Abstract
An HIV Env immunogen capable of eliciting broad immunity is critical for a successful vaccine. We constructed and characterized Adenovirus 5 host range mutant (Ad5hr) recombinants encoding HIVSF162 gp160 (subtype B) and HIVTV1 gp160 (subtype C). Immunization of mice with one or both induced cellular immunity to subtype B and C peptides by ELISpot, and antibody responses with high binding titers to HIV Env of subtypes A, B, C, and E. Notably, Ad5hr-HIVTV1 gp160 induced better cellular immunity than Ad5hr-HIVSF162 gp160, either alone or following co-administration. Thus, the TV1 Env recombinant alone may be sufficient for eliciting immune responses against both subtype B and C envelopes. Further studies of Ad5hr-HIVTV1 gp160 in rhesus macaques will evaluate the suitability of this insert for a future phase I clinical trial using a replication-competent Ad4 vector.
Keywords: replication-competent adenovirus, HIV envelope, vaccine
1. Introduction
Recently, the Centers for Disease Control and Prevention released the first estimates of HIV incidence in the United States from the nation’s new HIV incidence surveillance system [1]. This study revealed that 56,300 new HIV infections occurred in the United States in 2006, far higher than the previous estimate of 40,000 new infections annually. Whether this result reflects an actual increase, or simply better methods of prediction, it highlights the urgent need to halt the spread of the HIV/AIDS epidemic.
The most effective method for preventing new infections is vaccination. However, in the case of the HIV/AIDS pandemic, despite over 2 decades of research there are no prospects in the near future for a vaccine that would induce “sterilizing immunity”. The development of an Env immunogen able to elicit broad neutralizing antibody which completely blocks HIV entry and initial infection, thus providing sterilizing immunity, is ongoing and elusive. In the absence of potent neutralizing antibody, it is believed that protection against HIV will require both humoral and cellular immune responses at systemic and mucosal sites. Non-neutralizing antibodies (NNAbs) may contribute to control of infected cell foci in the brief window of time following HIV transmission prior to systemic spread of the virus [2]. By binding to Fc receptors on effector cells and targeting Env antigens on the surface of virus-infected cells, NNAbs can mediate several activities that eliminate the infected cells or inhibit viral replication, including antibody dependent cellular cytotoxicity (ADCC), antibody dependent cell-mediated viral inhibition (ADCVI), and opsonization [3-8]. In fact, vaccine-elicited NNAbs have been shown to correlate with reduced viral burdens following challenge in macaques [6-8]. On the other hand, T-cell responses also contribute to control of virus replication and progression to AIDS. Nevertheless, the elicited cellular immunity must be broadly reactive in order to protect against infection by the spectrum of HIV isolates across the diverse subtypes. Thus, additional vaccine components, including more conserved viral genes such as gag, pol, and nef, will likely be necessary. For our replicating Ad-recombinant approach, these are incorporated into vaccine regimens as separate recombinants. Here, we have investigated only the env component.
Both naked DNA and viral vectors encoding HIV genes of interest have been exploited in HIV vaccine design. Adenovirus vectors are among the most widely tested HIV vaccine vehicles. They are non-enveloped, double-stranded DNA viruses with an icosahedral structure made up of an array of a dozen different proteins, and are able to infect dividing and non-dividing cells. Their genome of approximately 36-kb is easily manipulated to generate recombinant constructs and can be grown to high titer. We have pursued a replication-competent Ad-recombinant vaccine approach [9, 10] with the rationale that a replicating vector will best mimic the highly successful live attenuated vaccines which give essentially life-long protection against diseases such as polio, yellow fever, smallpox, and measles. In fact, in the HIV field, a live attenuated SIV vaccine has exhibited the best efficacy in protecting macaques against SIV infection to date [11] yet due to eventual reversion to virulence is deemed too unsafe for use in people [12]. In addition to mimicking live attenuated SIV (or HIV) a replicating Ad-recombinant should also target mucosal inductive sites leading to induction of mucosal immune responses.
Adenoviruses are very species specific, with replication of human Ad restricted to people and chimpanzees. However, use of an Ad5 host range mutant vector (Ad5hr) which can replicate in cells of non-human primates [13] allows study of candidate replicating Ad-recombinant vectored vaccines in SIV or SHIV rhesus macaque monkey models. Replication competence is maintained by preserving the Ad E1 region and replacing the Ad E3-region with a transgene of interest for expression in target cells [14]. The Ad E3 region encodes proteins involved in evading host immunity and is non-essential for viral replication. We have shown that replication-competent Ad-HIV recombinants elicit better cellular immune responses and prime higher antibody titers compared to animals immunized with non-replicating Ad-HIV recombinants [15]. Further, in rhesus macaques, priming with replicating Ad recombinants and boosting with envelope protein has resulted in strong immune responses [16, 17] and potent protection against both SIV [18, 19] and SHIV [20-22] challenges.
