Abstract
In the course of lymphoid development, V(D)J recombination is subject to stringent locus-specific and temporal regulation. These constraints are ultimately responsible for several features peculiar to lymphoid development, including the lineage specificity of antigen receptor assembly, allelic exclusion and receptor editing. In addition, cell cycle phase-dependent regulation of V(D)J recombinase activity ensures that DNA rearrangement is completed by the appropriate mechanism of DNA repair. Regulation of V(D)J recombination involves interactions between the V(D)J recombinase – a heteromeric complex consisting of RAG-1 and RAG-2 subunits – and macromolecular assemblies extrinsic to the recombinase. This chapter will focus on those features of the recombinase itself – and in particular the RAG-2 subunit – that interact with extrinsic factors to establish patterns of temporal control and locus specificity in developing lymphocytes.
Keywords: V(D)J recombination, RAG-1, RAG-2, lymphocyte development, allelic exclusion, cell cycle, DNA repair, non-homologous end joining, homologous recombination, proteasomal degradation, cyclinA/Cdk2, Skp2-SCF, epigenetic control, DNA methylation, nucleosome phasing, histone acetylation, histone methylation, plant homeodomain, locus accessibility
Functional organization of RAG-1 and RAG-2
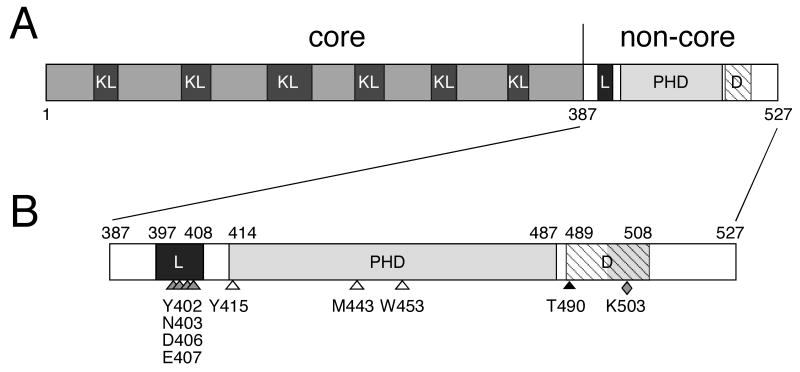
RAG-1 and RAG-2 are 1040 and 527 amino acid residues long, respectively. Residues 384 through 1008 of RAG-1 constitute the core fragment, which contains the catalytic site for DNA cleavage 1-3, mediates binding to recombination signal sequences (RSSs) 4-6 and makes contacts with the coding flanks 7, 8. The core RAG-2 fragment (Fig. 1), consisting of residues 1 through 387, extends interactions of RAG-1 with the RSS and is essential for helical distortion near the scissile bond, a possible prerequisite for transesterification 4-6, 9. Accordingly, mutations that impair recombinase-mediated cleavage and joining have been identified in core RAG-2 10.
Fig. 1.
Regulatory domains of RAG-2. A, schematic representation of RAG-2. Core and non-core regions are designated; amino acid residues are numbered below. KL, Kelch-like propeller domains; L, linker domain; PHD, plant homeodomain finger; D, domain governing programmed degradation and nuclear import of RAG-2. B, detailed representation of the non-core region. Amino acid residues at domain boundaries are numbered above. L (black rectangle), PHD (gray rectangle) and D (hatched rectangle) as defined in (A). The hatched interval denotes the extent of the domain governing cell cycle-dependent degradation of RAG-2; the shaded region within this interval marks the nuclear import signal that resides within the degradation domain. Shaded arrowheads, sites of mutations in the linker domain that impair V(D)J recombination. Open arrowheads, targets of mutations in the PHD domain that abolish H3K4me3 binding and impair V(D)J recombination. Black arrowhead, cyclinA/CDK2 phosphorylation site, essential for programmed degradation of RAG-2 at the G1-S transition. Shaded diamond, target of mutation that selectively impairs nuclear import of RAG-2.
Residues 387 through 527 of RAG-2 comprise the non-core region (Fig. 1) and are dispensable for DNA cleavage by the RAG proteins in vitro. Nonetheless, removal of this region reduces the efficiency of extrachromosomal recombination 11-16, increases production of hybrid joints 17, impedes endogenous VH-to-DJH joining 12, 18, 19 and promotes aberrant recombination 20. The mechanisms underlying these effects may be complex, as the non-core region includes multiple functional domains (Fig. 1B).
