Abstract
In protein synthesis initiation, the eukaryotic translation initiation factor (eIF) 2 (a G protein) functions in its GTP-bound state to deliver initiator methionyl-tRNA (tRNAiMet) to the small ribosomal subunit and is necessary for protein synthesis in all cells1,2. Phosphorylation of eIF2 [eIF2(αP)] is critical for translational control in diverse settings including nutrient deprivation, viral infection and memory formation3,4,5. eIF5 functions in start site selection as a GTPase accelerating protein (GAP) for the eIF2•GTP•tRNAiMet ternary complex (TC) within the ribosome bound pre-initiation complex6,7,8. Here we define new regulatory functions of eIF5 in the recycling of eIF2 from its inactive eIF2•GDP state between successive rounds of translation initiation. Firstly we show that eIF5 stabilizes the binding of GDP to eIF2 and is therefore a bi-functional protein that acts as a GDP dissociation inhibitor (GDI). We find that this activity is independent of the GAP function and identify conserved residues within eIF5 that are necessary for this role. In addition we show that eIF5 is a critical component of the eIF2(αP) regulatory complex that inhibits the activity of the guanine-nucleotide exchange factor (GEF) eIF2B. Together our studies define a new step in the translation initiation pathway, one that is critical for normal translational controls.
eIF2•GTP is one of multiple translation initiation factors that plays an essential role in facilitating precise ribosome-bound tRNAiMet recognition of initiator codons on mRNAs. A second factor, eIF5 binds eIF2 and accelerates GTP hydrolysis and release of both factors prior to small and large ribosomal subunit joining2. Modulating the activity of eIF2 via phosphorylation is a key control step in protein synthesis. eIF2α protein kinases respond to diverse cues and can mediate both general protein synthesis repression as well as translational activation of specific mRNAs including those bearing multiple short upstream open reading frames (uORFs) in their leaders. eIF2(αP) inhibits the GEF activity of eIF2B blocking the reactivation of eIF2•GTP1,2. Recent observations suggested that eIF5 has additional functions in translation. Both eIF2 and eIF5 are dissociated from ribosomes together9,10. In addition, in yeast, there is an abundant cellular fraction of eIF2/eIF5 complexes that greatly exceeds the detectable levels of TC11, suggesting that eIF5 may play a part in the recycling of eIF2•GDP. GDI proteins antagonise GEFs and have been described for some G proteins12, although not for translation initiation. We therefore assessed if eIF5 possesses GDI activity.
Yeast eIF2αβγ complexes were purified using his-tagged eIF2γ13. GDP release (Koff) was measured in a filter binding assay where eIF2•[3H]GDP complexes were dissociated in the presence of excess unlabelled GDP and magnesium ions (Fig.1a). Increasing [Mg2+] progressively stabilized GDP binding to eIF2, as expected, because Mg2+ helps coordinate the GDP-β phosphate-eIF2γ interaction14 (Fig.S1). The effect of increasing concentrations of recombinant eIF5 on the GDP dissociation rate was assessed using a glutathione S-transferase eIF5 fusion protein (GST-eIF5) purified from Escherichia coli (Figs.1b, S1, S2). A significant and progressive stabilization of GDP binding to eIF2 was observed with increasing eIF5 (Koff reduced from 0.12 to 0.077 min−1 with equimolar eIF2 and eIF5), which approached saturation at a 4:1 ratio. Importantly, GST alone did not have this activity (Fig.1b). We observed the GDP stabilization effect of eIF5 over a range of physiological magnesium concentrations. The data also show that Mg2+ is required for eIF5 GDI activity, as eIF5 has minimal GDI function in the absence of added Mg2+. However as Mg2+ concentration is increased within the physiological range, Mg2+ and eIF5 act together to stabilize GDP binding to eIF2 (Fig.S1). eIF5-FLAG, purified from yeast, behaved identically to GST-eIF5 (Fig.1c, #2& #10). These experiments demonstrate that eIF5 can act as a GDI factor for eIF2•GDP.
