Abstract
During the phase of overt tooth cytodifferentiation that occurs after birth in the mouse and using the 3.6Collagen1a-Cre, the BMP4 floxed and BMP4 knock-out mice, the BMP4 gene was deleted in early collagen producing odontoblasts around postnatal day 1. BMP4 expression was reduced over 90% in alveolar osteoblasts and odontoblasts. There was decreased rate of predentin to dentin formation and decreased mature odontoblast differentiation reflected in reduced DMP1 expression and proper dentinal tubule formation, as well as reduced Collagen type I and Osteocalcin expression. We observed mutant dysmorphogenic odontoblasts that failed to properly elongate and differentiate. The consequence of this failed differentiation process lead to permanent loss of dentin thickness, apparent enlarged pulp chambers in the molars and reduced bone supporting the tooth structures in mice as old as 10–12 months. Deletion of the BMP4 gene in odontoblasts also indirectly disrupted the process of enamel formation that persisted throughout life. The mechanism for this altered differentiation program in the absence of the BMP4 gene in odontoblasts is from decreased BMP signaling, and decreased expression of three key transcription factors, Dlx3, Dlx5, and Osterix. BMP signaling, as well as Dlx3 and Amelogenin expression, are also indirectly reduced in the ameloblasts of the odontoblast BMP4 cKO mice. This supports a key paracrine or endocrine role of odontoblasts derived BMP4 postnatally on the proper amelogenesis and formation of the enamel.
Introduction
A wealth of data that supports a role for BMP4 in tooth development in the embryo from the initial epithelial lamina to the late bell stage is using primarily the Msx1 knockout model. BMP4 expression has been demonstrated to be in both downstream and upstream of the Msx1 gene in the dental mesenchyme and epithelium [1–4]. Recombinant BMP4 (or presumably recombinant BMP2) can rescue most all of the Msx1 defects in tooth development, using a variety of epithelial–mesenchymal tissue grafts into the kidney capsule. Msx1 is transiently required in the dental mesenchyme to produce the BMP4 ligand at least up to the cap stage. These experiments clearly demonstrate that recombinant BMP4 can rescue the Msx1 mutant tooth phenotype and allows complete odontogenesis and amelogenesis to occur in these assays of kidney explants [3]. Again, using the kidney capsule assay with epithelial-mesenchymal implants from the tooth germ, Dr. Zhang’s laboratory demonstrates an early BMP activity requirement. This activity is achieved either by implantation of BMP4 beads or overexpression studies of a cDNA to BMP4 driven by the Msx1 promoter in the context of the Msx1 knockout animals for the formation of both dentin and enamel in this implant assay [4]. They demonstrate that both Dlx5 and Runx2 transcription factors are downstream of this BMP activity in early tooth development in this kidney capsule environment.
Classic experiments of Drs. Mina and Kollar, demonstrated that 1st pharyngeal arch ectoderm, that normally induces the underlying mesenchyme to produce teeth, is also capable of inducing some odontogenic differentiation with non-tooth related mesenchyme [5]. These experiments were extended to demonstrate that recombinant BMP4 could, in part, replace this ectodermal signal using non-tooth potential 2nd arch pharyngeal mesenchyme [6]. Thus, initial tooth development appears to require a BMP signal; most likely, BMP4 from the ectoderm activates BMP4 in the mesenchyme through an Msx1 dependent mechanism. All of these experiments with overexpression and addition of recombinant BMP4 protein, however, leave open questions that can only be addressed by a tissue specific deletion of the BMP4 gene.
Developmentally, the formation of the mandible and the tooth structure is a complex process and now has been conclusively demonstrated in a conditional knock out approach to require BMP4 [7]. Using a BMP4 floxed allele/Nkx2.5-Cre mouse which specifically activates the mandibular arch ectoderm and the pharyngeal arch endoderm, the deletion of the BMP4 gene resulted in a complete loss of the mandible. With this ectodermal deletion of the BMP4 gene, a large number of ectoderm genes are decreased in 10.5 dpc embryos; for example, the Isl1, Tlx1, and Dlx2 transcription factors. The expression of BMP4, Msx1, Msx2, and Alx4 is decreased in the adjacent mesodermal mesenchyme demonstrating conclusively that BMP4 from ectoderm can act in a paracrine manner.
Since the global knock-out of the BMP4 gene is an embryonic lethal [8], we have conditionally deleted the BMP4 gene in the mesenchymal precursors or preodontoblast stages, as well as the osteoblasts of the alveolar and other bone regions. We now address the consequences of the deletion of the BMP4 gene in the odontoblast stages during postnatal tooth cytodifferentiation. The Collagen type I gene rapidly turns on as the developing odontoblasts begin to make a collagen matrix. We used a Col1a1-Cre model that predominately activates Cre around the same time the collagen production is actively expressed in the odontoblasts. This activation time is just before birth through approximately 20 days after birth. The consequences of deleting the BMP4 gene in odontoblasts and surrounding osteoblasts result in permanent defects in tooth cytodifferentiation and permanent defects in the supporting periodontal apparatus. The physiological consequences of this new mutant model are discussed in relationship to related mouse and human tooth diseases. Some of the preliminary results related to these studies have been published [9].
Material and Methods
Animals
One day, 2 day, 8 day, 12 day, 18 day, one month, 9 month and 1 year old BMP4 cKO mice plus BMP4 control mice (wild type and heterozygotes BMP4fx/+), total of 58 animals were used for these studies. Control mice, both heterozygotes (BMP4fx/+) and wild type did not show any major difference in tooth or bone and were pooled for these studies and also in the statistical evaluation. BMP4 cKOs of odontoblasts and osteoblasts with loxP sites flanking exon 3 and 4 of the BMP4 gene were created by crossing the BMP4 floxed (BMP4-fx) mouse with a 3.6Col1a1-cre mouse [10]. The Cre line for deletion was a 3.6Col1a1-cre mouse model characterized by Dr. Liu [11]. We also used the BMP4null allele with the LacZ reporter [10]. BMP4-fx homozygote mice and the 3.6Col1a1-cre with either a BMP4-fx allele or the BMP4 null allele yielded viable BMP4 cKO mice (3.6Col1a1-cre; Bmp4null-lacZ/Bmp4fx) at about 20%. The mice have survived up to one and one-half years. All described procedures with animals performed in experiments were done with the UTHSCSA Institutional Animal Care and use Committee (IACUC) guidelines.
BMP4 cKO mice with deletion of Exon 3 and 4 were identified by Southern analysis and PCR of the DNA from the tails of weanlings. Total DNA was isolated using Proteinase K digestion in SDS buffer overnight followed by Phenol/Chloroform extraction. Primers specific for BMP4 Exon 4 were used. The primers used for detecting the wild and mutant alleles are: AGA CTC TTT AGT GAG CAT TTT CAA C and AGC CCA ATT TCC ACA ACT TC - 220bp (fx) and 180bp (wt), respectively; AGA CTC TTT AGT GAG CAT TTT CAA C and AGG TGA GCA GAG CTA AGA TG - 350bp (recombined).
X-ray analysis
Radiography was used as a non-invasive method to measure changes in teeth and bone without sacrificing the animal. Animals were radiographed using a Faxitron radiograph inspection unit, Model 8050-020, Field Emmission Corporation, Inc., with digital image capture. Digitized images were analyzed using the AnalySIS software to measure size and width of selected components in the 1st and 2nd molar teeth. Animals were anesthetized using Ketamine (Sigma-Aldrich, St Louis, MO).
In situ Hybridization
Tissue samples for in situ hybridization were processed RNase free with DEPC water in all reagents. The tissues were fixed in 4% paraformaldehyde/PBS overnight, decalcified in buffered 15% EDTA at 4°C for 4–7 weeks, embedded in paraffin, mounted on ProbeON Plus slides (Fisher Scientific, Pittsburgh, PA) and kept at 4°C. Procedures for in situ hybridization was done accordingly [12]. Briefly, all RNA probes for in situ hybridization were transcribed in vitro in the presence of Digoxigenin for production of antisense and sense probes with T3, T7 or SP6 polymerases, where appropriate. Sections were first deparaffinized and treated with Proteinase K. Hybridization was performed at 58°C overnight with probe concentration of 1μg/ml. After overnight hybridization, sections were treated with RNase, washed with 5XSSC, and with 50% formamide in 2XSSC. Detection of hybridization signal was done by adding alkaline phosphatase substrate (NTB/BCIP, Roche Diagnostic Corp, Dallas, TX) in detection buffer (10% polyvinyl alcohol 70–100kd Sigma, 100mM TRIS pH9, 100mM NaCl, 2mM Levamisole). The duration of hybridization signal development was from 1 hour to overnight at 30°C, depending on the probe and abundance of the transcript in the tissue. The sections were lightly stained with methyl green to give the best background level to barely see the tissue for quantitative analysis described below.