Temporal regulation of V(D)J recombination through interactions with the RAG-2 non-core region
The non-core region of RAG-2 supports the periodic destruction of RAG-2 protein. RAG-2 accumulates in quiescent cells and in dividing cells during the G1 phase; rapid degradation of RAG-2 begins at the G1-to-S transition and continues until the following entry into G1 21-23. Consequently, the appearance of recombination signal end intermediates 24, 25 and RAG-signal end complexes 26 is restricted to G0/G1. Destruction of RAG-2 is triggered by phosphorylation of threonine 490, which lies within a phylogenetically conserved cyclin-dependent kinase (Cdk) target site, and is also dependent on a lysine-rich interval spanning amino acid residues 499-508 21. Overlapping the RAG-2 degradation domain (Fig. 1B) is a non-canonical nuclear localization sequence that supports binding of importin 5 and nuclear import of RAG-2 27. At the G1-to-S transition, phosphorylation of RAG-2 by cyclinA/Cdk2 permits association of RAG-2 with the Skp2-SCF ubiquitin ligase. This phosphorylation-dependent interaction is mediated by the F-box protein Skp-2 and its associated protein Cks1. Upon polyubiquitylation of RAG-2 by Skp2-SCF, RAG-2 is subjected to proteasomal degradation 28.
The cell cycle dependence of V(D)J recombination may play a role in the coupling of DNA cleavage by the RAG complex to DNA repair. V(D)J recombination is normally completed by a form of DNA repair termed non-homologous end joining (NHEJ). NHEJ is active throughout the cell cycle, but an alternative mechanism for double-strand DNA repair, homologous recombination (HR), is nearly inactive during G1 29. In thymocytes of mice expressing RAG-2(T490A), aberrant recombinants resembling products of abortive homologous recombination are observed to accumulate 21. These observations suggest that restriction of RAG-2 accumulation to the G0 and G1 cell cycle phases promotes the correct repair of V(D)J recombination intermediates by NHEJ, perhaps by temporal sequestration of RAG activity from HR.
Locus specificity: general remarks
The V(D)J recombinase is directed toward particular sets of gene segments, depending on lymphoid lineage and developmental stage. Recent work has begun to provide a framework for understanding how this targeting is achieved. At the level of unchromatinized DNA, the V(D)J recombinase is targeted to antigen receptor gene segments by means of specific interactions with flanking RSSs, and this recognition does not require the non-core regions of RAG-1 or RAG-2. Not all RSSs support recombination with the same efficiency, because RSSs exhibit considerable sequence variation. Although sequence variation among RSSs can indeed affect gene segment usage 30, these differences cannot account for the dynamic shifts in locus specificity that accompany commitment to distinct lymphoid lineages and developmental transitions within lineages. Rather, ordered rearrangement of antigen receptor gene segments is associated with the imposition or relief of epigenetic marks. Specific chromatin modifications in the vicinity of RSSs are strongly associated with the presence or absence of ongoing rearrangement. The propensity of a particular locus to undergo rearrangement has been thought to be determined by accessibility to the RAG complex, a view that ascribes a passive role to the recombinase. Recent findings, however, indicate that the recombinase - through direct binding to modified chromatin - is an active partner in the epigenetic regulation of rearrangement. We discuss below how epigenetic marks interact with the V(D)J recombinase to promote locus-specific rearrangement.
Epigenetic modifications of possible relevance to V(D)J recombination
An alteration in gene function is termed epigenetic if it is maintained through cell division and does not involve a change in the DNA sequence. One extensively studied epigenetic mark is DNA methylation on cytosine, which in mammals occurs at most CpG dinucleotides. A far more complex set of epigenetic marks are associated with the protein components of chromatin. The basic unit of eukaryotic chromatin is the nucleosome. This consists of a histone core - two molecules each of the histones H2A, H2B, H3 and H4 - around which are wrapped about 146 base pairs of DNA. Histones are subject to a variety of posttranslational modifications including acetylation, methylation, phosphorylation, ubiquitylation and sumoylation. Differences in the degree and stereospecificity of modification contribute substantially to the complexity of these marks. Lysine, for example, can be mono-, di- or trimethylated, while arginine can be dimethylated symmetrically or asymmetrically. In addition to chemical modification, the register in which DNA is wrapped around the histone core - termed nucleosome phasing -can have profound effects on the accessibility of specific sequences to interacting factors. Observations relating these modes of epigenetic regulation to the activation or suppression of V(D)J recombination are summarized in turn below.