Figure 1. eIF5 has GDI activity.
a) Scheme for GDI activity assay. b) Increasing eIF5 stabilises GDP-binding to eIF2. Koff GDP from 60 pmol eIF2 with varying concentrations of GST-eIF5 (0-240 pmol, open circles) or GST alone (filled circle). Molar eIF2:GST-eIF5 protein ratios are indicated. c) Defining regions required for GDI activity. Mean Koff GDP (60 pmol eIF2) for indicated constructs derived from reactions with GST- or FLAG-eIF5 proteins (120 pmol). Black bars represent a significant reduction in Koff GDP (P<0.0001, unpaired Student's t-test). Errors show standard deviation (n>3). 2.9 mM Mg2+ was used in b) and c).
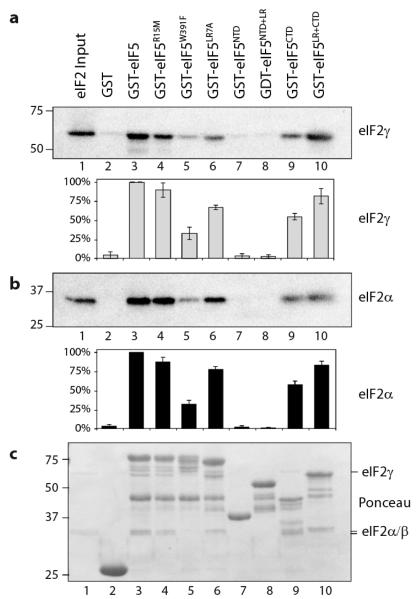
The invariant Arg15 residue in eIF5 is critical for eIF5-dependent GAP activity and the eIF5R15M mutation eliminates this function6-8. The R15M mutant retains full eIF5 GDI activity (Fig.1c, #3) demonstrating that GAP and GDI activities are independent. The tif5-7A allele is a well-characterised eIF5 mutant in which seven evolutionarily-conserved residues in the carboxy-terminal domain (CTD) are mutated to alanines15. These amino acid substitutions impair eIF5-eIF2 interactions11,15,16. We found that eIF57A-FLAG eliminated GDI function, as did a single conservative substitution within this motif at Trp391 (eIF5W391F; Fig.1c, #11& #4). GST-protein affinity chromatography was performed to examine interactions between purified eIF5 and eIF2. This confirmed that GST-eIF5W391F reduces the affinity of eIF5 for purified eIF2, similar to that described for the 7A mutant (Fig.2, lane 5)15 and shows that the eIF2 binding domain provided by the eIF5-CTD is necessary for GDI activity.
Figure 2. The CTD of eIF5 is critical for interaction with eIF2.
Affinity chromatography assay between eIF2 (110 pmol) and the indicated immobilized GST-eIF5 constructs. eIF2 was detected by immunoblotting using antibodies specific for a) eIF2γ or b) eIF2α. Representative blots are shown. Signal intensity was quantified (Adobe Photoshop) and the mean ± standard deviation (n=3) are shown below. c) Total protein in each sample stained with Ponceau S. Inputs (lanes 1) represent 10% of total.
To delineate regions of eIF5 necessary for its GDI activity we used sequence alignments and available structural information to define the ‘amino terminal domain’ (NTD, residues 1-152)17 and CTD (residues 241-405) of yeast eIF518. The intervening sequence (residues 153-240) is defined here as the ‘linker region’ (LR). GST-fusion proteins comprising these domains were assessed for eIF2 interaction and GDI activity (Figs. 1 and 2). Only GST-eIF5LR+CTD retained both activities, while the GST-eIF5CTD could bind to eIF2, but did not retain GDI function. These experiments suggested that residues within the LR are required in addition to the CTD for GDI activity. Sequence alignments from diverse organisms identified a highly-conserved 20 residue stretch within the LR which we have called the 'DWEAR' motif after the five absolutely conserved residues it contains (Fig.S3). A mutant allele was made within full-length eIF5 in which seven DWEAR motif residues were mutated to alanines; we term this the LR7A allele (eIF5LR7A). GST-eIF5LR7A retained eIF2-binding affinity (Fig.2, lane 6) but eliminated GDI function (Fig.1c, #5). These analyses separate eIF2 binding and GDI functions and identify conserved residues critical for GDI activity within the LR and CTD.