After hybridization, images were captured at various levels in the tissue using high resolution TIF files at 40X magnification and merged where necessary. Using ImageJ, the level of hybridization in each cell or tissue section was quantified, and the level of hybridization determined relative to the control level. For quantifying non-radioactive in situ hybridization signal, we used ImageJ measuring a purple-blue color from NBT formazan, which is a generated product of alkaline phosphatase reaction using BCIP and NTB substrate [12]. Areas for measurements were selected using “free hand selections” tool and corrected for background reading. The background was defined with average of 5 measured areas without hybridization signal. All obtained numbers are relative.
Immunocytochemistry
Paraffin sections of 8μm were prepared and deparaffinized with xylene and a graded ethanol series to 70% and rinsed in water. For antigen retrieval, sections were placed in 10mM sodium citrate buffer and microwaved for 5 minutes to boiling, with 2 minutes at boiling, then washed in water 3X, placed in 3% hydrogen peroxide for 10 minutes, washed in water 3X, and PBS for 5 minutes. After 30 minute treatment with blocking solution (10% non-immune goat serum or NGS, 0.6% BSA, 1% NaN3), the sections were treated with 1:50 dilution of primary P-Smad1/5/8 antibodies obtained from Cell Signaling Technology Inc. (Beverly, MA), in blocking solution overnight at 4°C. The sections were washed 3X with PBST (0.1% Tween 20 in PBS) and then treated with secondary antibody (goat biotinylated rabbit IgG antibody) at 1:200 dilution in blocking solution for 1 hour, followed by alkaline phosphatase (AP) conjugated avidin/biotin complex for 30 minutes, using VECTASTAIN ABC-AP kit (Vector Laboratories, Burlingame, CA). The alkaline phosphatase was developed using the Fast Red AP substrate (Roche Applied Science, Indianapolis, IN).
Statistical Methods
The data obtained from BMP4 cKO and control mice were correlated on similar sections and regions of the mandible and maxilla. Over the past 6 years, we have analyzed a large number of mice from appropriate crosses, with BMP4 cKO and control littermates. All animals were initially evaluated by X-rays. Groups of these different ages were evaluated with an n=3 to 5 per group for dental and enamel differences. Analysis of 4 independent mice for each genotype was carried out for μCT analysis. For gene expression studies and immunocytochemical experiments, we used at least 2 independent mice for each genotype and quantitation was preformed on at least one of these sets of animals. These analyses were followed by a two-tailed unpaired Student’s t-test where significant differences between control and BMP4 cKO animals were determined, with multiple measurements per section from each animal.
Results
BMP4 Expression In the 3.6Col1a1-cre; Bmp4null/Bmp4fx mouse model
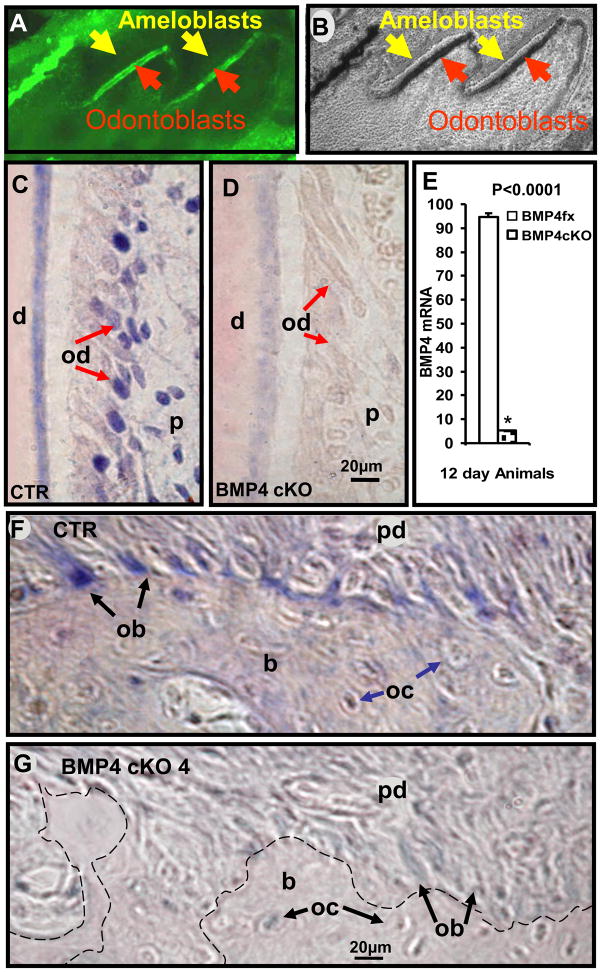
Previous studies with the 3.6Col1a1-cre model crossed with the Rosa26 reporter mice have shown that Cre activity correlates with Collagen type I expression predominately in osteoblasts and odontoblasts, as they begin to produce collagen matrix, and to a lesser extent in tendon [11]. Analysis of mandibles from newborn mice (P1) for deletion of BMP4 expression in odontoblasts was confirmed by the cross of 3.6Col1a1-cre line with ZEG-Reporter. The activated GFP expression observed in odontoblasts shows that Cre is already activated in odontoblasts after Cre-LoxP recombination at birth (Fig. 1A and B).
Fig. 1.
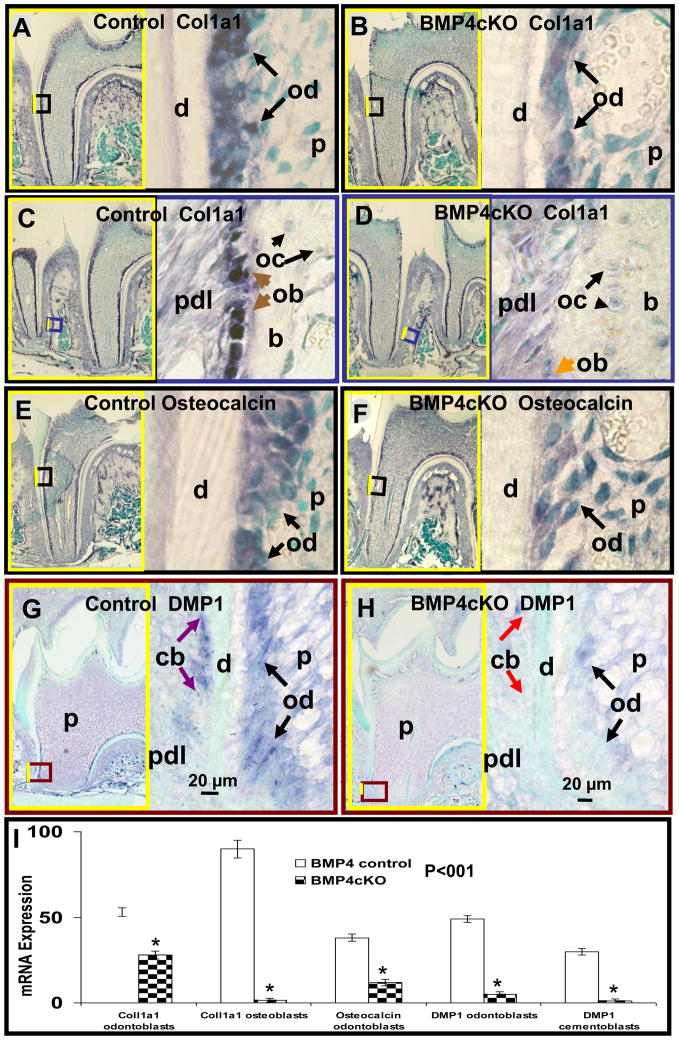
3.6Col1a1 targets odontoblasts at day 1 pp (P1) (A-fluorescent, B-light field microscopy) and in situ hybridization of BMP4 mRNA in representative sections of 12 day old mouse maxillary first molar odontoblasts (C, D) and osteoblasts (F, G) in BMP4 control (C, F) and BMP4 cKO mice (D, G). 3.6Col1a1-cre targeting at functional odontoblasts is confirmed by the cross of 3.6Col1a1-cre line with ZEG-Reporter. The activated GFP expression observed in odontoblasts of 1 day old mice (A-red arrows) shows that Cre is already activated in odontoblasts after Cre-LoxP recombination at birth. Note that GFP expression is not activated in ameloblasts (A-yellow arrows). The hybridization signal (blue color) in odontoblasts and osteoblasts is reduced 94% in BMP4 cKO mice compared to BMP4 control mice in this example with multiple measurements (P<0.0001 for the difference in CTR and BMP4 cKO) along the entire odontoblast area and confirmed visually in independent sets of control (CTR) and BMP4 cKO tissues at similar ages (data not shown) (E). d - dentin; od - odontoblasts (red arrows); ameloblasts (yellow arrows); p - dental pulp; ob - osteoblasts (black arrows); oc - osteocytes (blue arrows); b - bone; pd - periodontium. The dashed line (G) represents the boundary between the bone and periodontium in BMP4 cKO mice, which is clearly defined with blue stained osteoblasts in CTR mice.
