Abstract
MYCN amplification occurs in around 25% of neuroblastomas, and is associated with rapid tumor progression and poor prognosis. MYCN plays a paradoxical role in driving cellular proliferation and inducing apoptosis. We previously observed nuclear p53 accumulation in neuroblastoma and hypothesize that MYCN regulates p53 in neuroblastoma. Immunohistochemical analysis of 82 neuroblastoma tumors demonstrated an association between high p53 expression and MYCN expression and MYCN amplification. In a panel of 5 MYCN amplified and 5 non-amplified neuroblastoma cell lines and also the Tet21N regulatable MYCN expression system there was a correlation between p53 expression and MYCN expression. Knockdown of MYCN in 2 MYCN amplified cell lines led to a decrease in p53 expression. Tet21N MYCN+ cells expressed higher p53 mRNA and protein, and had greater p53 transcriptional activity, in comparison with Tet21N MYCN− cells. Using chromatin immunoprecipitation and reporter gene assays, MYCN was found to bind directly to an E-Box motif located close to the transcriptional start site within the p53 promoter and initiate transcription. Mutation of the E-Box led to a decrease in MYCN driven transcriptional activity. Microarray analysis of Tet21N MYCN+/− cells showed that several p53 regulated genes were upregulated in the presence of MYCN, including MDM2 and PUMA. Knockdown of MYCN and p53 in a MYCN amplified cell line led to reduced PUMA levels and other markers of apoptosis. We conclude that MYCN transcriptionally upregulates p53 expression in neuroblastoma and may be an important mechanism by which MYCN induces apoptosis.
Keywords: p53, MYCN, neuroblastoma, chromatin immunoprecipitation, microarray
INTRODUCTION
Neuroblastoma is an embryonal malignancy derived from precursor cells of the sympathetic nervous system. Despite significant advances in understanding the biology of this tumor, it currently still remains one of the most difficult childhood cancers to cure, with less than 40% of patients with high risk disease (stage 4 disease over 18 months of age or MYCN amplified disease) being long-term survivors 1, 2
Amplification of MYCN occurs in ~25% of neuroblastoma, and is associated with rapid tumor progression and a poor prognosis (reviewed by 3). MYCN belongs to the Myc family of basic-helix–loop–helix-leucine zipper (bHLH-LZ) transcription factors that have a critical role in cellular proliferation, differentiation, apoptosis, and oncogenesis. Members of this family function as heterodimers with Max, and exert transcriptional activity by specifically binding to consensus E-Box motifs (CA(C/T)GTG) located within the promoter regions of a diverse set of target genes (reviewed by 4). In contrast to c-MYC, which is expressed in a wide variety of embryonic and adult tissues, MYCN expression is limited to the developing nervous system and selected other sites.
Several genes have been identified as c-MYC transcriptional targets (http://www.myc-cancer-gene.org/site/mycTargetDB.asp) 5, however less is known about target genes of MYCN. Early studies found that several c-MYC target genes were expressed in some neuroblastoma cell lines with MYCN amplification, but not all, suggesting other cell specific factors may be important 6. Recent studies have reported significant overlap between c-MYC and MYCN regulated gene sets 7, 8. Enhanced ectopic expression of MYCN leads to both accelerated cell cycle progression and sensitization to apoptosis, therefore mechanisms which minimize or evade MYCN driven apoptosis are essential for tumor progression in neuroblastomas with MYCN amplification (reviewed by 9).
p53, often referred to as the “guardian of the genome”, is mutated in up to 60% of many human malignancies. In neuroblastoma, p53 is rarely mutated however protein accumulation is frequently observed in both neuroblastoma tumors and cell lines (reviewed by 10). The presence of accumulated p53 in neuroblastoma has been suggested to be due to the embryonic nature of these tumors, reflecting a failure of the precursor cells to mature 11. We and others have shown that the accumulated p53 is both predominantly nuclear and functional in neuroblastoma tumors and cell lines (reviewed by 12).
Early studies using reporter gene assays and electrophoretic gel mobility analysis reported that p53 was a direct target gene of c-MYC 13, 14. Furthermore, it was shown using quiescent fibroblasts that p53 directly mediates c-MYC induced apoptosis, and suggests that c-MYC driven p53 mediated apoptosis acts as a safeguard mechanism against aberrant oncogenic activation 15. The p53 promoter contains a non-canonical E-Box (CATGTG) located upstream of the transcription initiation site 13, 16 and is recognized by MYC-MAX heterodimers 17 which can bind and initiate transcription 16. Several studies have reported a positive correlation between c-MYC expression and p53 expression in both cell lines and tumors, and that inhibition of c-MYC expression using either antisense RNA or inhibitory peptides led to a decrease in p53 expression (reviewed by 18).
This study set out to test the hypothesis that p53 accumulation in neuroblastoma correlates with MYCN amplification status and MYCN expression, and that p53 is a direct transcriptional target of MYCN.
MATERIALS AND METHODS
Immunohistochemistry of Neuroblastoma Tumors
Eighty-two formalin-fixed, paraffin-embedded diagnostic, untreated neuroblastoma tumors were examined for p53 and MYCN by immunohistochemistry using antibodies and methods previously reported 12. MYCN hybridoma supernatant MYCN (NCMIX102) was used at a 1:4 dilution. Positive tissue controls included colonic adenocarcinoma for p53 and MYCN amplified NGP neuroblastoma cell cytoblocks for MYCN. Negative controls included incubations without primary antibodies and surrounding non-tumor tissue. The p53 and MYCN labeling indices were performed as previously described 12. MYCN amplification was performed routinely on 40 frozen tumors by Southern blot hybridization and fluorescent in-situ hybridization that were diagnosed after 1990.
Cell Lines
p53 wt neuroblastoma cell lines 19, 20 used were, MYCN amplified: IMR32, NBLW, CHLA136, LAN5, SMSKCNR and NGP, non-MYCN amplified: SHSY5Y, NB69, SKNRA, and SHEP, non-MYCN amplified high MYCN expressing: SJNB1, and NBLS, and the conditional MYCN expressing SHEP Tet21N system together with p53 null osteosarcoma SAOS2 and p53 mutant neuroblastoma SKNBE2C cells 21. Cells were cultured as previously described 22. To switch off MYCN expression, Tet21N cells were cultured in the presence of 1μg/ml of tetracycline (Sigma) for 48 hours. Cell lines were obtained from: the Children's Oncology Group Cell Culture and Xenograft Repository (LAN5), Sue Cohn (NBLW, NBLS), Linda Harris (SJNB1), Penny Lovat (SHSY5Y and IMR32), John Maris (NB69), Patrick Reynolds (SKNRA, SMSKCNR, CHLA136), Barbara Spengler (SKNBE2C), Manfred Schwab (SHEP, Tet21N), Rogier Versteeg (NGP) and Ximena Montano (SAOS2) between 1996 and 2007, and were validated upon receipt using cytogenetic analysis courtesy of Nick Bown, Institute of Human Genetics, Newcastle University.