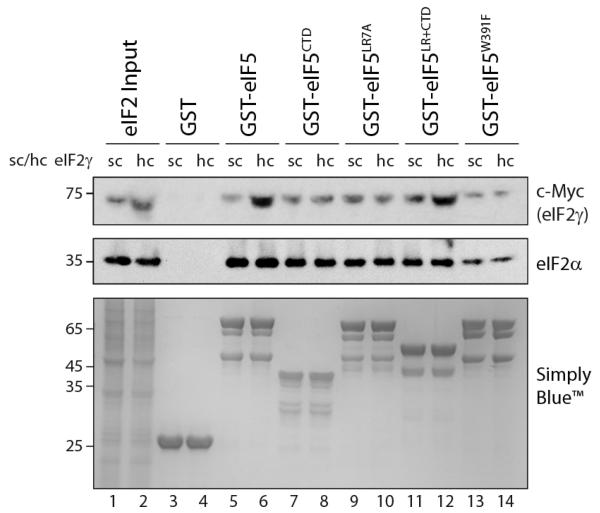
As GDP binds eIF2γ, we asked whether the eIF5LR+CTD could interact directly with eIF2γ to mediate GDI function. The eIF5 CTD was previously shown to interact with eIF2β15. In addition eIF2γ was found to bind a construct containing eIF5 residues 1-279 but not residues 280-40519 i.e. extending from the NTD through our LR into the CTD. We assessed the ability of GST-fusion proteins to interact with eIF2 from yeast cell extracts expressing c-Myc-6xHis tagged eIF2γ from either a single copy (sc) or high copy (hc) plasmid to provide a source both of eIF2αβγ complexes and excess free eIF2γ. As expected, GST-eIF5 could specifically interact with eIF2αβγ, as demonstrated by immunoblotting signals for the α and γ subunits, and with the excess eIF2γ when overexpressed (Fig.3, lanes 5-6). Consistent with Fig.2, eIF5 constructs with an intact CTD interacted with eIF2αβγ complexes in cell extracts similarly to full-length wild-type eIF5, while the W391F mutation weakened binding. However, among the mutants tested, only the eIF5LR+CTD construct containing an intact LR and CTD region stimulated binding to the excess eIF2γ (lane 12). Thus the LR7A allele interacts with eIF2 complexes but not excess free eIF2γ (lane 10). These results are consistent with eIF2β forming a major site for eIF5-CTD binding, while conserved residues within the LR region define a new region of eIF5 required for eIF2γ interaction and GDI function.
Figure 3. The Linker Region of eIF5 interacts with γ subunit of eIF2.
Affinity chromatography as in figure 2 between indicated immobilized GST-eIF5 constructs and total cell extracts (500 μg) expressing c-Myc-6xHis-eIF2γ from either a single copy (sc) or high copy (hc) plasmid. Immunoblots were developed with c-Myc and eIF2α antibodies. Total protein in each sample was stained with Ponceau S. Inputs (lane 1) represent 5% (blots) or 1% (stain) of total.
