Abstract
Approximately 20% of patients receiving liver transplants for end-stage hepatitis C rapidly develop severe allograph fibrosis within the first 24 months after transplant. Hepatitis C virus (HCV) variants were studied in 56 genotype 1-infected subjects with end-stage hepatitis C disease at the time before and 12-month after liver transplant, and post-transplant outcome was followed with serial liver biopsies. In 15 cases, pre-transplant HCV genetic diversity was studied in detail in liver (n=15), serum (n=15), peripheral blood mononuclear cells (n=13) and perihepatic lymph nodes (n=10). Our results revealed that pre-transplant HCV genetic diversity predicted the histological outcome of recurrent hepatitis C disease after transplant. Mild disease recurrence after transplant was significantly associated with higher genetic diversity, and greater diversity changes between the pre- and post-transplant time points (p=0.004). Meanwhile, pre-transplant genetic differences between serum and liver were related to a higher likelihood of development of mild recurrent disease after transplant (p=0.039).
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped, positive-strand RNA virus that circulates in vivo as a population of rapidly mutating yet closely related variants known as viral quasispecies. In nature, HCV infects only human, and disease manifestations are highly variable, ranging from self-limiting acute infections (~20%) to progressive chronic hepatitis C (~80%). Despite high levels of persistent viraemia during the majority of chronic infections, only 20–30% of such cases progress to cirrhosis, and progression typically occurs over several decades. The long disease incubation period and highly variable natural history are intriguing and also major impediments to hepatitis C pathogenesis studies.
Liver transplantation, a life saving intervention for many patients with end-stage hepatitis C disease, provides an extremely valuable model for studying HCV biology and disease progression over relatively short time intervals (Gretch et al., 1995; Gretch et al., 1996). In this model, de novo infection of liver allografts by HCV is virtually universal, and most patients have high titers of HCV viraemia by the second post-transplant week. Severe hepatitis C disease rapidly recurs in about 20% of cases, leading to accelerated cirrhosis in liver allografts within 5 years (Berenguer et al., 2000; Terrault, 2005). In contrast, approximately 50% of HCV genotype-matched cases have completely asymptomatic post-transplant infections despite persistence of high titers of HCV viraemia and potent anti-rejection immunosuppressive therapy. Although evidence suggests that pre-transplant HCV viral load (Berenguer et al., 2000; Charlton et al., 1998), HCV genotype (Charlton et al., 1998; Feray et al., 1995; Gane et al., 1996; Sugo et al., 2003), number of rejection episodes (Sheiner et al., 1995; Sugo et al., 2003), and type and degree of immunosuppression (Berenguer et al., 1998) are all associated with severity of HCV recurrence, solid conclusions about the determinants of recurrence outcome have not been available. HCV genetic heterogeneity in pre-transplant serum has also been proposed as a possible predictor of the outcome of recurrent disease (Gretch et al., 1996; Hassoba et al., 1999; Sullivan et al., 1998).
Although chronic hepatitis C is associated with a myriad of extrahepatic disease syndromes (Galossi et al., 2007), HCV replication in extrahepatic tissues remains poorly understood. Viral RNA has been detected in peripheral blood mononuclear cells (PBMCs), perihepatic lymph nodes or central nervous system with diversified variants distribution from serum (Laskus et al., 1998; Navas et al., 1998; Pal et al., 2006; Radkowski et al., 2002; Roque Afonso et al., 1999; Roque-Afonso et al., 2005; Vargas et al., 2002). Also, HCV variants within the liver have been observed to differ from the variants in the circulation, suggesting the existence of independent viral compartments (De Mitri et al., 1998; Laskus et al., 1998; Laskus et al., 2000; Okuda et al., 1999). Given the above evidence, extrahepatic tissues have been suggested to function as important HCV reservoirs and explain for the high rate of HCV therapeutic resistance. In post-transplant setting, the primary source of allograft reinfection is likely to come from the variants in circulation of the transplant recipient, while the variants derived from extrahepatic reservoirs, especially PBMCs, may also contribute to the circulating variants and serve as the origin of reinfection (Dahari et al., 2005; Laskus et al., 2002).
In the present study, genetic heterogeneity of pre-transplant HCV variants were analyzed in detail and related to severity of post-transplant disease recurrence. Genetic tools were used to explore and compare HCV variants in serum, liver and extrahepatic tissues of human subjects with end-stage hepatitis C disease. We tested the hypothesis that HCV genetics at the time of liver transplantation is predictive of the early post-transplant disease course, based on extremely heavy sampling of infected allografts for disease activity.
METHODS
Patients and sample collection
The study group comprises 56 patients who underwent orthotopic liver transplantation at the University of Washington Medical Center as described previously (Pal et al., 2006). Informed consent to participate in the study was obtained from all patients in accordance with the institutional review board requirements. All patients had HCV-related end-stage liver disease at the time of liver transplantation, and all developed recurrent HCV infection as demonstrated by the persistence of post-transplant HCV viremia. HCV genotypes were assigned using the restriction fragment length polymorphism analysis (RFLP) of the 5′-UTR region (Davidson et al., 1995) and confirmed by probe hybridization of the Core/E1 region (Li et al., 2008b). Serum samples and purified peripheral blood mononuclear cells (PBMCs) were obtained immediately prior to liver transplantation and stored at −80°C. For 15 patients, liver and perihepatic lymph node specimens were collected at the time of liver transplantation, immediately snap frozen in optimal cutting temperature (OCT) compound (Sakura Finetek, Torrance, CA) and stored at −80°C until further analysis. Serum samples were further obtained from 51 patients at around 12-month after transplant to define the genetic kinetics of recurrent HCV variants. Protocol liver biopsies were performed serially after transplant to monitor the recurrence of hepatitis C disease in the allograft. A median of 6 liver biopsies per patient was performed in a follow-up period of a median of 33 months, and disease severity was evaluated by an experienced transplant pathologist according to the Batts-Ludwig system (Batts and Ludwig, 1995).
