formation of stacked cisternae by low affinity protein interactions. J. Cell Biol. 2003;163:257–269. doi: 10.1083/jcb.200306020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapp E, Reinhart G, Bogert B, Lippincott-Schwartz J, Hegde R. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell Biol. 2004;164:997–1007. doi: 10.1083/jcb.200312079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L, Haugland RP. Energy transfer: A spectroscopic ruler. Proc. Natl. Acad. Sci. U.S.A. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rheenen J, Langeslag M, Jalink K. Correcting confocal acquisition to optimize imaging of fluorescence resonance energy transfer by sensitized emission. Biophys. J. 2004;86:2517–2529. doi: 10.1016/S0006-3495(04)74307-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotech. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Wouters FS, Verveer PJ, Bastiaens PI. Imaging biochemistry inside cells. Trends Cell Biol. 2001;11:203–211. doi: 10.1016/s0962-8924(01)01982-1. [DOI] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]