Selection of an appropriate env gene for insertion into a candidate Ad-HIV vaccine is critical. In order for the vaccine to be applicable to a global population, the ideal insert should elicit broad immune responses and recognize a spectrum of HIV isolates across subtypes. In addition, several lines of evidence have shown that initial HIV infections are established by macrophage tropic HIV isolates that utilize primarily CCR5 (R5) rather than CXCR4 (X4) co-receptors [23-25]. Hence, R5 strains of HIV are preferred for prophylactic HIV vaccine development. Previously, our preclinical vaccine studies evaluating HIV envelope immunogens made use of subtype B X4-tropic env inserts, including Ad4-, Ad5-, or Ad7-HIVIIIB or –HIVMN gp160 [15, 26, 27]. Similarly, for evaluating HIV Env in rhesus macaques we have used an Ad5hr recombinant expressing the subtype B dual-tropic HIV89.6P gp140 [20-22]. For this study we constructed replication-competent Ad5hr-HIV recombinants expressing the gp160 glycoprotein of subtype B (HIVSF162) and subtype C (HIVTV1) R5 strains. Subtype B HIV isolates are prevalent in North America, Latin America, the Caribbean, Europe, Japan, and Australia, while subtype C HIV isolates are prevalent in India and South Africa. Overall subtype C isolates are the most prevalent worldwide. The immunogenicity of both R5-tropic subtype B and C recombinants was evaluated in mice as a first step in selecting one or both env inserts for use in future clinical development of a replicating Ad-HIV vaccine. The Ad5hr-HIVTV1 recombinant was shown to be more immunogenic than Ad5hr-HIVSF162 and therefore a logical choice as an initial candidate immunogen. Recently initiated immunogenicity and protective efficacy studies in rhesus macaques will validate this selection.
2. Materials and methods
2.1. Ad5hr-HIV recombinants
Replication-competent Ad5hr recombinants carrying the HIVTV1 and HIVSF162 gp160 genes were constructed. The shuttle plasmid, pBRAd5ΔE3 containing the Ad5 sequence from 59.5 to 100 map units (mu) with a 78.8 to 85.7 mu deletion in the E3 region, and the plasmid carrying the Ad5 tripartite leader (pAd5tpl-18RD2) were obtained from Wyeth-Lederle Vaccines under a Cooperative Research and Development Agreement. The HIVTV1 and HIVSF162 gp160 genes, optimized for expression in mammalian cells, were obtained from Chiron Corporation, Emeryville, CA, as pCMVlink160mod-TV1 and pCMVlink160mod-SF162 plasmids, respectively.
A Kozak sequence (GCCACC) was inserted immediately upstream of the start codon of the HIVTVI gp160 gene. This was done by incorporating the sequence into the TV1.EcoRI.Kz.S primer (Table 1) which together with the TV1.DraIII.AS primer were used to amplify the first ~200bp of the gene. The synthetic PCR products were then digested with EcoRI and DraIII and used to replace the corresponding fragment in the original plasmid.
Table 1. Primers used in this study.
The translation initiation codon sequence is shown in bold. Enzyme recognition sites are underlined. Mutated nucleotide is indicated in white character on black background
| Name | Polarity | Sequence (5′→3′) |
|---|---|---|
| TV1.EcoRI.Kz.S | Sense | TATAGAATTCGCCACCATGCGCGTGATGGGCACCCAG |
| TV1.DraIII.AS | Antisense | TTCTCGGTCACGTTGCCCAGCA |
|
| ||
| 5-wt L01 | Sense | AATTCGCCACCATGCGCGTGAAGGGC |
| 5-wt L02 | Sense | ATCCGCAAGAACTACCAGCACCTGTGGCGCGGCGGCACCC |
| 5-wt L03 | Sense | TGCTGCTCCCGATGCTGATGATCTGCAGCGCCGTGGAGAA |
| 5-wt L04 | Sense | GCTGTGGGTGACCGTGTACTACGGCGTGCCCGTGTGGAAG |
| 5-wt L05 | Sense | GAGGCCACCACCACCCTGTTCTGCGCCAGCGACGCCAAG |
| 5-wt L06 | Sense | GCCTACGACACCGAGGTGCACAAC |
| 3-wt L01 | Antisense | GTGCACCTCGGTGTCGTAGGCCTTGGCGTCGCTGGCGCAGA |
| 3-wt L02 | Antisense | ACAGGGTGGTGGTGGCCTCCTTCCACACGGGCACGCCGT |
| 3-wt L03 | Antisense | AGTACACGGTCACCCACAGCTTCTCCACGGCGCTGCAGATC |
| 3-wt L04 | Antisense | ATCAGCATGCCCAGCAGCAGGGTGCCGCCGCGCCACAGGTG |
| 3-wt L05 | Antisense | CTGGTAGTTCTTGCGGATGCCCTTCACGCGCATGGTGGCG |
|
| ||
| SF162.BamHI.KO.S | Sense | AACCTGCTGCAGTACTGGATTCAGGAGCTGAAG |
| SF162.BamHI.KO.AS | Antisense | ATCCAGTACTGCAGCAGGTTGCCCCAGTAC |
|
| ||
| BGHpA.XhoI.S | Sense | TAACTCGAGCTGTGCCTTCTAGTTGCCAGC |
| BGHpA.XbaI.AS | Antisense | TAATCTAGACCATAGAGCCCACCGCATCCC |
|
| ||
| Ad5E3-P1 | Sense | TACGAGAGAACCTCTCCGAG |
| Ad5E3-P2 | Antisense | ACAGGCTGGCTCCTTAAAAT |
The tPA leader sequence in the pCMVlink160mod-SF162 plasmid was changed to an optimized wild type SF162 leader. Briefly, 11 oligonucleotides (5-wt L01 to 5-wt L06 and 3wt L01 to 3wt L05; Table 1) were phosphorylated with T4 polynucleotide kinase (Invitrogen) and boiled for 90 sec. The nucleotide mixture was allowed to cool down slowly to room temperature and subsequently ligated to the pCMVlink160mod-SF162 plasmid from which the EcoRI~DraIII fragment was already excised.
