Abstract
Cancer cells employ multiple mechanisms to evade tightly regulated cellular processes such as proliferation, apoptosis and senescence. Systems-wide analyses of tumors have recently identified receptor tyrosine kinase (RTK) coactivation as an important mechanism by which cancer cells achieve chemoresistance. This mini-review discusses our current understanding of the complex and dynamic process of RTK coactivation. We highlight how systems biology and computational modelling have been employed to predict integrated signalling outcomes and cancer phenotypes downstream of RTK coactivation. We conclude by providing an outlook on the feasibility of targeting RTK networks to overcome chemoresistance in cancer.
Keywords: Receptor Tyrosine Kinase, Signal Transduction, Systems Biology, Targeted Therapy
Introduction
The receptor tyrosine kinases (RTKs) have historically been the subject of intense investigation due to their widespread deregulation in cancer and the prospect of developing targeted therapeutics to these proteins. Since the first description of transphosphorylation between the epidermal growth factor receptor (EGFR) and the insulin receptor (IR) (1), an increasing number of studies have reported crosstalk between different members of the RTK superfamily. The introduction of large-scale high-throughput approaches for mapping cellular signaling networks have identified additional complexities in RTK network biology and led to the realization that RTK crosstalk is more prevalent than previously envisioned (2-4). This idea, referred to as “RTK coactivation”, is a process by which cancer cells simultaneously activate two or more RTKs in order to attain network robustness and increase the diversity of signaling outcomes that can be achieved using a limited repertoire of intracellular signaling components. Recent studies have shown that RTK coactivation plays critical roles in influencing tumor response to targeted therapeutics and manifestation of cancer phenotypes (2, 4). Furthermore, advances in network-based technologies and computational biology have enhanced our ability to evaluate these networks to the point of generating predictive computational models for cancer biology (5, 6). In this mini-review, we describe our evolving understanding of the complex and dynamic process of RTK coactivation. Using recent examples primarily focused on kinetic models, we will explore how cancer systems biology has been used to dissect RTK signaling networks. Finally, we offer a perspective on exploiting RTK coactivation networks as a means of designing therapeutic strategies targeting cancer.
Network robustness and chemoresistance
One advantage of RTK coactivation is that it imparts cancer cells with the ability to maintain network robustness in the face of acute perturbations, i.e. cells are able to preserve phenotypic outcomes despite disruption in one or more signaling elements. Many RTKs share common downstream effectors and activate similar signaling pathways, albeit to varying degrees (7) (Figure 1A). Using glioblastoma as a model, Stommel et al. have shown that RTK coactivation in tumors allows for ‘oncogene switching’ upon inhibition of specific RTKs (4). For instance, both EGFR and c-Met activate the PI3K pathway through the recruitment of the GAB1 adaptor protein. Under normal conditions, GAB1-PI3K preferentially associates with EGFR in glioblastoma cells expressing a constitutive mutant of EGFR (EGFRvIII). Pharmacological inhibition of EGFR leads to the compensatory recruitment of the GAB1-PI3K complex by c-Met, maintaining a robust PI3K signal and cell survival. Correspondingly, cancer cells that coactivate EGFR and c-Met fail to succumb to EGFR tyrosine kinase inhibitor monotherapy (2, 4). To overcome this resistance, combination strategies employing multiple RTK inhibitors have been proposed with the aim of shutting down signaling flux through key oncogenic nodes by simultaneously inactivating multiple upstream activators (Figure 1C). A cocktail of EGFR, c-Met and PDGFR inhibitors resulted in a synergistic decrease in cell viability compared to treatment with any single agent alone (4). This phenomenon has since been recapitulated in multiple cancer types, particularly in the context of acquired resistance to kinase inhibitors (8), suggesting that ‘oncogene switching’ as a result of RTK coactivation may be a general mechanism by which cancer cells achieve chemoresistance.
