Abstract
Though spermatozoon and egg contribute an equal share of nuclear DNA content to the newly formed embryo, there are inherent epigenetic differences between the paternal and maternal pronuclei in early cleavage stage embryos. Information about how to decipher sperm DNA in the embryo is established via sperm-specific DNA packaging that occurs during spermatogenesis. In addition to protamines, paternal factors that package sperm DNA distinctly from oocyte or somatic DNA include histones and their modifications, histone variants, chromatin-binding proteins, and non-coding RNAs. These evolutionarily conserved factors play interconnected roles in heterochromatin formation, gene regulation, and maintenance of genome integrity, which influence key processes after fertilization. This review focuses on recent developments from genomic and proteomic studies in model organisms showing that components closely associated with sperm DNA contribute to embryonic survival. These advances may reveal important insights into the treatment of infertility and use of assisted reproductive technologies.
Keywords: chromatin, embryogenesis, epigenetics, fertility, paternal, spermatogenesis
Introduction
Reproductive success does not begin and end with fertilization. Even when the union of sperm and egg is achieved in vitro using sophisticated assisted reproductive technologies, successful pregnancy rates are still less than 42% (CDC ART Report, 2004). Thus, improving reproductive outcomes is a daunting task. One way to surmount these challenges is to understand the role of evolutionarily conserved fertility factors. Because many proteins important for reproduction evolve quickly (Swanson and Vacquier, 2002), those that are conserved are likely to play fundamental roles in fertility (Hackstein et al., 2000; Xu EY et al., 2003; Chu et al., 2006). In addition, studying these components in model organisms is facilitated by the availability of experimental tools, like genomics, proteomics, and genetic mutations. Thus approaches using mice, zebrafish, Drosophila, and Caenorhabditis elegans that define elements important for germ cell formation and embryonic survival can accelerate the development of new avenues to improve reproductive outcomes.
Do spermatozoon and egg contribute equally to reproductive success? Proteins and RNAs in the oocyte are well-documented as essential for embryonic development (Stitzel and Se, 2007). However, few factors donated by the much smaller sperm cell are known. Sperm proteins with embryonic function include signalling proteins that trigger key events immediately after fertilization such as phospholipase C (PLCζ), which promotes cell cycle progression, and a Rho guanosine triphosphatase-activating protein (the CYK-4 protein in C. elegans), which establishes embryonic polarity (Krawetz, 2005; Jenkins et al., 2006; Swann et al., 2006). Although the zygotic genome is composed of equivalent shares of parental DNA, each chromosomal complement also contains non-equivalent epigenetic features that convey essential information about how maternal and paternal DNA is processed. DNA methylation is one well-documented mechanism of epigenetic regulation that alters dramatically in accordance with the developmental potential of the cell, which is reviewed elsewhere (Rousseaux et al., 2005; Trasler, 2006; Schaefer et al., 2007).
Proteins and RNAs directly associated with DNA also provide a considerable amount of epigenetic information to the new embryo. Spermatozoa carry a host of specific factors that package paternal DNA in a unique fashion. Subsequently, paternal DNA may contribute significantly more epigenetic information to the oocyte than was previously appreciated. This information is mediated by factors that include paternal proteins and nucleic acids such as histones and their modifications, histone variants, chromatin-binding proteins, and non-coding RNAs. This review will focus on recent exciting developments that demonstrate that components closely associated with sperm DNA before and after fertilization impact embryonic survival. These advances may reveal important insights into the applications and consequences of employing assisted reproduction treatment.
Packaging of the paternal genome is important for fertility
Because chromatin comprises a large proportion of each spermatozoon, proteins associated with DNA constitute a sizeable portion of the protein content donated to the embryo. Many of these proteins wrap the paternal DNA during sperm formation, resulting in a nuclear volume up to 40-fold less than that of somatic cells (Kimmins and Sassone-Corsi, 2005). The compaction of the sperm nucleus requires replacement of somatic histones with sperm nuclear basic proteins (Figure 1A). In mammals, this exchange occurs in a stepwise process. Histone proteins can be post-translationally modified or replaced by histone variants, which may be sperm- or testis-specific or enriched (Churikov et al., 2004; Caron et al., 2005). Next, modified histones or histone variants are largely replaced by transition proteins, which are then replaced by very small, highly basic proteins called protamine proteins. The shift from a histone-based chromatin architecture to one fundamentally based on protamines allows for the high degree of DNA compaction unique to sperm (Braun, 2001). Many organisms replace some or all somatic histones with a combination of histone variants and protamine-like proteins (Lewis et al., 2003). For example, human spermatozoa retain up to 15% of histone proteins (Gatewood et al., 1987). Histone variants such as histone H3.3 in C. elegans, CENP-A in bull, and H2AX and H2AZ in humans have also been detected in mature spermatozoa of various species (Gatewood et al., 1990; Palmer et al., 1990; Ooi et al., 2006).
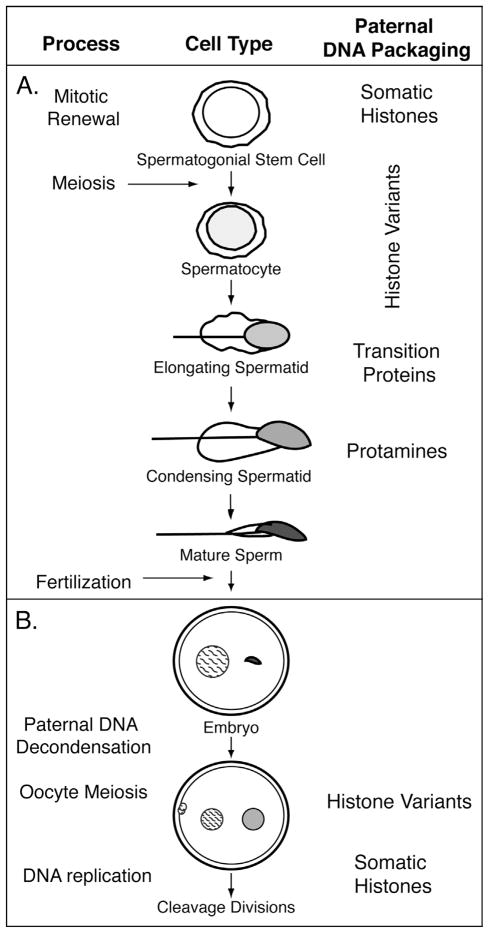
Figure 1.