RNA extraction, RT-PCR and sequencing
RNA was extracted from sera with the QIAamp Viral RNA mini kit (QIAGEN Inc., Valencia, CA) or from tissues with the RNeasy Mini kit (QIAGEN) according to the manufacture’s instructions. cDNA synthesis was carried out with the Moloney Murine Leukemia virus reverse transcriptase (Invitrogen, Carlsbad, CA) as described previously (Li et al., 2008a). Amplification of a 165-nucleotide sequence encompassing the HVR1 region was performed by nested PCR using two pairs of primers originally described by Weiner et al. (Weiner et al., 1991). Heteroduplex mobility analysis was applied to compare the difference of genetic diversity of HCV variants among the specimens from the same individuals (described below). For the 15 patients with multiple compartments available, the amplified fragments were further purified and ligated into the TA cloning vector (Invitrogen, Carlsbad, CA) and transfected into Escherichia coli TOP-10 competent cells. 18–23 colonies per specimen were then selected for clonal frequency analysis of unique variants (Wilson et al., 1995), and clones representing unique migration patterns during the gel electrophoresis were picked up for nucleotide sequencing in both directions.
Heteroduplex mobility analysis of HVR1 variants
HVR1 variants of pre- and post-transplant serum samples or tissue specimens were evaluated by heteroduplex mobility analysis (HMA) using specific probes generated from the major variant of the pre-transplant serum of each subject (Gretch et al., 1996; Polyak et al., 1997; Wilson et al., 1995). Probe labeling and hybridization were performed as described previously (Sullivan et al., 1998). Purified probes were end-labeled with T4 polynucleotide kinase plus 32P ATP (Amershan, Arlington Heights, IL) and then hybridized to HVR1 fragments derived by PCR amplification from each specimen. Hybridization mixtures were separated by electrophoresis on 6% polyacrylamide MDE gel (Cambrex, Rockland, ME) that was later vacuum dried and exposed to X-ray film. Hybridization to the unlabelled probe itself served as a marker for identification of homoduplexes. Heteroduplex mobility ratio (HMR) was estimated as the ratio of the shift distance of heteroduplex bands from the top of the gel compared to that of homoduplexes.
Phylogenetic and statistical analyses
HVR sequences were aligned using the CLUSTAL W program (Thompson et al., 1994) included in the MacVector 9.0 (Symantec Corp., Cupentino, CA). Genetic distances were calculated by the PHYLIP version 3.66 using the Kimura’s two-parameter model and an empirical transition/transversion ratio of 2 (Felsenstein, 1993). Synonymous (dS) and nonsynonymous (dN) substitutions were estimated for the HVR1 sequences by the Nei and Gojobori method (Nei and Gojobori, 1986). Phylogenetic trees were constructed with the neighbor-joining algorithm based on the nucleotide sequences of HVR1 variants (Saitou and Nei, 1987).
In the study including the 56 subjects, the Pearson Chi-square test was used to evaluate the gender and genotype according to groups of disease recurrence, while the Mann-Whitney U test was used to compare the other clinical characteristics and genetic diversity between the groups with the mild and severe disease outcomes. The Mann-Whitney U test was also used in the tissue reservoir studies of the 15 subjects to analyze difference of the nucleotide substitutions and genetic distances between the groups with mild and severe recurrence. The Mantel’s test was used to determine the genetic differences of the HCV variants, in other words, if the variants detected from a given compartment were genetically closer to each other than to variants detected from a different compartment (Ducoulombier et al., 2004; Zehender et al., 2005). The Pearson Chi-square test was used to compare the frequencies of genetic differences between the groups with mild and severe recurrence.
RESULTS
Characteristics of the study population
All the 56 patients were infected with HCV genotype 1 before transplant, with mixed or multiple genotype HCV infections ruled out using a highly sensitive Core/E1-based probe hybridization assay as recently described (Li et al., 2008b). A median of 6 follow-up liver biopsies was carried out per patient to evaluate the histological recurrence of hepatitis C disease after transplant, and the follow-up period ranged from a median of 36 months in the patients with mild recurrent disease to a median of 21 months in the patients with severe recurrent disease. Forty-eight patients were classified in the mild recurrent disease group based on the fibrosis stage during the initial 12 months after transplant (fibrosis stage 0 or 1), while eight patients were classified into the severe recurrent disease group with either bridging fibrosis or cirrhosis during the initial 12 months after transplant (fibrosis stage 3 or 4), or fibrosis stage 2 combined with severe, evolving inflammation defined histologically as bridging necrosis. The clinical characteristics of the two groups with mild versus severe disease recurrence were shown in Table 1. No significant differences existed between the two groups with respect to donor or recipient age, gender, race, immunosuppressive use, or allograft rejection episodes.
Table 1.
Clinical characteristics of the study groups of subjects that underwent liver transplantation and developed mild or severe post-transplant disease.
| Characteristic | Mild (n = 48) | Severe (n = 8) |
|---|---|---|
| Liver allograft donor | ||
| Median age (range) | 26 (10–62) | 31.5 (9–56) |
| Gender | ||
| Male | 37 | 7 |
| Female | 11 | 1 |
| Race | ||
| Caucasian | 41 | 7 |
| African | 3 | 0 |
| Hispanic | 0 | 0 |
| Others a | 4 | 1 |
| Liver transplant recipient | ||
| Median age (range) | 45 (35–69) | 47.5 (38–67) |
| Gender | ||
| Male | 40 | 5 |
| Female | 8 | 3 |
| Race | ||
| Caucasian | 42 | 7 |
| African | 3 | 0 |
| Hispanic | 3 | 1 |
| Others a | 0 | 0 |
| HCV genotype | ||
| 1a | 32 | 5 |
| 1b | 16 | 3 |
| Histologic biopsy follow-up | ||
| Median number (range) | 6 (2–15) | 6 (4–10) |
| Median length in months (range) b | 36 (12–163) | 21 (7–102) |
Note. The groups with mild and severe recurrent disease were compared using the Pearson Chi-square test for the gender, race and genotype or the Mann-Whitney U test for the other characteristics. The difference between the groups was not significant unless specifically indicated.