A BamHI site present in the HIVSF162 gp160 sequence was removed without changing the expressed amino acid sequence using the Gene Tailor Site-Directed Mutagenesis System (Invitrogen) according to the manufacturer’s instructions with some modification. Briefly, the target plasmid was methylated, followed by PCR amplification using AccuPrime Pfx Supermix (Invitrogen) with a pair of overlapping primers (SF162.BamHI.KO.S and SF162.BamHI.KO.AS; Table 1). The sense primer contained a single C→T nucleotide change. The amplification products were then transformed into One-Shot MAX Efficiency DH5α-T1 (Invitrogen) in which the methylated template DNA is digested by the host. Expression of gp160 by both TV1 and SF162 plasmids was confirmed by Western Blotting.
Using pCDNA3.1(−) plasmid (Invitrogen) as a template, the Bovine Growth Hormone (BGHpA) poly-A signal sequence was PCR-cloned using BGHpA.XhoI.S and BGHpA.XbaI.AS primers (Table 1) and inserted at the XhoI/XbaI site downstream of the HIV gp160 gene in both the TV1 and SF162 plasmids. The ~2.8kb SalI/XbaI fragments of the resulting plasmids were subsequently cloned downstream of Ad5tpl in the pAd5tpl-18RD2 plasmid to generate pAd5tpl-TV1-BGHpA or pAd5tpl-SF162-BGHpA plasmids, respectively. Finally, after removing the vector portion of these plasmids by NheI/XbaI digestion, the constructs were subcloned into the pBRAd5ΔE3 shuttle plasmid at the XbaI site. The resulting shuttle plasmids were confirmed by DNA sequencing.
Ad5hr-HIVgp160 recombinants were generated by homologous recombination as previously described [28]. In brief, DNA isolated from wild-type Ad5hr was digested with SpeI and separated on a 0.8% agarose gel. The larger fragment (~27 kbp) representing 0-75 mu was recovered from the gel using a Qiaex II Gel Extraction Kit (Qiagen). The shuttle plasmids were also digested with BamHI, separated on a 0.8% agarose gel and the larger fragments (~14 kbp) representing 59.5-100 mu and containing the HIV gp160 gene were isolated using the same method. Both Ad5hr and shuttle plasmid fragments were co-transfected into QBI-293 cells with lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Plaques were observed within 10-14 days. Ad5hr-HIVgp160 recombinant candidates were screened by PCR, and gp160 expression was evaluated by Western blotting. Potential contamination with full length Ad5 wild-type virus was excluded by the absence of the Ad5 E3 gene in PCR analysis using Ad5E3-P1 and Ad5E3-P2 primers (Table 1). The Ad5hr-HIVgp160 recombinant viruses were further purified three times by plaque purification.
2.2. Animals and Immunization
Sixty nine female Balb/c mice at 6 – 8 weeks of age were housed and maintained according to the standards of the American Association for Accreditation of Laboratory Animal Care. The animal protocol was reviewed and approved by the Animal Care and Use Committee prior to implementation. The immunization schedule is outlined in Table 2. Five mice were sacrificed on Day 0 to provide baseline data. The remaining 64 mice were divided into 4 groups of 16 mice. Ad5hr-recombinants were administered intraperitoneally at a dose of 1 × 108 infectious units (ifu). To balance group III which received both Ad5hr-HIV recombinants, the other groups received empty Ad5hr vector, Ad5hrΔE3, up to a total Ad dose of 2 × 108 ifu in a total volume of 500 μl phosphate-buffered saline (PBS). Five mice per group were sacrificed 2 weeks after the first immunization at week 0 and second immunization at week 4. The remaining mice were sacrificed at week 12. Spleens were collected to evaluate cellular immune responses. Mouse splenocytes were isolated by passing spleen fragments through a 70 μm nylon cell strainer. After lysis of erythrocytes, the splenocytes were resuspended in R-10 medium (RPMI-1640 containing 10% heat inactivated FCS, 2mM L-Glutamine, 50 μM β-mercaptoethanol, 10mM HEPES, 100 U/ml penicillin, 100 μg/ml streptomycin and 0.25μg/ml amphotericin B). Blood samples were also collected without anticoagulant. Clotted blood samples were centrifuged and sera of mice from the same group were pooled and stored at −70°C until being analyzed.