Schematic diagram of the progression of mammalian sperm cell nuclei through spermatogenesis and early post-fertilization stages. (A) In the testis, proliferating spermatogonial stem cells give rise to mature spermatozoa. During spermatogenesis, the chromatin of developing spermatozoa becomes successively more compact, with accompanying shifts in global chromosomal packaging by sperm nuclear basic proteins. This process is initiated during meiosis, with the bulk of the transition occurring in post-meiotic stages. Concurrently, epigenetic processes demarcate portions of the genome that undergo differential processing during post-meiotic stages (spermiogenesis) or post-fertilization. Developmental processes (stages) are indicated in the left column. Stage-specific incorporation of generalized relevant proteins that change nucleosomal composition are shown on the right. (B) Paternal chromatin (smooth shading) carries epigenetic marks established by chromatin factors during spermatogenesis that make it distinct from maternal chromatin (hatched shading) after fertilization and before the first cell division. Fertilization induces the oocyte to complete meiosis, resulting in one haploid maternal pronucleus and two extruded polar bodies. During this time, paternal chromatin delivered into the oocyte goes through extensive changes including the repackaging of sperm chromosomes with somatic histones, global decondensation, and imprinted transcriptional regulation, but maintains distinct epigenetic features.
Though many proteins have been identified that act as structural components to spool sperm DNA, little is known about additional roles for chromatin proteins. To address this lack of knowledge, a proteomic approach was used to identify proteins associated with C. elegans sperm DNA (Chu et al., 2006). Like mammalian DNA, C. elegans sperm DNA is highly compacted, though how this is achieved in C. elegans is unknown. In this large-scale analysis, specialized mass spectrometry known as Multidimensional Protein Identification Technology (McDonald and Yates, 2000) (MudPIT) identified 1099 proteins associated with chromatin purified from cells undergoing spermatogenesis. Sperm-specific and sperm-enriched proteins were found by subtracting factors in common with chromatin purified from oocytes isolated from fertilization-defective C. elegans mutant hermaphrodites (Chu et al., 2006). Mass spectrometric analysis also measured abundance, allowing prioritization of the most abundant 132 sperm-enriched proteins for further study.
Functional analysis of these abundant spermatogenic chromatin-associated proteins suggests they have roles in the germline and embryo (Chu et al., 2006). Taking advantage of the molecular and cytological tools available in C. elegans, phenotypes resulting from RNA interference (RNAi) against each of the 132 corresponding genes were assessed. Thirty-eight percent of these proteins were found to be important for germ cell formation or embryonic viability. The actual percentage is likely to be higher, as genes required for spermatogenesis are often RNAi resistant in C. elegans. Members of this large group of sperm chromatin-associated proteins are diverse, including putative DNA-binding proteins, RNA-binding proteins and signalling proteins. Significantly, 59% of the 132 proteins have human homologues and therefore potentially conserved functions in fertility.
Some of the identified proteins may function after fertilization. One such paternal protein required for C. elegans embryonic development, SPE-11, is associated with sperm chromatin (Hill et al., 1989; Browning and Strome, 1996). SPE-11 is not required to make functional spermatozoa; however, oocytes that are fertilized by spe-11 mutant spermatozoa fail to develop due to defects in oocyte meiotic progression (McNally and McNally, 2005). Although SPE-11 homologues are not found in mammals, higher organisms may use association with paternal chromatin to transmit factors to the embryo. Other studies discussed below provide evidence that evolutionarily conserved paternal proteins may indeed play roles after fertilization.
Distinguishing the paternal genome
One of the earliest events occurring after fertilization involves the transition of the paternally contributed chromosomes from a sperm-specific state to a somatic state (Figure 1B) (McLay and Clarke, 2003). Chromosome decondensation and the removal of protamines and other sperm nuclear basic proteins must occur prior to pronuclear fusion. Protamine removal is expected to be an active process, since protamines are anchored onto DNA by arginine rich domains (Brewer et al., 1999). Somatic histones must also be incorporated to maintain DNA integrity and chromatin structure. Sperm nuclear basic protein replacement begins prior to the first replication cycle, as recent studies have shown that paternal pronuclei are specifically targeted by oocyte factors for the replication-independent incorporation of the histone H3 variant, H3.3.
Global histone H3.3 incorporation into paternal DNA during decondensation before replication has been observed in worms, flies and mice (Loppin et al., 2005; van der Heijden et al., 2005; Ooi et al., 2006). Decondensation of paternal chromosomes is essential. Failure to decondense the paternal pronucleus in Drosophila due to a maternal-effect mutation, sesame, results in embryonic lethality (Loppin et al., 2000). Sesame encodes the fly homologue of the histone chaperone HIRA. Epitope-tagged histone H3.3 incorporates into paternal DNA during decondensation, a process that is dependant on fly HIRA (Loppin et al., 2005). The role of HIRA is probably conserved in mammals, as mouse HIRA associates exclusively with the male pronucleus prior to pronuclear fusion, implying H3.3 incorporation as well (van der Heijden et al., 2005). Furthermore, antibody staining against the major H3 histone, H3.1, demonstrated that unlike the maternal pronucleus, the paternal pronucleus is devoid of histone H3.1. Thus, in mice and flies paternal chromosomes are specifically targeted for the decondensation and repackaging steps that are necessary for subsequent development. It is still unclear which paternal proteins are initially recognized by oocyte factors for decondensation, though protamines are obvious suspects. Also unclear is whether HIRA directly recognizes paternal chromatin (Loppin et al., 2000). Another oocyte protein, nucleoplasmin, has also been implicated in targeting paternal DNA for decondensation in Xenopus (Philpott and Leno, 1992; Burns, 2003).
The significance of H3.3 incorporation in the newly formed embryo is so far unknown, but its presence does contribute to continued asymmetry of the paternal and maternal pronuclei (Ooi and Henikoff, 2007). Other epigenetic features are also observed to localize asymmetrically during this time period. Asymmetry in histone protein modifications exists immediately after fertilization, with some discrepancies between reports, generally attributed to differences in immunostaining procedures (Arney et al., 2002; Santos et al., 2005; van der Heijden et al., 2005; Yeo et al., 2005; Yoshida et al., 2007). In general, maternal chromatin appears to bear histone H3 mono-, di- and tri-methylation on lysines 4, 9, and 27 and lacks acetylation of histone 4 on lysine 12 before the four-cell stage. In contrast, paternal chromatin is devoid of histone tri-methylation and is acetylated on histone 4 on lysine 12 (Santos et al., 2005; van der Heijden et al., 2005; Yoshida et al., 2007).