DNA methylation
Methylation of CpG dinucleotides is normally associated with the suppression of transcription. Consistent with a general correlation of recombination with transcription, CpG methylation over antigen-receptor-gene segments is also associated with suppression of V(D)J recombination 31. Deletion of PDβ1, a promoter located 5′ to the Dβ1 gene segment or Eβ, an enhancer located 3′ to the TCRβ locus, is accompanied by increased CpG methylation in the Dβ1-Jβ1 region and defects in TCRβ1 rearrangement 32-34. Conversely, demethylation of DNA has been associated with activation of rearrangement. In developing B cells, for example, the Ig κ allele that is first activated for rearrangement is demethylated over the Jκ - Cκ region, while the opposite allele remains hypermethylated and is recruited to heterochromatin 35, 36.
Nucleosome phasing
Together, the core RAG-1 and RAG-2 fragments catalyze RSS-specific nicking and transesterification of DNA substrates in vitro. Efficient cleavage is not observed, however, when chromatinized nuclear substrates are used 37. RAG-mediated DNA cleavage in vitro is impeded when the target RSS is incorporated into a nucleosome 38-40; the degree of inhibition has been variously proposed to be dependent 38 or independent 39 of nucleosome phasing relative to the RSS. The resistance of mononucleosomal substrates to cleavage may result from inaccessibility of histone-associated DNA to the RAG complex as well as from helical distortion induced by wrapping of the DNA around the histone core 40, 41. The impediment to RAG-mediated DNA cleavage observed with mononucleosomal substrates in vitro can be relieved synergistically by histone acetylation and SWI/SNF-dependent remodeling, possibly as a result of alterations in chromatin structure that enhance accessibility of the RSS to the RAG complex 40, 41.
Histone acetylation
Acetylation of histones H3 and H4 is associated with active chromatin. A positive correlation between histone acetylation and active antigen receptor gene rearrangement has been widely documented. Decreased acetylation of H3 and H4 is associated with diminished germline transcription at unrearranged antigen receptor loci and is important for allelic exclusion 36, 42-45. During B cell development, diminished IL-7 signaling is associated with decreased histone acetylation and reduced accessibility to nucleases over distal VH segments 43. A similar relationship is observed over Vβ segments during the transition of intrathymic T cell progenitors from the CD4-CD8- to the CD4+CD8+ stage 46. Thus, decreases in histone acetylation are associated with diminished rearrangement. Consistent with this relationship, Ig κ alleles at which recombination is active exhibit increased acetylation of histone H3 36.
Histone H3 K9 methylation
Dimethylation of histone H3 at lysine 9 (H3K9me2), which is associated with silent chromatin, is positively correlated with inhibition of V(D)J recombination 47-49. Dimethyl marks at H3K9 are removed over VH segments at the pro-B to pre-B cell transition, at which stage VH-to-DJH joining occurs; H3K9 demethylation is dependent on expression of the transcription factor Pax5 in pro-B cells 47. A role for H3K9me2 in the control of V(D)J recombination was suggested in an experiment that targeted G9a, a histone H3K9 methyltransferase, to a TCRβ minilocus. In this setting, directed H3K9 methylation was found to inhibit both germline transcription and V(D)J recombination, overriding the presence of cis-acting accessibility control elements 49. An interpretation of these findings is complicated, because ablation of the G9a methyltransferase in mice had no significant effects on lymphoid development or stage specificity of V(D)J recombination, despite suppressive effects on λ light chain usage, B cell proliferation and plasma cell differentiation 50.