Table 2. Mouse immunization schedule.
| Group | Immunogens | # Mice sacrificed week 0 |
# Mice immunized week 0 |
# Mice sacrificed week 2 |
# Mice immunized week 4 |
# Mice sacrificed week 6 |
# Mice sacrificed week 12 |
|---|---|---|---|---|---|---|---|
| Baseline | 5 | 0 | 0 | 0 | 0 | 0 | |
| I | Ad5hr-HIVTV1 gp160 | 0 | 16 | 5 | 11 | 5 | 6 |
| II | Ad5hrHIVSF162 gp160 | 0 | 16 | 5 | 11 | 5 | 6 |
| III | Ad5hrHIVTV1 gp160 + Ad5hr-HIVSF162 gp160 | 0 | 16 | 5 | 11 | 5 | 6 |
| IV | Ad5hrΔE3 | 0 | 16 | 5 | 11 | 5 | 6 |
2.3. ELISpot assay
IFN-γ secretion in response to HIV envelope peptides was evaluated by ELISpot assay. Env peptides spanning each HIV-1 gp160 protein were pools of 15-mers with an 11 amino acid overlap. HIV-1 consensus subtype B and subtype C envelope peptides were obtained from the NIH AIDS Research & Reference Reagent Program, NIAID, NIH (Germantown, MD). HIVTV1 and HIVSF162 gp160 peptides were obtained from Advanced BioScience Laboratories, Inc. (Kensington, MD). The ELISpot assay was performed using commercial kits (Mabtech, Inc.) according to the manufacturer’s manual with slight modification. Briefly, activated 96-well PVDF membrane bottomed plates (MSIPS4510, Millipore) were coated overnight with 15 μg/ml anti-mouse IFN-γ monoclonal antibody, washed and blocked as described. Splenocytes were diluted in R-10 medium and transferred to triplicate wells so that each well contained 3 × 105 cells and 200 ng of each peptide in 100 μl total volume. Concanavalin A (Con A) (Sigma) at 5 μg/ml and R-10 alone were used as positive and negative controls, respectively. Following incubation at 37°C in 5% CO2 overnight, the cells were shaken-out and discarded. The plates were washed with PBS, reacted with 1 μg/ml Biotin conjugated anti mouse IFN-γ antibody in PBS-0.5% FCS and incubated for 2 hours at RT. After further washing, bound anti-IFN-γ antibody was reacted with Streptavidin-HRP for 1 hr at RT and washed again. Plates were incubated with EAC substrate (Vector Laboratories) until distinct spots emerged. The color reaction was stopped by washing extensively in deionized water. The plates were air-dried, and spots were counted using an ELISpot reader (Carl Zeiss MicroImaging GmbH).
2.4. Titration of Specific Binding Antibody
Sera from animals were pooled according to immunization groups and time points and assayed for specific binding antibody activity against HIV Env proteins of various subtypes by ELISA as previously described [29] with slight modification. Briefly, serially diluted sera were applied to a 96-well plate previously coated with 40 ng of HIV Env, blocked with skim milk, and incubated for 2 h at 37°C. After washing with PBS-Tween, the plate was reacted with peroxidase-conjugated goat anti-mouse IgG (H+L), incubated for another hour and washed again. A color reaction was performed by adding TMB (3,3′,5,5′-tetramethylbenzidine) peroxidase substrate solution. The reaction was stopped by adding 1M H3PO4 and the plate was read at 450 nm within 30 min. Antibody titer was defined as the reciprocal of the serum dilution at which the absorbance was twice that of sera of unimmunized mice at a 100× dilution.
2.5. Statistical analysis
Comparisons of HIV gp160 responses against the unimmunized controls were performed using the non-parametric exact two-tailed Wilcoxon rank sum test corrected by the Hochberg method for the three post-immunization times. Differences between immunized groups in IFN-γ-secreting cells were tested using repeated measures analysis of variance, with p values corrected for the four stimulating peptides. ELISpot counts were square-root transformed before analysis for consistency with the distributional assumptions of the method.