These early epigenetic marks are candidates to direct differences in transcriptional regulation of parental genomes in cleavage stage embryos. In mammals, androgenetic and gynogenetic embryos formed by pronuclear transplantation fail to develop, demonstrating that isogenic parental genomes are not interchangeable (Barton et al., 1984; McGrath and Solter, 1984; Surani et al., 1984). The global DNA demethylation of each parental genome, a process that occurs after fertilization, occurs at different stages (Haaf, 2006). Paternal DNA is demethylated before the first cell division. In contrast maternal DNA becomes demethylated between the 2- and 4-cell stage (Mayer et al., 2000). The male pronucleus is more transcriptionally active than the maternal pronucleus before the first cell division as assayed by incorporation of 5-bromouridine-5-triphosphate (Bouniol et al., 1995; Aoki et al., 1997; Liu et al., 2005). The early epigenetic marks established in the 1-cell embryo may act to protect the maternal genome from transcription or demethylation within the first cell divisions. Alternatively, they may also function to recruit factors to paternal chromatin to activate these processes.
Establishing paternal chromatin domains
Recent studies have implicated parentally differentiated epigenetic marks in additional embryonic functions. One lingering question is whether the shift from somatic histones to protamines during spermatogenesis necessitates re-establishment of subchromosomal regions after fertilization. Such domains include regions of heterochromatin that constitute centromeres and telomeres. Failure to do so could result in defects in gene expression or chromosome segregation during cell division in embryogenesis.
Recent work by Govin et al. has provided evidence that heterochromatin domains may not need to be re-established de novo (Govin et al., 2007). In their analysis, they found that chromatin at the pericentromere, a domain adjacent to centromeric DNA, changes composition during spermatogenesis in mice. In early germ cells, the pericentromere is distinguished by histone hypoacetylation, histone H3 tri-methylation at lysine 9, and binding of the heterochromatin protein, HP1. In post-meiotic stages of spermatogenesis (Figure 1A), pericentric heterochromatin undergoes a transition where histone H4 acetylation at lysines 8 or 12 and histone H3 tri-methylated at lysine 9 are retained in pericentric chromatin within elongating spermatids, then removed in condensing spermatids. This reorganization within the sperm nucleus coincides with the appearance of testis-specific histone variants. Proteomic analysis on acid extracted condensing nuclei in later stages of mouse spermatogenesis identified five new histone variants that are expressed exclusively during late spermiogenesis. Furthermore, immunolocalization experiments demonstrated that two of these histone variants, H2AL1 and H2AL2, demarcate pericentric heterochromatin regions in condensing spermatids where other marks of heterochromatin are now absent.
Immediately after fertilization in the one-cell embryo, paternal heterochromatic regions have different epigenetic marks than maternal heterochromatin. Heterochromatin on the maternal pronucleus is marked with HP1β, histone H3 mono- and tri-methylation at lysine 9, and histone H4 tri-methylation at lysine 20 (Santos et al., 2005; Probst et al., 2007). In contrast, paternal heterochromatin in the 1-cell embryo only shows histone H3 mono-methylation at lysine 9 and HP1β. Because tri-methylation of histone H3 at lysine 9 is thought to be required to establish a platform for HP1β (Grewal and Jia, 2007), it is not known how HP1β is recruited to paternal DNA in the one cell embryo. Speculation exists that the newly identified histone variants associated with heterochromatin domains in paternal DNA act as guides to identify paternal centromeres and telomeres of paternal chromosomes immediately after fertilization (Govin et al., 2007). Thus de-novo establishment of heterochromatin may be avoided by using histones, modified histones, or other chromatin proteins to mark these domains on sperm chromosomes before fertilization.
Inactivating paternal sex chromosomes in mammals
Differential regulation of paternal chromatin also occurs on the chromosome-wide level, both before and after fertilization. During spermatogenesis, the X and Y chromosomes undergo meiotic sex chromosome inactivation (MSCI) to form the sex body, which has unique chromatin properties that silence both sex chromosomes (Turner, 2007). During subsequent embryonic development in females, one of the two X chromosomes is silenced to achieve dosage compensation between XX females and XY males, a process called X chromosome inactivation (XCI). Initially, the paternally inherited X chromosome is preferentially inactivated in all tissues. During the blastocyst stage, cells in the embryo proper switch to the random mode of XCI, inactivating either the maternal or paternally derived X chromosome, which results in a mosaic female. In extra-embryonic tissues, including those that give rise to the placenta, preferential inactivation of the paternal X chromosome remains in place. The mechanism of MSCI is distinct from XCI. While XCI is dependent on the Xist non-coding RNA, MSCI is not (Marahrens et al., 1997; McCarrey et al., 2002; Turner et al., 2002). In addition, though some proteins that associate with the sex body are required for XCI, many are not, including proteins with dual functions in MSCI and processes like DNA repair (Turner, 2007).
Are there features of MSCI that lead to the exclusive choice of the paternal X for silencing during early embryonic XCI? On the one hand, this ‘preinactivation’ of the paternal X would be an efficient means to achieve dosage compensation in very early female embryos (Huynh and Lee, 2003). In support of this idea, recent work showed that the X and Y chromosomes are consistently silenced in post-meiotic stages, unlike autosomes (Namekawa et al., 2006; Turner et al., 2006). Correspondingly, the sex body is enriched for heterochromatin marks like histone H3 di-methylation at lysine 9 and HP1 binding. Microarray analysis of 676 X-linked genes demonstrated that the post-meiotic X is silenced to a comparable degree to that of the inactive human X (Carrel and Willard, 2005; Namekawa et al., 2006). On the other hand, evidence exists suggesting that the paternal X is not transcriptionally silent in two cell embryos, for expression of RNA pol II and X-specific transcripts have been detected at the paternal X (Okamoto et al., 2005). In addition, autosomal transgenes containing Xist that do not undergo MSCI in the male germline are still specifically targeted for XCI (Okamoto et al., 2005), suggesting the dispensability of MSCI for XCI. Due to the highly condensed nature of mature sperm chromatin, the fate of histone modifications on the X chromosome after spermatid elongation is difficult to assess. It is possible that epigenetic features of MSCI could remain on the paternal X, helping to implement some degree of dosage compensation in the earliest stages of embryogenesis, providing additional marks for paternal X chromosome for XCI, or modulating X chromosome-wide repression in early cleavage stage embryos.