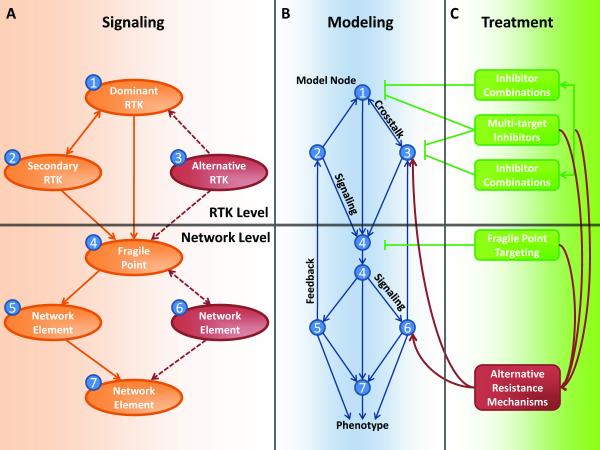
Figure 1. Features of RTK coactivation networks.
(A) RTK-mediated signalling pathways share multiple elements and inhibiting the dominant RTK often results in the compensatory recruitment of downstream components by secondary RTKs. Examples of dominant RTKs include EGFR (2, 4, 10) and ErbB2 (15), while secondary RTKs such as c-Met, PDGFR and IGF-1R (4, 9, 15) have been reported. These RTK coactivation events converge on a number of fragile points in the network such as AKT (6). (B) Kinetic modelling approaches simplify complex signalling networks and identify fragile points that have disproportionate large effects on signalling and phenotypic output. (C) Effective treatment strategies (green) include targeting of multiple RTKs or fragile points determined by the implementation of network models. Alternative resistance mechanisms may arise in response to these therapeutic interventions (red) which may include activation of alternative RTKs (e.g. Axl (16)) or other intracellular network elements.
An elaboration of this concept has been put forward by Pillay et al., where a dominant kinase sits on top of a hierarchy of RTKs and inhibition of this dominant RTK results in the elevation of a secondary RTK to the primary position (9) (Figure 1A). In the glioblastoma example described earlier, EGFRvIII is the dominant kinase while c-Met acts as the secondary kinase. Interestingly, the dominant RTK may itself be responsible for the activation of secondary RTKs. Utilizing high-throughput microwestern arrays combined with Bayesian network modeling, Ciacco et al. derived a model depicting the network architecture of eight RTKs activated by EGFR in A431 epidermoid carcinoma cells (10). Mechanisms for secondary RTK coactivation include (a) autocrine/paracrine growth factor secretion (11), (b) direct transphosphorylation by the dominant RTK (9), (c) indirect phosphorylation through a signaling intermediate (e.g. Src) (12) or (d) transcriptional regulation (13). Phosphoproteomic studies have also shown that the stimulation of different dominant/initiator RTKs result in the coactivation of distinct secondary RTK profiles. For instance, EGF activation of EGFR-HER2 heterodimers in HER2-overexpressing human mammary epithelial cells (HMECs) led to the preferential activation of c-Met, Axl, and EphA2 compared to heregulin-driven HER2-HER3 dimers (14). However, both EGFR-HER2 and HER2-HER3 dimers activated IGF-1R to the same degree (14). These observations would suggest that inhibition of the dominant RTK could result in significant rewiring of the web of RTKs it activates and ultimately the resultant downstream signaling network. Understanding this process of differential signal utilization remains a significant challenge in developing targeted therapeutics to overcome network robustness.