Histone H3 K4 methylation
Methylation of histone H3 lysine 4 (H3K4) is a phylogenetically conserved modification that has been linked to transcriptional activation in yeast and metazoans 51. The relationship between histone H3K4 methylation and V(D)J recombination has been the subject of much recent study 36, 48, 52, 53. Dimethylated histone H3K4 (H3K4me2) 48, 53 and H3K4me3 (54, 55 exhibit distinct patterns of enhancement within the D-JH cluster in pro-B cells poised to undergo D-to-JH rearrangement. Moreover, the recombinationally active Ig κ allele in pre-B cells is marked by hypermethylation of H3K4 36.
Monoubiquitylation of histone H2B at lysine 123 (ubH2B) promotes histone H3K4 methylation in yeast 56-58. UbH2B is associated with transcriptionally active chromatin both in yeast 59-63 and in mammalian cells 60, 64. Patterns of ubH2B deposition have yet to be extensively mapped. As H2B ubiquitylation appears to be a prerequisite for H3K4 hypermethylation, it will be of interest to know whether the density of ubH2B is enhanced at sites of active V(D)J recombination, possibly extending the chain of causation one step upstream.
Direct recognition of modified histone H3 by the V(D)J recombinase
The observations outlined above, while essential to an understanding of epigenetic control, do not in themselves provide mechanistic insight into how histone modification is linked mechanistically to V(D)J recombination. Building on recent progress in the understanding of how histone methylation patterns are read, several studies have combined biochemical, structural and genetic approaches to outline how one such linkage is established.
A variety of protein domains are capable of binding the N-terminal region of histone H3 when this is hypermethylated at lysine 4. These include the chromodomains of CHD1 65, 66, the double tudor domain of JMJD2A 67 and the plant homeodomain (PHD) fingers of ING2 68-71, BPTF 68, 71 and Yng1 72. Crystallographic analysis reveals that the PHD fingers of ING2 69, BPTF 68 and Yng1 72 all contain an aromatic cage that mediates binding to methyl-lysine, a feature shared by other methyl-lysine-binding domains 73. The structural basis of H3K4me2 or H3K4me3 binding by the PHD finger is of particularly broad significance, because this recognition domain is present in many chromatin-associated proteins that carry out histone modification 74, 75.
The ability of the PHD finger to mediate binding to H3K4me2 and H3K4me3 led several groups to examine the function of a similar domain that earlier had been identified within residues 419 through 481 of the non-core region of RAG-2 76. This non-canonical PHD finger (Fig. 1B) was shown to mediate direct binding of RAG-2 to histone H3 di- or trimethylated at K4, with a preference for H3K4me3 54, 55. Mutations that abolish binding of the RAG-2 PHD finger to H3K4me3 (Fig. 1B) were found to impair V(D)J recombination both within extrachomosomal substrates and at endogenous loci 54, 55. Moreover, the association of the RAG-2 PHD finger with chromatin across the immunoglobulin heavy chain locus is positively correlated with the density of H3K4me3 54. Mutations that disrupt H3K4me3 binding or Zn++ coordination by the RAG-2 PHD finger had been associated earlier with combined hereditary immunodeficiencies in humans 77-81, underscoring the physiologic importance of these interactions.
The crystal structures of complexes between the RAG-2 PHD finger and modified H3 peptides have shown that this domain, while functionally related to its canonical cousins, exhibits the unusual ability to integrate epigenetic marks 82. In the complex with a peptide bearing K4me3, the trimethyl ammonium group of K4 is buried in an “aromatic cage” similar to that of other methyl-lysine-binding domains. An important difference between the PHD finger of RAG-2 and other H3K4me3-binding domains, however, was observed: an enhanced affinity for a doubly modified histone – namely, H3 bearing both K4Me3 and a symmetrically dimethylated arginine at position 2 (R2Me2s). This is possible because the RAG-2 PHD finger lacks a side chain carboxylate that in homologous domains forms salt bridges with unmodified R2. In RAG-2 this is replaced by tyrosine, which mediates interactions with H3R2me2s 82. An important consequence is that binding of RAG-2 to an H3 peptide bearing K4me3 is enhanced by the presence of R2Me2s 82. While the differential affinities of RAG-2 for singly and doubly modified histone H3 could in principle contribute to locus discrimination by the V(D)J recombinase, the physiological relevance of this property remains unclear, because symmetric methylation of histone H3 R2 has as yet not been detected in vivo.