Others include Native Americans, Pacific Islanders, and races unknown.
Mild vs. severe, p= 0.031.
Heteroduplex mobility analysis of pre- and post-transplant serum variants
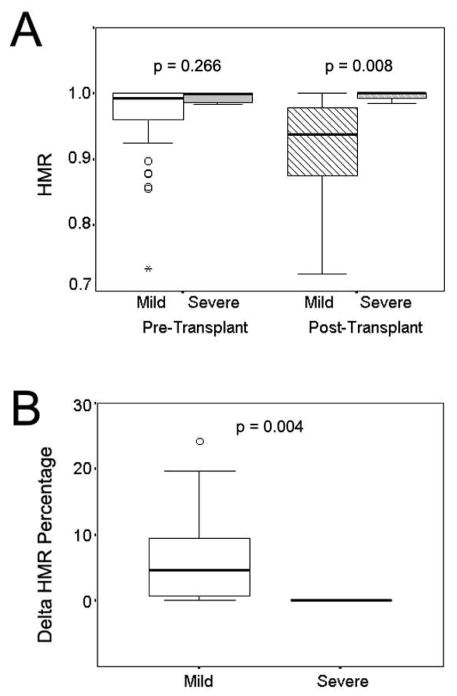
HCV variants in pre-transplant and 12-month post-transplant serum samples were evaluated with heteroduplex mobility analysis (HMA) targeting the hypervariable region 1 (HVR1) of the envelope E2 gene (Gretch et al., 1996). As demonstrated in previous studies, the shift distance of heteroduplex and homoduplex bands from the top of the gel gave a precise indication of nucleotide sequence similarities between the probe and target fragments (Polyak et al., 1997; Wilson et al., 1995). Analysis of heteroduplex mobility ratio (HMR) demonstrated greater viral diversity at both pre-transplant and post-transplant time points in patients with mild recurrent disease compared to those with rapid severe recurrent disease (Figure 1A). The median HMR at the time of pre-transplant was 0.992 and 0.995 for the groups with mild and severe recurrent diseases, respectively. The HMR of the mild group showed great diversification 12 months after transplant (median 0.937), while the HMR of the severe group remained relatively stable (median 0.995). The difference of post-transplant HMR was statistically significant between the two groups (p=0.008). Figure 1B further shows that the rate of change in viral diversity over time (delta HMR) between the pre- and post-transplant samples differed significantly between the two groups, with greater changes in those with mild disease recurrence (median 4.6% and 0% for the mild and severe groups, respectively, p=0.004).
Figure 1.
Evaluation of HVR1 genetic heterogeneity with heteroduplex mobility analysis. Panel A, heteroduplex mobility ratios in subjects with mild and severe recurrent disease at pre- and post-transplant time points; Panel B, the percentage of change in heteroduplex mobility ratios in subjects with mild and severe recurrent disease between the pre- and post-transplant time points.
Pre-transplant HCV genetic diversity correlated with recurrent disease
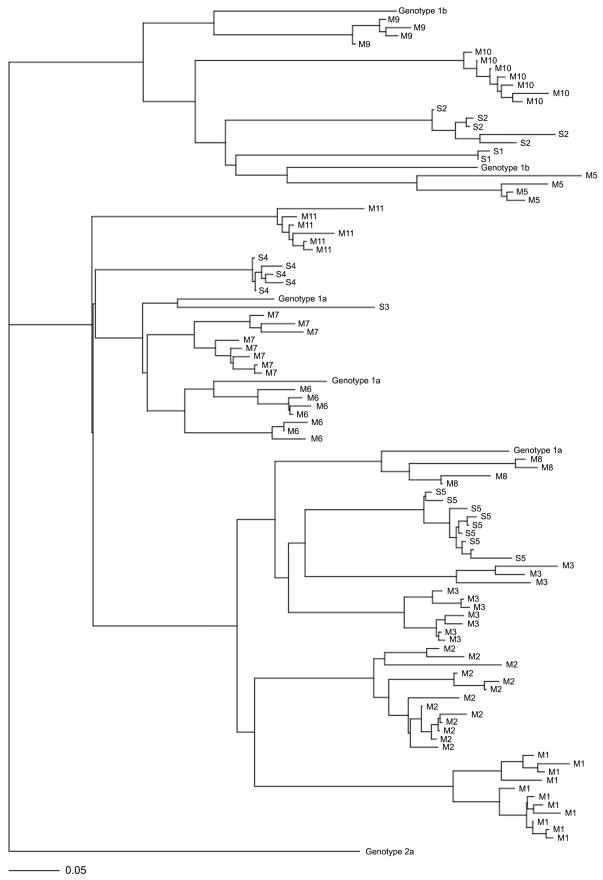
Specimens from tissue reservoirs, including liver, perihepatic lymph node and peripheral blood mononuclear cells (PBMCs), were available from a subset of 15 patients (10 mild and 5 severe cases) at the pre-transplant time point. HCV variants were detected in all 15 liver specimens, 10 of the lymph nodes and 13 of the PBMCs (5 of the lymph node samples and 2 of the PBMC samples were HCV RNA negative). Using the clonal frequency analysis previously described (Wilson et al., 1995), a total of 1035 variants (18 to 23 variants per sample) were characterized by gel electrophoresis, and a total of 282 variants were identified as unique variants and subjected to nucleotide sequencing (GenBank Accession No. GU169906-GU169994, GU170008-GU170200). Figure 2 shows the phylogenetic tree of the unique sequences of serum specimens from each of the patient, along with the reference genotype 1a and 1b sequences and a genotype 2a sequence as an outgroup. As shown in the figure, the variants formed two major clades on the phylogenetic tree based on the HCV subgenotypes (1a and 1b). The variants segregated only with others from the same individual host, and intermingling of variants from subjects with various disease outcomes was observed for each clade.
Figure 2.