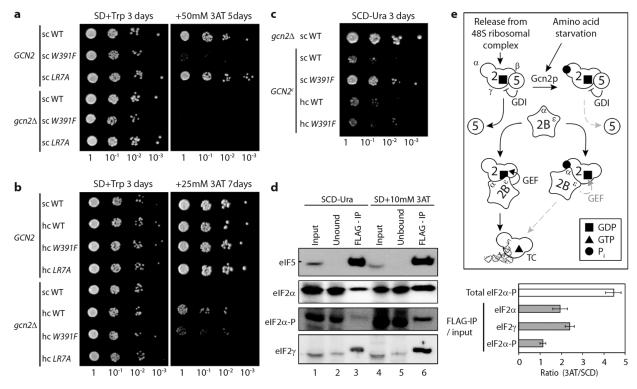
To examine the effects of mutations on eIF5 function in vivo, the R15M, W391F and LR7A mutations were introduced into eIF5-FLAG on both sc and hc plasmids, then transformed into an eIF5 deleted (tif5Δ) yeast strain by plasmid shuffling so that each mutant became the sole source of eIF5. The R15M GAP mutant was lethal, despite appropriate expression (Fig.S4), while W391F and LR7A mutants grew well (Fig.4a). These data demonstrate GAP activity is the essential function of eIF5 and imply that these mutations eliminating GDI function have minimal impact on GAP activity or growth under standard conditions.
Figure 4. GDI activity antagonises eIF2B and affects GCN4 activation in vivo.
a-c) Strains expressing single (sc) or high copy (hc) eIF5 plasmids as the source of eIF5 and co-transformed with plasmids expressing GCN2, vector alone (gcn2Δ) [a, b] or the constitutively active mutant GCN2M788V,E1591K (GCN2c) [c] were grown as stated. d) Left, immunoblots following FLAG-eIF5 immune precipitation of protein complexes from cells grown in nutrient sufficient conditions (SCD) and following starvation (SD+3AT). Right, quantification ± standard deviation (n=3). e) Model for recycling and regulation of eIF2, incorporating eIF5 GDI activity. Dashed grey arrows indicate steps that limit eIF2 recycling.
Translation of GCN4 mRNA is highly-sensitive to eIF2 activity and is widely used as a reporter for the activity of translation initiation factors20. Derepression of GCN4 translation can be scored by monitoring cell growth in the presence of the HIS3 inhibitor 3-amino-triazole (3AT). Growth of wild-type cells on media containing 3AT is dependent upon activation of Gcn2p, the eIF2α kinase that responds to amino acid starvation (Fig.4a, gcn2Δ), which inhibits eIF2B GEF activity lowering TC abundance. This enables ribosomes to bypass inhibitory uORFs in GCN4 mRNA that normally repress GCN4 translation20. As the GDI deficient mutants exhibit different eIF2 binding activities in vitro (Figs. 2 and 3) we anticipated that both mutants could provide complementary tools to probe GDI function in vivo. hc eIF5 bypasses the requirement for eIF2(αP) for GCN4 activation, conferring resistance to 3AT in gcn2Δ cells11 (Fig.4b, row 6). An increase in the concentration of an eIF2/eIF5 complex in cells, which antagonises eIF2B GEF activity, explains this phenotype11. The GDI mutant, hc eIF5LR7A, fails to grow on 3AT medium (Fig.4b, row 8), consistent with reduced eIF2B antagonism in vivo. In agreement with this, immune precipitation of hc FLAG-eIF5LR7A failed to enhance binding with eIF2 over that bound to sc eIF5 (Fig.S5a). Therefore these data are consistent with the idea that loss of GDI function in eIF5LR7A can diminish competition between eIF5 and eIF2B (GEF) for eIF2.