Evidence for allosteric regulation of V(D)J recombinase activity by histone H3 trimethylated at lysine 4
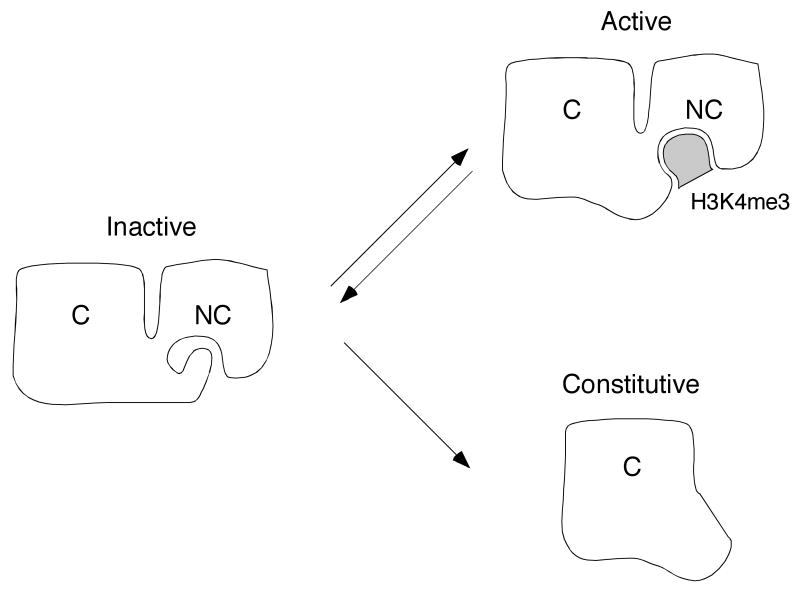
The engagement of histone H3K4me3 by the RAG-2 PHD finger provides a bridge between one chemical mark of active chromatin and the V(D)J recombinase machinery. Paradoxically, while V(D)J recombination is profoundly impaired by a point mutation that abolishes H3K4me3 binding by the RAG-2 PHD finger, complete removal of the non-core region, including the entire PHD finger, has only a modest debilitating effect 54, 55. To reconcile these observations it has been proposed that an inhibitory domain resides within the non-core region of RAG-2, and that suppression of recombinase activity by this domain is relieved upon engagement of the PHD finger by H3K4me3 (Fig. 2). Consistent with this proposal is a crystal structure in which the RAG-2 PHD finger - in the absence of an H3K4me3 ligand - is occupied by an amino-terminal peptide encoded by the expression construct 82. It may be that hypermethylated H3K4 does not simply act as a docking site for the recombinase but rather plays a more active role as an allosteric trigger of RAG catalysis.
Fig. 2.
A model for allosteric activation of the RAG complex by modified histone H3. White figures represent RAG-2; C and NC denote core and non-core regions. respectively. Shaded object represents trimethylated lysine 4 of histone N3 (H3K4me3). In a hypothetical inactive conformation (left), the aromatic channel of the RAG-2 PHD finger is occupied by an inhibitory domain residing elsewhere in the non-core region. In the hypothetical active conformation (upper right), the PHD finger is bound by histone H3K4me3 and the putative inhibitory domain is released. The RAG-2 core fragment (lower right) lacks both the PHD finger and the putative inhibitory domain. In this configuration RAG-2 is proposed to assume an active configuration constitutively. For further discussion, see text.
Future directions: deposition and integration of epigenetic signals controlling V(D)J recombination
The link between transcriptional activation and locus-specificity of V(D)J recombination has long suggested that transcription and V(D)J recombination are controlled by shared epigenetic mechanisms. Progress in understanding these mechanisms has awaited the chemical characterization of epigenetic marks and the development of methods by which the genomic distribution of these marks could be mapped. These approaches have begun to provide a detailed view of epigenetic change at antigen receptor genes as a function of development. Several important questions will continue to dominate the field.