A phylogenetic tree of the unique serum-derived variants from the 15 patients with multiple compartments. The tree was constructed with the neighbor-joining method based on the nucleotide sequences using the PAUP software. Five known sequences of genotypes 1a or 1b (1a.US.JL_77, 1a.US.PEN_77, 1a.H77, 1b.KR.HCV-L2, 1b.JP.11-4) were included as genotype references along with a genotype 2a sequence (2a.JP.JFH-1) as an outgroup. The variants were labeled with patient numbers (M1–M3, M5–M11, S1–S5) with M and S indicating the mild or severe post-transplant outcomes, respectively. The scale bar indicates the horizontal branch length corresponding to 5 substitutions per 100 nucleotide sites.
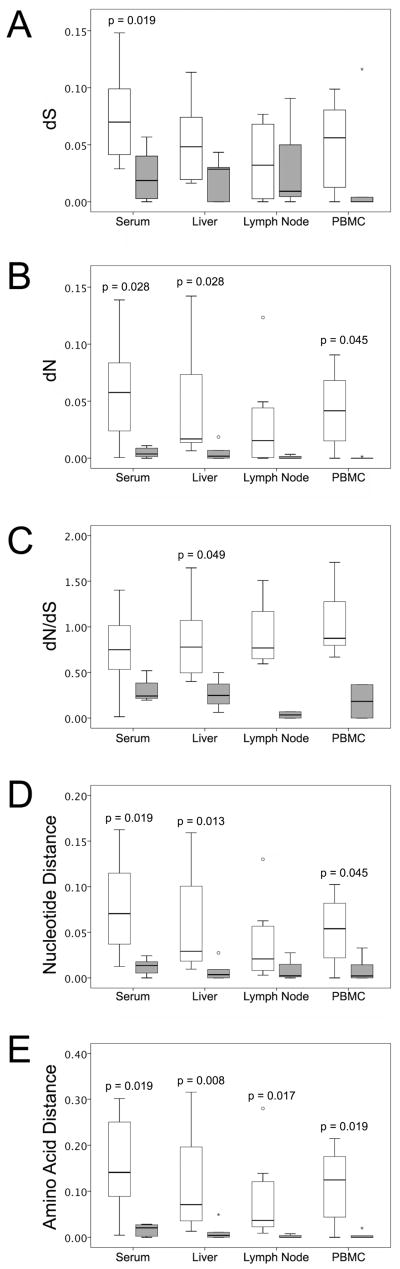
When comparing HVR genetic diversity across the tissue samples, serum variants had the highest genetic heterogeneity, while lower heterogeneity was found in liver, lymph node and PBMC compartments. Compared to the subjects in the mild post-transplant disease category, the group with severe recurrent disease showed significant lower mean values of synonymous and non-synonymous substitutions of the serum-derived variants (p=0.019 for dS and p=0.028 for dN, respectively, Figure 3A and 3B). Lower mean values of dS and dN were also observed within the viral variants isolated from the pre-transplant liver, lymph node and PBMC specimens of subjects with severe hepatitis C recurrence. In all four compartments, the mean intra-population dN/dS ratio also tended to be lower in the patients with severe recurrent disease, although significant difference was only found in the liver compartment (p=0.049, Figure 3C). HVR1 genetic distances, evaluated at the nucleotide and amino acid levels, are summarized in Figure 3D and 3E. Higher genetic distances were associated with mild recurrent disease, and statistical significance was found with respect to the nucleotide distances of serum, liver and PBMC variants, and the amino acid distances of the variants from all the four compartments. When comparing across the compartments, the serum-derived variants had a mean nucleotide distance of 0.052 and a mean amino acid distance of 0.100, representing the highest genetic diversity of the four compartments.
Figure 3.
Comparison of the genetic parameters of multiple compartments between patients with mild and severe recurrent disease. Panel A, the mean values of synonymous nucleotide substitutions (dS); Panel B, the mean values of non-synonymous nucleotide substitutions (dN); Panel C, the mean values of dN/dS ratios; Panel D, the mean values of nucleotide genetic distances; Panel E, the mean values of amino acid genetic distances. The Mann-Whitney U test was used to compare differences between the groups with mild and severe recurrent disease.
Pre-transplant HCV genetic differences in multiple compartments correlate with recurrent disease
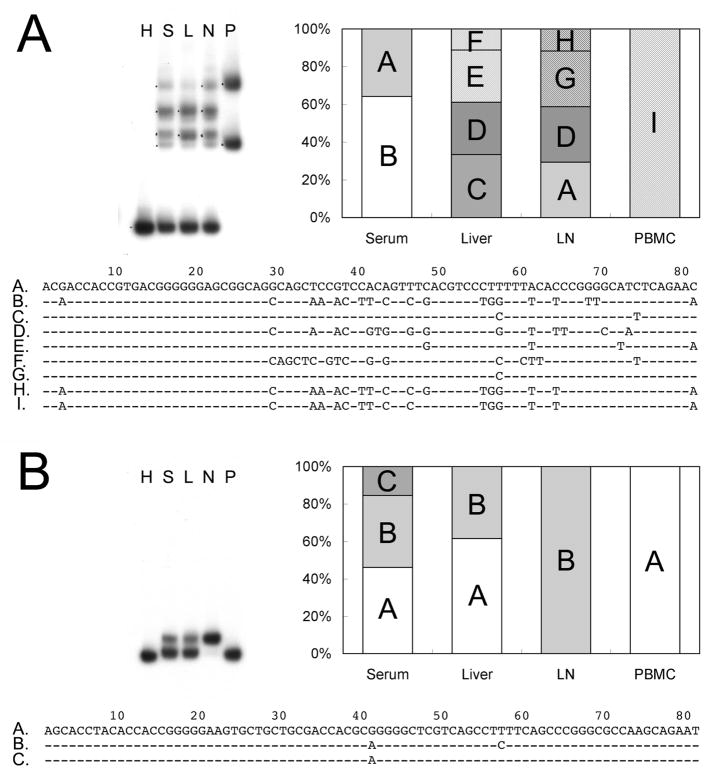
The heteroduplex mobility analysis, while applied to the specimens from multiple compartments of the same individual, provided direct evidence of genetic diversity of the HCV variants. Figure 4 shows the HMA gel shifts of the variants derived from serum, liver, lymph node and PBMCs from two representative subjects (Panel A, M5 with mild recurrent hepatitis and Panel B, S5 with severe recurrent disease) along with the bar figure of major variant composition and the alignment of variant nucleotide sequences. As shown in Panel A, each of the compartments displayed multiple bands of heteroduplexes that migrated at a retarded distance compared to the homoduplex band representing the shifting distance of the major variant in serum. Nucleotide sequencing further confirmed that the compartments harbored unique variants other than the major variant in serum. For the subject S5 that developed severe recurrent disease (Panel B), the migration distances of the hereroduplex bands appeared to be highly similar to the homoduplex control on the HMA gel shift. Only minor difference was found among the compartments of serum, liver, lymph node and PBMCs, and the difference was associated with only one or two nucleotide inconsistence (see the alignment of nucleotide sequences). Overall, the HMA gel shifts were performed in 13 out of the 15 subjects and revealed differences of heteroduplex migration patterns among the compartments in 7 out 8 subjects with mild recurrent disease (M2, M3, M5, M6, M7, M10, M11) and 3 out of 5 subjects with severe recurrent disease (S2, S4, S5). Interestingly, minor differences of heteroduplex migration patterns were found in all the three cases with severe disease outcomes.