We assessed whether eIF5 GDI was required for the normal response to eIF2 phosphorylation in GCN2 cells and found that W391F mutant cells fail to grow on 3AT medium, showing they cannot induce GCN4 translation (Fig.4a, row 2). This Gcn− phenotype is shared with other eIF5-CTD mutations21, and was previously ascribed to a defect within the 48S initiation complex causing enhanced leaky-scanning of uORF1 within the GCN4 mRNA leader21. However, this explanation does not take into account the eIF5 GDI function. Because the eIF5W391F mutation reduces levels of the eIF2/eIF5 complex (Figs 2, 3, S5) and eliminates GDI activity, we postulated that eIF5 binding to eIF2•GDP is necessary to inhibit eIF2B GEF function via eIF2(αP) and that the observed reduced affinity of eIF5W391F for eIF2 was sufficient to alter the sensitivity of cells to eIF2(αP), thus causing the 3AT sensitivity. To test whether eIF5W391F could affect global translational sensitivity to eIF2(αP), we introduced constitutively active GCN2c alleles that hyper-phosphorylate eIF2α to high-levels independent of amino acid starvation and down-regulate global translation initiation conferring severely reduced rates of growth22 (Fig.4c, row 2). The eIF5W391F allele reverted slow-growth phenotypes associated with two different GCN2c alleles of varying phenotypic severity (Fig.4c, row 3 and Fig.S5b). Immunoblotting analysis showed that the eIF5 mutation did not alter the ability of Gcn2p to phosphorylate eIF2α (Fig.S5c). The Gcn− and GCN2c growth-reversion phenotypes reported here cannot be explained by enhanced leaky-scanning and are identical to those conferred by mutations in eIF2B subunits which alter the ability of eIF2B to respond to eIF2(αP)13,23 and by the overexpression of eIF2B or eIF224. Thus in W391F mutant cells, translation is significantly less sensitive to the inhibitory effects of eIF2(αP).
We found that over-expression of eIF5W391F restored eIF2/eIF5 complex levels to those found in wildtype cells (Fig.S5a). Consistent with this, hc eIF5W391F reversed the growth enhancement associated with GCN2c alleles (Fig.4c, row 5) as well as the Gcn− phenotype (compare Fig.4a, row 2 and Fig.4b, row 3) and partially reverted the Gcd− phenotype caused by hc eIF5 (compare Fig.4b, rows 6 and 7). Although we assume that the W391F mutation, in common with other CTD mutations21, has a minor impact upon pre-initiation complex formation and GAP function, the W391F phenotypes and their reversion are consistent with reduced eIF2/eIF5 complex formation and GDI activity promoting eIF2 nucleotide exchange and disrupting the ability of cells to regulate translation appropriately in response to eIF2(αP) (Fig.S6b).
These data suggested that the eIF2/eIF5 complex was a new critical component of the regulatory circuit that inhibits eIF2B GEF function and regulates translation by diverse means. To test this prediction, we immune precipitated FLAG-eIF5 from wild-type cells grown in amino acid complete medium and following starvation when eIF2 phosphorylation was enhanced ~4-fold (Fig.4d). A significantly greater fraction of eIF2 was found associated with the precipitated FLAG-eIF5 following amino acid starvation (2-2.5 fold), indicating that the affinity of eIF5 for eIF2 is enhanced, or that the release of eIF5 from eIF2 is retarded, when the latter is phosphorylated. However we do not suggest that eIF5 interacts directly with eIF2α because the increase in precipitated eIF2(αP) observed following amino acid starvation is in proportion with its higher total abundance (Fig.4d).
Taken together our data demonstrate that eIF5-GDI is a novel component of the inhibitory eIF2(αP) pathway that restricts the recycling of eIF2 to TC and acts in addition to the well defined inhibition of eIF2B whereby eIF2α phosphorylation promotes its association with the eIF2Bαβδ subunits25, to impair eIF2Bε subunit-mediated nucleotide exchange26. Thus our data are consistent with a model where eIF5 is a multifunctional protein with separate GDI and GAP activities, both necessary for controlled translation initiation in eukaryotes and we propose an altered scheme (Fig.4e, Fig.S6) to account for eIF5-GDI activity and the eIF2•GDP/eIF5 complex in modulating the recycling of eIF2 in response to eIF2 phosphorylation between rounds of translation initiation.