Eliminating paternal meiotic errors
Investigations into the role of germline sex chromosome silencing in various organisms have identified specific aspects of meiosis critical for producing functional spermatozoa. During meiotic prophase I, replicated homologous chromosomes must pair, synapse, undergo recombination, and repair the double-stranded breaks routinely generated during recombination. Embryonic lethality can result if the male pronucleus contains gross defects, like compromised genomic integrity, aneuploidy of chromosomes, or mis-established epigenetic information. In the germline, culling of defective germ cells through mechanisms such as apoptosis helps guard against the transmission of such defects. For example, failure of sex chromosomes to undergo MSCI results in spermatogonial apoptosis and also male infertility (Turner, 2007), potentially revealing the kinds of problems that can arise during spermatogenesis.
Why does failure to implement MSCI result in germline apoptosis? One hypothesis is that lingering DNA damage triggers the apoptotic pathway since many proteins required for MSCI also function in DNA repair. For example, disruption of the mouse breast cancer related protein BRCA1 shows pleiotropic defects including embryonic lethality. A hypomorphic allele of Brca1 in a p53/+ genetic background, however, revealed defects in DNA recombination repair and failed formation and silencing of the sex body that led to apoptosis and male infertility (Xu X et al., 2003b; Turner et al., 2004). Milder phenotypes of knockout mice in two DNA repair proteins, PARP-1 and H2AX, were observed. PARP-1, which can add poly-(ADP-ribose) polymers to histone proteins, functions in base excision repair (Dantzer et al., 2006a). Phosphorylated H2AX, called γH2AX, localizes to sites of DNA damage (Fernandez-Capetillo et al., 2004). Disruption of either of these genes in mice also caused male infertility associated with failure of MSCI, apoptosis with some defects in localization of DNA damage proteins and slower resolution of double-stranded breaks (Celeste et al., 2002; Fernandez-Capetillo et al., 2003; Dantzer et al., 2006b). Thus, due to the dual functions of such proteins in DNA repair and MSCI, failure to implement MSCI may elicit removal of nuclei through mechanisms that sense DNA damage and therefore disrupt genome integrity.
A second link between failure to implement MSCI and germline apoptosis is revealed by recent work showing that MSCI is one consequence of a more general process called meiotic silencing of unpaired chromatin (MSUC). The X chromosome is much larger than the Y chromosome, its pairing partner during spermatogenesis. Whereas asynapsed regions between the X and Y chromosomes in the male germline specifically trigger the process of MSCI, unsynapsed chromosomes in general are targeted for repression in both sexes via MSUC (Baarends et al., 2005; Turner et al., 2006). The two processes are mechanistically linked, for male mice carrying a chromosomal translocation between chromosomes X and 16, T(X;16)16H, exhibit silencing of unpaired chromatin, which accumulates the same proteins that associate with the XY body (Baarends et al., 2005; Turner et al., 2005). Cells containing chromosomes that fail to synapse are specifically targeted for elimination during pachytene (Turner et al., 2005). Recent work in C. elegans has shown that homologous chromosome synapsis is required to avoid culling by apoptosis (Bhalla and Dernburg, 2005). This work was significant because the authors demonstrated that the requirement for synapsis is DNA repair-independent by abolishing double-stranded break generation using a mutation of spo-11, the meiotic endonuclease. Because unsynapsed chromatin and/or failure to execute MSCI induces apoptosis, one model is that MSCI has evolved to prevent the activation of the apoptotic pathway in normal male germlines, where unsynapsed X and Y chromosomes are found in every germ cell nucleus.
It is also possible that failure to silence unsynapsed chromosomes results in inappropriate expression levels of genes located within those regions (Turner, 2007). Failure to inactivate the sex chromosomes through MSCI, thus permitting their expression, may result in toxicity to developing spermatogonia. This may be because some X- or Y-linked genes required in one dose have duplicate copies on autosomes that are normally active during spermatogenesis to compensate for the silenced X or Y copy (Handel, 2004; Maciejowski, 2005). Conversely, nuclei containing translocations of autosomes that do not undergo synapsis may exhibit inappropriate repression of essential genes that map to the translocation, causing cell death. It is not known whether these putative transcriptional effects directly cause cell death, or whether they could also activate a checkpoint response and eliminate defective cells via apoptosis.
These studies clearly illustrate that several kinds of errors that would produce potentially defective spermatozoa are eliminated through apoptosis during meiosis, thus providing one barrier to transmitting meiotic errors to offspring.
Protecting integrity of paternal genome organization
Significant roles in maintaining genomic integrity during spermatogenesis have also been recently identified for non-coding RNAs and their associated proteins, which also function in heterochromatin formation and transcriptional repression (Bernstein and Allis, 2005). Sperm RNAs themselves have been identified in the perinuclear theca, an electron-dense layer tightly surrounding the condensed nucleus of mammalian spermatozoa (Wykes et al., 1997). The importance of RNA associated with spermatogenic chromatin is supported by the finding that a significant proportion (30/132) of spermatogenic chromatin-enriched C. elegans proteins are potentially associated with RNA (Chu et al., 2006).
Among these proteins are RNA binding proteins such as the Argonaute family members PIWI/PAZ proteins, germline-specific components of RNAi machinery that possess RNase H-like nuclease activity. Ultra-high throughput cloning and sequencing technology has allowed the identification of a distinct class of small RNAs called piRNAs, which are 26–30 base pairs in length and associate with Piwi proteins (Girard et al., 2006; Brennecke et al., 2007; Lin, 2007; O’Donnell and Boeke, 2007). Tens to hundreds of thousands of piRNA sequences are known and map to a limited number of loci. Interestingly, these include loci that give rise to Drosophila repeat-associated RNAs (rasiRNAs) and regions that encode transposons or control transposon activity (Aravin et al., 2004; Saito et al., 2006; Brennecke et al., 2007). Piwi proteins are thought to implement silencing of transposable elements through targeting by piRNAs (O’Donnell et al., 2007), a key process since inappropriate activation of transposons in germ cells could result in alterations in gene structure or expression.