3. Results
3.1. Construction of Ad5hr-recombinants encoding HIV gp160 genes
Both Ad5hr-recombinants containing the HIVSF162 or HIVTV1 gp160 genes were constructed as described in Materials and Methods and outlined in Figure 1. After insertion of the Kozak sequence upstream of the start codon in the HIVTV1 env sequence and substitution of the tPA leader sequence for the wild type SF162 leader and removal of the BamHI site in the SF162 plasmid, the expression of both HIV gp160 proteins was confirmed by Western blot following transfection of the resulting plasmids into QBI 293 cells (Fig. 1A). The expression pattern of both envelope proteins is in accord with the previously published observations [30].
Figure 1. Construction of Ad5hr HIV-1 gp160 recombinants.
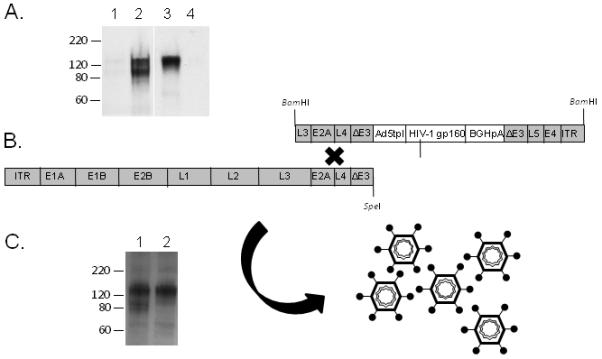
(A) Western blot of QBI 293 cell lysate after being transfected with plasmid expressing HIVTV1 gp160 (lane 2) or HIVSF162 gp160 (lane 3). The Env band was not observed in mock transfected cells (lane 1) or untransfected cells (lane 4). (B) Schematic diagram of the Ad5hr-shuttle plasmid and the left hand Ad5hr fragment used to create replication-competent Ad5hr-recombinant virus by homologous recombination. (C) Expression of HIV-1 gp160 in QBI 293 cells after being transduced with 1 MOI of Ad5hr-HIVTV1 (lane 1) or Ad5hr-HIVSF162 (lane 2).
In the shuttle plasmid, the transgene cassette-consisting of the Ad5 tripartite leader (TPL) sequence, HIV gp160, and BGHpA signal, was placed in the deleted E3 region of the Ad5 shuttle plasmid (Fig 1B, upper right strand). After removing the vector portion of the shuttle by BamHI digestion, the DNA fragment was co-transfected with the left hand part of SpeI digested Ad5hr wt genomic DNA (Fig 1B, left lower strand) into QBI 293 cells to produce Ad5hr-HIV gp160 recombinants by homologous recombination (Fig 1B). Each clone was tested for the presence of the inserted HIV gene by PCR, and positive clones were evaluated for protein expression by Western blotting. Clones showing the best gp160 expression were selected and underwent three successive plaque purifications, followed by propagation and research-grade purification for vaccination purposes. Before inoculating into animals, the propagated recombinants were titrated and the expression of HIV gp160 was confirmed again by Western blotting following transduction into QBI 293 cells (Fig. 1C). As shown, the final Ad5hr-HIVTV1 and –HIVSF162 recombinants expressed equivalent amounts of gp160 protein as determined by measuring band densities using ImageJ software (http://rsbweb.nih.gov/ij/).
Immunogenicity of the recombinants was evaluated in Balb/C mice. Ad5hr recombinants are able to infect mice, but due to host range restriction, replication does not occur. Nevertheless, the inserted gene products are expressed on a one-time basis. The mouse immunization protocol is outlined in Table 2 and detailed in Materials and Methods. Immune responses were evaluated 2 weeks after each immunization and again at 8 weeks after the second immunization in order to evaluate persistence of the induced immunity.
3.2. HIV gp160 specific cellular immune responses
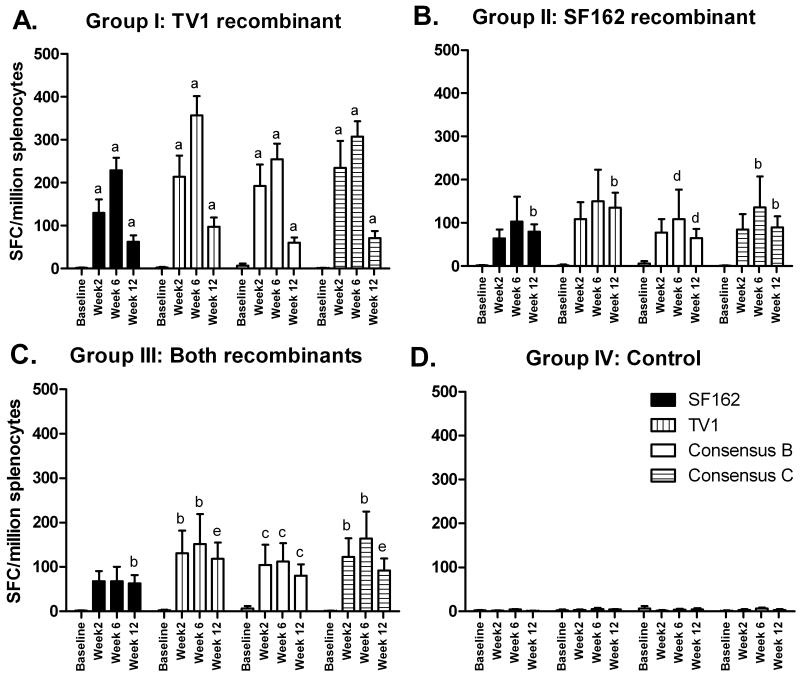
HIVenvelope-specific cellular immunity elicited by the Ad5hr-recombinant vaccines was evaluated by IFN-γ ELISPOT assay (Fig. 2). Both recombinants were immunogenic, inducing IFN-γ-secreting cells in response to stimulation with both SF162 and TV1 Env peptides. To evaluate whether the Ad5hr-recombinants would elicit cellular immune responses against a broader range of HIV envelopes from subtype B and C isolates, consensus subtype B [31] and consensus subtype C [32] envelope peptides were also tested. Cross-reactive responses were elicited by both recombinants to the consensus B and C peptides (Fig. 2).