In the maxillary 1st molar at postnatal day 12, BMP4 expression is reduced over 90%, as quantitated using these and similar images shown in Figure 1C, D, and E. Quantitation of the expression levels is carefully done using areas of the tissue with only the light counter stain color for determining background. The level of expression is determined using ImageJ and capturing the area of odontoblasts showing the alkaline phosphatase dependent purple color. Previously, using a dot blot technique with increasing mRNA samples and the same concentrations of biotinylated probe, secondary antibody and development reagents, we established a linear relationship with mRNA levels and extent of purple stain in the range used for all in situ hybridization experiments [12]. In a similar manner, the expression of BMP4 in the BMP4 cKO mice is also reduced over 90% in the osteoblasts on the surface of the alveolar bone adjacent to the periodontal ligament (Figure 1F and G). Similar results were obtained with an independent set of mice with the same age and appropriate phenotypes (data not shown).
Dentin Thickness is reduced in BMP4 cKO mice from 12 days to 9 months of age in both the Coronal and Root Region
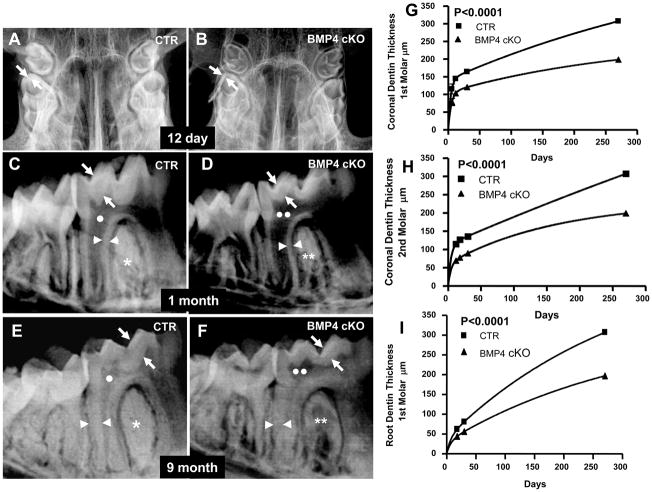
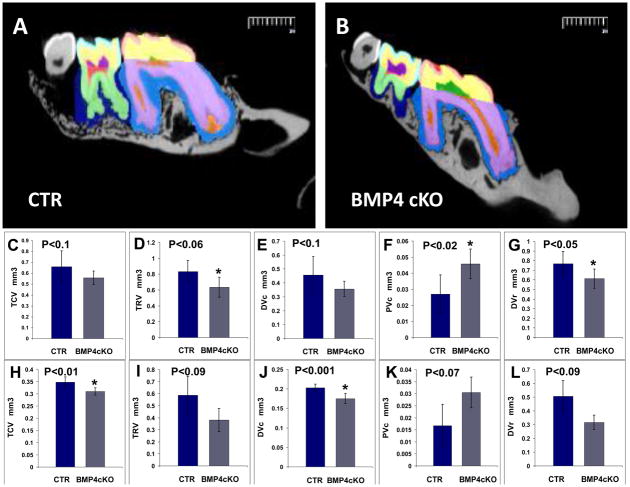
We first initiated a longitudinal age study of both the 1st and 2nd molars by high resolution digital X-ray analysis, followed by histological analysis. Figure 2a demonstrates that the BMP4 cKOs have a severe dentin phenotype, with reduced dentin thickness and enlarged pulp chambers (taurodontism). Both the dentin around the crown of 1st and 2nd molars (coronal) and the dentin layer in the root region are reduced 30 to 40% beginning as early as 6–12 days of age and continuing up to 9 months of age. The deletion of the BMP4 gene in differentiating odontoblasts leads to the phenotype shown in Figure 2aA and B with enlarged pulp chamber and decreased dentin-enamel thickness at 12 days of age. We have followed the same phenotype to 12 months of age (data not shown). The enamel radioopacity also decreased in the BMP4 cKO mice compared to control mice. At 1 month of age (after tooth cytodifferentiation greatly decreased in a normal mouse), the decreased dentin persists, and of note is the decreased alveolar bone between the teeth and roots in absence of BMP4 in the odontoblasts and alveolar osteoblasts (Fig. 2a-panel C and D). Even at 9 months, the phenotype persists, as shown in Figure 2a-panel E and F. Figures 2a-panel G, H, and I show the quantitation from the X-ray analysis of the coronal (1st and 2nd molars) and root dentin thickness (1st molar only) in at least 2 mice of each genotype at each age, with over 20 measurements per tooth per animal. The age related curves were relative to an exponential associated equation with R values greater than 0.95. The difference in the phenotypes between control and BMP4 cKO mice are highly significant with a P<0.0001. Figure 2b shows μCT analysis of n=4 animals of the control and BMP4 cKOs at 1 year of age for both 1st molar (Figure 2b-Panels C–G), and 2nd molar (Figure 2b-Panel H–L). Figure 2b-Panel A and B shows an example cross section from the μCT and the color coded legend for the volumes that were quantitated in the μCT analysis. There are significant decreases in the total crown volume (TCV), total root volume (TRV), dentin volume in the crown (DVc), pulp volume in the crown (PVc-predentin levels could be in these measurements, although predentin excess in the BMP4 cKO mice was not seen by histology in these older animals), and dentin volume in the root (DVr). The μCT data show similar trends in changed tooth parameters in both 1st and 2nd molars but at least one of them did not reach significance at a P<0.05, as noted.
Fig. 2.
Fig. 2a. High Resolution Digital X-ray analysis and estimation of dentin thickness in control (Panels A, C, E) and BMP4 cKO (Panels B, D, F) mice. These are representative X-rays from 12 day mice (Panel A, B), 1 month (Panel C, D) and 9 month (Panel E, F) mice. The dentin thickness in crown or coronal region and the root region was quantitated in 1 to 2 animals of each genotype at each age, from 12 days to 9 months. The values were then fit to an exponential associated curve using Prism-GraphPad software program (Panel G, H, I). This equation gave R values >0.95 for best curve of analysis. The significance of the difference in the genotype measures between CTR and BMP4 cKO was P<0.0001. Thus there was significantly reduced coronal (arrows) and root (arrowheads) dentin thickness (30 to 35%), as well as apparent enlarged pulp chambers (two white circles-Panel D and F) in BMP4 cKO mice compared to control mice (one white circle-Panel C and D). BMP4 cKO mice have also reduced radio-opacity (two white asterisks-Panel D and F) and display osteopenia like phenotype in the alveolar bone compared to the wild type control mice (one white asterisks-Panel C and E).