Methods summary
Plasmid and strain construction employed standard methods27. Plasmid, oligonucleotide and yeast construction details are given in Tables S1-S3. eIF2 and eIF5 proteins were purified as previously described13,28. The eIF5 GDI activity assay is a variant of an assay used to measure eIF2B GEF activity13. Full methods are provided in the electronic version of this paper.
Methods
Yeast Genetics
Yeast strains were grown in standard media as described27. Plasmid transformations used the lithium acetate method29 and plasmid shuffling employed 5-fluoro-orotic acid as described27. Cells were grown to exponential phase then diluted to A600=0.1 and ten-fold serially diluted. 2μl of each dilution was spotted onto the indicated medium and incubated at 30°C.
Plasmid constructions
Individual mutants were generated by overlapping primer site directed mutagenesis (Stratagene) or using standard PCR cloning methods. The L7RA mutant was commercially synthesised (Mr Gene GmbH) then sub-cloned.
SDS PAGE/ Immunoblotting
This was performed as described previously26 using specific antibodies for eIF5, eIF2α and 2γ (custom antibodies), c-Myc-HRP (9E10, Santa Cruz Biotechnology), FLAG (M2, Sigma) and eIF2α–P (9721, Cell Signaling Technology). HRP secondary chemiluminescent detection as per manufacturer's instructions (Perkin Elmer).
Protein purification
eIF2 was purified as described previously13 using strain GP3511. For purification of GST-tagged eIF5 wt and mutant constructs, ArcticExpress™ BL21 DE3 RIL E. coli cells (Stratagene) were transformed with recombinant pGEX-4T-1 plasmids and then grown in LB medium to an A600 of 0.7. Expression was induced using 1 mM of isopropyl-β-d-thiogalactopyranoside (IPTG) and cells were then grown overnight at 12oC. Cells were harvested and lysed with BugBuster® (EMDBiosciences) with protease inhibitors added [1 × complete EDTA-free protease inhibitor tablet (Roche)] before clearing by centrifugation (16,000 × g, 20 minutes, 4°C). The soluble lysate was then mixed for 2hrs at 4°C with glutathione-Sepharose beads (GE Healthcare) equilibrated in 1 × PBS. The beads were then washed thrice, then suspended in glutathione elution buffer (50 mM Tris-HCl [pH 9.0], 500 mM NaCl, 50 mM glutathione, 1 mM DTT, 0.1 % Triton X-100) and incubated overnight at 4°C. Eluates were dialysed into storage buffer (30 mM HEPES [pH 7.5], 100 mM KCl, 0.1 mM MgCl2, 10 % glycerol, 1 mM DTT) and stored at −80°C. Quantification was performed using the Bradford Protein Assay (Bio-Rad).
To purify FLAG tagged eIF5 from yeast, strains expressing hc eIF5-FLAG (GP5424) or eIF5-7A-FLAG (GP5423) were grown in SCD medium (2% dextrose) but without leucine to A600 of 3-5. Cells were harvested, washed in ice-cold water and then resuspended in high-salt lysis buffer (100mM Tris-HCl [pH 8.0], 500 mM KCl, 5 mM MgCl2, 5 mM NaF, 2.5 mM PMSF, 10% glycerol, 0.1% Triton X-100 with 1 μg/ml pepstatin A, 1 μg/ml leupeptin, 5 μg/ml aprotinin, and 1 × complete EDTA-free protease inhibitor tablet [Roche]). Lysis was performed by grinding with a pestle and mortar under liquid nitrogen and lysates were then cleared by centrifugation (16,000 × g, 20 minutes, 4°C). The extract was then incubated for 2 hours at 4°C with EZview™ Red ANTI-FLAG® M2 Affinity Gel (Sigma) equilibrated in high-salt lysis buffer, washed once with high-salt lysis buffer then twice with low-salt lysis buffer (as high-salt lysis buffer but with only 100 mM KCl). Bound protein was then released by incubation for 30 minutes in the presence of 0.1 mg/ml of 3xFLAG peptide (Sigma) and dialysed into storage buffer. Quantification was performed using the Bradford Assay (Bio-Rad).