Kinetic models and RTK networks
Perhaps a less well-studied aspect is the effect of RTK coactivation on integrated signaling networks and tumor phenotypes. An analysis by Macbeath and co-workers demonstrated that 6 distinct RTKs differentially phosphorylated common downstream signaling molecules and displayed disparate phosphotyrosine binding domain affinities (7). This study raises the important question of whether an integrated network downstream of coactivated RTKs is the result of a simple signal amplification of these unique RTK signatures or do novel (or possibly antagonistic) functions arise from RTK coactivation? A recent study on trastuzumab (an ErbB2 specific therapeutic antibody) resistance illustrates the complexity of this problem (15). The authors found that a heterotrimer of activated ErbB2, ErbB3 and IGF-IR was responsible for conferring trastuzumab resistance in breast cancer cell lines. Depletion of either ErbB3 or IGF-IR did not alter the interactions between the remaining two RTKs but nonetheless sensitized cells to trastuzumab treatment. However, this drug sensitivity was achieved through different mechanisms. Knockdown of ErbB3 decreased Akt activation but had no effect on MAPK signaling while removal of IGF-IR reduced Src and MAPK activation levels with minimal effect on Akt and ErbB3 phosphorylation. It is plausible that some of the features (e.g. tratuzumab resistance) observed in patients may be attributed to shared RTK co-activation signaling nodes while others (e.g. tumor cell invasion) may be due to the action of specific RTKs. “Combination indices” for RTK co-activation must be established to determine if these signaling and phenotypic outcomes are additive or synergistic compared to those arising from individual RTKs. Such experiments combined with computational biology approaches are key to addressing some of the complexity inherent in RTK signaling networks.
The attractiveness of computational biology lies in its ability to generate models that are capable of distilling how RTK interactions impinge on tumor phenotypes and signal transduction. The challenge of generating such integrated models is the identification and incorporation of points of crosstalk between individual contributing growth factor signaling networks (Figure 1B). Additionally, for computational models to be informative, it is critical that suitable experiments are performed to include the appropriate signaling space and avoid inaccuracies that may result in incorrect predictions or relationships. Borisov et al. generated an ODE (ordinary differential equation)-based model that merges points of crosstalk and integrates the feedback mechanisms inherent in both the EGFR and IR signaling networks (5). Using a combination of experiments and computational simulations, the authors determined that while insulin weakly activates the Erk pathway compared to EGF, co-stimulation of HEK293 cells with a low dose of EGF with insulin leads to a synergistic activation of the Erk pathway. This observation is due to the increase in the levels of PIP3 as a result of EGF and insulin co-activation of PI3K that ultimately facilitates the membrane recruitment of GAB1 and increased activation of the Ras pathway. Consistent with this hypothesis, addition of wortmannin (a PI3K inhibitor) abrogated this synergy. Interestingly, the addition of insulin appears to confer robustness to EGF stimulated pErk in GAB1 knockdown cells which suggests that co-stimulated cells use multiple redundant pathways to activate critical signaling nodes. The identification and incorporation of these redundant pathways into next generation kinetic models will allow for more accurate predictions of signaling outcomes in disease. Additionally, the inclusion of additional RTK inputs, such as c-Met or PDGFR, will undoubtedly enrich our understanding of mechanisms of crosstalk and redundancy in RTK coactivation networks.
Exploiting RTK coactivation networks for therapy
Strategies to overcome RTK coactivation broadly fall into two classes. As described above, the first strategy involves therapeutically targeting multiple RTKs simultaneously in order to shut down oncogenic RTK signaling and overcome compensatory mechanisms (Figure 1C). More recently, single agents targeting multiple RTKs have been developed and show promise in preclinical models of disease (16, 17). An example is foretinib, a small molecule inhibitor of Axl, c-Met and VEGFR, which reduced tumor burden in a mouse model of lung metastasis and overcame lapatinib (an EGFR/ErbB2 inhibitor)-induced resistance in BT474 breast cancer cells (16, 17). While these multi-target approaches are effective in pre-clinical models, it remains to be seen if additional resistance mechanisms, either driven by the activation of alternative RTKs or by other processes, will arise in cancer cells treated in this manner (Figure 1C).