PIWI protein homologues in several organisms function in silencing transposons in the germline. Drosophila piwi mutant male germlines inappropriately activate the copia and mdg1 retrotransposon elements and the endogenous retrovirus gypsy (Sarot et al., 2004; Kalmykova et al., 2005). Retrotransposons are also inappropriately activated in mutants of the mouse PIWI homologues MIWI, MILI, and MIWI2 (Grivna et al., 2006; Aravin et al., 2007; Carmell et al., 2007) and zebrafish ZIWI mutants (Houwing et al., 2007). The activation of transposable elements in PIWI homologue mutants coincides with defects in germ cell progression through different stages of gamete formation. Drosophila Piwi is required for spermatogenesis and stem cell maintenance in the germline (Cox et al., 2000). Consistent with the observation that piRNAs in mammals have thus far only been identified in testis (Girard et al., 2006; Watanabe et al., 2006), MIWI, MILI, and MIWI2 mutants are male infertile, exhibiting loss of germ cells in different stages of meiosis (Lin and Spradling, 1997; Deng and Lin, 2002; Kuramochi-Miyagawa et al., 2004). The zebrafish homologue ZIWI is expressed in male and female germ cells, but not required for germline stem cell specification. Mutation of ZIWI results in loss of germ cells and germline apoptosis (Houwing et al., 2007). Despite some common defects, differences in germline phenotypes have been observed, indicating that the homologues may have distinct or overlapping essential functions in meiotic progression.
In any organism studied thus far, it has not yet been shown that elimination of germ cells in mutant testes is a direct result of transposon activity. However, these recent findings have clearly implicated PIWI proteins in the silencing of male germline transposon activity, a process that is important for maintaining genomic integrity (O’Donnell and Boeke, 2007). Protecting the genome integrity in developing gametes is critical because unlike mutation of somatic cells, genes disrupted by rogue sequences in the germline will be passed on to the next generation.
Conclusion
As discussed above, proteins and RNAs associated with sperm DNA mediate fundamental processes that affect development of the embryo. These include early events such as targeting the male pronucleus for decondensation or establishing heterochromatin domains required for early cell divisions. Additionally, implementing differential gene expression of paternal DNA in early embryos and of sex chromosomes is essential. Germline mechanisms have also evolved to monitor and protect spermatocytes against meiotic errors, mediated by chromatin-associated factors. This review emphasizes that wide-ranging processes contribute to successful reproduction, and that embryonic effects specified by either maternal or paternal chromatin factors are not trivial.
The advent of new technologies has brought about an explosion in the amount and access to new information about reproduction. With the sequencing of the human genome complete, researchers are now actively cataloguing the entire protein and RNA content of reproductive components like human spermatozoa (Johnston et al., 2005; Ostermeier et al., 2005; Pilch and Mann, 2006). An imposing question looms: what next? As demonstrated by the studies discussed above, model organisms like worms, flies, fish, and mice are tremendously useful in characterizing the roles of evolutionarily conserved proteins in fertility and reproduction. Thus, the combination of large-scale genomic or proteomic analyses with the use of basic model organisms to dissect molecular function represents a powerful approach to the study of fundamental processes. Information garnered from model organisms should prove highly useful for human fertility studies, since most of the factors discussed have human homologues.
The knowledge gained from genomic and proteomic approaches, discussed here and elsewhere, has indicated that overlapping mechanisms have evolved, probably to prevent reproductive failure (Bazer and Spencer, 2005). On the genomic level, functional redundancy or gene duplication of reproductive genes may help ensure fertility. For example, several protein families with multiple members were identified that associate to C. elegans spermatogenic chromatin (Chu et al., 2006). Other organisms also have large gene families that function in the germline like Argonaut (Parker and Barford, 2006), nanos (Suzuki et al., 2007), and DAZ (Reynolds and Cooke, 2005), as well as histone variants (Govin et al., 2004) and tyrosine kinases (Kierszenbaum, 2006). Though some family members have distinct function, others often have overlapping or redundant function. On the RNA level, RNA transcripts can be processed extensively for testis-specific function. In mammals, alternative splicing of genes is routinely employed to generate multiple protein isoforms. Genome-wide analysis of expressed sequence tags (EST) in humans indicates the testis is enriched for tissue-specific transcripts (Xu et al., 2002; Huang et al., 2005). Germ cells in particular produce mRNA isoforms of varying length throughout development, suggesting RNA processing during gamete formation is highly regulated (Eddy, 2002). In addition, RNAs can be cleaved into non-coding small RNAs, with either strand used for subsequent targeting of germline gene silencing (Munroe, 2006). The examples cited above indicate that functional redundancy may be a conserved mechanism to maintain fertility, regardless of organismal complexity.
In fact, few causes of human infertility are known to arise from mutation of a single gene. Though functional redundancy afforded by gene families may be a contributing factor, there are likely other mechanisms working in combination to ensure reproductive success. Therefore, the most common effect of single gene loss may be predisposition towards infertility. In this scenario, elimination of multiple genes, which individually have no obvious affect, could result in additive or synergistic subfertility. This has been observed in mice where infertility resulted only from the combination of several mutations in genes from multiple families. Triple mutants in various combinations of transition protein 2 (Tnp2), proacrosin (Acr), histone H1.1 (H1.1), histone H1t (H1t), and sperm mitochondria-associated cysteine-rich protein (Smcp) showed infertility, even though single mutants and other double and triple mutant combinations were fertile (Nayernia et al., 2003, 2005). Because complex mutational studies are more feasible in model organisms, future work to identify potential synergistic fertility gene networks can be greatly facilitated by their use.
When mechanisms like functional redundancy fail to protect the chromatin content of germ cells, defective gametes can be eliminated via germline apoptosis. Germline elimination of spermatocytes can result in complete sterility in extreme cases, as shown by studies on PIWI proteins and MSCI (Turner et al., 2006; Carmell et al., 2007; Houwing et al., 2007). Thus, excessive programmed cell death signifies that a fundamental process is being compromised. However, in some cases, a subset of germ cells can escape this culling and proceed to fertilize oocytes despite the impairment of critical processes such as MSCI or DNA repair, as observed in Parp-2 deficient mice (Dantzer et al., 2006b). Therefore, germ cells produced by subfertile males may carry chromosomal defects such as aneuploidy or loss of genome integrity, whose consequences may not be apparent until after sperm–egg fusion. This is highly relevant with regards to human fertility, because spermatozoa from subfertile males with oligospermia or spermatozoa extracted from azoospermic males are used in assisted reproduction procedures such as intracytoplasmic sperm injection (Buffat et al., 2006; Georgiou et al., 2006; Nikolettos et al., 2006; Verpoest and Tournaye, 2006).
