Abstract
Powassan virus (POWV, Flaviviridae: Flavivirus) is the sole North American member of the tick-borne encephalitis complex and consists of two distinct lineages that are maintained in ecologically discrete enzootic transmission cycles. The underlying genetic mechanisms that lead to niche partitioning in arboviruses are poorly understood. Therefore, intra- and interhost genetic diversity was analyzed to determine if POWV exists as a quasispecies in nature and quantify selective pressures within and between hosts. In contrast to previous reports for West Nile virus (WNV), significant intrahost genetic diversity was not observed. However, pN (0.238) and dN/dS ratios (0.092) for interhost diversity were similar to those of WNV. Combined, these data suggest that purifying selection and/or population bottlenecks constrain quasispecies diversity within ticks. These same selective and stochastic mechanisms appear to drive minor sequence changes between ticks. Moreover, Powassan virus populations seem not to be structured as quasispecies in naturally infected adult deer ticks.
Keywords: Powassan virus, TBE, quasispecies, Ixodes scapularis, genetic diversity
Introduction
Powassan virus (POWV, Flaviviridae: Flavivirus) is the sole North American member of the tick-borne encephalitis (TBE) complex which is otherwise geographically distributed across temperate Eurasia (Gritsun 2003). Many of these viruses, including POWV, have been identified as the causative agents of severe central nervous system infections in humans (Gritsun 2003; McLean 1959). Historically, there have been few confirmed human cases of POWV; however, recent evidence suggests that there has been an apparent increase in incidence (Hinten et al. 2008; Tavakoli et al. 2009). Molecular epidemiological studies have established that POWV consists of two distinct lineages (Ebel et al. 2001; Kuno et al. 2001). Lineage I, the “prototype” lineage, is maintained in an enzootic transmission cycle between Ixodes cookei and Ix. marxi and small and medium-sized woodland mammals (Main et al. 1979; McLean and Chernesky 1969; McLean and Larke 1963). Lineage II, the “Deer tick virus” (DTV) lineage, is apparently maintained in an enzootic transmission cycle between Ix. scapularis and Peromyscus leucopus (white-footed mice) (Ebel et al. 2000; Telford et al. 1997). While the two lineages are maintained in ecologically discrete transmission cycles and share only 84% nucleotide and 94% amino acid sequence identity, they are serologically indistinguishable and thus classified as the same viral species (Beasley et al. 2001; Kuno et al. 2001). Analysis of variation within each lineage revealed that the DTV-lineage has greater genetic variation and therefore is most likely the ancestral lineage (Ebel et al. 2001). Furthermore, analysis of synonymous and non-synonymous variation suggests that the two lineages diverged as a result of positive natural selection which may have arisen as a result of their distinct transmission cycles (Ebel et al. 2001). However, the underlying mechanisms facilitating this virus’ adaptation to two unique transmission cycles remain unknown.
Powassan virus, as with all RNA viruses, is thought to exist in nature as a diverse population of competing mutants which vary in their degree of nucleotide divergence from a master sequence (Domingo et al. 1998; Eigen et al. 1988). These assorted populations are referred to as quasispecies and arise from the error-prone replication associated with virally encoded RNA-dependent RNA-polymerases (RdRp) which lack 3’ to 5’ exonucleolytic proofreading activity (Holland et al. 1982). In addition, RNA viruses typically have short generation times and large populations, both of which contribute to their observed quasispecies structure (Domingo and Holland 1997; Duffy et al. 2008). It is thought that the genetic plasticity of RNA viruses is advantageous in the presence of novel or changing environments. For example, the quasispecies structure has been identified as an important determinant in viral evasion of the host immune response and the development of resistance to antiviral drugs and therapies (Essajee et al. 2000; Farci and Purcell 2000; Farci et al. 2002). We recently suggested that the high genetic diversity typical of mosquito-associated WNV populations may present a more complex target to the primary invertebrate antiviral pathway, RNA interference, promoting the persistence of virus within these hosts (Brackney et al. 2009). Therefore, the mutational diversity of RNA viruses facilitates their persistence at the cellular, organismal and population levels.
RNA viruses have the potential for rapid evolution and high levels of genetic diversity due to high mutational diversity. However, these features are not static and can differ significantly between viral families. In fact, significant differences in the level of genetic diversity and the rate of evolution have been observed between single-host viruses and the two-host arthropod-borne viruses (arboviruses) (Duffy et al. 2008; Jenkins et al. 2002; Weaver et al. 1992). For instance, dengue virus (DENV) has mutation rates ranging from 5.11 × 10−4 to 9.01 × 10−4 substitutions per site per year, whereas influenza A virus and HIV-1 have rates of 1.62 × 10−3 and 1.95 × 10−3, respectively (Gojobori et al. 1990; Gorman et al. 1990; Jenkins et al. 2002; Twiddy et al. 2003). This is typical among arboviruses and may be the result of having to adapt to and persist in two hosts as divergent as arthropods and vertebrates (Weaver 2006; Weaver et al. 1992). This alternating replication scheme may generate negative fitness trade-offs in which viral traits favored in one host are purified in another (Holmes 2003). Despite the bottlenecking effect of a two host transmission cycle, arboviruses continually adapt to new ecological niches as demonstrated by the diversification of multiple lineages of eastern equine encephalitis virus (EEEV) and POWV and the ability of each lineage to adapt to a unique transmission cycle (Brault et al. 1999; Ebel et al. 2001). Similarly, the ongoing adaptation of West Nile virus (WNV) to North American Culex pipiens mosquitoes demonstrates the evolutionary potential of arboviruses (Davis et al. 2005; Moudy et al. 2007). To date, only one study has examined the interhost variation of a tick-borne virus population, however, this study did not investigate intrahost variation or the relevance of quasispecies to naturally occurring tick-borne virus populations (Casati et al. 2006). Therefore, the hypothesis that POWV is maintained in nature as a quasispecies was evaluated. Specifically, lineage II POWV (i.e. DTV) was sampled from naturally infected Ix. scapularis ticks and intrahost genetic diversity was quantified within each host. We determined whether positive or purifying selection is occurring in the invertebrate host and compared our findings to the previously published data of the mosquito-borne flavivirus, WNV (Jerzak et al. 2005).