Quantitative Reverse Transcription PCR (qRT-PCR)
qRT-PCR was performed as previously described 22. All primers and probes were Inventoried TaqMan Gene Expression Assays (Applied Biosystems, Foster City, CA). RNA expression values were normalized to actin. All experiments were performed in triplicate.
Western Analysis
Western blotting was performed using previously described methods 21. Primary antibodies used were p53 DO-7 (Novocastra Laboratories Ltd., Newcastle upon Tyne, UK) at 1:1000, MYCN (NCMII100) at 1:100, actin (Sigma, St. Louis, MO) at 1:500, MDM2 (Calbiochem, Cambridge, MA) at 1:100, p21WAF1 (Calbiochem) at 1:100, Phospho-p53 (Ser-15) at 1:1000 and cleaved caspase 3 (Cell Signaling Technology Inc., Danvers, MA) at 1:1000 and PUMA (Calbiochem.
Secondary goat anti-mouse/rabbit HRP conjugated antibody (Dako, Glostrup, Denmark) was used at 1:500. Densitometry was performed as previously described 22 and expression normalized to actin.
Plasmids and Luciferase Assays
Plasmids used were the p53-dependent pGL3-P2 reporter construct 23, pCMV-β-galactosidase (Stratagene, La Jolla, CA), pRenilla-TK vector (Promega, Madison, WI) and p53 promoter reporter constructs pGL2-200bp and pGL2-356bp originally described by Wang & El-Deiry 24 and purchased from Addgene (Addgene, Cambridge, MA). pGL2-Δ200bp and pGL2-Δ356bp were generated by replacing the E-Box CATGTG with CTGCAG, and confirmed by sequencing. The pCMV14-MYCN expression plasmid was generated by cloning the CDS of MYCN into the p3XFLAG-CMV™-14 vector (Sigma). Cells were transfected using FuGENE®6 (Roche, Basel, Switzerland) or Polyethyleneimine (PEI) (Sigma). Luciferase activity was measured as previously described 25 or using the Dual-Luciferase Reporter Assay (Promega). To control for transfection efficiency, luciferase activity was normalized to β-galactosidase activity or Renilla luciferase activity. All experiments were performed in triplicate.
p53 DNA Binding Assay
The TransAM™ ELISA based p53 transcription factor assay (Active Motif, Carlsbad, CA) was used according to the manufacturer's protocol. Nuclear extracts were prepared using previously described methods 12, and 5μg of protein used per well. Samples were prepared in triplicate and assayed in duplicate. Absorption was read at 450nm with a reference wavelength of 655nm.
Gene Expression Microarray
Cells were harvested from Tet21N MYCN− and Tet21N MYCN+ cells in triplicate and analyzed using Affymetrix U133 plus 2.0 oligonucleotide arrays. Arrays were performed at the CRUK facility (Paterson Institute, Manchester, UK). Microarray data was analyzed using GeneSpring GX (Agilent Technologies Inc., Santa Clara, CA). Selected genes were validated using qRT-PCR. Array data is deposited in ArrayExpress, accession number E-MEXP-2340.
RNA Interference
siRNAs were transfected into cells using Lipofectamine (Invitrogen, Paisley, UK) to concentrations as indicated. siRNAs were purchased from Eurogentec (Eurogentec Ltd., Seraing, Belgium), using previously described sequences for p53 26 and MYCN 22. All experiments were performed in triplicate.
Irradiation Induced DNA Damage and Caspase Assays
Cells were treated with 4Gy X-irradiation from a RS320 irradiation system (Gulmay Medical, Surrey, UK). Cells were harvested at the indicated times post irradiation for protein analysis or caspase analysis. Experiments were performed in triplicate. Caspase-3/7 activity was measured using the Caspase-Glo 3/7 assay (Promega) according to the manufacturer's instructions. Experiments were performed in triplicate.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as previously described 27, using 5μg of anti-MYCN B8.4B (Santa Cruz Biotechnology Inc., Santa Cruz, CA), anti-Max C-17 (Santa Cruz) or IgG (Santa Cruz) as a negative control. Samples were analyzed using SYBR green quantitative PCR with specific primers. Nucleolin was used as a positive control. Experiments were performed in triplicate. All primer sequences are listed in Supplementary Information Table S1.
Actinomycin D and Cycloheximide Treatment
Tet21N MYCN+ and MYCN− cells were treated with either 1μg/ml of Actinomycin D (Sigma), or 25μM of Cycloheximide (Calbiochem). Cells were harvested at the indicated times after Actinomycin D treatment for qRT-PCR analysis of mRNA expression, and after Cycloheximide treatment for protein analysis. Experiments were performed in triplicate.
Statistical Analyses
Statistical tests were performed using Minitab (Minitab Inc., State College, PA.) or GraphPad Prism (GraphPad Software Inc., La Jolla, CA). All statistical tests were two-sided. If the expected frequency of a χ2 test was less than 5, Fisher's exact test was performed using Graph Pad Prism.