The findings discussed in this review underscore the importance of a comprehensive approach to studying the root causes of human infertility. It is possible that developmental defects in embryos are in actuality due to faulty spermatogenic chromatin, and may contribute to infertility and the moderate success rate of assisted reproduction. In fact, a consistent correlation between sperm chromatin structure and pregnancy rate from assisted reproduction is building (Barri et al., 2005; Boe-Hansen et al., 2006; Cebesoy et al., 2006; Bungum et al., 2007). In addition, environmental factors can also perturb male germ cell content and fertility (Aitken and De Iuliis, 2007). Studies assessing the protein and RNA content of spermatozoa before using assisted reproduction, like intracytoplasmic sperm injection, has identified molecular markers that are related to reproductive outcome (Steger, 2003; Nasr-Esfahani et al., 2004). The development of further assays to assess protein and RNA components of sperm chromatin, in conjunction with existing assays for sperm quantity and quality, may prove useful in the quest to improve the outcomes for couples using assisted reproduction.
Acknowledgments
Thanks to Bill Kelly, Chun Tsai, Susan Mirsoian and Needhi Bhalla for helpful discussions. This work was supported by the NIH MBRS SCORE S06 GM52588 grant to DC.
Biographies
 Tammy Wu is a post-doctoral fellow in the laboratory of Diana Chu at San Francisco State University. She obtained her PhD in molecular and cell biology at the University of California, Berkeley and her BA from Johns Hopkins University. Her research interests include using genetic and molecular approaches to study basic developmental processes. Currently, she is examining chromatin-mediated regulation of gene expression during spermatogenesis, using the nematode worm Caenorhabditis elegans as a model system.
Tammy Wu is a post-doctoral fellow in the laboratory of Diana Chu at San Francisco State University. She obtained her PhD in molecular and cell biology at the University of California, Berkeley and her BA from Johns Hopkins University. Her research interests include using genetic and molecular approaches to study basic developmental processes. Currently, she is examining chromatin-mediated regulation of gene expression during spermatogenesis, using the nematode worm Caenorhabditis elegans as a model system.
 Diana Chu is an Assistant Professor at San Francisco State University. She obtained a BA in Biochemistry at the University of California at Berkeley and a PhD in Molecular Biology at the University of California, Los Angeles. Dr Chu conducted post-doctoral work at the University of California at Berkeley studying the molecular mechanisms of dosage compensation and sex determination using the model organism C. elegans. Her current work focuses on understanding how chromatin composition affects gene expression and chromosome dynamics during spermatogenesis. She is interested in characterizing evolutionarily conserved factors important for male fertility.
Diana Chu is an Assistant Professor at San Francisco State University. She obtained a BA in Biochemistry at the University of California at Berkeley and a PhD in Molecular Biology at the University of California, Los Angeles. Dr Chu conducted post-doctoral work at the University of California at Berkeley studying the molecular mechanisms of dosage compensation and sex determination using the model organism C. elegans. Her current work focuses on understanding how chromatin composition affects gene expression and chromosome dynamics during spermatogenesis. She is interested in characterizing evolutionarily conserved factors important for male fertility.
References
- Aitken RJ, De Iuliis GN. Origins and consequences of DNA damage in male germ cells. Reproductive BioMedicine Online. 2007;14:727–733. doi: 10.1016/s1472-6483(10)60676-1. [DOI] [PubMed] [Google Scholar]
- Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental Biology. 1997;181:296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, et al. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, et al. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Molecular and Cellular Biology. 2004;24:6742–6750. doi: 10.1128/MCB.24.15.6742-6750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arney KL, Bao S, Bannister AJ, et al. Histone methylation defines epigenetic asymmetry in the mouse zygote. International Journal of Developmental Biology. 2002;46:317–320. [PubMed] [Google Scholar]
- Baarends WM, Wassenaar E, van der Laan R, et al. Silencing of unpaired chromatin and histone H2A ubiquitination in mammalian meiosis. Molecular and Cellular Biology. 2005;25:1041–1053. doi: 10.1128/MCB.25.3.1041-1053.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barri PN, Vendrell JM, Martinez F, et al. Influence of spermatogenic profile and meiotic abnormalities on reproductive outcome of infertile patients. Reproductive BioMedicine Online. 2005;10:735–739. doi: 10.1016/s1472-6483(10)61117-0. [DOI] [PubMed] [Google Scholar]
- Barton SC, Surani MA, Norris ML. Role of paternal and maternal genomes in mouse development. Nature. 1984;311:374–376. doi: 10.1038/311374a0. [DOI] [PubMed] [Google Scholar]
- Bazer FW, Spencer TE. Reproductive biology in the era of genomics biology. Theriogenology. 2005;64:442–456. doi: 10.1016/j.theriogenology.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD. RNA meets chromatin. Genes and Development. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- Bhalla N, Dernburg AF. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- Boe-Hansen GB, Fedder J, Ersboll AK, Christensen P. The sperm chromatin structure assay as a diagnostic tool in the human fertility clinic. Human Reproduction. 2006;21:1576–1582. doi: 10.1093/humrep/del019. [DOI] [PubMed] [Google Scholar]
- Bouniol C, Nguyen E, Debey P. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Experimental Cell Research. 1995;218:57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- Braun RE. Packaging paternal chromosomes with protamine. Nature Genetics. 2001;28:10–12. doi: 10.1038/ng0501-10. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- Brewer LR, Corzett M, Balhorn R. Protamine-induced condensation and decondensation of the same DNA molecule. Science. 1999;286:120–123. doi: 10.1126/science.286.5437.120. [DOI] [PubMed] [Google Scholar]
- Browning H, Strome S. A sperm-supplied factor required for embryogenesis in C. elegans. Development. 1996;122:391–404. doi: 10.1242/dev.122.1.391. [DOI] [PubMed] [Google Scholar]
- Buffat C, Patrat C, Merlet F, et al. ICSI outcomes in obstructive azoospermia: influence of the origin of surgically retrieved spermatozoa and the cause of obstruction. Human Reproduction. 2006;21:1018–1024. doi: 10.1093/humrep/dei418. [DOI] [PubMed] [Google Scholar]
- Bungum M, Humaidan P, Axmon A, et al. Sperm DNA integrity assessment in prediction of assisted reproduction technology outcome. Human Reproduction. 2007;22:174–179. doi: 10.1093/humrep/del326. [DOI] [PubMed] [Google Scholar]