FLAG immune precipitations
These were performed similarly to purifications but using buffers containing 80 mM KCl to maintain interactions.
GDI activity
eIF2•[3H]GDP binary complexes were formed in 75 mm × 12 mm soda-lime glass tubes (Fisher). Typically using eIF2 (60 pmol) and 0.25 μCi [3H]GDP (10-15 Ci/mmol) in assay buffer (30 mM HEPES pH 7.5, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 2 mg/ml CPK). Complex formation was carried out for 10 minutes at room temperature before stabilisation by addition of MgCl2 to 2.9 mM and incubation for a further 2 minutes at room temperature. Purified eIF5 (120 pmol) or sample buffer was then bound to the eIF2 binary complex for 30 minutes at 10°C. Dissociation of the binary complex was initiated by addition of a >100-fold excess of unlabelled GDP (2 nmol). 12 μl samples were taken immediately (t=0) and at 2, 4, 6, 8 and 10 minutes. At each time point, samples were added to 2.5 ml ice-cold Stop buffer (30 mM HEPES [pH 7.5], 100 mM KCl, 0.1 mM EDTA, 5 mM MgCl2), filtered through Whatman 0.45 μm 25mm cellulose nitrate filters using a Millipore vacuum manifold then washed twice with 2.5 ml of ice-cold Stop buffer. Filters were dried at 65°C then counted by liquid scintillation in Ultima Gold™ F (Perkin Elmer). Experimental data were fitted to exponential dissociation curves to obtain the dissociation rate constant (Koff).
GST affinity chromatography
1. Using purified eIF2
Purified GST-eIF5 proteins or GST alone (140 pmol) and eIF2 (110 pmol) were incubated in 400 μl of interaction buffer (30 mM HEPES [pH 7.5], 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 2.5 mM MgCl2, 0.05% Triton X-100) with 50 μl of glutathione-Sepharose beads for 2 hours at 4°C. The beads were then washed 3 times with 1 ml of interaction buffer and the bound proteins eluted by boiling in sample buffer. Bound eIF2 was then resolved by SDS-PAGE separation and detected immunoblot analysis using the indicated antibodies.
2. Using cell extracts
Strains GP5463 and GP5661 expressing sc and hc c-Myc-tagged GCD11 (respectively) were grown in SC-Leu-Trp medium to A600 of 3-5. Cells were harvested, washed in water and then resuspended in 1ml low-salt lysis buffer. Lysis was performed using 1.0 mm zirconia/silica beads (Thistle Scientific) and the FastPrep 24 homogeniser at 4°C (MP Biomedicals). Lysates were cleared by centrifugation (16,000 × g, 20 minutes, 4°C) and proteins quantified using Bradford assay (Bio-Rad). Total protein lysate (500 μg) was incubated with purified GST-eIF5 (140 pmol) or GST alone and 40 μl of glutathione-Sepharose beads in a total volume of 1 ml of low-salt lysis buffer for 2 hrs at 4°C. The resin was washed twice with 1 ml of low-salt lysis buffer and the bound proteins eluted by boiling in sample buffer. Bound eIF2α and γ was assayed by immunoblot analysis.
Supplementary Material
Acknowledgements
We thank Katsura Asano for plasmids and yeast strains, and helpful discussions. Mark Ashe and members of the Pavitt and Ashe labs for helpful discussions. This work was supported by grants BB/E002005/1 and BB/H010599/1 from the BBSRC to GDP.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
The authors declare that they have no competing financial interests.