Materials and Methods
Specimen Collection
Three sites in Hayward and Spooner, Wisconsin were chosen for specimen collection based on the presence of POWV positive ticks previously reported from these locales (Ebel et al. 2000; Ebel et al. 1999). Sites A and B are located in Hayward and are ~1.5 miles apart. The third site (C) is located in Spooner about 30 miles southwest from sites A and B. Adult Ix. scapularis ticks were collected by dragging a 1 m2 flannel cloth over emergent vegetation in October of 2007 and May 2008. Live ticks were transported back to the laboratory and sorted by site, sex and date.
Powassan Virus Detection
POWV infections were detected according to methods described previously (Brackney et al. 2008; Ebel et al. 2000). Briefly, Ix. scapularis tick cuticles were compromised with a sterilized glass pestle in 50 µl of phosphate buffered saline (PBS) with 20% fetal calf serum (FCS) and RNA extracted from pools of two to 10 ticks. The presence of POWV RNA was determined by reverse transcription polymerase chain reaction (RT-PCR) as described previously (Brackney et al. 2008). The forward (POWV 1274 F) and reverse primers (POWV 3180 R) were designed to amplify a ~2 kilobase (kb) region spanning the E/ NS1 junction (Table 1). Subsequently, RNA was extracted from individual ticks from positive pools and screened by RT-PCR as described above.
Table 1.
List of Primers.
| Primer Name | 5’- Sequence - 3’ |
|---|---|
| POWV 1274 F | GTGCCAAGTTTGAATGCGAGGAAG |
| POWV 3180 R | GAACGGGGCCCAGCGAGAGTGAC |
| POWV 8652 F | AATGGCCATGACAGACACAACAGC |
| POWV 9509 R | CCTTCCATCATGCGGATTAGTTGG |
| POWV 8783 F | GTGACTGGCTATTTGAGCACCTT |
| POWV 8842 R | TGGATCTAACCTTCGCTATGAATTC |
| POWV NS5 PROBE | 6FAM-CGAGCCAGGGTGA-MGBNFQ |
| POWV 1622 Fa | ATCAGGACTGGAACAGTGTGG |
| POWV 2427 Fa | ATGTGCAGTTGATCCAGAGAGG |
| POWV 2678 Ra | ATCTGTCTTGTCTACCACGATGG |
| M13 Fa | GTAAAACGACGGCCAG |
| M13 Ra | CAGGAAACAGCTATGAC |
sequencing primers
Quantification of Viral Genomic Equivalents
Quantification of DTV genomic equivalents was performed using Quantitative-RT-PCR (Q-RT-PCR). Briefly, a ~1 kb fragment from the POWV NS5 gene was amplified using the POWV 8652 forward and POWV 9509 reverse primers (Table 1). The amplicon was cloned into the multiple cloning site downstream of the T7 promoter in the pCR4-TOPO vector (Invitrogen, Carlsbad, CA). The recombinant vector was linearized with Pst I and purified. RNA of the insert was produced by in vitro transcription using the T7 Megascript kit according the manufacturer’s instructions (Ambion, Austin, TX). RNA was quantified, diluted and used as a standard control for the Q-RT-PCR. Viral RNA copy numbers were determined for each tick using the TaqMan ® One-Step RT-PCR Master Mix Reagent (Applied Biosystems, Foster City, CA) with the POWV NS5 probe and the POWV 8783 F and POWV 8842 R primers on the ABI Prism 7000 Sequence Detection System (Applied Biosystems) (Table 1) (Ebel and Kramer 2004).
Determination of Powassan Virus Genetic Diversity
Intrahost genetic diversity was determined according to methods described elsewhere (Jerzak et al. 2005). For these studies, five micoliters of total RNA were reverse transcribed using the High-Fidelity Accuscript Reverse transcription (RT) kit (Stratagene, Cedar Creek, TX) according to manufacturer’s specifications. Subsequently, the cDNA samples were PCR amplified with the high fidelity Pfu Ultra polymerase (Stratagene). The PCR parameters were 94°C for 10 min. followed by 40 cycles of 94°C for 30 sec., 56°C for 30 sec. and 72°C for 2 min. after which a 10 min. final extension at 72°C was applied. The primers used for both the RT and PCR reactions were POWV 1274 forward and POWV 3180 reverse. Amplicons were gel purified and cloned into the pCR Script Amp SK(+) vector (Stratagene). Clones were sequenced at the University of New Mexico’s School of Medicine DNA Services laboratory using an ABI 3130XL automated sequencer (Applied Biosystems). The sequencing primers were chosen so as to provide two-fold coverage of the amplicons and are listed in Table 1.
Sequence Analysis
Seventeen to twenty-one clones from each individual tick were sequenced. Sequences were aligned and analyzed for genetic diversity using DNAStar’s SeqMan module (DNASTAR Inc., Madison, WI). Twofold redundancy throughout each clone was required for sequence data to be considered complete with the exception of the first 350 nt. which only had single coverage. Signals from this region were consistently very strong and only in the case of mutations within this region were the clones (two clones in total) re-sequenced. Consensus sequences for each sample were determined and each clone was compared to the consensus. The percentage of nucleotide mutations (total number of mutations divided by the total number of sequenced nucleotides) and the percentage of mutant clones were used as an indicator of genetic diversity.