- Burns KH, Viveiros MM, Ren Y, et al. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300:633–636. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Developmental Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Caron C, Govin J, Rousseaux S, Khochbin S. How to pack the genome for a safe trip. Progress in Molecular Subcellular Biology. 2005;38:65–89. doi: 10.1007/3-540-27310-7_3. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Cebesoy FB, Aydos K, Unlu C. Effect of sperm chromatin damage on fertilization ratio and embryo quality post-ICSI. Archives of Andrology. 2006;52:397–402. doi: 10.1080/01485010600666953. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu DS, Liu H, Nix P, et al. Sperm chromatin proteomics identifies evolutionarily conserved fertility factors. Nature. 2006;443:101–105. doi: 10.1038/nature05050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churikov D, Zalenskaya IA, Zalensky AO. Male germline-specific histones in mouse and man. Cytogenetic Genome Research. 2004;105:203–214. doi: 10.1159/000078190. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127:503–514. doi: 10.1242/dev.127.3.503. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Ame JC, Schreiber V, et al. Poly(ADP-ribose) polymerase-1 activation during DNA damage and repair. Methods in Enzymology. 2006a;409:493–510. doi: 10.1016/S0076-6879(05)09029-4. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Mark M, Quenet D, et al. Poly(ADP-ribose) polymerase-2 contributes to the fidelity of male meiosis I and spermiogenesis. Proceedings of the National Academy of Sciences of the USA. 2006b;103:14854–14859. doi: 10.1073/pnas.0604252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Developmental Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Male germ cell gene expression. Recent Progress in Hormone Research. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair. 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Mahadevaiah SK, Celeste A, et al. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Developmental Cell. 2003;4:497–508. doi: 10.1016/s1534-5807(03)00093-5. [DOI] [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, et al. Isolation of four core histones from human sperm chromatin representing a minor subset of somatic histones. Journal of Biological Chemistry. 1990;265:20662–20666. [PubMed] [Google Scholar]
- Gatewood JM, Cook GR, Balhorn R, et al. Sequence-specific packaging of DNA in human sperm chromatin. Science. 1987;236:962–964. doi: 10.1126/science.3576213. [DOI] [PubMed] [Google Scholar]
- Georgiou I, Syrrou M, Pardalidis N, et al. Genetic and epigenetic risks of intracytoplasmic sperm injection method. Asian Journal of Andrology. 2006;8:643–673. doi: 10.1111/j.1745-7262.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. Journal of Cell Biology. 2007;176:283–94. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Caron C, Lestrat C, et al. The role of histones in chromatin remodelling during mammalian spermiogenesis. European Journal of Biochemistry. 2004;271:3459–3469. doi: 10.1111/j.1432-1033.2004.04266.x. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nature Reviews Genetics. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proceedings of the National Academy of Sciences of the USA. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T. Methylation dynamics in the early mammalian embryo: implications of genome reprogramming defects for development. Current Topics in Microbiology and Immunology. 2006;310:13–22. doi: 10.1007/3-540-31181-5_2. [DOI] [PubMed] [Google Scholar]
- Hackstein JH, Hochstenbach R, Pearson PL. Towards an understanding of the genetics of human male infertility: lessons from flies. Trends in Genetics. 2000;16:565–572. doi: 10.1016/s0168-9525(00)02140-5. [DOI] [PubMed] [Google Scholar]
- Handel MA. The XY body: a specialized meiotic chromatin domain. Experimental Cell Research. 2004;296:57–63. doi: 10.1016/j.yexcr.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Hill DP, Shakes DC, Ward S, Strome S. A sperm-supplied product essential for initiation of normal embryogenesis in Caenorhabditis elegans is encoded by the paternal-effect embryonic-lethal gene, spe-11. Developmental Biology. 1989;136:154–166. doi: 10.1016/0012-1606(89)90138-3. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Huang X, Li J, Lu L, et al. Novel development-related alternative splices in human testis identified by cDNA microarrays. Journal of Andrology. 2005;26:189–196. doi: 10.1002/j.1939-4640.2005.tb01085.x. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- Jenkins N, Saam JR, Mango SE. CYK-4/GAP provides a localized cue to initiate anteroposterior polarity upon fertilization. Science. 2006;313:1298–1301. doi: 10.1126/science.1130291. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Wooters J, Kopf GS, et al. Analysis of the human sperm proteome. Annals of the New York Academy of Sciences. 2005;1061:190–202. doi: 10.1196/annals.1336.021. [DOI] [PubMed] [Google Scholar]
- Kalmykova AI, Klenov MS, Gvozdev VA. Argonaute protein PIWI controls mobilization of retrotransposons in the Drosophila male germline. Nucleic Acids Research. 2005;33:2052–2059. doi: 10.1093/nar/gki323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum, et al. Tyrosine protein kinases and spermatogenesis: truncation matters. Molecular Reproduction and Development. 2006;73:399–403. doi: 10.1002/mrd.20456. [DOI] [PubMed] [Google Scholar]
- Kimmins S, Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- Krawetz SA. Paternal contribution: new insights and future challenges. Nature Reviews Genetics. 2005;6:633–642. doi: 10.1038/nrg1654. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Song Y, de Jong ME, et al. A walk though vertebrate and invertebrate protamines. Chromosoma. 2003;111:473–482. doi: 10.1007/s00412-002-0226-0. [DOI] [PubMed] [Google Scholar]
- Lin H. piRNAs in the germ line. Science. 2007;316:397. doi: 10.1126/science.1137543. [DOI] [PubMed] [Google Scholar]
- Lin H, Spradling AC. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development. 1997;124:2463–2476. doi: 10.1242/dev.124.12.2463. [DOI] [PubMed] [Google Scholar]
- Liu HL, Hara KT, Aoki F. Role of the first mitosis in the remodeling of the parental genomes in mouse embryos. Cell Research. 2005;15:127–132. doi: 10.1038/sj.cr.7290277. [DOI] [PubMed] [Google Scholar]
- Loppin B, Bonnefoy E, Anselme C, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature. 2005;437:1386–1390. doi: 10.1038/nature04059. [DOI] [PubMed] [Google Scholar]
- Loppin B, Docquier M, Bonneton F, Couble P. The maternal effect mutation sesame affects the formation of the male pronucleus in Drosophila melanogaster. Developmental Biology. 2000;222:392–404. doi: 10.1006/dbio.2000.9718. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, Ahn JH, Cipriani PG, et al. Autosomal genes of autosomal/X-linked duplicated gene pairs and germ-line proliferation in Caenorhabditis elegans. Genetics. 2005;169:1997–2011. doi: 10.1534/genetics.104.040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes and Development. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, et al. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- McCarrey JR, Watson C, Atencio J, et al. X-chromosome inactivation during spermatogenesis is regulated by an Xist/Tsix-independent mechanism in the mouse. Genesis. 2002;34:257–266. doi: 10.1002/gene.10163. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Yates JR., 3rd Proteomic tools for cell biology. Traffic. 2000;1:747–754. doi: 10.1034/j.1600-0854.2000.011001.x. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McLay DW, Clarke HJ. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125:625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans. Developmental Biology. 2005;282:218–230. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- Munroe SH, Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell and Molecular Life Sciences. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, et al. Postmeiotic sex chromatin in the male germline of mice. Current Biology. 2006;16:660–667. doi: 10.1016/j.cub.2006.01.066. [DOI] [PubMed] [Google Scholar]