Figure 2. HIV-1 gp160 specific ELISpot responses following immunization with Ad5hr recombinants.
Spleen cells of immunized mice were harvested at the indicated time points and tested for IFN-γ secretion after stimulation with the indicated HIV-1 gp160 peptides. Significant differences in comparison to the baseline control are indicated as follows: a, p <0.01; b, p <0.02; c, p <0.03; d, p < 0.04; e, p <0.05.
Compared to unimmunized mice, mice immunized with Ad5hr-HIVTV1 showed significantly higher HIV gp160 responses to all Env peptides at weeks 2, 6, and 12 (p=0.0079 for SF162, TV1 and consensus C; p=0.0089 for consensus B). In contrast, mice immunized with Ad5hr-HIVSF162 developed responses more slowly. Significantly higher responses to consensus B and C peptides in comparison to baseline controls were not seen until weeks 6 and 12 (p<0.032 and p<0.016, respectively for both time points), while significant responses to SF162 and TV1 peptides were seen only at week 12 (p=0.013 for both). Co-administration of Ad5hr-HIVSF162 and Ad5hr-HIVTV1 gave an intermediate outcome. Significant responses were seen at all three timepoints to consensus B peptides (p<0.026 for all 3 times) as well as to TV1 and consensus C peptides (p=0.016, for both at weeks 2 and 6; p=0.0498 for both at week 12). However, reactivity to SF162 peptides only reached significance at week 12 (p=0.013). Control mice receiving empty Ad5hrΔE3 did not develop cellular responses to HIV gp160 peptides.
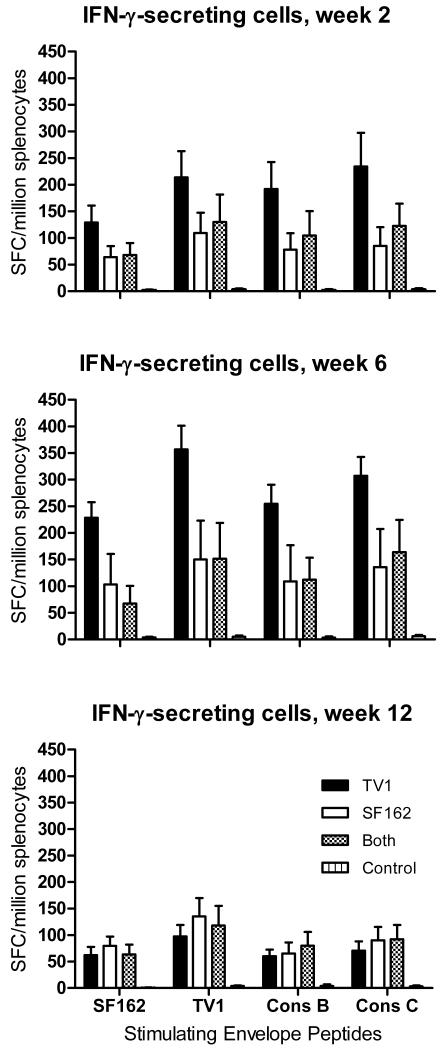
While Ad5hr-HIVTV1 and Ad5hr-HIVSF162 exhibited similar levels of expression of gp160 protein (Fig. 1C), overall the TV1 recombinant elicited stronger cellular immune responses. Mean ELISPOT responses for mice immunized with Ad5hr-HIVTV1 were higher than those of mice immunized with Ad5hr-HIVSF162 or both recombinants for all 4 groups of Env peptides at weeks 2 and 6, although lower at week 12 (Fig. 3). When the data over the 3 time points were analyzed for each pool of stimulating peptides, the Ad5hr-HIVTV1 immunogen elicited higher responses to TV1, consensus B and consensus C peptides (p = 0.04 for each) and marginally higher to SF162 peptides (p = 0.07) compared to the Ad5hr-HIVSF162 immunogen. When the data were analyzed for the combined group of 4 peptide pools, the Ad5hr-HIVTV1 immunogen still elicited greater numbers of IFN-γ-secreting cells compared to the Ad5hr-HIVSF162 immunogen (p = 0.033). Similar analysis for the Ad5hr-HIVTV1 immunization group versus group III that received both immunogens shows marginally nonsignificant differences (p = 0.06 for each of the peptide pools individually; p = 0.085 combined). No significant difference was seen between the groups of mice that received the Ad5hr-HIVSF162 immunogen compared to the group that received both recombinants.