The first is to define precisely the structural features that confer locus specificity to the V(D)J recombinase. While recognition of histone H3K4me3 by RAG-2 provides a link between active chromatin and V(D)J recombination, it is obvious that H3K4me3 – a general mark of transcriptionally active chromatin – is too broadly distributed to act alone in directing the recombinase to specific sites of action. Clearly other modes of regulation must contribute to locus specificity of recombinase activity. While it seems likely that this will involve a combinatorial summation of chromatin modifications and DNA sequence elements, the answer is far from clear. A related question concerns the direct role of modified chromatin in regulating RAG activity. The proposal that the recombinase is allosterically activated upon binding of the RAG-2 PHD finger to modified chromatin will need to be tested, and the relative contributions of modifications at H3K4, H3R2 and elsewhere will need to be defined. Regions of the RAG-2 other than the PHD finger may also mediate functional interactions with chromatin. The RAG-2 linker region, which lies at the amino-terminal side of the PHD finger (Fig. 1B), has been reported to bind core histones and mutations within this region were found to impair VH-to-DJH joining 83; the basis for this apparently selective effect is unclear. A third question concerns how developmental signals, such as those that emanate from the pre-BCR, govern deposition and removal of epigenetic marks at antigen receptor loci. A resolution of these outstanding issues will provide a starting point from which to address the larger problem of allelic exclusion.
Contributor Information
Yun Liu, Email: yliu39@jhmi.edu, Department of Molecular Biology and Genetics and Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD 21210, USA.
Li Zhang, Email: lzhang32@jhmi.edu, Department of Molecular Biology and Genetics and Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD 21210, USA.
Stephen Desiderio, Email: sdesider@jhmi.edu, Department of Molecular Biology and Genetics and Institute for Cell Engineering, The Johns Hopkins University School of Medicine, Baltimore, MD 21210, USA.
References
- 1.Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell. 2000;5:97–107. doi: 10.1016/s1097-2765(00)80406-2. [DOI] [PubMed] [Google Scholar]
- 2.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akamatsu Y, Oettinger MA. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol Cell Biol. 1998;18:4670–4678. doi: 10.1128/mcb.18.8.4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Difilippantonio MJ, McMahan CJ, Eastman QM, Spanopoulou E, Schatz DG. RAG1 mediates signal sequence recognition and recruitment of RAG2 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 6.Swanson PC, Desiderio S. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity. 1998;9:115–125. doi: 10.1016/s1074-7613(00)80593-2. [DOI] [PubMed] [Google Scholar]
- 7.Eastman QM, Villey IJ, Schatz DG. Detection of RAG protein-V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol. 1999;19:3788–3797. doi: 10.1128/mcb.19.5.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson PC, Desiderio S. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol. 1999;19:3674–3683. doi: 10.1128/mcb.19.5.3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 10.Qiu JX, Kale SB, Yarnell Schultz H, Roth DB. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol Cell. 2001;7:77–87. doi: 10.1016/s1097-2765(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 11.Cuomo CA, Oettinger MA. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirch SA, Rathbun GA, Oettinger MA. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. Embo J. 1998;17:4881–4886. doi: 10.1093/emboj/17.16.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahan CJ, Difilippantonio MJ, Rao N, Spanopoulou E, Schatz DG. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol Cell Biol. 1997;17:4544–4552. doi: 10.1128/mcb.17.8.4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sadofsky MJ, Hesse JE, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadofsky MJ, Hesse JE, McBlane JF, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19:3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekiguchi JA, Whitlow S, Alt FW. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol Cell. 2001;8:1383–1390. doi: 10.1016/s1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 18.Akamatsu Y, Monroe R, Dudley DD, et al. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc Natl Acad Sci U S A. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 20.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res. 2004;32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Z, Dordai DI, Lee J, Desiderio S. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity. 1996;5:575–589. doi: 10.1016/s1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 22.Lin WC, Desiderio S. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science. 1993;260:953–959. doi: 10.1126/science.8493533. [DOI] [PubMed] [Google Scholar]