Analysis of Divergence and Selection
Analyses of the intrahost and interhost POWV populations were conducted as previously described (Jerzak et al. 2005). Briefly, nucleotide alignments of the 17–21 clones for each of the 10 POWV specimens (intrahost) or the consensus sequences from all 10 specimens (interhost) were used to determine the mean pairwise nucleotide divergence (Π) using DnaSP (Librado and Rozas 2009). Additionally, pN, the proportion of mutations in each alignment that were non-synonymous, and the mean number of synonymous and non-synonymous sites per sequence were determined in DnaSP using the Nei-Gojobori method. Using these values the ratio of non-synonymous to synonymous mutations per site (dN/dS) were computed in Microsoft Excel. The dN/dS ratios for alignments with only non-synonymous mutations were defined as 1.000.
The values determined within and between the POWV populations were compared to those previously published for WNV (Jerzak et al. 2005). Values for WNV intrahost variation were taken directly from the manuscript; however, the values for interhost variation were determined from the unpublished consensus sequences from all 10 mosquito specimens as described above.
Phylogenetic Analysis
All ten POWV consensus sequences and each unique haplotype within each population were subjected to a Neighbor-Joining analysis using the Maximum Composite Likelihood model for nucleotide substitutions in MEGA4. An un-rooted 50% majority rule consensus tree of 1000 bootstrap replicates was subsequently rendered.
Statistical Analyses
Statistical analyses on the proportion of mutations that resulted in amino acid substitutions, pN, in the intrahost and interhost populations were performed with a two-tailed Fisher’s Exact test.
Results
POWV-infected samples were taken from Ix. scapularis, the primary vector of the enzootic transmission cycle of DTV. POWV-positive samples were initially identified by RT-PCR and subsequently confirmed by viral isolation (Brackney et al. 2008). Viral isolate identifiers are as follows; first position is either (F) female or (M) male followed by the site (A, B, or C) and finally the collection date and tick number (Table 2). Examination of POWV RNA copy numbers demonstrated the presence of high RNA copy numbers (range 5.85 to 7.19 log10 RNA copies per 10 µl tissue suspension) in all of the specimens analyzed (8 of 8) (Table 2). RNA copy numbers were not determined for two of the samples, FA512 #40 and MA512 #40, because the RNA had been exhausted during the cloning process.
Table 2.
Tick specimens included in this study: source, nucleotide diversity, intrahost variation and infection status.
| Percentage with Mutations |
|||||||
|---|---|---|---|---|---|---|---|
| Specimen | Nucleotides | Clones | Haplotypes | Π | pN | dN/dS | RNA Copies |
| DTV FB1015 #5 | 0.003a | 5 | 2 | ND | ND | ND | 7.17e5 |
| DTV MB1015 #38 | 0.005 | 10 | 3 | 0.00011 | 1.000 | 1.000 | 1.05e6 |
| DTV FB1017 #177 | 0.003 | 9.5 | 2 | 0.0001 | 1.000 | 1.000 | 1.33e6 |
| DTV MA1017 #254 | 0.003 | 4.7 | 2 | 0.00005 | 1.000 | 1.000 | 1.93e6 |
| DTV FA512 #40 | 0.006 | 12 | 3 | 0.00016 | 1.000 | 1.000 | ND |
| DTV FB513 #1 | 0.003 | 5 | 2 | 0.00005 | 1.000 | 1.000 | 1.57e7 |
| DTV MB512 #27 | 0.003 | 6 | 2 | 0.00006 | 1.000 | 1.000 | 2.10e6 |
| DTV MA512 #40 | 0.008a | 20 | 4 | 0.00011 | 1.000 | 1.000 | ND |
| DTV MB512 #73 | 0.003 | 5.3 | 2 | 0.00006 | 1.000 | 1.000 | 9.49e5 |
| DTV MC514 #32 | 0.006 | 10.5 | 3 | 0.00011 | 0.500 | 0.302 | 1.20e6 |
| Mean | 0.0043 | 8.8 | 2.1 | 0.00009 | 0.944 | 0.922 | 3.12e6 |
specimens included one insertion or deletion mutant which were used in determining the nucleotide and clone diversities and were included in the total haplotype count, but were not used to calculate Π, pN and dN/dS ratios.
Analysis of the intrahost genetic diversity within individual ticks revealed that POWV is maintained in adult Ix. scapularis as a relatively homogenous population. Overall, the percentage of individual clones within each population differing from the consensus sequence ranged from 4.7% to 20% with an average of 8.8% (Table 2). Similarly, the percentage of nucleotides with mutations was extremely low. Overall, there were 15 mutations in 360,367 nucleotides sequenced (0.0043%) with a range of 0.003% to 0.008% (Table 2). The majority of the mutations were point mutations with one clone containing a 143 nucleotide insertion and one with a single nucleotide deletion. Interestingly, 94% of the nucleotide substitutions resulted in amino acid changes (Table 2). Previously published results from a control population derived from the WNV infectious cDNA clone was used as a proxy to estimate the rate of misincorporations introduced by our experimental approach (Jerzak et al. 2005). While using the same techniques and reagents, the previous study found a misincorporation rate of 0.004% with 7.7% of the clones differing from the consensus sequence. Furthermore, all of the identified mutations in the WNV control resulted in amino acid changes. These results are strikingly similar to the results found in our POWV-infected tick samples.
Patterns of genetic divergence and non-synonymous variation were examined in intra-and interhost POWV populations. The mean genetic distance (Π) was extremely low in all of the intrahost alignments (0.00009) and was 30-fold higher in the interhost alignments (0.0027) (Table 3a and 3b). Analysis of the proportion of mutations that resulted in amino acid substitutions, pN, revealed that our intrahost populations had surprisingly higher values (0.944) than the expected ~70% if mutations occurred at random and most likely represent experimentally introduced errors (Table 3a). These proportions are significantly higher than those found in the interhost populations (0.238) (p = 0.0002) (Table 3b). Similarly, dN/dS ratios were approaching 1.000 in the intrahost populations (0.922), but were 10-fold lower in the interhost populations (0.092) (Table 3a and Table 3b).