References
- 1.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scheuner D, et al. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 4.Costa-Mattioli M, et al. eIF2alpha phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell. 2007;129:195–206. doi: 10.1016/j.cell.2007.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohr I. Neutralizing innate host defenses to control viral translation in HSV-1 infected cells. Int. Rev. Immunol. 2004;23:199–220. doi: 10.1080/08830180490265600. [DOI] [PubMed] [Google Scholar]
- 6.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Das S, Ghosh R, Maitra U. Eukaryotic translation initiation factor 5 functions as a GTPase-activating protein. J. Biol. Chem. 2001;276:6720–6726. doi: 10.1074/jbc.M008863200. [DOI] [PubMed] [Google Scholar]
- 8.Paulin FE, et al. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr. Biol. 2001;11:55–59. doi: 10.1016/s0960-9822(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 9.Cheung YN, et al. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh CR, et al. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J. 2006;25:4537–4546. doi: 10.1038/sj.emboj.7601339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DerMardirossian C, Bokoch GM. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 2005;15:356–363. doi: 10.1016/j.tcb.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev. 1998;12:514–526. doi: 10.1101/gad.12.4.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt E, Blanquet S, Mechulam Y. The large subunit of initiation factor aIF2 is a close structural homologue of elongation factors. EMBO J. 2002;21:1821–1832. doi: 10.1093/emboj/21.7.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Asano K, et al. Conserved bipartite motifs in yeast eIF5 and eIF2Bepsilon, GTPase- activating and GDP-GTP exchange factors in translation initiation, mediate binding to their common substrate eIF2. EMBO J. 1999;18:1673–1688. doi: 10.1093/emboj/18.6.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asano K, et al. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conte MR, et al. Structure of the eukaryotic initiation factor (eIF) 5 reveals a fold common to several translation factors. Biochemistry. 2006;45:4550–4558. doi: 10.1021/bi052387u. [DOI] [PubMed] [Google Scholar]
- 18.Wei Z, Xue Y, Xu H, Gong W. Crystal structure of the C-terminal domain of S. cerevisiae eIF5. J. Mol. Biol. 2006;359:1–9. doi: 10.1016/j.jmb.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 19.Alone PV, Dever TE. Direct binding of translation initiation factor eIF2gamma-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2B epsilon. J. Biol. Chem. 2006;281:12636–12644. doi: 10.1074/jbc.M511700200. [DOI] [PubMed] [Google Scholar]
- 20.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 21.Singh CR, et al. Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol. Cell. Biol. 2005;25:5480–5491. doi: 10.1128/MCB.25.13.5480-5491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramirez M, et al. Mutations activating the yeast eIF-2 alpha kinase GCN2: isolation of alleles altering the domain related to histidyl-tRNA synthetases. Mol. Cell. Biol. 1992;12:5801–5815. doi: 10.1128/mcb.12.12.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavitt GD, Yang W, Hinnebusch AG. Homologous segments in three subunits of the guanine nucleotide exchange factor eIF2B mediate translational regulation by phosphorylation of eIF2. Mol. Cell. Biol. 1997;17:1298–1313. doi: 10.1128/mcb.17.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dever TE, et al. Modulation of tRNAiMet, eIF-2 and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNAiMet ternary complexes. Mol. Cell. Biol. 1995;15:6351–6363. doi: 10.1128/mcb.15.11.6351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishnamoorthy T, et al. Tight binding of the phosphorylated alpha subunit of initiation factor 2 (eIF2α) to the regulatory subunits of guanine nucleotide exchange factor eIF2B is required for inhibition of translation initiation. Mol. Cell. Biol. 2001;21:5018–5030. doi: 10.1128/MCB.21.15.5018-5030.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohammad-Qureshi SS, et al. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2beta and -2gamma mediate guanine nucleotide exchange. Mol. Cell. Biol. 2007;27:5225–5234. doi: 10.1128/MCB.00495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast genetics: A Cold Spring Harbor Laboratory Course Manual. 1997 Edition Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. [Google Scholar]
- 28.Phan L, et al. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 1998;18:4935–4946. doi: 10.1128/mcb.18.8.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
Additional Reference
- 29.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.