- 23.Lin WC, Desiderio S. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc Natl Acad Sci U S A. 1994;91:2733–2737. doi: 10.1073/pnas.91.7.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Desiderio S, Lin WC, Li Z. The cell cycle and V(D)J recombination. Curr Top Microbiol Immunol. 1996;217:45–59. doi: 10.1007/978-3-642-50140-1_4. [DOI] [PubMed] [Google Scholar]
- 25.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 26.Jiang H, Ross AE, Desiderio S. Cell cycle-dependent accumulation in vivo of transposition-competent complexes between recombination signal ends and full-length RAG proteins. J Biol Chem. 2004;279:8478–8486. doi: 10.1074/jbc.M311219200. [DOI] [PubMed] [Google Scholar]
- 27.Ross AE, Vuica M, Desiderio S. Overlapping signals for protein degradation and nuclear localization define a role for intrinsic RAG-2 nuclear uptake in dividing cells. Mol Cell Biol. 2003;23:5308–5319. doi: 10.1128/MCB.23.15.5308-5319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang H, Chang FC, Ross AE, et al. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell. 2005;18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Takata M, Sasaki MS, Sonoda E, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. Embo J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feeney AJ. Factors that influence formation of B cell repertoire. Immunol Res. 2000;21:195–202. doi: 10.1385/IR:21:2-3:195. [DOI] [PubMed] [Google Scholar]
- 31.Nakase H, Takahama Y, Akamatsu Y. Effect of CpG methylation on RAG1/RAG2 reactivity: implications of direct and indirect mechanisms for controlling V(D)J cleavage. EMBO Rep. 2003;4:774–780. doi: 10.1038/sj.embor.embor904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitehurst CE, Chattopadhyay S, Chen J. Control of V(D)J recombinational accessibility of the D beta 1 gene segment at the TCR beta locus by a germline promoter. Immunity. 1999;10:313–322. doi: 10.1016/s1074-7613(00)80031-x. [DOI] [PubMed] [Google Scholar]
- 33.Mathieu N, Hempel WM, Spicuglia S, Verthuy C, Ferrier P. Chromatin remodeling by the T cell receptor (TCR)-beta gene enhancer during early T cell development: Implications for the control of TCR-beta locus recombination. J Exp Med. 2000;192:625–636. doi: 10.1084/jem.192.5.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitehurst CE, Schlissel MS, Chen J. Deletion of germline promoter PD beta 1 from the TCR beta locus causes hypermethylation that impairs D beta 1 recombination by multiple mechanisms. Immunity. 2000;13:703–714. doi: 10.1016/s1074-7613(00)00069-8. [DOI] [PubMed] [Google Scholar]
- 35.Mostoslavsky R, Singh N, Kirillov A, et al. Kappa chain monoallelic demethylation and the establishment of allelic exclusion. Genes Dev. 1998;12:1801–1811. doi: 10.1101/gad.12.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldmit M, Ji Y, Skok J, et al. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 37.Stanhope-Baker P, Hudson KM, Shaffer AL, Constantinescu A, Schlissel MS. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 38.Kwon J, Imbalzano AN, Matthews A, Oettinger MA. Accessibility of nucleosomal DNA to V(D)J cleavage is modulated by RSS positioning and HMG1. Mol Cell. 1998;2:829–839. doi: 10.1016/s1097-2765(00)80297-x. [DOI] [PubMed] [Google Scholar]
- 39.Golding A, Chandler S, Ballestar E, Wolffe AP, Schlissel MS. Nucleosome structure completely inhibits in vitro cleavage by the V(D)J recombinase. Embo J. 1999;18:3712–3723. doi: 10.1093/emboj/18.13.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwon J, Morshead KB, Guyon JR, Kingston RE, Oettinger MA. Histone acetylation and hSWI/SNF remodeling act in concert to stimulate V(D)J cleavage of nucleosomal DNA. Mol Cell. 2000;6:1037–1048. doi: 10.1016/s1097-2765(00)00102-7. [DOI] [PubMed] [Google Scholar]
- 41.Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. Embo J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin mu heavy chain gene locus. Embo J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury D, Sen R. Transient IL-7/IL-7R signaling provides a mechanism for feedback inhibition of immunoglobulin heavy chain gene rearrangements. Immunity. 2003;18:229–241. doi: 10.1016/s1074-7613(03)00030-x. [DOI] [PubMed] [Google Scholar]
- 44.Johnson K, Calame K. Transcription factors controlling the beginning and end of B-cell differentiation. Curr Opin Genet Dev. 2003;13:522–528. doi: 10.1016/j.gde.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 45.McMurry MT, Krangel MS. A role for histone acetylation in the developmental regulation of VDJ recombination. Science. 2000;287:495–498. doi: 10.1126/science.287.5452.495. [DOI] [PubMed] [Google Scholar]