Table 3.
Intra- and Interhost Comparisons with West Nile virus.
| A | Intrahost (Virus: Vector) |
Percentage with Mutations |
||||
|---|---|---|---|---|---|---|
| Nucleotides | Clones | Π | pN | dN/dS | ||
| POWV and Ix. scapularis |
0.0043 | 8.8 | 0.00009 | 0.944 | 0.922 | |
| WNV and Cx. Pipiens (Jerzak et. al. 2005) |
0.0205 | 23.5 | 0.00034 | 0.419 | 0.334 | |
| B | Interhost (Virus: Vector) |
Percentage with Mutations |
||||
|---|---|---|---|---|---|---|
| Nucleotides | Clones | Π | pN | dN/dS | ||
| POWV and Ix. scapularis |
0.113 | NA | 0.0027 | 0.238 | 0.092 | |
| WNV and Cx. Pipiens (Adapted from Jerzak et. al. 2005) |
0.098 | NA | 0.00226 | 0.191 | 0.072 | |
Comparison of the genetic diversity of POWV in Ix. scapularis to those previously reported for WNV populations isolated from the North American vector, Culex pipiens, reveals strikingly different profiles of intrahost variability between these two flaviviruses (Jerzak et al. 2005). Estimates of WNV intrahost genetic diversity derived from mosquitoes determined that the percentage of substituted bases (0.0205%) and mutant clones (23.5%) as well as the mean pairwise distance (Π = 0.00034) were all considerably higher than in our POWV populations (Table 3a). Conversely, when comparing the proportion of mutations resulting in amino acid substitutions (pN) and the dN/dS ratios, the values determined from the POWV populations were significantly higher (pN; p = 0.027) (Table 3a). However, upon inspection of the interhost populations, by comparison the two results were very similar with a slight increase in all measurable parameters in the POWV populations (Table 3b).
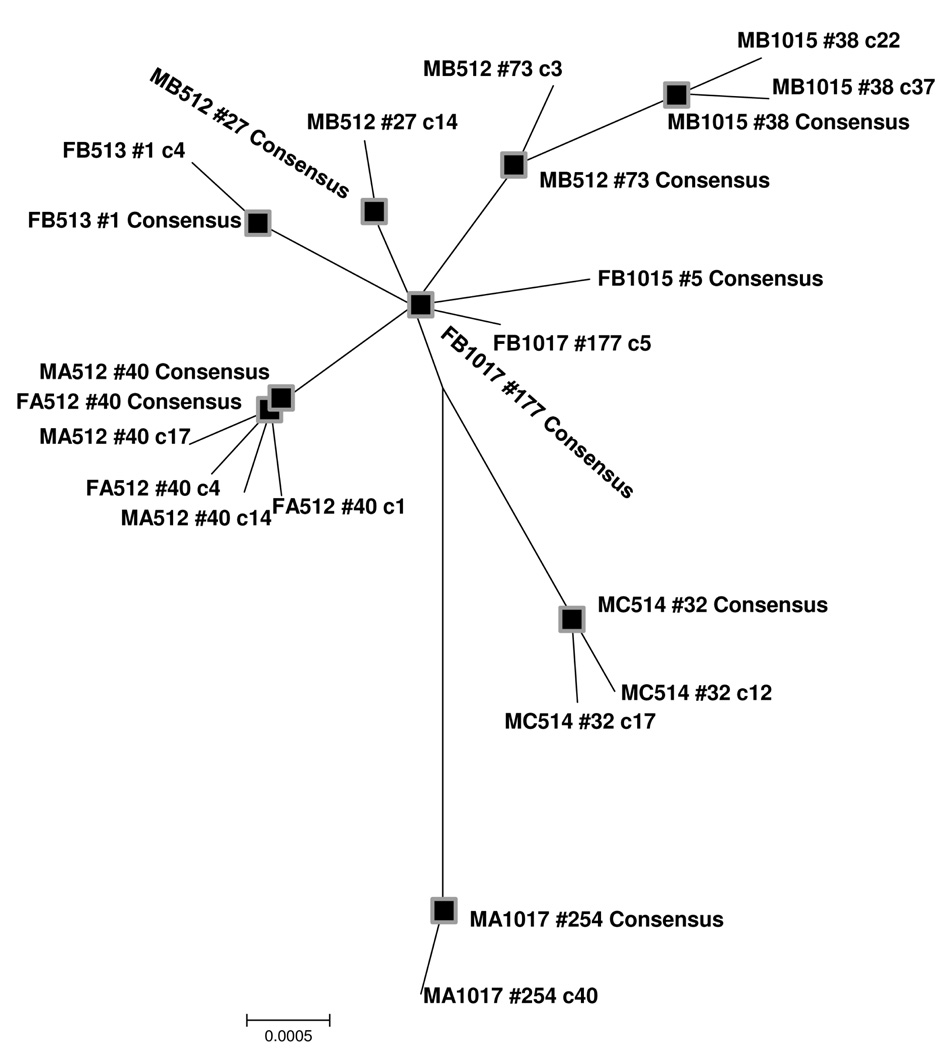
All ten POWV isolate consensus sequences and each individual haplotype were subjected to a Neighbor-Joining analysis. It was determined that genetic diversity is not shared among ticks (Figure 1). Furthermore, it was tentatively observed that viral isolates seemed to cluster according to geographic distribution.
Figure 1. Phylogenetic analysis of all consensus and haplotype sequences.