The second approach involves identifying and targeting fragile points that exist downstream of RTK co-activation networks (Figure 1B). Due to the inherent plasticity of oncogenic networks, it remains a challenge to determine a priori signaling nodes that sensitize cancer cells when perturbed therapeutically. A recent study demonstrates that sensitivity analysis in ODE-based models may be a promising technique towards achieving this goal of predictive cancer biology. Employing an ErbB network-based ODE model, Schoeberl and co-workers investigated the effects of EGFR ligand betacellulin and ErbB3 ligand heregulin on multiple downstream ErbB signaling outputs (6). By performing species-specific sensitivity analyses (varying the levels of each species in the model and determining its effect on the model output), they predicted that regardless of the ligand administered; ErbB3 is the most sensitive node in the network that regulates AKT phosphorylation. The authors experimentally validated this prediction using an ErbB3-specific monoclonal antibody MM-121 and showed that they could accurately simulate the dose response of ErbB and AKT phosphorylation levels upon antibody treatment. While prior studies have largely been limited to single cell types or groups of closely related cell lines, the authors were able to extend the predictive capability of the model to two additional cell types and three distinct ErbB targeting antibodies/inhibitors through the incorporation of cell-/drug-specific parameters such as receptor abundance or kinetic rate constants. It has previously been shown that that distinct cell types mediate cell-specific phenotypes through the use of common effector processing (18), supporting the idea that computational models may be extended to other cell types or stimuli through additional data generation and model refinement. While RTKs have traditionally been considered as signal initiators or transducers, it would be interesting to establish if specific RTKs (e.g. ErbB3) may themselves be part of the common effector processing machinery in the context of an RTK coactivation network and thus be amenable to therapeutic intervention. The study by Schoeberl et al. highlights the power of sensitivity analyses as a means of identifying fragile points in networks and ODE-models as tools for predicting network-activity responses to therapeutics.
Conclusions
The emerging picture of RTK coactivation suggests that cancer cells have evolved this mechanism to adapt and respond to challenges presented by exogeneous or intrinsic perturbations. The integrated signaling networks and phenotypes arising from RTK coactivation are poorly annotated and will require additional high-density signaling and biochemical datasets to better inform existing computational modeling efforts. Early examples of implementing kinetic models as well as additional computational approaches such as statistical modeling (10, 19) have shown promise in facilitating network simulations and generating testable predictions pertinent to RTK interactions. The challenge moving forward is to extend these models to include diverse cell and tissue types that incorporate complex influences such as the tumor microenvironment or cell-tissue interactions in vivo. The success of these efforts hinges on the development of new technologies that are capable of generating quantitative in vivo signaling assessments with spatial resolution (20) that will refine existing computational models that have been constructed using in vitro data. We are optimistic that these systems-based approaches will be useful in identifying fragile points that are inherent in RTK coactivation networks and will have a positive impact in the development of network-based therapeutics for translation into the clinic.
Acknowledgements
Grant support: PHH is supported by the Wellcome Trust and The Institute of Cancer Research. The authors do not have any conflicting interests involving this work.
The authors thank Nathan Tedford for helpful discussions on the manuscript.