- Nasr-Esfahani MH, Salehi M, Razavi S, et al. Effect of protamine-2 deficiency on ICSI outcome. Reproductive BioMedicine Online. 2004;9:652–658. doi: 10.1016/s1472-6483(10)61776-2. [DOI] [PubMed] [Google Scholar]
- Nayernia K, Drabent B, Meinhardt A, et al. Triple knockouts reveal gene interactions affecting fertility of male mice. Molecular Reproduction and Development. 2005;70:406–416. doi: 10.1002/mrd.20227. [DOI] [PubMed] [Google Scholar]
- Nayernia K, Meinhardt A, Drabent B, et al. Synergistic effects of germ cell expressed genes on male fertility in mice. Cytogenetic Genome Research. 2003;103:314–320. doi: 10.1159/000076819. [DOI] [PubMed] [Google Scholar]
- Nikolettos N, Asimakopoulos B, Papastefanou IS. Intracytoplasmic sperm injection–an assisted reproduction technique that should make us cautious about imprinting deregulation. Journal of the Society for Gynecologic Investigation. 2006;13:317–328. doi: 10.1016/j.jsgi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Boeke JD. Mighty Piwis defend the germline against genome intruders. Cell. 2007;129:37–44. doi: 10.1016/j.cell.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Arnaud D, Le Baccon P, et al. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature. 2005;438:369–373. doi: 10.1038/nature04155. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Henikoff S. Germline histone dynamics and epigenetics. Current Opinions in Cell Biology. 2007;19:1–9. doi: 10.1016/j.ceb.2007.04.015. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Priess JR, Henikoff S. Histone H3.3 variant dynamics in the germline of Caenorhabditis elegans. PLoS Genetics. 2006;2:e97. doi: 10.1371/journal.pgen.0020097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Moldenhauer JS, et al. A suite of novel human spermatozoal RNAs. Journal of Andrology. 2005;26:70–74. [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Margolis RL. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma. 1990;100:32–36. doi: 10.1007/BF00337600. [DOI] [PubMed] [Google Scholar]
- Parker JS, Barford D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends in Biochemical Sciences. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Philpott A, Leno GH. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell. 1992;69:759–767. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- Pilch B, Mann M. Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biology. 2006;7:R40. doi: 10.1186/gb-2006-7-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst AV, Santos F, Reik W, et al. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma. 2007;116:403–415. doi: 10.1007/s00412-007-0106-8. [DOI] [PubMed] [Google Scholar]
- Reynolds N, Cooke HJ. Role of the DAZ genes in male fertility. Reproductive BioMedicine Online. 2005;10:72–80. doi: 10.1016/s1472-6483(10)60806-1. [DOI] [PubMed] [Google Scholar]
- Rousseaux S, Caron C, Govin J, et al. Establishment of male-specific epigenetic information. Gene. 2005;345:139–153. doi: 10.1016/j.gene.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, et al. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes and Development. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, et al. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Developmental Biology. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics. 2004;166:1313–1321. doi: 10.1534/genetics.166.3.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CB, Ooi SK, Bestor TH, Bourc’his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- Steger K. Possible predictive factors for ICSI? Molecular biology techniques in combination with therapeutic testicular biopsies. Andrologia. 2003;35:200–208. doi: 10.1046/j.1439-0272.2003.00581.x. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Tsuda M, Saga Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development. 2007;134:77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- Swann K, Saunders CM, Rogers NT, Lai FA. PLCzeta(zeta): a sperm protein that triggers Ca2+ oscillations and egg activation in mammals. Seminars in Cell and Developmental Biology. 2006;17:264–273. doi: 10.1016/j.semcdb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nature Reviews Genetics. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Trasler JM. Gamete imprinting: setting epigenetic patterns for the next generation. Reproduction, Fertility and Development. 2006;18:63–69. doi: 10.1071/rd05118. [DOI] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Ellis PJ, et al. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Developmental Cell. 2006;10:521–529. doi: 10.1016/j.devcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, et al. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature Genetics. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- Turner JM, Aprelikova O, Xu X, et al. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Current Biology. 2004;14:2135–2142. doi: 10.1016/j.cub.2004.11.032. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Elliott DJ, et al. Meiotic sex chromosome inactivation in male mice with targeted disruptions of Xist. Journal of Cell Science. 2002;115:4097–4105. doi: 10.1242/jcs.00111. [DOI] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AA, et al. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mechanisms of Development. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Verpoest W, Tournaye H. ICSI: hype or hazard? Human Fertility (Cambridge) 2006;9:81–92. doi: 10.1080/14647270500422158. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Takeda A, Tsukiyama T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes and Development. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykes SM, Visscher DW, Krawetz SA. Haploid transcripts persist in mature human spermatozoa. Molecular Human Reproduction. 1997;3:15–19. doi: 10.1093/molehr/3.1.15. [DOI] [PubMed] [Google Scholar]
- Xu EY, Lee DF, Klebes A, et al. Human BOULE gene rescues meiotic defects in infertile flies. Human Molecular Genetics. 2003;12:169–175. doi: 10.1093/hmg/ddg017. [DOI] [PubMed] [Google Scholar]
- Xu Q, Modrek B, Lee C. Genome-wide detection of tissue-specific alternative splicing in the human transcriptome. Nucleic Acids Research. 2002;30:3754–3766. doi: 10.1093/nar/gkf492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Aprelikova O, Moens P, et al. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development. 2003;130:2001–2012. doi: 10.1242/dev.00410. [DOI] [PubMed] [Google Scholar]
- Yeo S, Lee KK, Han YM, Kang YK. Methylation changes of lysine 9 of histone H3 during preimplantation mouse development. Molecules and Cells. 2005;20:423–428. [PubMed] [Google Scholar]
- Yoshida N, Brahmajosyula M, Shoji S, et al. Epigenetic discrimination by mouse metaphase II oocytes mediates asymmetric chromatin remodeling independently of meiotic exit. Developmental Biology. 2007;301:464–477. doi: 10.1016/j.ydbio.2006.08.006. [DOI] [PubMed] [Google Scholar]