Neighbor-joining analysis of consensus sequences (black squares) and all unique haplotypes detected in POWV populations infecting adult Ixodes scapularis. Presented is the 50% majority rule consensus tree of 1000 bootstrap replicates. The FB1015 #5 insertion haplotype and the MA512 #40 deletion haplotype were not included in this analysis.
Discussion
The presence of intrahost genetic diversity has been described for numerous RNA viruses in the laboratory and in nature (Alves et al. 2002; Bonneau et al. 2001; Domingo et al. 1992; Farci et al. 2002; Plyusnin et al. 1996). Specifically, multiple studies have found that mosquito-borne flaviviruses, such DENV and WNV, exist as true quasispecies (Beasley et al. 2003; Davis et al. 2003; Jerzak et al. 2005; Lin et al. 2004; Wang et al. 2002). However, drawing direct parallels to tick-borne flavivirus populations may be tenuous provided the evolutionary divergence between the invertebrate vectors and the marked differences in their life cycles. To date, few studies have examined the genetic variation associated with tick-borne flaviviruses and no studies have evaluated the quasispecies structure in the laboratory or nature. Therefore, we examined the intra- and interhost variability of the tick-borne flavivirus, POWV, in order to determine whether POWV exists as a quasispecies in its enzootic transmission cycle.
To ensure that any observable differences in intrahost genetic diversity were not biased due to variation in viral loads we quantified viral RNA copies in eight of ten samples. The RNA copy number values were very similar among all samples tested (Table 2) and were about 10-fold higher than those previously reported for POWV-infected adult Ix. scapularis (Costero and Grayson 1996). However, this previous study ascertained viral titers (viral titers ≥ 5.2 log10) from experimentally infected ticks by the suckling mouse ID50 method. While it may be difficult to directly compare these two approaches, evidence suggests that flaviviruses typically have 100–1000 times more RNA copies than infectious virions (Takhampunya et al. 2006). Therefore, these values seem to be concordant with previously published literature.
Approximately 0.0043% of bases sequenced and 8.8% of clones differed from the consensus (Table 2). These estimates of nucleotide diversity are similar to the misincorporation rates previously reported for a control WNV infectious cDNA clone used to estimate the error rate of the experimental approach (Jerzak et al. 2005). Furthermore, the proportion of non-synonymous to synonymous mutations between the intrahost populations (pN = 0.944) and the experimental control (pN = 1) were nearly identical. While differences in the time and place of these experiments as well as differences in the sequencing platforms and human variability in the analysis of the sequence data may make a direct comparison problematic, extensive efforts were made to limit these effects. Specifically, the polymerase enzyme (Pfu Ultra), the cloning vector (pCR Script Amp SK(+)), and bacterial cells (XL10-Gold Ultracompetent cells) were identical in both studies. Therefore, we used WNV-derived estimates for the rate of misincorporations introduced by our experimental methods in this study. Comparing WNV-derived estimates of experimental error with the genetic diversity detected in our field-derived POWV populations allows us to conclude that POWV populations within adult Ix. scapularis have undetectable levels of genetic diversity.
The low genetic diversity observed in POWV-infected ticks is strikingly different from WNV in Cx. pipiens mosquitoes (Table 3a) (Jerzak et al. 2005). Several factors may contribute to these differences. The WNV genetic diversity study examined virus populations isolated from adult mosquitoes during the peak of the transmission season. Under this scenario, WNV is primarily transmitted horizontally between mosquitoes and birds with only minor contributions from vertical and consequently transtadial (between life-stages) or co-feeding modes of transmission (Anderson et al. 2008; McGee et al. 2007). Therefore, the likelihood that bottlenecking events influencing the observed WNV genetic diversity arose from vertical or transtadial transmission would be very rare. Conversely, the POWV populations isolated from adult Ix. scapularis may have been subject to bottlenecks during horizontal/ transtadial or vertical/ transtadial transmission. Previous studies have demonstrated that POWV can be transmitted both transovarially and transtadially (Costero and Grayson 1996). Considering, our samples were collected from unattached questing adult ticks it is possible that the POWV populations examined arose from vertical transmission. Furthermore, it is reasonable to assume that POWV populations isolated from adult deer ticks have been subject to at least one round of transtadial transmission since they most likely acquired infection as immatures. Interestingly, one study has demonstrated that POWV titers dramatically increase during the life-stage in which the infectious bloodmeal is acquired and level off in subsequent life-stages (Costero and Grayson 1996). These findings seem to be independent of the life-stage in which the infectious bloodmeal was consumed. This could be explained with two equally plausible scenarios. POWV replication may proceed logarithmically in the exposed life-stage until reaching a threshold titer after which viral infections become ‘dormant’ and remain stable in subsequent life-stages. Alternatively, virus replication may be cyclical with regards to transtadial transmission with threshold titers being achieved in the exposed life-stage, titers dropping during molting, and replication being reinitiated in the subsequent life-stage, potentially introducing bottlenecks during molting. To date, it is unknown whether transovarial and/ or transtadial transmission serve as a natural genetic bottleneck and further studies are warranted. Virological factors may also contribute to the observed difference. While both POWV and WNV are flaviviruses, they are genetically quite divergent. It is possible that this divergence may have resulted in differences in the basic fidelity of their RdRps as even differences in the RdRp fidelity between closely related DENV serotypes have been reported (Twiddy et al. 2003).