Fig. 2b. MicroCT analysis of 1st and 2nd molars of CTR and BMP4 cKOs of 1 year old animals. A set of 4 CTR and 4 BMP4 cKO jaws from 1 year old mice were subjected to μCT analysis by Numira, Inc (www.numirabio.com) using parameters that we developed with them to quantitate various volumetric aspects of the tooth structure. Panel A and B show cross-sections from the μCT analysis representing colored parameters to be measured in the μCT analysis. In the 1st molar, pink represents the volume measure for the enamel (EV-see Figure 6). Yellow represents the volume of the crown mineralized dentin (DVc). Green represents the pulp volume in the crown that would include any predentin (PVc). The light purple represents the dentin volume in the root region (DVr). The red region represents the pulp volume in the root, including any predentin (PVr). Total crown volume (TCV) and total root volume were also measured in these 1 year animals. In a similar manner, color coding for the 2nd molar is light blue for the enamel volume, EV (see Figure 6G), yellow for the dentin volume of the crown region (DVc), dark purple for the pulp volume of the crown (PVc), and light green for the dentin volume in the root (DVr). The outer blue region around the molar teeth represents the periodontal region of unmineralized tissue and showed no significant difference in CTR and BMP4 cKO animals. Panel C to F shown quantitation and t-test for the 1st molar, with significant increase in PVc and decrease in DVr. There were decreases in the other parameters that showed a tread but were not significant. Panels H to L represent the data and P values for the 2nd molar with significant decreases in TCV and DVc. There was an increase in PVc that had a P=0.07 and less significant decreases in TRV and DVr. In at least one of the molars there was a significant decrease in all the volume measurements that were observed.
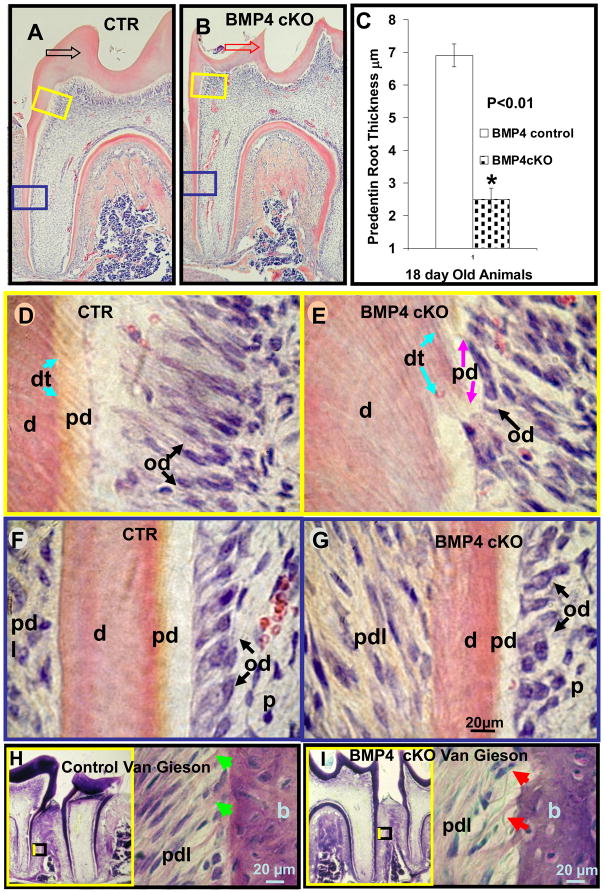
Histological evaluation of the surrounding periodontium was carried out at several stages with the 1st and 2nd molars. As shown as an example in 18 day mouse 1st molars, sections near the mid region of the tooth shown in Figure 3A and B have reduced dentin thickness greater than 30% in the coronal area and over 40% in the developing root dentin. At higher magnification, shown in Figure 3D and E for the coronal region, the odontoblasts are retarded and delayed in their differentiation with very little polarized properties. Very little predentin and much thinner dentinal tubules are noted in the BMP4 cKO mice compared to controls. In the root region where odontoblasts are less mature and dentin formation is actively developing at this stage in the root, the odontoblasts are less elongated and the dentin is much thinner with much less active predentin being laid down. The quantitative coronal and root dentin thickness, as well as the predentin thickness results are summarized in Figure 2a-Panel G, H, I and Figure 3C, respectively. These measurements are derived from at least 2 animals of each genotype and quantitation from 2–3 different sections of each animal. When the tooth sections are stained with Van Gieson which stains predominately collagen fibers, less staining is shown in the dentin and in the alveolar bone between the roots of BMP4cKO mice (Figure 3H, I). Of note is the decreased and disorganized collagen staining pattern in the periodontal ligament region in the BMP4 cKO animals compared to the control animals.
Fig. 3.
Histological evaluation of 1st molar of 18 day old control (A, D, F, H) and BMP4 cKO (B, E, G, I) mice. BMP4 cKO mice have a reduced root predentin thickness of 50% as measured in this set of animals (C and G) compared to the control mice (C and F). From another set of CTR and BMP4 cKO animals of a different litter of the same age, similar reductions in predentin thickness were obtained as in the BMP4 cKO mice (C, G) and as compared to control wild type mice (C, F) (data not shown). There is also less apparent predentin in the coronal area of BMP4 cKO molars (B, E) than in controls (A, D). The predentin of BMP4 cKO mice is disorganized and patchy in character; pink-purple arrows (E). These histological observations were observed in two independent sets of similar aged animals. The dentinal tubules appear thinner and disorganized in BMP4 cKO mice; blue arrows (E) compared to the control mice (D). The boxed areas (yellow) in A and B represent positions and origins of coronal dentin and predentin thickness shown with higher magnifications in D and E. The boxed areas with blue lines in A and B represent positions and origins of root dentin and predentin thickness shown with higher magnifications in F and G. Note the difference in thickness and shape of coronal dentin between CTR and BMP4 cKO mice with hollow arrows in A-black (CTR) and B-red (BMP4 cKO). The coronal and root dentins were also reduced 30 to 35%, as shown in Figure 2G and 2I. Van Gieson stain for collagen (H, I) in periodontium of BMP4 cKO mice have a marked decrease in the characteristic pink stain for collagen - red arrows (I) with the light pink stain representing less mature compact collagen fibers, as seen in normal Sharpie fibers in control mice, green arrows (H). On the left side of (H) and (I) surrounded with yellow frame is the smaller magnification of each picture which contains the smaller boxed area representing positions and origins of higher magnifications shown on the right site. d - dentin; pd - predentin; od - odontoblasts (black arrows); pdl - periodontium; dt - dentin tubules (blue arrows); p - dental pulp; b - bone.
Deletion of BMP4 Gene in Odontoblasts Inhibits Their Maturation to Dentin Matrix Formation and Dentinal Tubule Formation Stages
In order to begin to investigate the mechanism of the observed phenotypes in this BMP4 cKO model, we first carried out key gene expression studies of several dentin matrix and dentinal tubule related gene products by in situ hybridization. Figure 4A–F and I demonstrate that there are 50 to 70% reduction in Collagen type 1 and Osteocalcin expression in odontoblasts, as well as 98% Collagen type 1 reduction in periodontal osteoblasts, of the BMP4 cKO 1st molars of 18 day animals. Hybridization was carried out on two animals per genotype with similar results. Quantitation of expression levels was determined in one set of control and BMP4 cKO slides.
Fig. 4.
Deletion of BMP4 in odontoblasts suppresses expression of dentin matrix and dentinal tubule genes: Collagen type I (Col1a1) (B), Osteocalcin (F) and DMP1 (H) in 12–18 day old BMP4 cKO (B, F, H) mice compared to the control (A, E, G). An example in situ hybridization data are shown from various sections from the same set of control and BMP4 cKO animals. All hybridizations have been repeated in independent set of animals with similar results. Here we show quantitation of signal level from this example and shown in Figure 4I. In situ hybridization expression (blue color), in odontoblasts of BMP4 cKO mice, of Col1a1 mRNA (B), Osteocalcin mRNA (F) and DMP1 (H) was reduced by 47%, 68% and 90% as compared to control mice (A, E, G), respectively (I). We noted a 97% reduced mRNA expression in BMP4 cKO mice of DMP1 in cementoblasts (H-cb, red arrows) and of Col1a1 in periodontal osteoblasts (D-ob, orange arrow) compared to the controls (G-cb, purple arrows and C-ob, brown arrows). On the left side of (A through H) surrounded with yellow frame is the smaller magnification of each picture which contains the smaller boxed area representing positions and origins of higher magnifications shown on the right side. d - dentin; od - odontoblasts; pdl - periodontium; p - dental pulp; pdl - periodontium; b - bone; oc - alveolar bone osteocytes; ob - periodontal osteoblasts; cb - cementoblasts.