Figure 3. Mean vaccine-induced IFN-γ-secreting cells by immunization group.
SFC are plotted as a function of time post-immunization, in response to the indicated Env peptide stimuli. Mean values and standard errors of the mean are shown.
3.3. HIV specific humoral immune responses
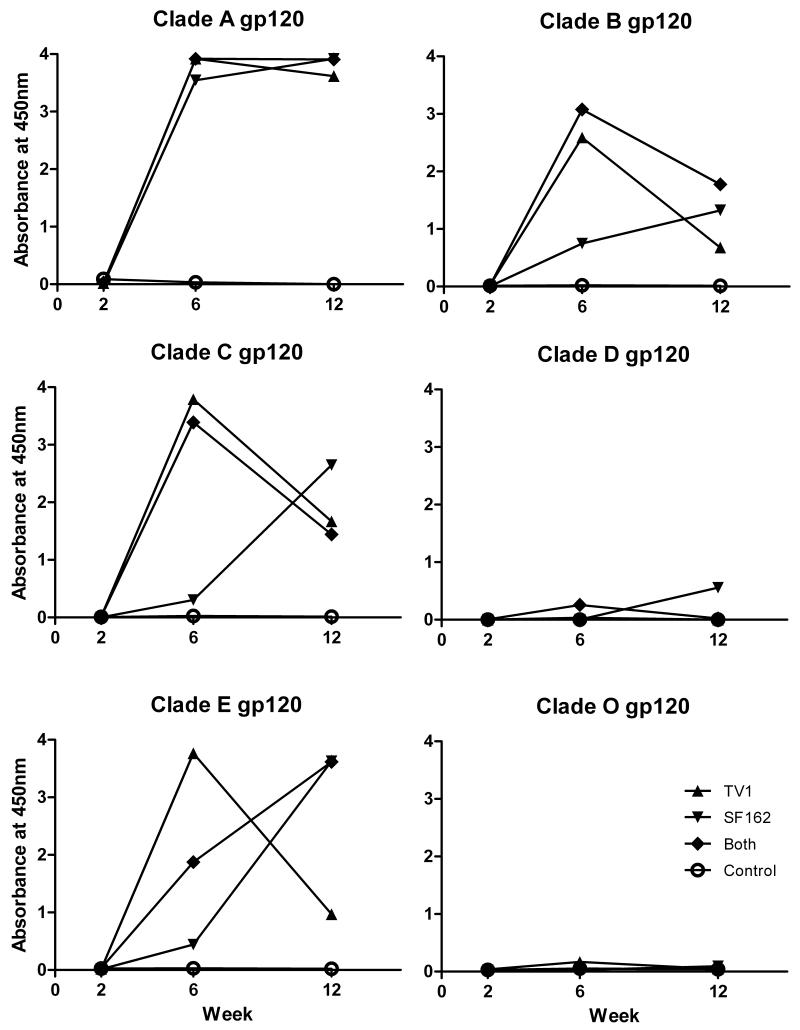
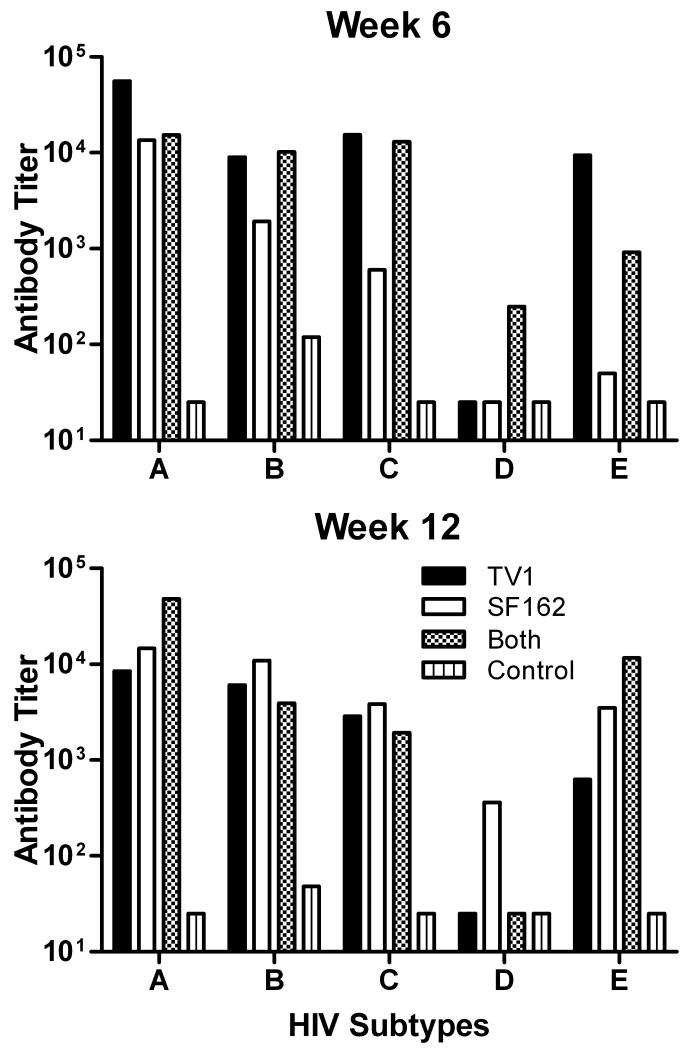
Immunizations with vectored recombinant vaccines are intended primarily to induce cellular immune responses as the inserted gene is expressed in infected cells, processed, and presented via class I MHC molecules to CD8+ T cells. We have seen however, that immunization with Ad-recombinant vaccines can also elicit antibody responses to expressed gene products and when coupled with subsequent protein booster immunizations lead to potent humoral immunity. The current protocol was not designed with an envelope protein boost. Nevertheless, we assessed the potency and breadth of antibody responses elicited by the recombinants on their own. As shown in Figure 4, two successive Ad recombinant immunizations were needed to elicit significant binding antibody responses to HIV Env proteins. Subsequently, HIV envelope proteins of subtypes A, B, C, and E were recognized in general, while subtypes D and O were poorly or not at all recognized. As shown in Figure 5, at week 6 (2 weeks after the second Ad-recombinant immunization) mice immunized with Ad5hr-HIVTV1 exhibited the highest titers against HIV subtype A and E envelopes and titers equivalent to those elicited in mice that received both Ad recombinants against subtype B and C envelopes. Statistical analysis was not possible as pooled samples were compared. The Ad5hr-HIVSF162 immunogen elicited the lowest antibody titers across the 5 different HIV envelope subtypes. All three immunization groups exhibited similarly persistent titers across subytpes by week 12 post-immunization.
Figure 4. Anti-HIV Env binding antibody in immunized mouse sera.
Sera collected from immunized mice in each group at indicated time points were pooled and tested for specific binding to HIV Env proteins of several different HIV-1 subtypes. Results are the average of 2 independent measurements of 450nm absorbance at serum dilutions of 1:50.