The combined intra- and interhost genetic variability data permitted examination of the selective pressures acting on POWV within and between hosts. The mean pairwise distance for within host alignments was 30-fold lower than was observed in the interhost alignments (Table 3a and Table 3b). Therefore, POWV sequences are more diverse between hosts than within an individual host. These results are not surprising and are consistent with other examinations of within and between host genetic variations (Jerzak et al. 2005). The pN and dN/dS ratios among the intrahost alignments were close to 1.000 thus suggesting neutral selection; however, with nucleotide diversity levels below background, these conclusions are tenuous at best. On the other hand, the pN and dN/dS ratios among the interhost alignments were low suggesting strong purifying selection. Values for pN and dN/dS ratios are slightly higher than those observed in WNV populations among mosquito hosts and may suggest that constraints on POWV sequence variation may be looser in the POWV transmission cycle than in the WNV transmission cycle (Table 3b). Interestingly, our values are much more similar to those in WNV than observed in the closely related western European TBEV subtype (W-TBEV) collected from Ix. ricinus (Casati et al. 2006). Numerous factors may account for this discrepancy in interhost variation. The authors used Taq polymerase during the PCR step which has a significantly higher error rate than the proofreading Pfu polymerase used in these studies (Malet et al. 2003). This may account for the almost 10-fold increase in estimates of nucleotide diversity. In addition, different regions within the genome were analyzed. It may be that the fitness costs are greater in the E/ NS1 region analyzed here than in the 5’-NCR/ capsid region analyzed by Casati et. al. and thereby reducing the observed variation. Finally, differences in the viruses and vectors analyzed between the studies may attribute to the lack of concordance. Nevertheless, these findings support the previous observations that arbovirus sequence variation is tightly constrained.
The phylogenetic analysis of the consensus/ unique haplotype alignment revealed that POWV haplotypes are not shared among ticks (Figure 1). Whether these findings are a result of our inability to detect genetic variants within hosts or a result of tight population bottlenecks during some phase of the virus/ tick lifecycle remains to be determined. The Neighbor-Joining analysis did reveal some clustering of viral isolates according to geography. Two of the three samples isolated from site A are closely grouped and all samples from site B are grouped and both are more closely related to one another (~1.5 miles apart) than either is to the sample from site C (~30 miles away). In fact, the two closely related isolates from site A, MA512 #40 and FA512 #40, have identical consensus sequences (Figure 1). It is possible that these two isolates were obtained from ticks that had shared a common vertebrate host or the ticks represent transovarially infected siblings. One isolate from site A did seem to group more closely with the sole isolate from site C than to the other site A specimens. This may be due to transportation of a POWV-infected tick from site C to site A while attached to its mammalian host(s). This scenario is conceivable due to the broad host feeding preference of nymphal and adult deer ticks and the lack of large physical barriers impeding gene flow between locales.
This is the first report examining the intrahost genetic variability of a tick-borne flavivirus either in the laboratory or nature. While evidence for strong purifying selection of POWV was observed between hosts, there seems to be very limited intrahost genetic variation. Without greater sequencing depth, such as those afforded by massively parallel sequencing, we were unable to determine whether POWV exists as a quasispecies in adult deer ticks. Overall, these findings highlight the need for further investigation into the quasispecies structure of other tick-borne viruses and the examination of the potential bottlenecking effects of vertical and transtadial transmission on arbovirus populations.
Acknowledgements
The authors would like to thank Kendra Pesko for technical assistance and her thoughtful comments on the manuscript. Funding for this project was provided in part by a grant from that National Institute of Allergy and Infectious Disease, National Institutes of Health, under grant number AI067380, the Ruth L. Kirschstein National Research Service Award under grant T32 AI007538-11, and by the University of New Mexico School of Medicine, Department of Pathology. Ivy K. Brown was supported by the University of New Mexico Initiative to Maximize Student Diversity, which is funded by the National Institute of General Medical Sciences, NIH, under grant GM060201. Sanger sequencing was performed at the UNM DNA research facility.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alves K, Canzian M, Delwart EL. HIV type 1 envelope quasispecies in the thymus and lymph nodes of AIDS patients. Aids Research and Human Retroviruses. 2002;18:161–165. doi: 10.1089/08892220252779700. [DOI] [PubMed] [Google Scholar]
- Anderson JF, Main AJ, Delroux K, Fikrig E. Extrinsic incubation periods for horizontal and vertical transmission of West Nile virus by Culex pipiens pipiens (Diptera : Culicidae) Journal of Medical Entomology. 2008;45:445–451. doi: 10.1603/0022-2585(2008)45[445:eipfha]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Beasley DWC, Davis CT, Guzman H, Vanlandingham DL, da Rosa A, Parsons RE, Higgs S, Tesh RB, Barrett ADT. Limited evolution of West Nile virus has occurred during its southwesterly spread in the United States. Virology. 2003;309:190–195. doi: 10.1016/s0042-6822(03)00150-8. [DOI] [PubMed] [Google Scholar]