Since deletion of the DMP1 gene in mice leads to major defects in dentinal tubule formation and expanded pulp chamber with decreased total dentin thickness [13] similar to the BMP4 cKO phenotype, we examined DMP1 expression in rapidly developing odontoblasts of 12 day 1st molars near the root region and as shown in Figure 4G and H. There was over a 90% reduction of DMP1 in this example where expression is observed in both the cementoblasts and in the odontoblasts, supporting a role of BMP4 in cementogenesis as well as odontogenesis (Figure 4I). Similar results were obtained in sets of animals from other litters of similar age. As cementoblasts differentiate and produce cementum that contain Collagen type I and the BMP4 gene is expressed in cementoblasts, the BMP4 gene was likely deleted in these cementoblasts as well, but we have no direct proof at this time.
Key Transcription Factors and BMP Signaling are Reduced in Odontoblasts of the BMP4 cKO Animals
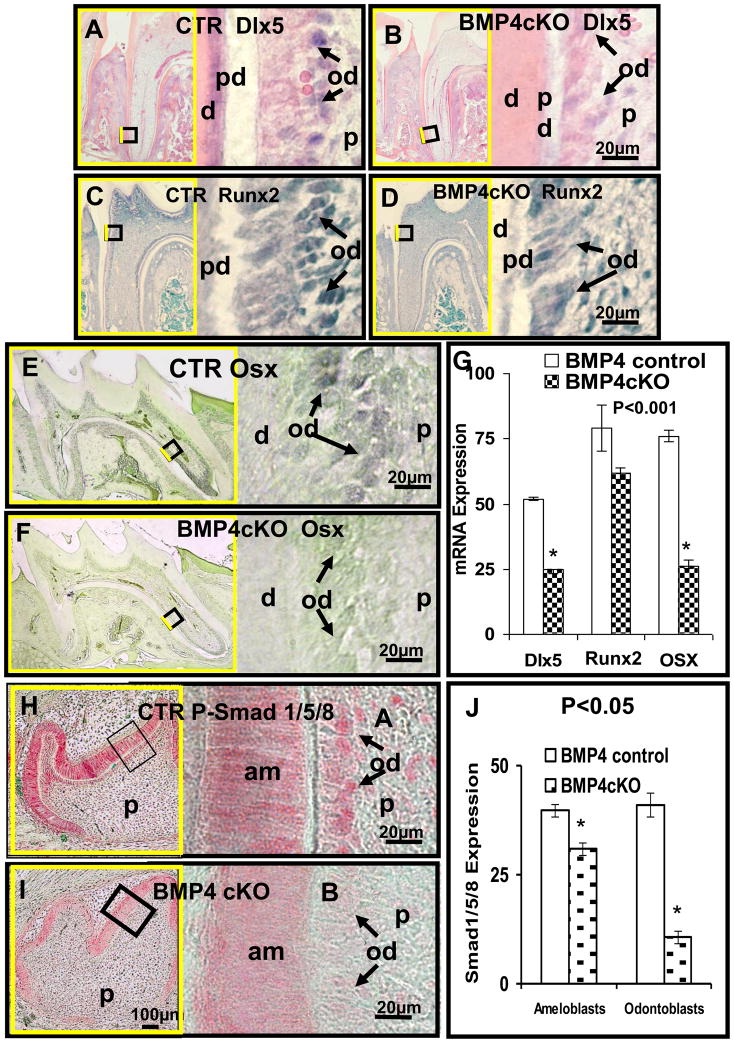
Several transcription factors, Dlx5, Runx2 and Osterix, regulated by BMP signaling and expressed in osteoblasts, chondrocytes, and odontoblasts have been shown when deleted globally to have major affects on the process of tooth formation, where the animals at least survived until birth [14–16]. Their specific roles in postnatal tooth cytodifferentiation are less clear. There is also a suggested hierarchy in that in osteoblasts, at least, BMP signaling induces Osterix expression and is mediated by Dlx5 and independent of Runx2 [17]. We explored expression of these three key transcription factors in odontoblasts in the BMP4 cKO model and found that both Dlx5 and Osterix expression was reduced over 50%, as determined in two independent animals with multiple measurements per animal.
A modest decrease in Runx2 expression was observed (Figure 5A–G) by in situ hybridization. These results suggest the mechanism of action of BMP4 in odontoblasts is at least autocrine for odontoblasts and that the BMP4 induced Osterix expression is required for proper differentiation of odontoblasts. This is supported with deletions of Osterix in embryonic (by conventional knockout approach) or postnatal stage (by conditional knockout approach) which both led to a dentin defect with low mineral content and thin dentin [16].
Fig. 5.
Deleted BMP4 in odontoblasts of 18 day old BMP4 cKO mice molars suppresses expression of transcription factors: Dlx5 (A, B), Runx2 (C, D) and Osterix (E, F) as well as P-Smad1/5/8 representing BMP signaling (H, I). Quantitation shown in Figure G and J was determined in multiple sections in one set of control and BMP4 cKO animals with similar in situ hybridizations done in another set of animals at the same age. Quantitative in situ hybridization (G) shows reduced expression of Dlx5 (B), Runx2 (D) and Osx (F) mRNA in BMP4 cKO odontoblasts of 52%, 22%, 66% compared to control (A, C, E), respectively, in this example. The immunostaining signal (red color) of P-Smad1/5/8, in odontoblasts and ameloblasts of 2 day postnatal BMP4 control mice (H, J) is reduced by 74% in odontoblasts but only 22% in ameloblasts in BMP4 cKO animals (I, J). Similar results were obtained from independent sets animals at age of 2 and 18 days. d - dentin; pd - predentin; od - odontoblasts (black arrows); p - dental pulp; am - ameloblasts.
To determine if there was altered BMP signaling in odontoblasts after deletion of BMP4, we carried out immunocytochemistry with an antibody to phospho-Smad158, a measure of early phosphorylation from the ligand activated BMP receptor II-I complex [18]. As shown with these sections of 1st molars from 12 day animals, as an example, there is a dramatic decrease in P-Smad158 levels of over 70% in the odontoblasts with specific deletion of the BMP4 gene (Figure 5H, I, J). Quantitation was determined in one set of animals and confirmed in an independent set of control and BMP4 cKO sections using similar age animals.
Surprisingly, a reduced level of P-Smad158 immunoreactivity in the ameloblasts (20 to 30%) was observed, suggesting a paracrine role of odontoblast BMP4 on the process of amelogenesis and enamel formation. We have also noted a delay in enamel formation in the BMP4 cKO mice compared to control littermates (Figure 6).
Fig. 6.
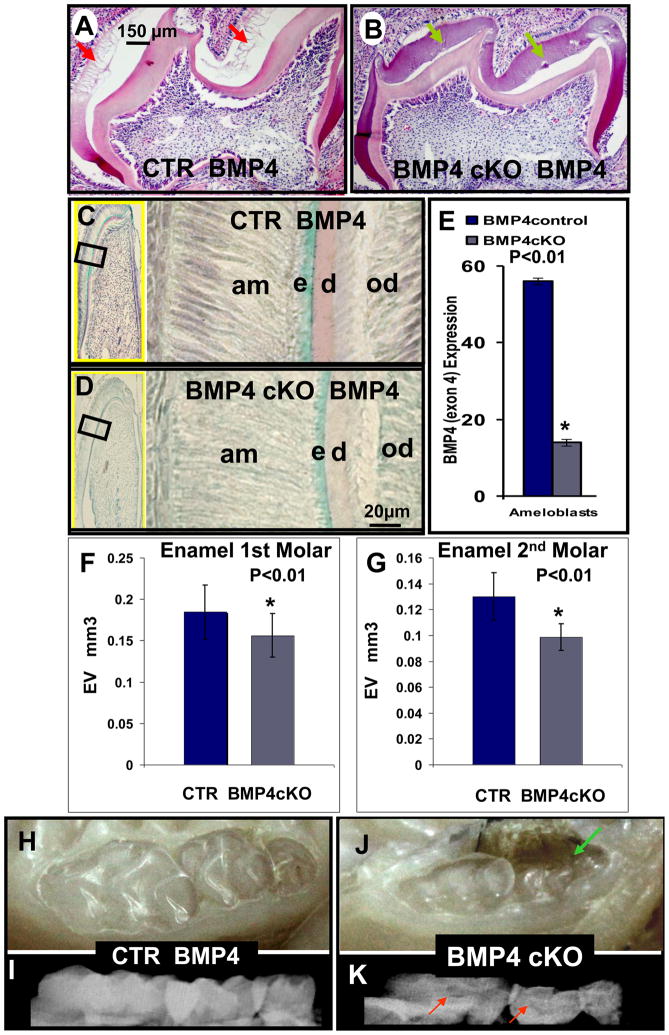
Hematoxylin - eosin staining (A, B) and in situ hybridization of BMP4 expression (C, D) in CTR (A, C) and BMP4 cKO (B, D) mice at 8–12 days; Photographs (H and J) and X-rays (I and K) of CTR (H and I) and BMP4 cKO (J and K) mice at 1 year. Hematoxylin – eosin staining, after demineralization of wild type mice, shows that tooth immature enamel matrix was not present as the enamel has matured to >90% mineral phase with little matrix proteins (A - red arrows); however, this immature enamel matrix was still present in BMP4 cKO mice (B - green arrows). These observations were seen in 2 independent sets of animals. BMP4 expression is also reduced in ameloblasts (C and D) of BMP4cKO mice and quantitated in multiple sections and measurements in CTR and BMP4 cKO animals as shown in (E). By quantitative μCT analysis with 4 independent CTR and BMP4 cKO 1 year animals, a 20 to 30% reduction in the enamel volume (EV) as shown in F and G for the 1st and 2nd molars was observed. Also, the BMP4 cKO mice have more transparent teeth (J) with unequal distribution of mineral on the X-ray image, red arrows (K) in comparison to control animals (H and I). We note in some BMP4 cKO animals an apparent periodontal loss that needs further investigation, green arrow (J).