Figure 4.
Genetic diversity of HCV variants among multiple compartments shown with heteroduplex mobility analysis. The HMA gel shift presented the migration distances of heteroduplex bands along with the homoduplex control after electropheresis. The lanes on the gel shift were labeled with H for homoduplex, S for serum, L for liver, N for lymph node and P for PBMCs. The composition of the major variants in each compartment (equal or more than two clones in a single compartment) was shown in the bar figure with unique variants labeled with letters. The alignment of the nucleotide sequences was shown right below. Panel A, variants from a representative patient with mild post-transplant outcome (M5); Panel B, variants from a representative patient with severe post-transplant outcome (S5).
With sequences available, the genetic differences of HVR1 variants were quantitatively evaluated using the phylogenetic analysis and the Mantel’s statistical test (Ducoulombier et al., 2004; Zehender et al., 2005). The Mantel’s test, based on comparison of matrixes of genetic and phenotypic distances, determines whether the sequences in one compartment are more similar to each other than to the sequences in other compartments. A total of 70 inter-compartment comparisons were made using the sequences from the various compartments (Table 2). The Mantel’s test revealed 14 out of 15 subjects with significant genetic differences between serum and at least one of the tissue compartments at the pre-transplant time point. It is noticeable that genetic differences between serum and liver were detected in 9 out of 10 subjects with mild recurrent disease compared to 2 out of 5 with severe recurrent disease, suggesting a correlation between serum versus liver genetic differences and a higher probability of developing severe recurrent hepatitis C disease after transplant (p=0.039).
Table 2.
Results of the Mantel’s test performed between the pairwise compartments in the patients with mild and severe recurrent hepatitis. P value of less than 0.05 in the Mantel’s test is considered as an indication of significant genetic differences.
| Mild | Severe | P value | |||
|---|---|---|---|---|---|
| Significant | Non-Significant | Significant | Non-Significant | ||
| Serum v.s. | |||||
| Liver | 9 | 1 | 2 | 3 | 0.039 |
| LN | 4 | 3 | 1 | 2 | 0.490 |
| PBMC | 6 | 2 | 2 | 3 | 0.207 |
| Liver v.s. | |||||
| LN | 3 | 4 | 2 | 1 | 0.490 |
| PBMC | 6 | 2 | 2 | 3 | 0.207 |
| LN v.s. | |||||
| PBMC | 4 | 2 | 1 | 2 | 0.343 |
Note. The Pearson Chi-square test was used to compare the frequency of genetic differences between the two groups with mild and severe recurrent disease.
DISCUSSION
Recurrent HCV infection remains to be a universal problem in patients who are actively infected with HCV and undergo liver transplantation. Besides, the natural history of HCV disease is accelerated in the immunosuppressed liver-transplant recipients compared to immunocompetent patients in the acute infection setting. In general, the circulating viral titer is one log higher after transplant than before transplant (Gretch et al., 1995), and severe graft damage may occur in the early years after transplant (Berenguer et al., 2000; Gane et al., 1996). The accelerated disease progression in liver-transplant recipients allows precise definition of disease outcomes within a relatively short period of follow-up. Thus, the liver transplantation model provides a unique opportunity to understand the pathogenesis of HCV-related liver disease.
Our study investigated the correlation between pre-transplant HCV genetics and the clinical and histological recurrent hepatitis in liver-transplant recipients. We analyzed HVR1 of the E2 gene since the region has been shown to play a role in cell attachment and to be the main target of antibody-mediated neutralization. In the present study, we observed that the genetic diversity of HCV variants is significantly associated with the recurrence of mild hepatitis. Heteroduplex mobility analysis of pre- and post-transplant serum specimens demonstrated a clear, direct correlation between HVR1 homogeneity and the onset of severe recurrent hepatitis C disease by 12 months after transplant. Investigation of different reservoirs further demonstrated that pre-transplant genetic distances were universally higher in the patients who subsequently developed mild recurrent disease compared to those with severe disease. Higher dN/dS ratios, averaged over the entire HVR1, were found in the patients with mild recurrent disease, which may reflect a protective aspect of stronger host immune pressure before transplant in the mild disease cases. HCV genetic differences between serum and tissues are commonly observed in the allograft recipients before transplant, and genetic differences between serum and liver are associated with higher likelihood of mild disease recurrence at statistical significance.