RESULTS
p53 protein expression is associated with MYCN amplification and expression in neuroblastoma tumors
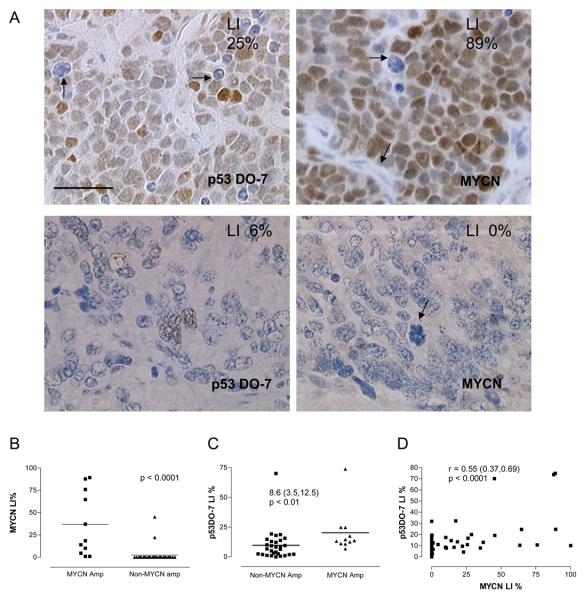
p53 expression, localization and relationship with histology in this set of neuroblastoma tumors has been previously reported 12. p53 was found to be predominantly nuclear and expressed at high levels in undifferentiated and poorly differentiated neuroblastoma (Figure 1A, and 12). MYCN expression was nuclear, heterogeneous and expressed at higher levels in undifferentiated, poorly differentiated and International Neuroblastoma Pathology Classification unfavorable histology tumors (Figure 1A and data not shown). MYCN was not expressed in Schwannian stroma (Figure 1A). MYCN expression was significantly higher in MYCN amplified tumors as expected and only three non-MYCN amplified tumors expressed MYCN detectable by immunohistochemistry (Figure 1A, 1B and Table 1). Using the median Labeling Index for p53 and MYCN in all 82 tumors as a cut off between positive and negative expression, increased p53 expression was significantly associated with MYCN amplification with two of the p53 antibodies used (Table 1 and Figure 1A & C), and with MYCN expression with all three p53 antibodies (Table 1 and Figure 1A & D). For DO-7 the median difference in labeling index (LI) between MYCN amplified and non-amplified neuroblastoma was 8.6%, median p53DO-7 LI for non-MYCN amplified tumors was 6.7% and for MYCN amplified tumors was 13.5% (Figure 1C).
Figure 1. p53 expression is associated with MYCN amplification and MYCN protein expression in untreated neuroblastoma tumors.
A) Upper panel Immunohistochemistry of a stroma-poor, undifferentiated, MYCN amplified, stage 4 primary neuroblastoma, using (Left) p53DO-7 antibody showing heterogeneous nuclear immunostaining, absent in apoptotic cells (arrowed) and (Right) MYCN antibody showing heterogeneous nuclear immunostaining with negative stroma and negative mitotic and apoptotic cells (arrowed). Lower Panel Immunohistochemistry of a stroma-poor nodule of a non-MYCN amplified, stage 4 primary nodular ganglioneuroblastoma, using (Left) p53DO-7 antibody showing low level nuclear immunostaining and (Right) MYCN antibody showing absent immunostaining and a mitotic cell (arrowed). Labeling Indices (LI) for antibodies are also shown in top right of images. Scale bar 50μM. B & C Scatter plots of Labeling Indices (LI) showing median expression (two tailed Mann-Whitney tests used) B) MYCN amplification vs MYCN LI, and C) MYCN amplification vs p53DO-7 LI (median difference shown and 95% Confidence Interval). D) Correlation between p53DO-7 LI and MYCN LI (Spearman Correlation and 95% Confidence Interval).
Table 1.
MYCN amplification and expression is associated with p53 protein expression in 82 neuroblastomas
| Median LI % (range) |
MYCN n = 82 |
MNA1 n = 40 |
|||||
|---|---|---|---|---|---|---|---|
| − | + | p | + | − | p | ||
| MYCN | >0 (0,100) | − | 0 | 26 | <0.001 | ||
| + | 11 | 3 | |||||
| p53 DO-7 | 8, (0, 75) | − 35 | 6 | <0.001 | 1 | 14 | <0.05 |
| + 17 | 24 | 10 | 15 | ||||
| p53 DO-1 | 7 (0, 89) | − 34 | 5 | < 0.001 | 1 | 15 | < 0.05 |
| + 17 | 25 | 10 | 13 | ||||
| p53 1801 | 2, (0, 20) | − 38 | 10 | < 0.001 | 2 | 14 | 0.2 |
| + 14 | 20 | 9 | 15 | ||||
Fisher's exact test two-sided. All other parameters χ2 used. MNA = MYCN amplification. The Median Labeling Index (LI) for all 82 tumours was used as a cut-off for positive and negative expression (range).
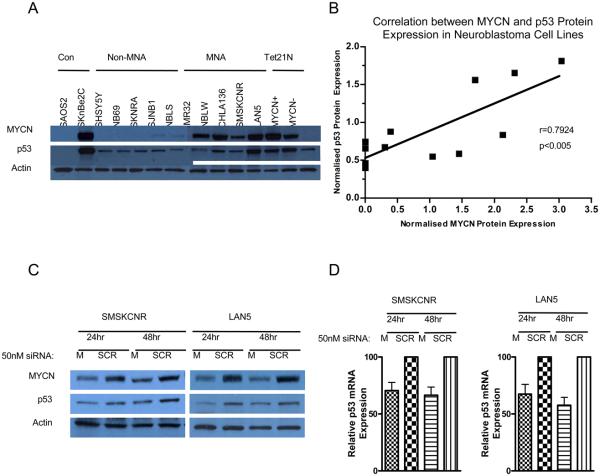
p53 protein expression correlates with MYCN expression in neuroblastoma cell lines
p53 protein levels were analyzed in a panel of ten p53 wt neuroblastoma cell lines, conditional MYCN expressing SHEP Tet21N cells, and two negative control cell lines, p53 mutant SKNBE2C, and p53 null SAOS2 cells (Figure 2A). Induced levels of MYCN in Tet21N MYCN+ cells were comparable with MYCN amplified neuroblastoma cell lines (Figure 2A). p53 expression was found to correlate with MYCN expression (p<0.005, r=0.7924, Spearman Correlation) (Figure 2A and B), but not with MYCN amplification (data not shown).
Figure 2. p53 expression correlates with MYCN status and expression in neuroblastoma cell lines.
A) Western blot showing p53 and MYCN protein expression in neuroblastoma cell lines Con = control. B) Correlation between p53 protein and MYCN protein expression in neuroblastoma cell lines (Spearman Correlation, r=0.7924, p<0.005). p53 and MYCN expression was determined using densitometry, and normalized to actin. Knockdown of MYCN expression using siRNA (50nM) (M) in two MYCN amplified neuroblastoma cell lines, SMSKCNR and LAN5 compared with scrambled siRNA (SCR) led to decreased C) p53 protein expression and D) mRNA expression in both cell lines.
To further investigate p53 expression in relation to MYCN expression, MYCN expression was inhibited using siRNA in p53 wt MYCN amplified SMSKCNR and LAN5 cells. This led to decreased p53 protein (Figure 2C) and mRNA expression (Figure 2D) in both cell lines. At 24 hours after MYCN knockdown LAN5 cells exhibited a ~40% reduction in p53 mRNA expression (Figure 2D) and ~50% reduction in p53 protein expression (p<0.05, paired t-test) (Figure 2C), whereas the SMSKCNR cell line exhibited a ~30% (Figure 2D) and ~40% (Figure 2C) reduction in p53 mRNA and protein expression respectively.