BMP4 from Odontoblasts Plays a Paracrine Role in stimulating BMP4, Amelogenin and Dlx3 expression in Ameloblasts, as well as Maturation of the Enamel
After decalcification, sectioning and staining with Hematoxylin-eosin in teeth from mature Wt or control animals, we observed a blank space where the mature enamel normally exists. As noted in Figure 6A and B, the enamel matrix in BMP4 cKO animals is delayed in its maturation process, as indicated by the presence of the immature enamel matrix proteins. We then explored the removal of BMP4 in odontoblasts and how this would alter BMP4 expression in the ameloblasts. This observation suggests a paracrine role of odontoblast BMP4 on amelogenesis. As shown in Figure 6C, D and E, from results of in situ hybridization, BMP4 expression is reduced 70% in the ameloblasts in this example from one set of animals. From μCT analysis of n=4 of control and n=4 of BMP4 cKO mice, a significant decrease of 20–30% in the enamel volume (EV), as shown in Figure 6F (1st molar) and 6G (2nd molar) was observed. Visual inspection of the teeth in 1 year control (6H) and BMP4 cKO (6J) animals showed less apparent enamel (Figure 6H and J) and unequally distributed mineral opacity on the surface of the X-rayed teeth (Figure 6I and K), supporting the μCT analysis of 4 independent animals of each genotype from different litters, 1 year of age.
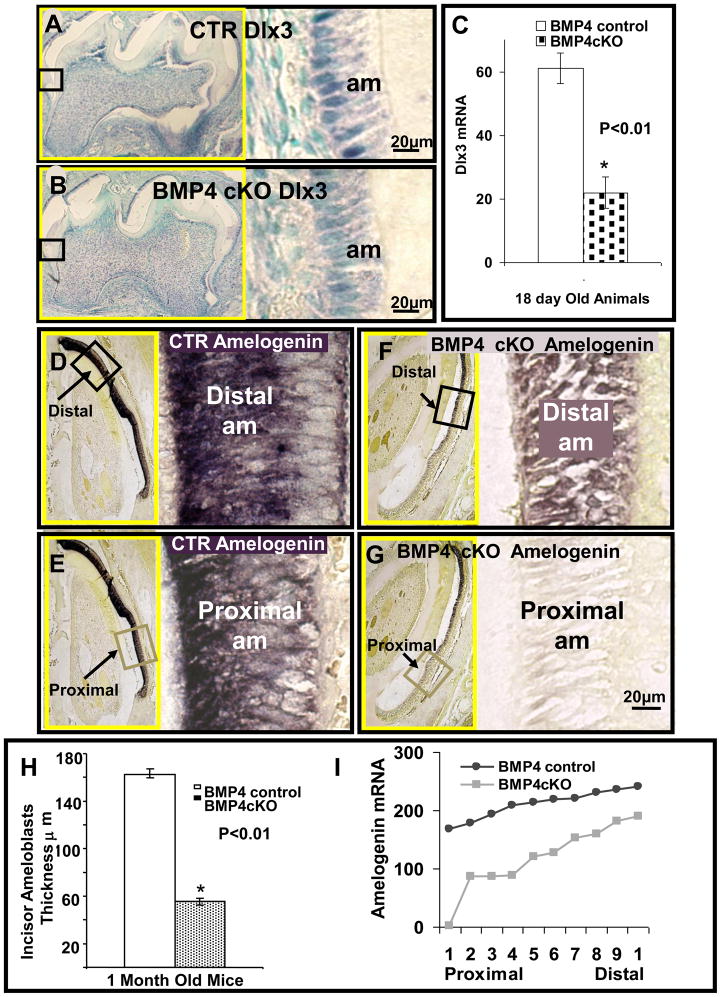
Dlx3 is highly expressed in ameloblasts and controls many of the downstream genes involved in amelogenesis, such as Ameloblastin and Amelogenin. Figure 7A and B demonstrate from this example of 12 day animals that Dlx3 expression in ameloblasts of 1st molars is reduced in BMP4 cKO animals and correlates to less elongated ameloblasts. Quantitation of Dlx3 in this example shows a 64% reduction of Dlx3 expression in the BMP4 cKO mice. Similar results were observed in another set of control and BMP4 cKOs (data not shown). In Figures 7D–G and I, we demonstrate that Amelogenin expression is also decreased 50% in this example of BMP4 cKO mice compared to control mice at this 12 day stage. As noted in other sets of animals, the ameloblasts are not nearly as elongated in the BMP4 cKOs as the control littermates, both in the proximal and distal regions. These expression patterns of Amelogenin in control and BMP4 cKO mice were confirmed in independent sets of animals of similar age (data not shown).
Fig. 7.
Figure 7 shows in situ hybridization of Dlx3 (A–C) and Amelogenin (D–G and I) in ameloblasts of control (A, D, E) and BMP4 cKO (B, F, G) mice. An example of the hybridization signal for Dlx3 is shown in Figure A and B for one set of CTR and BMP4 cKO animals, with quantitation from this example shown in Figure 7C from multiple sections. Similar results have been seen with an independent set of CTR and BMP4 cKO animals of similar age (data not shown). Representative sections for Dlx3 were from molars (A, B) and for Amelogenin from incisors (D–G), in 18–30 day old mice. The expressions of Dlx3 mRNA in control mice (A) are reduced in BMP4 cKO (B) mice by 64% (C) in this example. Amelogenin mRNA hybridization signal (tones of dark purple to black color) in control ameloblasts (D, E) is higher on average 59% (I) as compared to BMP4 cKO ameloblasts (F, G). In control mice, expression of Amelogenin mRNA of proximal less mature incisor ameloblasts is 27% lower (E) from more mature distal ameloblasts (D). In BMP4 cKO mice, expression of Amelogenin mRNA in proximal incisor ameloblasts is barely detectable (G) compared to distal incisor ameloblasts (F). Distal expression of Amelogenin in control mice (D) is increased by 58% compared to BMP4 cKO mice (F, I) in this example. Proximal expression of Amelogenin in BMP4 cKO mice (G) decreased to an undetectable level compared to control mice (E, I). A similar incisor gradient of Amelogenin expression was observed in an independent set of animals of similar age. Ameloblast (am) thickness in incisors was also reduced by 65% in BMP4 cKO mice compared to the control mice (H), as determined in this set of CTR and BMP4 cKO animals. On the left side of A, B and D–G, surrounded with yellow frame is the smaller magnification of each picture which contains the smaller boxed area representing positions and origins of higher magnifications shown on the right side.