Although the pathogenesis mechanism of recurrent hepatitis C is still not clear, the correlation between increased HCV genetic diversity and mild post-transplant hepatitis has been reported in numerous studies. The degree of relatedness between pre- and post-transplant HCV genetics tended to be higher in patients with severe compared to those with mild disease recurrence (Gretch et al., 1996; Sanchez-Fueyo et al., 2001). In other studies, direct sequencing indicated that intra-sample genetic distances and amino acid diversification were inversely related to the histological severity of HCV recurrence (Lyra et al., 2002; Sanchez-Fueyo et al., 2001). Furthermore, the greater genetic divergence was observed in multiple regions over the whole HCV genome in patients with mild post-transplant disease than in those with severe disease (Gretch et al., 1996; Sullivan et al., 1998). The dN/dS ratio, which generally decreased in the broadly immunosuppressed patients after transplant, was reported to be lower in the patients with severe than in those with mild or moderate HCV infection recurrence (Gretch et al., 1996; Sanchez-Fueyo et al., 2001). Thus, a stronger selective pressure is associated with a better outcome in the post-transplant setting.
When assessed with statistical methods, genetic differences were found among HCV variants from the pre-transplant serum, liver, lymph node and PBMC specimens in the allograft recipients. HCV variants circulating in serum are considered as the main source of allograft reinfection after transplant, whereas lymphocytes and macrophages also enter the allograft soon after transplant. In theory, extrahepatic strains in the serum or lymphocytes may compete with liver-derived variants during reinfection of hepatocytes, allowing selective population to dominate within the allograft during the first few hours after reperfusion (Hughes et al., 2004). In the current study, we found that the patients with heterogeneous HCV variants, especially those with genetic differences between liver and serum, were more likely to develop mild post-transplant disease. This observation suggests that liver-derived and extrahepatic variants may both contribute to reinfection of the naive allograft and influence the development of recurrent hepatitis. In contrast, when pre-transplant genetic differences are not observed, allograft reinfection may be dominated by liver-derived variants that are the major variants in circulation as well as in extrahepatic compartments.
The association between HCV genetic diversity and progression of post-transplant disease suggests two possible explanations. First, the histological switch to severe recurrent disease might simply be a consequence of re-infection with highly pathogenic variants that dominate at the pre-transplant stage. The pathogenic variants rapidly adapt to the naive allograft to propagate a large amount of homogeneous descendants followed with the recurrence of severe liver disease. The analogous model of acute infection seems to support the enhanced inherent virulence of specific variants, when extreme high levels of HCV titer was observed with the lowest degree of genetic diversity in patients with fulminant hepatitis (Farci et al., 2000). In the transplantation setting, it is noteworthy that multiple factors such as age of recipients, severity of hyperbilirubinemia, variable immunosuppression level, and HLA mismatch between donors and recipients may also contribute to the outcome of recurrent disease along with the factor of HCV genetics, which makes the situation even complicated. Second, the HCV genetic diversity at both pre- and post-transplant stages might be a consequence of host antiviral immune defense, which drives viral mutation and protects the liver from the development of severe disease. In such case, the HCV genetic diversity is rather a co-occurrence during the development of post-transplant liver disease. Our current data presented higher dN/dS ratio in the patients with mild recurrent disease, suggesting that host immune pressure is more prevalent at the pre-transplant stage in the mild cases. Immunology data in a study by Rosen et al. (Rosen et al., 1999) also demonstrated a statistically significant correlation between markers of HCV-specific CD4+ T cell immunity during the early post-transplant infection period and mild disease phenotypes.
When assessing viral heterogeneity from tissue compartments and serum, a major concern is sampling bias. This can occur if the viral genome variants are non-uniformly distributed throughout the infected tissue. A second type of sampling bias, referred to as “resampling bias” (Liu et al., 1996), occurs when genome template input copy number is very low. Regarding the first issue, extensive tissue sampling was performed on two whole explanted HCV-infected livers at 10 independent and spatially separated sites per liver (data not shown). HCV genetic heterogeneity was very close or identical for all samples, suggesting that HCV genome distribution is most likely uniform in liver at end stage disease. Regarding the issue of “resampling bias”, which has been documented in the setting of low titer HIV infections (Liu et al., 1996), we found no evidence of resampling in the few cases where repeat testing was performed, although the possibility of this amplification error may not be with confidence excluded during our study of HCV heterogeneity in tissue reservoirs (data not shown).
In summary, our data suggest that HCV genetics at the pre-transplant stage predicts hepatitis C disease severity in transplanted allograft. Viral predictors of poor outcome include low genetic diversity in circulation, and decreased genetic differences between serum and tissue specimens. Longitudinal studies are still going on in our lab to further define the HCV pathogenic mechanism that determines the outcome of hepatitis C disease after transplant.