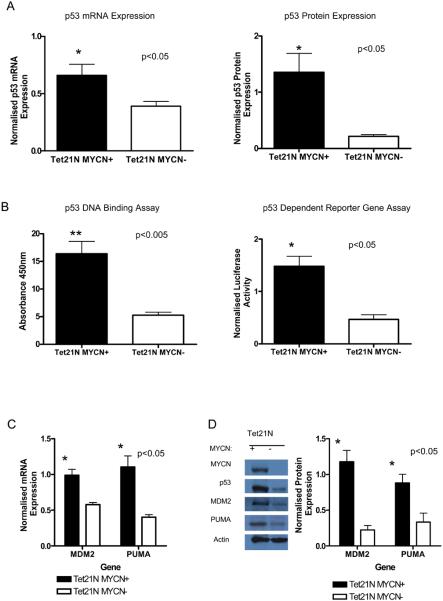
p53 expression, transcriptional activity and induction of p53 regulated genes in SHEP Tet21N cells
Tet21N MYCN+ cells were found to express significantly higher p53 mRNA (p<0.05, paired t-test) (Figure 3A) and protein (p<0.05, paired t-test) (Figure 2A and 3A) compared with Tet21N MYCN− cells. Consistent with higher p53 transcript and protein levels, Tet21N MYCN+ cells exhibited significantly greater p53 DNA binding capacity (p<0.005, paired t-test) and transcriptional activity (p<0.05, paired t-test) (Figure 3B) in comparison with Tet21N MYCN− cells. Furthermore, higher mRNA (p<0.05, paired t-test) (Figure 3C) and protein (p<0.05, paired t-test) (Figure 3D) expression of two endogenous p53 target genes, MDM2 and PUMA were observed in Tet21N MYCN+ cells compared with Tet21N MYCN− cells, confirming the results of the reporter gene and DNA binding assays, indicating that the accumulated p53 in Tet21N MYCN+ cells is transcriptionally active.
Figure 3. p53 mRNA and protein expression, and DNA binding and transcriptional activity in Tet21N cells.
A) p53 (Left) mRNA and (Right) protein expression in the presence of MYCN, (paired t-test, p<0.05). B) (Left) p53 DNA binding capacity (paired t-test, p<0.005) and (Right) p53-dependent reporter gene activity (paired t-test, p<0.05) in the presence of MYCN. The luciferase activity of the p53-dependent pGL3-P2 reporter construct was normalized to β-galactosidase activity of pCMV-β-galactosidase plasmid construct. C) Bar chart showing higher mRNA expression of p53 target genes MDM2 and PUMA in the presence of MYCN (paired t-test, p<0.05). D) (Left) Western blot and (Right) densitometry analysis normalized to actin showing higher protein expression of p53 target genes MDM2 and PUMA in the presence of MYCN (paired t-test, p<0.05).
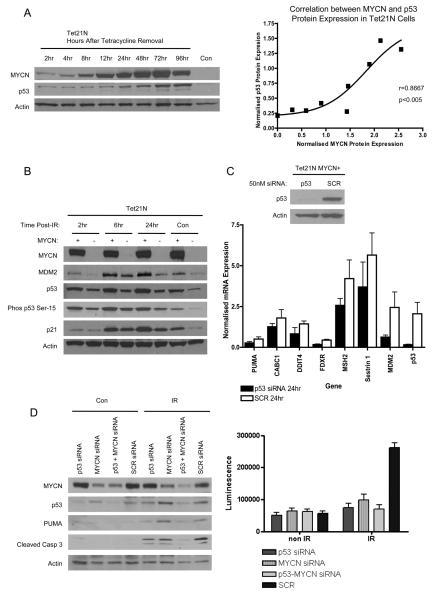
Modulation of MYCN expression in Tet21N cells by continuous culture in the presence of tetracycline for two weeks followed by removal of tetracycline from the growth media resulting in the induction of MYCN expression, showed that p53 mRNA (data not shown) and protein expression positively correlated with MYCN protein expression (p<0.005, r=0.8667, Spearman Correlation) (Figure 4A), lending further support for MYCN regulation of p53 in this system.
Figure 4. p53 expression in the presence of MYCN is functional after DNA damage, and necessary for apoptosis.
A) (Left) Western blot and (Right) representative graph showing the positive correlation between MYCN protein and p53 protein expression in Tet21N cells harvested after tetracycline removal from growth media (Spearman Correlation, r=0.7924, p<0.005). Con, cells cultured continuously in the presence of tetracycline. B) Western analysis showing p53 is functional in both Tet21N MYCN+ and MYCN− cells after DNA damage, leading to induction of target genes p21WAF1 and MDM2. Con, non-irradiated samples. C) qRT-PCR analysis of 7 p53 regulated genes identified using microarray analysis, upregulated in the presence of MYCN and showed a decrease in expression after p53 siRNA treatment (50nM) for 24 hours. D) MYCN amplified NGP cells treated with MYCN siRNA (40nM), p53 siRNA (50nM), p53 and MYCN siRNA (35nM of each siRNA) or SCR siRNA (70nM) for 48 hours prior to irradiation induced DNA damage. 24 hours post irradiation cells were harvested and analyzed for the expression of apoptosis mediators cleaved caspase 3 and PUMA (Left) and Capsase-3/7 activity (Right).
To determine whether p53 is functional after DNA damage in Tet21N MYCN+/− cells, cells were subjected to 4Gy of X-irradiation and analyzed for expression of p21WAF1 and MDM2. Following DNA damage there was an increase in serine -15 phosphorylated p53 as well as induction of p53, p21WAF1 and MDM2 in Tet21N MYCN+ and MYCN− cells (Figure 4B). The basal and induced levels of p53, phosphorylated p53 at serine-15, MDM2 and p21WAF1 were all higher in Tet21N MYCN+ cells compared with Tet21N MYCN− cells (Figure 4B) consistent with higher levels of p53 activity in the presence of MYCN. The above data confirms that p53 is functional both in the presence and absence of MYCN in this system.