- Beasley DWC, Suderman MT, Holbrook MR, Barrett ADT. Nucleotide sequencing and serological evidence that the recently recognized deer tick virus is a genotype of Powassan virus. Virus Res. 2001;79:81–89. doi: 10.1016/s0168-1702(01)00330-6. [DOI] [PubMed] [Google Scholar]
- Bonneau KR, Mullens BA, MacLachlan NJ. Occurrence of genetic drift and founder effect during quasispecies evolution of the VP2 and NS3/NS3A genes of bluetongue virus upon passage between sheep, cattle, and Culicoides sonorensis. Journal of Virology. 2001;75:8298–8305. doi: 10.1128/JVI.75.17.8298-8305.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney DE, Beane JE, Ebel GD. RNAi Targeting of West Nile Virus in Mosquito Midguts Promotes Virus Diversification. PLoS Pathog. 2009;5:e1000502. doi: 10.1371/journal.ppat.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackney DE, Nofchissey RA, Fitzpatrick KA, Brown IK, Ebel GD. Stable Prevalence of Powassan Virus in Ixodes scapularis in a Northern Wisconsin Focus. American Journal of Tropical Medicine and Hygiene. 2008;79:971–973. [PMC free article] [PubMed] [Google Scholar]
- Brault AC, Powers AM, Chavez CLV, Lopez RN, Cachon MF, Gutierrez LFL, Kang WL, Tesh RB, Shope RE, Weaver SC. Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. American Journal of Tropical Medicine and Hygiene. 1999;61:579–586. doi: 10.4269/ajtmh.1999.61.579. [DOI] [PubMed] [Google Scholar]
- Casati S, Gern L, Piffaretti JC. Diversity of the population of Tick-borne encephalitis virus infecting Ixodes ricinus ticks in an endemic area of central Switzerland (Canton Bern) Journal of General Virology. 2006;87:2235–2241. doi: 10.1099/vir.0.81783-0. [DOI] [PubMed] [Google Scholar]
- Costero A, Grayson MA. Experimental transmission of Powassan virus (Flaviviridae) by Ixodes scapularis ticks (Acari:Ixodidae) American Journal of Tropical Medicine and Hygiene. 1996;55:536–546. doi: 10.4269/ajtmh.1996.55.536. [DOI] [PubMed] [Google Scholar]
- Davis CT, Beasley DWC, Guzman H, Raj P, D'Anton M, Novak RJ, Unnasch TR, Tesh RB, Barrett ADT. Genetic variation among temporally and geographically distinct West Nile virus isolates, United States, 2001, 2002. Emerging Infectious Diseases. 2003;9:1423–1429. doi: 10.3201/eid0911.030301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CT, Ebel GD, Lanciotti RS, Brault AC, Guzman H, Siirin M, Lambert A, Parsons RE, Beasley DWC, Novak RJ, Elizondo-Quiroga D, Green EN, Young DS, Stark LM, Drebot MA, Artsob H, Tesh RB, Kramer LD, Barrett ADT. Phylogenetic analysis of North American West Nile virus isolates, 2001–2004: Evidence for the emergence of a dominant genotype. Virology. 2005;342:252–265. doi: 10.1016/j.virol.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Domingo E, Baranowski E, Ruiz-Jarabo CM, Martin-Hernandez AM, Saiz JC, Escarmis C. Quasispecies structure and persistence of RNA viruses. Emerg Infect Dis. 1998;4:521–527. doi: 10.3201/eid0404.980402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Escarmis C, Martinez MA, Martinezsalas E, Mateu MG. Foot and Mouth Disease virus Populations are Quasispecies. Current Topics in Microbiology and Immunology. 1992;176:33–47. doi: 10.1007/978-3-642-77011-1_3. [DOI] [PubMed] [Google Scholar]
- Domingo E, Holland JJ. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- Duffy S, Shackelton LA, Holmes EC. Rates of evolutionary change in viruses: patterns and determinants. Nat Rev Genet. 2008;9:267–276. doi: 10.1038/nrg2323. [DOI] [PubMed] [Google Scholar]
- Ebel G, Campbell E, Goethert H, Spielman A, Telford SR., 3rd Enzootic transmission of deer tick virus in New England and Wisconsin sites. Am J Trop Med Hyg. 2000;63:36–42. doi: 10.4269/ajtmh.2000.63.36. [DOI] [PubMed] [Google Scholar]
- Ebel GD, Foppa I, Spielman A, Telford SR. A focus of deer tick virus transmission in the northcentral United States. Emerging Infectious Diseases. 1999;5:570–574. doi: 10.3201/eid0504.990423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel GD, Kramer LD. Short report: Duration of tick attachment required for transmission of Powassan virus by deer ticks. American Journal of Tropical Medicine and Hygiene. 2004;71:268–271. [PubMed] [Google Scholar]
- Ebel GD, Spielman A, Telford SR., III Phylogeny of North American Powassan virus. J Gen Virol. 2001;82:1657–1665. doi: 10.1099/0022-1317-82-7-1657. [DOI] [PubMed] [Google Scholar]
- Eigen M, McCaskill J, Schuster P. Molecular quasispecies. J Phys Chem. 1988;92:6881–6891. [Google Scholar]
- Essajee SM, Pollack H, Rochford G, Oransky I, Krasinski K, Borkowsky W. Early changes in quasispecies repertoire in HIV-infected infants: Correlation with disease progression. Aids Research and Human Retroviruses. 2000;16:1949–1957. doi: 10.1089/088922200750054675. [DOI] [PubMed] [Google Scholar]
- Farci P, Purcell RH. Clinical significance of hepatitis C virus genotypes and quasispecies. Semin Liver Dis. 2000;20:103–126. [PubMed] [Google Scholar]
- Farci P, Strazzera R, Alter HJ, Farci S, Degioannis D, Coiana A, Peddis G, Usai F, Serra G, Chessa L, Diaz G, Balestrieri A, Purcell RH. Early changes in hepatitis C viral quasispecies during interferon therapy predict the therapeutic outcome. Proc Nat Acad Sci USA. 2002;99:3081–3086. doi: 10.1073/pnas.052712599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojobori T, Moriyama EN, Kimura M. Molecular clock of viral evolution, and the neutral theory. Proc Nat Acad Sci USA. 1990;87:10015–10018. doi: 10.1073/pnas.87.24.10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman OT, Bean WJ, Kawaoka Y, Webster RG. Evolution of the nucleoprotein gene of influenza-a virus. Journal of Virology. 1990;64:1487–1497. doi: 10.1128/jvi.64.4.1487-1497.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gritsun TS, Nuttal PA, Gould Ernest A. Tick-borne Flaviviruses. In: Monath TJCaTP., editor. The Flaviviruses: Detection, Diagnosis and Vaccine Development. Vol. 61. San Diego: Elsevier Academic Press; 2003. pp. 317–371. [Google Scholar]