Acknowledgments
The authors acknowledge Matt Maria for assistance in manuscript preparation. The work was supported by NIH grants R01-DK-98-017, R01 AI 66209-01 and R01 AI 49168-06.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Batts KP, Ludwig J. Chronic hepatitis. An update on terminology and reporting. Am J Surg Pathol. 1995;19:1409–1417. doi: 10.1097/00000478-199512000-00007. [DOI] [PubMed] [Google Scholar]
- Berenguer M, Ferrell L, Watson J, Prieto M, Kim M, Rayon M, Cordoba J, Herola A, Ascher N, Mir J, Berenguer J, Wright TL. HCV-related fibrosis progression following liver transplantation: increase in recent years. J Hepatol. 2000;32:673–684. doi: 10.1016/s0168-8278(00)80231-7. [DOI] [PubMed] [Google Scholar]
- Berenguer M, Prieto M, Cordoba J, Rayon JM, Carrasco D, Olaso V, San-Juan F, Gobernado M, Mir J, Berenguer J. Early development of chronic active hepatitis in recurrent hepatitis C virus infection after liver transplantation: association with treatment of rejection. J Hepatol. 1998;28:756–763. doi: 10.1016/s0168-8278(98)80224-9. [DOI] [PubMed] [Google Scholar]
- Charlton M, Seaberg E, Wiesner R, Everhart J, Zetterman R, Lake J, Detre K, Hoofnagle J. Predictors of patient and graft survival following liver transplantation for hepatitis C. Hepatology. 1998;28:823–830. doi: 10.1002/hep.510280333. [DOI] [PubMed] [Google Scholar]
- Dahari H, Feliu A, Garcia-Retortillo M, Forns X, Neumann AU. Second hepatitis C replication compartment indicated by viral dynamics during liver transplantation. J Hepatol. 2005;42:491–498. doi: 10.1016/j.jhep.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Davidson F, Simmonds P, Ferguson JC, Jarvis LM, Dow BC, Follett EA, Seed CR, Krusius T, Lin C, Medgyesi GA, et al. Survey of major genotypes and subtypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76 ( Pt 5):1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- De Mitri MS, Mele L, Chen CH, Piccinini A, Chianese R, D’Errico A, Alberti A, Pisi E. Comparison of serum and liver hepatitis C virus quasispecies in HCV-related hepatocellular carcinoma. J Hepatol. 1998;29:887–892. doi: 10.1016/s0168-8278(98)80115-3. [DOI] [PubMed] [Google Scholar]
- Ducoulombier D, Roque-Afonso AM, Di Liberto G, Penin F, Kara R, Richard Y, Dussaix E, Feray C. Frequent compartmentalization of hepatitis C virus variants in circulating B cells and monocytes. Hepatology. 2004;39:817–825. doi: 10.1002/hep.20087. [DOI] [PubMed] [Google Scholar]
- Farci P, Shimoda A, Coiana A, Diaz G, Peddis G, Melpolder JC, Strazzera A, Chien DY, Munoz SJ, Balestrieri A, Purcell RH, Alter HJ. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science. 2000;288:339–344. doi: 10.1126/science.288.5464.339. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP - Phylogeny Inference Package Version 3.5. Department of Genetics, University of Washington; Seattle, WA: 1993. [Google Scholar]
- Feray C, Gigou M, Samuel D, Paradis V, Mishiro S, Maertens G, Reynes M, Okamoto H, Bismuth H, Brechot C. Influence of the genotypes of hepatitis C virus on the severity of recurrent liver disease after liver transplantation. Gastroenterology. 1995;108:1088–1096. doi: 10.1016/0016-5085(95)90207-4. [DOI] [PubMed] [Google Scholar]
- Galossi A, Guarisco R, Bellis L, Puoti C. Extrahepatic manifestations of chronic HCV infection. J Gastrointestin Liver Dis. 2007;16:65–73. [PubMed] [Google Scholar]
- Gane EJ, Portmann BC, Naoumov NV, Smith HM, Underhill JA, Donaldson PT, Maertens G, Williams R. Long-term outcome of hepatitis C infection after liver transplantation. N Engl J Med. 1996;334:815–820. doi: 10.1056/NEJM199603283341302. [DOI] [PubMed] [Google Scholar]
- Gretch DR, Bacchi CE, Corey L, dela Rosa C, Lesniewski RR, Kowdley K, Gown A, Frank I, Perkins JD, Carithers RL., Jr Persistent hepatitis C virus infection after liver transplantation: clinical and virological features. Hepatology. 1995;22:1–9. [PubMed] [Google Scholar]
- Gretch DR, Polyak SJ, Wilson JJ, Carithers RL, Jr, Perkins JD, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassoba HM, Bzowej N, Berenguer M, Kim M, Zhou S, Phung Y, Grant R, Pessoa MG, Wright TL. Evolution of viral quasispecies in interferon-treated patients with chronic hepatitis C virus infection. J Hepatol. 1999;31:618–627. doi: 10.1016/s0168-8278(99)80340-7. [DOI] [PubMed] [Google Scholar]
- Hughes MG, Jr, Rudy CK, Chong TW, Smith RL, Evans HL, Iezzoni JC, Sawyer RG, Pruett TL. E2 quasispecies specificity of hepatitis C virus association with allografts immediately after liver transplantation. Liver Transpl. 2004;10:208–216. doi: 10.1002/lt.20060. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang LF, Jang SJ, Vargas H, Rakela J. Hepatitis C virus quasispecies in patients infected with HIV-1: correlation with extrahepatic viral replication. Virology. 1998;248:164–171. doi: 10.1006/viro.1998.9269. [DOI] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wang LF, Nowicki M, Rakela J. Uneven distribution of hepatitis C virus quasispecies in tissues from subjects with end-stage liver disease: confounding effect of viral adsorption and mounting evidence for the presence of low-level extrahepatic replication. J Virol. 2000;74:1014–1017. doi: 10.1128/jvi.74.2.1014-1017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskus T, Radkowski M, Wilkinson J, Vargas H, Rakela J. The origin of hepatitis C virus reinfecting transplanted livers: serum-derived versus peripheral blood mononuclear cell-derived virus. J Infect Dis. 2002;185:417–421. doi: 10.1086/338635. [DOI] [PubMed] [Google Scholar]
- Li H, McMahon BJ, McArdle S, Bruden D, Sullivan DG, Shelton D, Deubner H, Gretch DR. Hepatitis C virus envelope glycoprotein co-evolutionary dynamics during chronic hepatitis C. Virology. 2008a;375:580–591. doi: 10.1016/j.virol.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Thomassen LV, Majid A, McMahon BJ, Bruden D, McArdle S, Bano N, Chung M, Carithers RL, Perkins JD, Sullivan DG, Gretch DR. Investigation of putative multisubtype hepatitis C virus infections in vivo by heteroduplex mobility analysis of core/envelope subgenomes. J Virol. 2008b;82:7524–7532. doi: 10.1128/JVI.02220-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SL, Rodrigo AG, Shankarappa R, Learn GH, Hsu L, Davidov O, Zhao LP, Mullins JI. HIV quasispecies and resampling. Science. 1996;273:415–416. doi: 10.1126/science.273.5274.415. [DOI] [PubMed] [Google Scholar]