To determine whether the greater induction of p21WAF1 and MDM2 in Tet21N MYCN+ cells is dependent on functional p53, p53 was knocked down using siRNA and the cells treated with 4Gy of X-irradiation prior to analysis of p21WAF1 and MDM2 expression. Inhibition of p53 using siRNA led to complete abrogation of p21WAF1 and MDM2 induction following DNA damage further confirming that p53 is functional and that induction of p21WAF1 and MDM2 is dependent on functional p53 (data not shown). In addition, following knockdown of p53, basal levels of MDM2 and p21WAF1 were much lower than in the cells treated with scrambled (SCR) control siRNA (data not shown).
Gene expression microarray analysis of p53 and p53 regulated gene expression in the presence of ectopic MYCN
Affymetrix gene expression microarray analysis of SHEP Tet21N MYCN+ and MYCN− cells was performed to determine whether there was upregulation of p53 and/or p53 regulated genes in the presence of MYCN. qRT-PCR was performed on selected genes to validate microarray results (data not shown), and the results were consistent. Several previously reported putative MYCN target genes were found to be differentially expressed in the presence of MYCN (Supplementary Information, Table S2). In agreement with qRT-PCR analysis, p53 transcript expression detected by microarray analysis was upregulated 1.9 fold in the presence of MYCN (Supplementary Information, Table S3). In addition, many p53 regulated genes were also found to be differentially upregulated in the presence of MYCN (Supplementary Information, Table S3 and Figure S1). To confirm that the increased expression of p53 regulatable genes in the presence of MYCN was p53-dependent, p53 expression in the Tet21N MYCN+ cells was inhibited using siRNA, and selected p53 regulated genes analyzed by qRT-PCR (Figure 4C). All genes tested showed a decrease in transcript expression in the presence of p53 siRNA compared with SCR control, confirming that the observed increase in expression of p53 regulatable genes is at least in part dependent on higher levels of p53 present in Tet21N MYCN+ cells (Figure 4C).
Irradiation induced apoptosis in MYCN amplified cells is dependent on p53
To determine whether higher levels of apoptosis in MYCN amplified neuroblastoma cells is dependent on higher levels of p53 in the presence of MYCN, MYCN amplified NGP cells were treated with MYCN and/or p53 siRNA or SCR siRNA prior to irradiation induced DNA damage (Figure 4D). NGP cells were chosen because we have previously reported high levels of apoptosis following irradiation in this cell line (21). Apoptosis was determined by analysis of expression of apoptosis mediator PUMA and cleavage of caspase 3 together with quantification of caspase 3/7 activity. Twenty-four hours after irradiation, inhibition of MYCN or p53 led to decreased caspase-3/7 activity, compared to cells treated with SCR siRNA (Figure 4D). Furthermore, inhibition of MYCN and p53 led to a slightly greater reduction in caspase-3/7 activity than inhibition of MYCN or p53 alone (Figure 4D). Similarly, inhibition of MYCN and p53 led to a greater reduction in cleavage of caspase 3 and less induced PUMA than inhibition of MYCN or p53 alone (Figure 4D).
p53 regulation is predominantly at the level of transcription
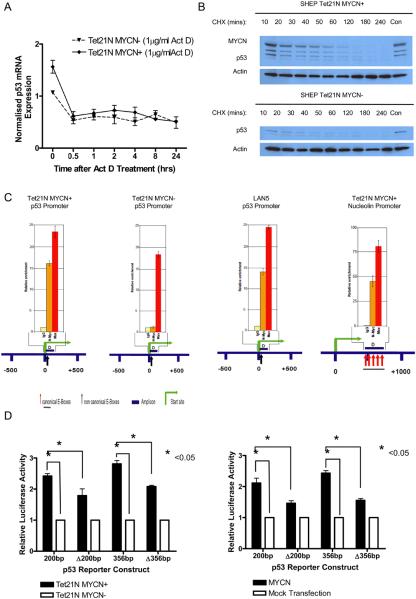
To determine whether the increased p53 expression in the presence of MYCN is due to transcriptional regulation or protein stabilization through post-translational modifications, Tet21N cells were treated with either 1μg/ml of actinomycin D or 25μM of cycloheximide. Actinomycin D (Figure 5A) and cycloheximide (Figure 5B) treatment led to decreases in p53 mRNA and protein levels, respectively, in both Tet21N MYCN+ and MYCN− cells. p53 was found to have a half life of 30-40 mins in this system. MYCN levels also decreased following cycloheximide treatment (Figure 5B). The above data suggests that the increased expression of p53 in the presence of MYCN is predominantly via transcriptional regulation rather than p53 stabilization by post-translational modifications.
Figure 5. MYCN predominantly regulates p53 at the transcriptional level, via direct binding to the E-Box motif within the p53 promoter.
A) qRT-PCR analysis of p53 mRNA expression in Tet21N MYCN+ and MYCN− cells after treatment with 1μg/ml Actinomycin D, shows a decrease in p53 mRNA expression, suggesting that MYCN regulates p53 transcription. B) Western blot showing p53 and MYCN protein expression in (Top) Tet21N MYCN+ and (Bottom) MYCN− cells harvested after treatment with 25μM of Cycloheximide (CHX), demonstrates a decrease in p53 expression suggesting that p53 is not predominantly post-translationally stabilized. C) MYCN ChIP analysis of the p53 promoter in (Left) Tet21N MYCN+ cells, (second from left) Tet21N MYCN− cells and (second from right) MYCN amplified LAN5 cells. (Right) As a positive control direct binding of MYCN to the nucleolin promoter in Tet21N MYCN+ cells is shown. D) Relative luciferase activity of p53 promoter constructs (pGL2-200bp, pGL2-Δ200bp, pGL2-356bp, pGL2-Δ356bp) transfected into (Left) Tet21N MYCN+ and MYCN− cells, and (Right) co-transfected with pCMV14-MYCN expression plasmid into SHEP cells.
p53 is a direct transcriptional target of MYCN
MYCN ChIP was performed in two MYCN amplified neuroblastoma cell lines and the Tet21N system to determine whether MYCN can directly bind to an E-Box motif within the p53 promoter in vivo. It was found that MYCN binds directly to the non-canonical E-Box motif located close to the transcriptional start site of p53 in Tet21N MYCN+ cells (Figure 5C) and MYCN amplified neuroblastoma cell lines LAN5 (Figure 5C) and SKNBE2C (Supplementary Information, Figure S2). As expected, no MYCN binding was observed in Tet21N MYCN− cells (Figure 5C). MYCN binding to the E-Box within the nucleolin promoter in the Tet21N MYCN+ cells was used as a positive control (Figure 5C).