Surprisingly, we found in 1 month old incisors which are continually undergoing amelogenesis and enamel formation that Amelogenin expression, a late marker in the process of amelogenesis, is greatly reduced in the proximal and the less mature region in the BMP4 cKO animals. There is less of a difference in the distal more mature region compared to control mice. These results are quantitated in Figure 7I from one set of animals at 12 days of age. The overall thickness of the ameloblast layer is reduced 60 to 70% in the BMP4 cKO incisors from this example and observed in similar sets of littermates as shown in Figure 7H. Thus, the deletion of the BMP4 gene in odontoblasts leads to disruption of the process of amelogenesis and enamel formation. These observations in young animals are most likely related to the phenotype observed in older 1 year old animals in which the enamel appears hypomineralized and thinner in nature and may represent another form of amelogenesis imperfecta (AI).
Discussion
These studies demonstrate a requirement of BMP4 for proper tooth cytodifferentiation in the period from birth to approximately 20 days after birth, as well as for tooth maturation up to 1 year of age. We used the 3.6Col1a1-cre mouse model for early deletion of BMP4 in early Collagen type I producing odontoblasts. Deletion of the BMP4 gene was activated in early odontoblasts of the 1st molar at E18 to P0 when Collagen type I begins to be expressed. We found both odontogenesis and amelogenesis disrupted, with the final consequence of enlarged pulp chambers, less dentin in both the crown and root region, and thinner and hypomineralized enamel. This combined amelogenesis and dentinogenesis imperfecta phenotype resulted in teeth morphology found in a variety of human tooth diseases, such as mutations of the Dlx 3 gene and others [19–24]. It is likely that mutations in the BMP4 gene and signaling pathways are involved in human tooth disorders, but to date none have been well documented.
Previous studies have shown that BMP4 is highly expressed in both the odontoblasts and ameloblasts during postnatal tooth cytodifferentiation [25]. We have shown in these studies that deletion of the BMP4 gene early in the odontogenic process leads to down-regulation of Dlx5 and Osterix in odontoblasts. Dlx5 has been shown to regulate Collagen type I transcription; thus, this could explain the decreased col1a1 expression seen in both the odontoblasts and associated osteoblasts in the periodontium [26]. DMP1 expression in odontoblasts is decreased in this model and can explain the disrupted dentinal tubule formation [13]. Dlx5 is thought to directly regulate the Osterix gene, and decreased expression could help explain the decreased Osterix expression in odontoblasts [27]. As part of the mechanism for the disrupted and delayed odontogenesis and dentin formation, we have shown a dramatic decrease in BMP signaling, using the P-Smad158 immunocytochemistry. After observing the decreased BMP signaling in the ameloblasts as well as odontoblasts, we determined that BMP4 gene expression in ameloblasts is decreased and correlates to reduced Dlx3, Amelogenin, and Ameloblastin gene expression (data not shown). The phenotype is permanent and persistent and not just a delay in the early cytodifferentiation phase. As noted in Figure 2a, the molar dentin continues to expand as does the jaw with age up to at least 9 months of age; however, by 9 months of age, we observed a 30% decrease in the dentin thickness and volume in the BMP4 cKO animals (monitored at 1 year of age by μCT). There is an apparent enlargement of pulp chambers in the molars and less volume of dentin and enamel that was significant in at least one of the molars. Clearly, in odontoblasts, BMP4 is a critical ligand for proper tooth cytodifferentiation and continued postnatal development up to 1 year of age.
What is remarkable in these studies is the potential role of BMP4 in these processes and how similar the process is to overt tooth development in the embryo starting at E10.5. At this stage, epithelial BMP4 stimulates the underlying mesenchyme to produce BMP4 in an Msx1 dependent manner [28]. The mesenchymal BMP4 then signals in a paracrine manner to the epithelium to continue growing and developing. BMP4 from the mesenchymal component also functions in an autocrine manner to drive the mesenchymal development process, leading to a well organized bud stage of the tooth germ. Similarly, our data suggests that the odontoblast BMP4 is functioning in both a paracrine and autocrine manner to coordinate the simultaneous cytodifferentiation of the mesenchymal dentin and the epithelial derived enamel. This hypothesis has long been suggested by a variety of experiments and observations [29, 30]. Perhaps the high levels of BMP4 in the preameloblasts initiate this process as in the embryonic paradigm. BMP4 and possibly other BMPs activate the preodontoblasts to increase BMP4 production that drives the odontogenic process and feeds back on the ameloblasts to continue their cytodifferentiation process. Recent experiments show Osterix, downstream of BMP4, was specifically deleted in the differentiating odontoblasts using a DMP1-cre model [31]. Not only was odontogenesis disrupted but the process of amelogenesis was also altered, again supporting the connection of odontogenesis and amelogenesis. Specific deletion of BMP4 in the ameloblasts and analysis of both amelogenesis and odontogenesis should help test some of these ideas. A BMP4 enhancer that controls expression only in ameloblasts is now being used to create a Cre model to conditionally remove the BMP4 gene in this cell lineage [25].
Previous studies have suggested a role of BMP4 in tooth cytodifferentiation and that recombinant BMP4 protein or overexpression of BMP4 driven by a cDNA to the mRNA can in fact drive the whole tooth formation process to well formed dentin and enamel. Most of these studies have been carried out in an ectopic kidney capsule site [3, 4, 6]. However, overexpression studies do not rigorously demonstrate that the causative gene is BMP4 itself. This requires specific deletion of the BMP4 gene, as shown in these studies. The first specific deletion of the BMP4 gene, using the Nkx2.5-cre model, in which Cre is targeted to the mandibular epithelial and endothelial components, clearly demonstrates a role of the BMP4 gene in the process of mandibular development and tooth formation. These data again suggest that an initial epithelial BMP4 signal may be very important in the whole process of tooth cytodifferentiation and will be tested in the future. In our studies, we have demonstrated that at least the preodontoblast BMP4 is required for continued stages of tooth cytodifferentiation.
Recent studies in our lab have also uncovered a role of the BMP2 gene in postnatal tooth development. Using the same 3.6Col1a1-cre and floxed BMP2 gene, we noted a delay in both dentin formation and enamel. However, the phenotype is different in several ways. First, DMP1 expression is actually increased in the BMP2 cKO mice, but late markers such as Amelogenin and Ameloblastin are dramatically decreased. This suggests a hierarchal use of BMP2 and BMP4 in odontogenesis, in which the action of BMP4 precedes BMP2 in odontogenesis, at least with the 3.6Col1a1-cre model. Deletion of BMP2 in odontoblasts leads to an accumulation of altered or dimorphic odontoblasts at this dentinal tubule stage and failure to mature to the later stages. We have also deleted BMP2 at an earlier stage in tooth development using the Sp7 (Osterix)-cre model. Dental pulp cells as well as odontoblasts express this Sp7-cre, as assayed using an EGFP reporter linked to the Cre cassette. These mice are under analysis, but the phenotype appears more like the 3.6Col1a1-cre; BMP4 model with dramatic decrease in DMP1 and BMP4 expression. Thus the BMP2 gene may play a very early role and be important in activation of BMP4. This activated BMP4 then plays a critical role in activating BMP2 in later stage odontoblasts that drive them to the mature stage and coupling to amelogenesis. Further analysis is underway to sort out these possible interactions.
In summary, we have deleted the BMP4 gene at early stages in the mesenchymal derived preodontoblasts and shown a permanent development of defective teeth. We show that, in odontoblasts, BMP4 is acting in both a paracrine and autocrine fashion to coordinate both the processes of dentin and enamel formation.
Acknowledgments
Kulessa H and Hogan BLM are acknowledged for the development of BMP4 floxed mouse.