- Hinten SR, Beckett GA, Gensheimer KF, Pritchard E, Courtney TM, Sears SD, Woytowicz JM, Preston DG, Smith RP, Rand PW, Lacombe EH, Holman MS, Lubelczyk CB, Kelso PT, Beelen AP, Stobierski MG, Sotir MJ, Wong S, Ebel G, Kosoy O, Piesman J, Campbell GL, Marfin AA. Increased Recognition of Powassan Encephalitis in the United States, 1999–2005. Vector-Borne and Zoonotic Diseases. 2008;8:733–740. doi: 10.1089/vbz.2008.0022. [DOI] [PubMed] [Google Scholar]
- Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, Vandepol S. Rapid Evolution of RNA Genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Patterns of Intra- and Interhost Nonsynonymous Variation Reveal Strong Purifying Selection in Dengue Virus. J. Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Rambaut A, Pybus OG, Holmes EC. Rates of molecular evolution in RNA viruses: A quantitative phylogenetic analysis. J Mol Evol. 2002;54:156–165. doi: 10.1007/s00239-001-0064-3. [DOI] [PubMed] [Google Scholar]
- Jerzak G, Bernard KA, Kramer LD, Ebel GD. Genetic variation in West Nile virus from naturally infected mosquitoes and birds suggests quasispecies structure and strong purifying selection. Journal of General Virology. 2005;86:2175–2183. doi: 10.1099/vir.0.81015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G, Artsob H, Karabatsos N, Tsuchiya KR, Chang GJJ. Genomic sequencing of deer tick virus and phylogeny of powassan-related viruses of North America. American Journal of Tropical Medicine and Hygiene. 2001;65:671–676. doi: 10.4269/ajtmh.2001.65.671. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lin SR, Hsieh SC, Yueh YY, Lin TH, Chao DY, Chen WJ, King CC, Wang WK. Study of sequence variation of dengue type 3 virus in naturally infected mosquitoes and human hosts: Implications for transmission and evolution. Journal of Virology. 2004;78:12717–12721. doi: 10.1128/JVI.78.22.12717-12721.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Main AJ, Carey AB, Downs WG. Powassan Virus in Ixodes cookei and Mustelidae in New England. Journal of Wildlife Diseases. 1979;15:585–591. doi: 10.7589/0090-3558-15.4.585. [DOI] [PubMed] [Google Scholar]
- Malet I, Belnard M, Agut H, Cahour A. From RNA to quasispecies: a DNA polymerase with proofreading activity is highly recommended for accurate assessment of viral diversity. J Virol Method. 2003;109:161–170. doi: 10.1016/s0166-0934(03)00067-3. [DOI] [PubMed] [Google Scholar]
- McGee CE, Schneider BS, Girard YA, Vanlandingham DL, Higgs S. Nonviremic transmission of West Nile virus: Evaluation of the effects of space, time, and mosquito species. American Journal of Tropical Medicine and Hygiene. 2007;76:424–430. [PubMed] [Google Scholar]
- McLean DM, Chernesky MA. Tick Vectors of Powassan Virus. Fed Proc. 1969;28:430. [Google Scholar]
- McLean DM, Larke RP. Powassan and Silverwater Viruses - Ecology of 2 Ontario Arboviruses. Can Med Assoc J. 1963;88:182. [PMC free article] [PubMed] [Google Scholar]
- McLean DMaWD. Powassan virus: Isolation of virus from a fatal case of encephalitis. Can Med Assoc J. 1959;80:708–711. [PMC free article] [PubMed] [Google Scholar]
- Moudy RM, Meola MA, Morin LLL, Ebel GD, Kramer LD. A newly emergent genotype of west Nile virus is transmitted earlier and more efficiently by Culex mosquitoes. Am J Trop Med Hyg. 2007;77:365–370. [PubMed] [Google Scholar]
- Plyusnin A, Cheng Y, Lehvaslaiho H, Vaheri A. Quasispecies in wild-type Tula hantavirus populations. Journal of Virology. 1996;70:9060–9063. doi: 10.1128/jvi.70.12.9060-9063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takhampunya R, Ubol S, Houng HS, Cameron CE, Padmanabhan R. Inhibition of dengue virus replication by mycophenolic acid and ribavirin. Journal of General Virology. 2006;87:1947–1952. doi: 10.1099/vir.0.81655-0. [DOI] [PubMed] [Google Scholar]
- Tavakoli NP, Wang H, Dupuis M, Hull R, Ebel GD, Gilmore EJ, Faust PL. Brief Report: Fatal Case of Deer Tick Virus Encephalitis. New England Journal of Medicine. 2009;360:2099–2107. doi: 10.1056/NEJMoa0806326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telford SR, Armstrong PM, Katavolos P, Foppa I, Garcia ASO, Wilson ML, Spielman A. A new tick-borne encephalitis-like virus infecting New England deer ticks, Ixodes dammini. Emerging Infectious Diseases. 1997;3:165–170. doi: 10.3201/eid0302.970209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiddy SS, Holmes EC, Rambaut A. Inferring the rate and time-scale of dengue virus evolution. Mol Biol Evol. 2003;20:122–129. doi: 10.1093/molbev/msg010. [DOI] [PubMed] [Google Scholar]
- Wang WK, Lin SR, Lee CM, King CC, Chang SC. Dengue type 3 virus in plasma is a population of closely related genomes: Quasispecies. Journal of Virology. 2002;76:4662–4665. doi: 10.1128/JVI.76.9.4662-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC. Evolutionary influences in arboviral disease. Quasispecies: Concept and Implications for Virology. 2006;Vol. 299:285–314. doi: 10.1007/3-540-26397-7_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver SC, Rico-Hesse R, Scott TW. Genetic diversity and slow rates of evolution in new world alphaviruses. Curr Top Microbiol Immunol. 1992;176:99–117. doi: 10.1007/978-3-642-77011-1_7. [DOI] [PubMed] [Google Scholar]