To confirm functional regulation of p53 by MYCN, p53 reporter constructs incorporating the wt E-Box motif, CATGTG, (pGL2-200bp and pGL2-356bp), or the mutated E-Box motif, CTGCAG, (pGL2-Δ200bp and pGL2-Δ356bp) were transfected into Tet21N MYCN+ and Tet21N MYCN− cells, and also co-transfected with the pCMV14-MYCN expression plasmid into parental SHEP cells. In line with the above ChIP data, p53 promoter constructs incorporating the wt E-Box motif exhibited significantly greater luciferase activity in the presence of MYCN (p<0.05, paired t-test) (Figure 5D) in Tet21N cells, and also in SHEP cells co-transfected with pCMV14-MYCN (p<0.05, paired t-test) (Figure 5D). Mutation of the E-Box motif led to significantly reduced luciferase activity (p<0.05, paired t-test) in Tet21N MYCN+ cells (Figure 5D) and also in SHEP cells co-transfected with pCMV14-MYCN (p<0.05, paired t-test) (Figure 5D).
DISCUSSION
Like c-MYC, MYCN plays a paradoxical role in driving both cellular proliferation 28 and inducing apoptosis 29-31. This is observed histologically by a high mitosis-karyorrhexis index (MKI) in both human MYCN amplified neuroblastoma tumors 32-35 and MYCN transgenic mouse neuroblastoma tumors 36. In addition, in transgenic mouse tumors we reported tingible body macrophages reflecting high levels of apoptosis within these tumors 36. Studies using MYCN amplified cell lines, MYCN transgenic mouse tumors and ectopic MYCN expressing Tet21N cells have demonstrated higher p53 levels in the presence of MYCN 22, 37, 38 and increased levels of apoptosis in response to chemotherapeutic agents 29, 39-41, and irradiation 22.
We and others have previously observed wt nuclear functional p53 accumulation in neuroblastoma cell lines (reviewed by 10) and tumors 12. p53 has been shown to be a direct target gene of c-MYC, in which the p53 promoter has been reported to contain an E-Box related sequence (CATGTG) through which the Myc gene family exert transcriptional activity 13, 14. We hypothesized that p53 accumulation in neuroblastoma is related to MYCN amplification and expression and that p53 is a direct transcriptional target of MYCN in neuroblastoma.
Here we show that p53 protein expression correlates with MYCN protein expression in both primary neuroblastoma tumors and neuroblastoma cell lines. Using a regulatable MYCN expression system, p53 was found to be functional and exhibit greater transcriptional activity in the presence of MYCN leading to increased expression of several p53 target genes. Using quantitative MYCN ChIP, MYCN was found to bind directly to the non-canonical E-Box motif (CATGTG) within the p53 promoter in vivo, and using reporter gene assays direct functional upregulation of transcription from the p53 promoter by MYCN was demonstrated. Furthermore, deletion of this E-box led to reduced transcriptional activity from the p53 promoter by MYCN. This data provides strong support to the model that this specific E-Box is responsible for MYCN mediated regulation of p53. The binding of MYCN to an E-Box within the p53 promoter has very recently been reported using ChIP-chip arrays 7, however this study did not include functional upregulation of p53. Taken together these findings provide an important and direct mechanism by which MYCN is able to sensitize cells for p53 mediated apoptosis.
Our data is both consistent with and provides a direct mechanism to account for previous studies which have shown that MYCN amplified neuroblastoma tumors expressed significantly higher levels of p53 RNA in comparison with non-amplified tumors 7, 42, 43. It is also possible that the association of p53 with undifferentiated, highly proliferative neuroblastoma we previously reported is mediated through MYCN 12, and that MYCN may be involved in other studies linking p53 RNA expression with unfavorable neuroblastoma 44.
Our data is also in line with work in the TH-MYCN transgenic mouse model which demonstrated that MYCN driven tumor formation had higher penetrance and reduced latency in p53 halpoinsufficient mice, and that chemotherapy induced apoptosis was shown to be p53-dependent, where apoptosis was significantly reduced in TH-MYCN p53 +/− tumors compared to p53+/+ tumors 41.
Mechanisms by which members of the Myc family induce apoptosis are not fully understood (reviewed by 45). One mechanism is via p14ARF mediated increased p53 expression, stability, and activity 46, although we have previously reported that SHEP cells are homozygously deleted for p14ARF 19, and this was further confirmed in the present study (data not shown). However, p14ARFmay be involved in other neuroblastoma cell lines in addition, or instead of, direct regulation of the p53 gene shown in the present study.
MYCN and p53 are both expressed in the normal embryonic developing nervous system, during the phase of precursor cell expansion prior to the onset of differentiation. In the context of normal embryonic development, the present findings therefore provide a mechanism for MYCN driven p53-dependent apoptosis, that may be important in eliminating any rapidly proliferating neuroblasts exposed to potential teratogens, to prevent deregulated proliferation and aberrations during development.
Our findings may help to explain why neuroblastoma are initially chemosensitive, however despite an initial response, more than half of all cases relapse with chemoresistant disease suggesting that MYCN amplified tumors eventually develop mechanisms to evade MYCN driven apoptosis (reviewed by 9). There is evidence from cell line studies that MYCN amplified cell lines may circumvent MYCN driven p53-dependent apoptosis by selecting for cells with aberrations in the p53/MDM2/p14ARF pathway. Analysis of neuroblastoma cell lines reported to date with aberrations in the p53/MDM2/p14ARF pathway demonstrates that 25/34 (74%) of cell lines are MYCN amplified (Supplementary Information, Table S4) and predominantly established following previous therapy at relapse 19, when abnormalities of the p53 pathway in neuroblastoma tumors have been previously reported (reviewed by 10). A study examining the p53 pathway in relapsed MYCN amplified neuroblastomas is warranted.
Further illustrating the complexity of MYCN's paradoxical role in proliferation and apoptosis, MDM2 the critical negative regulator of p53, has previously been reported to be a direct target of MYCN 47. In the present study siRNA mediated inhibition of p53 led to a decrease in MDM2 expression (data not shown), suggesting that increased expression of MDM2 in the presence of MYCN may also be dependent on the higher expression of p53 and not solely due to direct upregulation by MYCN.