- Lyra AC, Fan X, Lang DM, Yusim K, Ramrakhiani S, Brunt EM, Korber B, Perelson AS, Di Bisceglie AM. Evolution of hepatitis C viral quasispecies after liver transplantation. Gastroenterology. 2002;123:1485–1493. doi: 10.1053/gast.2002.36546. [DOI] [PubMed] [Google Scholar]
- Navas S, Martin J, Quiroga JA, Castillo I, Carreno V. Genetic diversity and tissue compartmentalization of the hepatitis C virus genome in blood mononuclear cells, liver, and serum from chronic hepatitis C patients. J Virol. 1998;72:1640–1646. doi: 10.1128/jvi.72.2.1640-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Okuda M, Hino K, Korenaga M, Yamaguchi Y, Katoh Y, Okita K. Differences in hypervariable region 1 quasispecies of hepatitis C virus in human serum, peripheral blood mononuclear cells, and liver. Hepatology. 1999;29:217–222. doi: 10.1002/hep.510290117. [DOI] [PubMed] [Google Scholar]
- Pal S, Sullivan DG, Kim S, Lai KK, Kae J, Cotler SJ, Carithers RL, Jr, Wood BL, Perkins JD, Gretch DR. Productive replication of hepatitis C virus in perihepatic lymph nodes in vivo: implications of HCV lymphotropism. Gastroenterology. 2006;130:1107–1116. doi: 10.1053/j.gastro.2005.12.039. [DOI] [PubMed] [Google Scholar]
- Polyak SJ, Faulkner G, Carithers RL, Jr, Corey L, Gretch DR. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J Infect Dis. 1997;175:1101–1107. doi: 10.1086/516448. [DOI] [PubMed] [Google Scholar]
- Radkowski M, Wilkinson J, Nowicki M, Adair D, Vargas H, Ingui C, Rakela J, Laskus T. Search for hepatitis C virus negative-strand RNA sequences and analysis of viral sequences in the central nervous system: evidence of replication. J Virol. 2002;76:600–608. doi: 10.1128/JVI.76.2.600-608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque Afonso AM, Jiang J, Penin F, Tareau C, Samuel D, Petit MA, Bismuth H, Dussaix E, Feray C. Nonrandom distribution of hepatitis C virus quasispecies in plasma and peripheral blood mononuclear cell subsets. J Virol. 1999;73:9213–9221. doi: 10.1128/jvi.73.11.9213-9221.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roque-Afonso AM, Ducoulombier D, Di Liberto G, Kara R, Gigou M, Dussaix E, Samuel D, Feray C. Compartmentalization of hepatitis C virus genotypes between plasma and peripheral blood mononuclear cells. J Virol. 2005;79:6349–6357. doi: 10.1128/JVI.79.10.6349-6357.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen HR, Hinrichs DJ, Gretch DR, Koziel MJ, Chou S, Houghton M, Rabkin J, Corless CL, Bouwer HG. Association of multispecific CD4(+) response to hepatitis C and severity of recurrence after liver transplantation. Gastroenterology. 1999;117:926–932. doi: 10.1016/s0016-5085(99)70352-5. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sanchez-Fueyo A, Gimenez-Barcons M, Puig-Basagoiti F, Rimola A, Sanchez-Tapias JM, Saiz JC, Rodes J. Influence of the dynamics of the hypervariable region 1 of hepatitis C virus (HCV) on the histological severity of HCV recurrence after liver transplantation. J Med Virol. 2001;65:266–275. doi: 10.1002/jmv.2029. [DOI] [PubMed] [Google Scholar]
- Sheiner PA, Schwartz ME, Mor E, Schluger LK, Theise N, Kishikawa K, Kolesnikov V, Bodenheimer H, Emre S, Miller CM. Severe or multiple rejection episodes are associated with early recurrence of hepatitis C after orthotopic liver transplantation. Hepatology. 1995;21:30–34. [PubMed] [Google Scholar]
- Sugo H, Balderson GA, Crawford DH, Fawcett J, Lynch SV, Strong RW, Futagawa S. The influence of viral genotypes and rejection episodes on the recurrence of hepatitis C after liver transplantation. Surg Today. 2003;33:421–425. doi: 10.1007/s10595-002-2537-5. [DOI] [PubMed] [Google Scholar]
- Sullivan DG, Wilson JJ, Carithers RL, Jr, Perkins JD, Gretch DR. Multigene tracking of hepatitis C virus quasispecies after liver transplantation: correlation of genetic diversification in the envelope region with asymptomatic or mild disease patterns. J Virol. 1998;72:10036–10043. doi: 10.1128/jvi.72.12.10036-10043.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrault NA. Treatment of recurrent hepatitis C in liver transplant recipients. Clin Gastroenterol Hepatol. 2005;3:S125–131. doi: 10.1016/s1542-3565(05)00709-3. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas HE, Laskus T, Radkowski M, Wilkinson J, Balan V, Douglas DD, Harrison ME, Mulligan DC, Olden K, Adair D, Rakela J. Detection of hepatitis C virus sequences in brain tissue obtained in recurrent hepatitis C after liver transplantation. Liver Transpl. 2002;8:1014–1019. doi: 10.1053/jlts.2002.36393. [DOI] [PubMed] [Google Scholar]
- Weiner AJ, Brauer MJ, Rosenblatt J, Richman KH, Tung J, Crawford K, Bonino F, Saracco G, Choo QL, Houghton M, et al. Variable and hypervariable domains are found in the regions of HCV corresponding to the flavivirus envelope and NS1 proteins and the pestivirus envelope glycoproteins. Virology. 1991;180:842–848. doi: 10.1016/0042-6822(91)90104-j. [DOI] [PubMed] [Google Scholar]
- Wilson JJ, Polyak SJ, Day TD, Gretch DR. Characterization of simple and complex hepatitis C virus quasispecies by heteroduplex gel shift analysis: correlation with nucleotide sequencing. J Gen Virol. 1995;76 ( Pt 7):1763–1771. doi: 10.1099/0022-1317-76-7-1763. [DOI] [PubMed] [Google Scholar]
- Zehender G, De Maddalena C, Bernini F, Ebranati E, Monti G, Pioltelli P, Galli M. Compartmentalization of hepatitis C virus quasispecies in blood mononuclear cells of patients with mixed cryoglobulinemic syndrome. J Virol. 2005;79:9145–9156. doi: 10.1128/JVI.79.14.9145-9156.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]