Abbreviations
- RTK
Receptor Tyrosine Kinase
- EGFR
Epidermal Growth Factor Receptor
- IR
Insulin Receptor
References
- 1.Lammers R, Van Obberghen E, Ballotti R, Schlessinger J, Ullrich A. Transphosphorylation as a possible mechanism for insulin and epidermal growth factor receptor activation. J Biol Chem. 1990;265:16886–90. [PubMed] [Google Scholar]
- 2.Huang PH, Mukasa A, Bonavia R, et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A. 2007;104:12867–72. doi: 10.1073/pnas.0705158104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 4.Stommel JM, Kimmelman AC, Ying H, et al. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287–90. doi: 10.1126/science.1142946. [DOI] [PubMed] [Google Scholar]
- 5.Borisov N, Aksamitiene E, Kiyatkin A, et al. Systems-level interactions between insulin-EGF networks amplify mitogenic signaling. Mol Syst Biol. 2009;5:256. doi: 10.1038/msb.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoeberl B, Pace EA, Fitzgerald JB, et al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci Signal. 2009;2:ra31. doi: 10.1126/scisignal.2000352. [DOI] [PubMed] [Google Scholar]
- 7.Gordus A, Krall JA, Beyer EM, et al. Linear combinations of docking affinities explain quantitative differences in RTK signaling. Mol Syst Biol. 2009;5:235. doi: 10.1038/msb.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guix M, Faber AC, Wang SE, et al. Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest. 2008;118:2609–19. doi: 10.1172/JCI34588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pillay V, Allaf L, Wilding AL, et al. The plasticity of oncogene addiction: implications for targeted therapies directed to receptor tyrosine kinases. Neoplasia. 2009;11:448–58. doi: 10.1593/neo.09230. 2 p following 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciaccio MF, Wagner JP, Chuu CP, Lauffenburger DA, Jones RB. Systems analysis of EGF receptor signaling dynamics with microwestern arrays. Nat Methods. 2010;7:148–55. doi: 10.1038/nmeth.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena NK, Taliaferro-Smith L, Knight BB, et al. Bidirectional crosstalk between leptin and insulin-like growth factor-I signaling promotes invasion and migration of breast cancer cells via transactivation of epidermal growth factor receptor. Cancer Res. 2008;68:9712–22. doi: 10.1158/0008-5472.CAN-08-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto N, Mammadova G, Song RX, Fukami Y, Sato K. Tyrosine phosphorylation of p145met mediated by EGFR and Src is required for serum-independent survival of human bladder carcinoma cells. J Cell Sci. 2006;119:4623–33. doi: 10.1242/jcs.03236. [DOI] [PubMed] [Google Scholar]
- 13.Reznik TE, Sang Y, Ma Y, et al. Transcription-dependent epidermal growth factor receptor activation by hepatocyte growth factor. Mol Cancer Res. 2008;6:139–50. doi: 10.1158/1541-7786.MCR-07-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf-Yadlin A, Kumar N, Zhang Y, et al. Effects of HER2 overexpression on cell signaling networks governing proliferation and migration. Mol Syst Biol. 2006;2:54. doi: 10.1038/msb4100094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X, Gao L, Wang S, et al. Heterotrimerization of the growth factor receptors erbB2, erbB3, and insulin-like growth factor-I receptor in breast cancer cells resistant to herceptin. Cancer Res. 2010;70:1204–14. doi: 10.1158/0008-5472.CAN-09-3321. [DOI] [PubMed] [Google Scholar]
- 16.Liu L, Greger J, Shi H, et al. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69:6871–8. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- 17.Qian F, Engst S, Yamaguchi K, et al. Inhibition of tumor cell growth, invasion, and metastasis by EXEL-2880 (XL880, GSK1363089), a novel inhibitor of HGF and VEGF receptor tyrosine kinases. Cancer Res. 2009;69:8009–16. doi: 10.1158/0008-5472.CAN-08-4889. [DOI] [PubMed] [Google Scholar]
- 18.Miller-Jensen K, Janes KA, Brugge JS, Lauffenburger DA. Common effector processing mediates cell-specific responses to stimuli. Nature. 2007;448:604–8. doi: 10.1038/nature06001. [DOI] [PubMed] [Google Scholar]
- 19.Janes KA, Lauffenburger DA. A biological approach to computational models of proteomic networks. Curr Opin Chem Biol. 2006;10:73–80. doi: 10.1016/j.cbpa.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Giampieri S, Manning C, Hooper S, Jones L, Hill CS, Sahai E. Localized and reversible TGFbeta signalling switches breast cancer cells from cohesive to single cell motility. Nat Cell Biol. 2009;11:1287–96. doi: 10.1038/ncb1973. [DOI] [PMC free article] [PubMed] [Google Scholar]