In conclusion, this study provides several lines of evidence that p53 is a direct transcriptional target of MYCN in neuroblastoma and is likely to be a mechanism for MYCN induced p53-dependent apoptosis. However the various selection pressures on MYCN amplified tumors either spontaneously or via chemotherapy may lead to selection for those cells that can evade apoptosis by various mechanisms which result in MYCN amplified neuroblastoma progressing to become such a severe phenotype.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the following for providing cell lines: Sue Cohn, Linda Harris, Penny Lovat, John Maris, Patrick Reynolds, Barbara Spengler, Manfred Schwab and Ximena Montano. We would also like to thank Naohiko Ikegaki for the MYCN antibodies, Ximena Montano for the p53 DO-1 antibody and Novocastra, Newcastle upon Tyne, for the p53DO-7 and Pab1801 antibodies used for immunohistochemistry. We are also grateful to Nick Bown, Angela Baker and Julian Board for MYCN analyzes, Allison Carpenter, Christine Estlin, Janet Thompson and Susan Wood for pathology technical support, Donatella Manzoni for assistance with cell culture and Dan Swan for bioinformatics support.
Sources of Support: The North of England Children's Cancer Research Fund, the U.K Department of Health and Cancer Research U.K. Italian Association for Cancer Research (AIRC) and Italian Ministry of Research and Education.
Abbreviations
- LI
labeling index
- wt
wild type
- SCR
scrambled
- qRT-PCR
Quantitative Reverse Transcription PCR
- ChIP
Chromatin Immunoprecipitation
- CHX
Cycloheximide
- MKI
mitosis-karyorrhexis index
Footnotes
Conflicts of Interest: None
REFERENCES
- 1.Matthay KK, Reynolds CP, Seeger RC, et al. Long-term results for children with high-risk neuroblastoma treated on a randomized trial of myeloablative therapy followed by 13-cis-retinoic acid: a children's oncology group study. Journal of Clinical Oncology. 2009;27:1007–13. doi: 10.1200/JCO.2007.13.8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pearson AD, Pinkerton CR, Lewis IJ, Imeson J, Ellershaw C, Machin D. High-dose rapid and standard induction chemotherapy for patients aged over 1 year with stage 4 neuroblastoma: a randomised trial. The lancet oncology. 2008;9:247–56. doi: 10.1016/S1470-2045(08)70069-X. [DOI] [PubMed] [Google Scholar]
- 3.Cohn SL, Tweddle DA. MYCN amplification remains prognostically strong 20 years after its “clinical debut”. European Journal of Cancer. 2004;40:2639–42. doi: 10.1016/j.ejca.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 4.Grandori C, Eisenman RN. Myc target genes. Trends in Biochemical Sciences. 1997;22:177–81. doi: 10.1016/s0968-0004(97)01025-6. [DOI] [PubMed] [Google Scholar]
- 5.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome biology. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Yosef T, Yanuka O, Halle D, Benvenisty N. Involvement of Myc targets in c-myc and N-myc induced human tumors. Oncogene. 1998;17:165–71. doi: 10.1038/sj.onc.1201939. [DOI] [PubMed] [Google Scholar]
- 7.Westermann F, Muth D, Benner A, et al. Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome biology. 2008;9:R150. doi: 10.1186/gb-2008-9-10-r150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laurenti E, Varnum-Finney B, Wilson A, et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–24. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogarty MD. The requirement for evasion of programmed cell death in neuroblastomas with MYCN amplification. Cancer letters. 2003;197:173–9. doi: 10.1016/s0304-3835(03)00103-4. [DOI] [PubMed] [Google Scholar]
- 10.Tweddle DA, Pearson AD, Haber M, et al. The p53 pathway and its inactivation in neuroblastoma. Cancer letters. 2003;197:93–8. doi: 10.1016/s0304-3835(03)00088-0. [DOI] [PubMed] [Google Scholar]
- 11.Davidoff AM, Pence JC, Shorter NA, Iglehart JD, Marks JR. Expression of p53 in human neuroblastoma- and neuroepithelioma-derived cell lines. Oncogene. 1992;7:127–33. [PubMed] [Google Scholar]
- 12.Chen L, Malcolm AJ, Wood KM, et al. p53 is nuclear and functional in both undifferentiated and differentiated neuroblastoma. Cell Cycle. 2007;6:2685–96. doi: 10.4161/cc.6.21.4853. [DOI] [PubMed] [Google Scholar]
- 13.Reisman D, Elkind NB, Roy B, Beamon J, Rotter V. c-Myc trans-activates the p53 promoter through a required downstream CACGTG motif. Cell Growth & Differentiation. 1993;4:57–65. [PubMed] [Google Scholar]
- 14.Roy B, Beamon J, Balint E, Reisman D. Transactivation of the human p53 tumor suppressor gene by c-Myc/Max contributes to elevated mutant p53 expression in some tumors. Molecular & Cellular Biology. 1994;14:7805–15. doi: 10.1128/mcb.14.12.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–3. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- 16.Roy B, Beamon J, Balint E, Reisman D. Transactivation of the human p53 tumor suppressor gene by c-Myc/Max contributes to elevated mutant p53 expression in some tumors. Molecular and cellular biology. 1994;14:7805–15. doi: 10.1128/mcb.14.12.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blackwell TK, Huang J, Ma A, et al. Binding of myc proteins to canonical and noncanonical DNA sequences. Molecular & Cellular Biology. 1993;13:5216–24. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reisman D, Loging WT. Transcriptional regulation of the p53 tumor suppressor gene. Seminars in Cancer Biology. 1998;8:317–24. doi: 10.1006/scbi.1998.0094. [DOI] [PubMed] [Google Scholar]
- 19.Carr J, Bell E, Pearson AD, et al. Increased frequency of aberrations in the p53/MDM2/p14(ARF) pathway in neuroblastoma cell lines established at relapse. Cancer research. 2006;66:2138–45. doi: 10.1158/0008-5472.CAN-05-2623. [DOI] [PubMed] [Google Scholar]
- 20.Tweddle DA, Malcolm AJ, Bown N, Pearson AD, Lunec J. Evidence for the development of p53 mutations after cytotoxic therapy in a neuroblastoma cell line. Cancer research. 2001;61:8–13. [PubMed] [Google Scholar]