The work was supported by National Institute of Health research grants NIDCR DE16949 and DE018865, NIAMS AR054616 and AR46798.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vainio S, Karavanova I, Jowett A, Thesleff I. Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell. 1993;75:45–58. [PubMed] [Google Scholar]
- 2.Maas R, Bei M. The genetic control of early tooth development. Critical Reviews in Oral Biology & Medicine. 1997;8:4–39. doi: 10.1177/10454411970080010101. [DOI] [PubMed] [Google Scholar]
- 3.Bei M, Kratochwil K, Maas RL. BMP4 rescues a non-cell-autonomous function of Msx1 in tooth development. Development. 2000;127:4711–8. doi: 10.1242/dev.127.21.4711. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Song Y, Zhang X, Tang J, Chen J, Chen Y. Msx1/Bmp4 genetic pathway regulates mammalian alveolar bone formation via induction of Dlx5 and Cbfa1. Mechanisms of Development. 2003;120:1469–79. doi: 10.1016/j.mod.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Mina M, Kollar EJ. The induction of odontogenesis in non-dental mesenchyme combined with early murine mandibular arch epithelium. Archives of Oral Biology. 1987;32:123–7. doi: 10.1016/0003-9969(87)90055-0. [DOI] [PubMed] [Google Scholar]
- 6.Ohazama A, Tucker A, Sharpe PT. Organized tooth-specific cellular differentiation stimulated by BMP4. Journal of Dental Research. 2005;84:603–6. doi: 10.1177/154405910508400704. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, Schwartz RJ, Maxson R, Furuta Y, Martin JF. Threshold-specific requirements for Bmp4 in mandibular development. Developmental Biology. 2005;283:282–93. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 8.Winnier G, Blessing M, Labosky PA, Hogan BL. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes & Development. 1995;9:2105–16. doi: 10.1101/gad.9.17.2105. [DOI] [PubMed] [Google Scholar]
- 9.Gluhak-Heinrich J, Guo D, Yang W, Martinez L, Harris M, Kulessa H, Lichtler AC, Kream B, Zhang J, Feng JQ, Harris SE. Role of Bone Morphogenetic Protein 4 (BMP-4) in Tooth Development. In: Vukicevic S, Sampath KT, editors. Bone Morphogenetic Proteins: From Local to Systemic Therapeutics. Vol. 199. Basel, Switzeland: Birkhauser Verlag; 2008. p. 211. [Google Scholar]
- 10.Kulessa H, Hogan BL. Generation of a loxP flanked bmp4loxP-lacZ allele marked by conditional lacZ expression. Genesis: the Journal of Genetics & Development. 2002;32:66–8. doi: 10.1002/gene.10032.abs. [DOI] [PubMed] [Google Scholar]
- 11.Liu F, Woitge HW, Braut A, Kronenberg MS, Lichtler AC, Mina M, Kream BE. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. International Journal of Developmental Biology. 2004;48:645–53. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- 12.Gluhak-Heinrich J, Yang W, Harris M, Harris SE. Quantitative In Situ Hybridization with Enhanced sensitivity in Soft, Bone and Tooth Tissue Using Digoxigenin Tagged RNA Probes. Boiochemia Medica. 2008;18:59–80. [Google Scholar]
- 13.Ye L, MacDougall M, Zhang S, Xie Y, Zhang J, Li Z, Lu Y, Mishina Y, Feng JQ. Deletion of dentin matrix protein-1 leads to a partial failure of maturation of predentin into dentin, hypomineralization, and expanded cavities of pulp and root canal during postnatal tooth development. J Biol Chem. 2004;279:19141–19148. doi: 10.1074/jbc.M400490200. [DOI] [PubMed] [Google Scholar]
- 14.D’Souza RN, Aberg T, Gaikwad JS, Cavender A, Owen M, Karsenty G, Thesleff I. Cbfa1 is required for epithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–2920. doi: 10.1242/dev.126.13.2911. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Marijanovic I, Kronenberg MS, Erceg I, Stover ML, Velonis D, Mina M, Heinrich JG, Harris SE, Upholt WB, Kalajzic I, Lichtler AC. Expression and function of Dlx genes in the osteoblast lineage. Developmental Biology. 2008;316:458–70. doi: 10.1016/j.ydbio.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapadia H, Xie Y, Zhou X, Crombrugghe BD, D’Souza RN, Feng JQ. Osterix is critical for odontogenesis and condyle formation. 87 General session of the IADR; Miami FL. 2009. [Google Scholar]
- 17.Yagi K, Tsuji K, Nifuji A, Shinomiya K, Nakashima K, DeCrombrugghe B, Noda M. Bone morphogenetic protein-2 enhances Osterix gene expression in chondrocytes. Journal of Cellular Biochemistry. 2003;88:1077–83. doi: 10.1002/jcb.10467. [DOI] [PubMed] [Google Scholar]
- 18.Massague J. Integration of Smad and MAPK pathways: a link and a linker revisited. Genes & Development. 2003;17:2993–2997. doi: 10.1101/gad.1167003. [DOI] [PubMed] [Google Scholar]
- 19.Dong J, Amor D, Aldred MJ, Gu T, Escamilla M, MacDougall M. DLX3 mutation associated with autosomal dominant amelogenesis imperfecta with taurodontism. American Journal of Medical Genetics Part A. 2005;133A:138–41. doi: 10.1002/ajmg.a.30521. [DOI] [PubMed] [Google Scholar]
- 20.Lee SK, Lee ZH, Lee SJ, Ahn BD, Kim YJ, Lee SH, Kim JW. DLX3 mutation in a new family and its phenotypic variations. Journal of Dental Research. 2008;87:354–7. doi: 10.1177/154405910808700402. [DOI] [PubMed] [Google Scholar]
- 21.Wright JT, Hong SP, Simmons D, Daly B, Uebelhart D, Luder HU. DLX3 c.561_562delCT mutation causes attenuated phenotype of tricho-dento-osseous syndrome. American Journal of Medical Genetics Part A. 2008;146:343–9. doi: 10.1002/ajmg.a.32132. [DOI] [PubMed] [Google Scholar]
- 22.Lee SK, Lee KE, Jeon D, Lee G, Lee H, Shin CU, Jung YJ, Lee SH, Hahn SH, Kim JW. A novel mutation in the DSPP gene associated with dentinogenesis imperfecta type II. Journal of Dental Research. 2009;88:51–5. doi: 10.1177/0022034508328168. [DOI] [PubMed] [Google Scholar]
- 23.Haruyama N, Sreenath TL, Suzuki S, Yao X, Wang Z, Wang Y, Honeycutt C, Iozzo RV, Young MF, Kulkarni AB. Genetic evidence for key roles of decorin and biglycan in dentin mineralization. Matrix Biology. 2009;28:129–36. doi: 10.1016/j.matbio.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chae YM, Heo SH, Kim JY, Lee JM, Ryoo HM, Cho JY. Upregulation of smpd3 via BMP2 stimulation and Runx2. BMB reports. 2009;42:86–90. doi: 10.5483/bmbrep.2009.42.2.086. [DOI] [PubMed] [Google Scholar]
- 25.Feng JQ, Zhang J, Tan X, Lu Y, Guo D, Harris SE. Identification of cis-DNA regions controlling Bmp4 expression during tooth morphogenesis in vivo. Journal of Dental Research. 2002;81:6–10. doi: 10.1177/002203450208100103. [DOI] [PubMed] [Google Scholar]
- 26.Tadic T, Erceg I, Stover ML, Rowe DW, Lichtler AC. Dlx5 induces expression of COL1A1 promoter contained in a retrovirus vector. Croatian Medical Journal. 2001;42:436–9. [PubMed] [Google Scholar]
- 27.Chen S, Gluhak-Heinrich J, Wang YH, Wu YM, Chuang HH, Chen L, Yuan GH, Dong J, Gay I, MacDougall M. Runx2, Osx and Dspp expression in tooth development. Journal of Dental Research. 2009 doi: 10.1177/0022034509342873. in press manuscript # 08-0369RRR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bei M, Maas R. FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development. 1998;125:4325–33. doi: 10.1242/dev.125.21.4325. [DOI] [PubMed] [Google Scholar]
- 29.Veis A. Amelogenin gene splice products: potential signaling molecules. Cellular & Molecular Life Sciences. 2003;60:38–55. doi: 10.1007/s000180300003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iacob S, Veis A. Identification of temporal and spatial expression patterns of Amelogenin isoforms during mouse molar development. European Journal of Oral Sciences. 114(Suppl 1):194–200. doi: 10.1111/j.1600-0722.2006.00287.x. discussion 201–2. [DOI] [PubMed] [Google Scholar]
- 31.Lu Y, Xie Y, Zhang S, Dusevich V, Bonewald LF, Feng JQ. DMP1-targeted Cre expression in odontoblasts and osteocytes. Journal of Dental Research. 2007;86:320–5. doi: 10.1177/154405910708600404. [DOI] [PubMed] [Google Scholar]