- 46.Tripathi R, Jackson A, Krangel MS. A change in the structure of Vbeta chromatin associated with TCR beta allelic exclusion. J Immunol. 2002;168:2316–2324. doi: 10.4049/jimmunol.168.5.2316. [DOI] [PubMed] [Google Scholar]
- 47.Johnson K, Pflugh DL, Yu D, et al. B cell-specific loss of histone 3 lysine 9 methylation in the V(H) locus depends on Pax5. Nat Immunol. 2004;5:853–861. doi: 10.1038/ni1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A. 2003;100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Osipovich O, Milley R, Meade A, et al. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol. 2004;5:309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 50.Thomas LR, Miyashita H, Cobb RM, et al. Functional analysis of histone methyltransferase g9a in B and T lymphocytes. J Immunol. 2008;181:485–493. doi: 10.4049/jimmunol.181.1.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sims RJ, 3rd, Reinberg D. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 2006;20:2779–2786. doi: 10.1101/gad.1468206. [DOI] [PubMed] [Google Scholar]
- 52.Perkins EJ, Kee BL, Ramsden DA. Histone 3 lysine 4 methylation during the pre-B to immature B-cell transition. Nucleic Acids Res. 2004;32:1942–1947. doi: 10.1093/nar/gkh523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakraborty T, Chowdhury D, Keyes A, et al. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 54.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matthews AG, Kuo AJ, Ramon-Maiques S, et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dover J, Schneider J, Tawiah-Boateng MA, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- 57.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 58.Lee JS, Shukla A, Schneider J, et al. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 59.Henry KW, Wyce A, Lo WS, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003;17:2733–2740. doi: 10.1101/gad.1156403. [DOI] [PubMed] [Google Scholar]
- 61.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xiao T, Kao CF, Krogan NJ, et al. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol Cell Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 64.Kirmizis A, Santos-Rosa H, Penkett CJ, et al. Arginine methylation at histone H3R2 controls deposition of H3K4 trimethylation. Nature. 2007;449:928–932. doi: 10.1038/nature06160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Flanagan JF, Mi LZ, Chruszcz M, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 66.Sims RJ, 3rd, Chen CF, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–41792. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang Y, Fang J, Bedford MT, Zhang Y, Xu RM. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Ilin S, Wang W, et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pena PV, Davrazou F, Shi X, et al. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi X, Hong T, Walter KL, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wysocka J, Swigut T, Xiao H, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 72.Taverna SD, Ilin S, Rogers RS, et al. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol Cell. 2006;24:785–796. doi: 10.1016/j.molcel.2006.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 74.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 75.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 76.Callebaut I, Mornon JP. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci. 1998;54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gomez CA, Ptaszek LM, Villa A, et al. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol Cell Biol. 2000;20:5653–5664. doi: 10.1128/mcb.20.15.5653-5664.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Noordzij JG, de Bruin-Versteeg S, Verkaik NS, et al. The immunophenotypic and immunogenotypic B-cell differentiation arrest in bone marrow of RAG-deficient SCID patients corresponds to residual recombination activities of mutated RAG proteins. Blood. 2002;100:2145–2152. [PubMed] [Google Scholar]
- 79.Schwarz K, Gauss GH, Ludwig L, et al. RAG mutations in human B cell-negative SCID. Science. 1996;274:97–99. doi: 10.1126/science.274.5284.97. [DOI] [PubMed] [Google Scholar]
- 80.Villa A, Sobacchi C, Notarangelo LD, et al. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 81.Villa A, Santagata S, Bozzi F, et al. Partial V(D)J recombination activity leads to Omenn syndrome. Cell. 1998;93:885–896. doi: 10.1016/s0092-8674(00)81448-8. [DOI] [PubMed] [Google Scholar]
- 82.Ramon-Maiques S, Kuo AJ, Carney D, et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A. 2007;104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.West KL, Singha NC, De Ioannes P, et al. A direct interaction between the RAG2 C terminus and the core histones is required for efficient V(D)J recombination. Immunity. 2005;23:203–212. doi: 10.1016/j.immuni.2005.07.004. [DOI] [PubMed] [Google Scholar]