- 21.Tweddle DA, Malcolm AJ, Cole M, Pearson AD, Lunec J. p53 cellular localization and function in neuroblastoma: evidence for defective G(1) arrest despite WAF1 induction in MYCN-amplified cells. The American journal of pathology. 2001;158:2067–77. doi: 10.1016/S0002-9440(10)64678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell E, Premkumar R, Carr J, et al. The role of MYCN in the failure of MYCN amplified neuroblastoma cell lines to G1 arrest after DNA damage. Cell Cycle. 2006;5(22):2639–47. doi: 10.4161/cc.5.22.3443. [DOI] [PubMed] [Google Scholar]
- 23.Liang H, Lunec J. Characterisation of a novel p53 down-regulated promoter in intron 3 of the human MDM2 oncogene. Gene. 2005;361:112–8. doi: 10.1016/j.gene.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 24.Wang S, El-Deiry WS. p73 or p53 directly regulates human p53 transcription to maintain cell cycle checkpoints. Cancer research. 2006;66:6982–9. doi: 10.1158/0008-5472.CAN-06-0511. [DOI] [PubMed] [Google Scholar]
- 25.Lu X, Vogt PK, Boger DL, Lunec J. Disruption of the MYC transcriptional function by a small-molecule antagonist of MYC/MAX dimerization. Oncology reports. 2008;19:825–30. [PubMed] [Google Scholar]
- 26.Armstrong JL, Veal GJ, Redfern CP, Lovat PE. Role of Noxa in p53-independent fenretinide-induced apoptosis of neuroectodermal tumours. Apoptosis. 2007;12:613–22. doi: 10.1007/s10495-006-0020-1. [DOI] [PubMed] [Google Scholar]
- 27.Weinmann AS, Farnham PJ. Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods. 2002;26:37–47. doi: 10.1016/S1046-2023(02)00006-3. [DOI] [PubMed] [Google Scholar]
- 28.Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene. 1996;13:803–12. [PubMed] [Google Scholar]
- 29.Fulda S, Lutz W, Schwab M, Debatin KM. MycN sensitizes neuroblastoma cells for drug-induced apoptosis. Oncogene. 1999;18:1479–86. doi: 10.1038/sj.onc.1202435. [DOI] [PubMed] [Google Scholar]
- 30.Aubry S, Charron J. N-Myc shares cellular functions wiht c-Myc. DNA and cell biology. 2000;19:353–64. doi: 10.1089/10445490050043326. [DOI] [PubMed] [Google Scholar]
- 31.van Noesel MM, Pieters R, Voute PA, Versteeg R. The N-myc paradox: N-myc overexpression in neuroblastomas is associated with sensitivity as well as resistance to apoptosis. Cancer letters. 2003;197:165–72. doi: 10.1016/s0304-3835(03)00101-0. [DOI] [PubMed] [Google Scholar]
- 32.Goto S, Umehara S, Gerbing RB, et al. Histopathology (International Neuroblastoma Pathology Classification) and MYCN status in patients with peripheral neuroblastic tumors: a report from the Children's Cancer Group. Cancer. 2001;92:2699–708. doi: 10.1002/1097-0142(20011115)92:10<2699::aid-cncr1624>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 33.Shimada H, Stram DO, Chatten J, et al. Identification of subsets of neuroblastomas by combined histopathologic and N-myc analysis. Journal of the National Cancer Institute. 1995;87:1470–6. doi: 10.1093/jnci/87.19.1470. [DOI] [PubMed] [Google Scholar]
- 34.Shimada H, Ambros IM, Dehner LP, Hata J, Joshi VV, Roald B. Terminology and morphologic criteria of neuroblastic tumors: recommendations by the International Neuroblastoma Pathology Committee. Cancer. 1999;86:349–63. [PubMed] [Google Scholar]
- 35.Altungoz O, Aygun N, Tumer S, Ozer E, Olgun N, Sakizli M. Correlation of modified Shimada classification with MYCN and 1p36 status detected by fluorescence in situ hybridization in neuroblastoma. Cancer genetics and cytogenetics. 2007;172:113–9. doi: 10.1016/j.cancergencyto.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Moore HC, Wood KM, Jackson MS, et al. Histological profile of tumours from MYCN transgenic mice. Journal of clinical pathology. 2008;61:1098–103. doi: 10.1136/jcp.2007.054627. [DOI] [PubMed] [Google Scholar]
- 37.Sugihara E, Kanai M, Saito S, et al. Suppression of centrosome amplification after DNA damage depends on p27 accumulation. Cancer research. 2006;66:4020–9. doi: 10.1158/0008-5472.CAN-05-3250. [DOI] [PubMed] [Google Scholar]
- 38.Cui H, Li T, Ding HF. Linking of N-Myc to death receptor machinery in neuroblastoma cells. Journal of Biological Chemistry. 2005;280:9474–81. doi: 10.1074/jbc.M410450200. [DOI] [PubMed] [Google Scholar]
- 39.Fulda S, Lutz W, Schwab M, Debatin KM. MycN sensitizes neuroblastoma cells for drug-triggered apoptosis. Medical & Pediatric Oncology. 2000;35:582–4. doi: 10.1002/1096-911x(20001201)35:6<582::aid-mpo19>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 40.Paffhausen T, Schwab M, Westermann F. Targeted MYCN expression affects cytotoxic potential of chemotherapeutic drugs in neuroblastoma cells. Cancer letters. 2007;250:17–24. doi: 10.1016/j.canlet.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 41.Chesler L, Goldenberg DD, Collins R, et al. Chemotherapy-induced apoptosis in a transgenic model of neuroblastoma proceeds through p53 induction. Neoplasia. 2008;10:1268–74. doi: 10.1593/neo.08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berwanger B, Hartmann O, Bergmann E, et al. Loss of a FYN-regulated differentiation and growth arrest pathway in advanced stage neuroblastoma. Cancer cell. 2002;2:377–86. doi: 10.1016/s1535-6108(02)00179-4. [DOI] [PubMed] [Google Scholar]
- 43.Raschella G, Negroni A, Giubilei C, et al. Transcription of N-myc and proliferation-related genes is linked in human neuroblastoma. Cancer letters. 1991;56:45–51. doi: 10.1016/0304-3835(91)90192-k. [DOI] [PubMed] [Google Scholar]
- 44.Ohira M, Oba S, Nakamura Y, et al. Expression profiling using a tumor-specific cDNA microarray predicts the prognosis of intermediate risk neuroblastomas. Cancer cell. 2005;7:337–50. doi: 10.1016/j.ccr.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 45.Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nature Reviews Molecular Cell Biology. 2005;6:635–45. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- 46.Zindy F, Eischen CM, Randle DH, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes & development. 1998;12:2424–33. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Slack A, Chen Z, Tonelli R, et al. The p53 regulatory gene MDM2 is a direct transcriptional target of MYCN in neuroblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:731–6. doi: 10.1073/pnas.0405495102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.