Figure 5. Antibody binding titers against HIV Env proteins of different subtypes.
Results of sera pooled according to immunization group and time post-immunization. Means of each HIV-Env subtype are shown.
4. Discussion
Our previous studies using replication-competent Ad recombinants expressing HIV or SIV envelope as one component of our vaccination regimen have shown strong elicitation of cellular and humoral immune responses in vaccinated rhesus macaques and chimpanzees. Here we constructed two new Ad5hr recombinants expressing full length HIV-1 gp160 of subtype B (HIVSF162) and subtype C (HIVTV1). Both strains are R5-tropic HIV-1 isolates.
HIV-1SF162 was originally isolated by cocultivating PBMC from seronegative donors with cerebrospinal fluid of an HIV-1 seropositive patient with toxoplasmosis [33]. The virus is “neurotropic” and has distinguishing characteristics including macrophage tropism, relative inability to infect established T-cell lines and down modulate the CD4 receptor, and resistance to serum neutralization [33, 34]. HIVTV1 was derived from the peripheral blood of an infected individual in South Africa. The molecular clone of the envelope sequence was selected for high expression and the ability to mediate cell-to-cell fusion, indicating functionality [35].
Efficient expression of HIV structural proteins (Gag and Env) requires the presence of the HIV Rev protein for exporting the relevant unspliced mRNAs from the host nucleus into the cytoplasm. Adenoviruses also replicate in the host nucleus and thus when recombinants are used for expressing HIV structural proteins the presence of Rev is also required. However, when genes expressing HIV gp160 are codon optimized [36], the presence of Rev mRNA nuclear export is no longer necessary. Both HIV gp160 sequences used here were already modified, using codons from highly expressed mammalian genes. After transfection of the plasmid constructs, both env glycoprotein gene products were highly expressed in cell culture under the strong CMV early promoter. Strong expression was also observed in cell lines infected with the recombinants, where expression was driven by the Ad major late promoter, confirming that Rev was no longer required for efficient expression of the HIV Env protein in these recombinants. The insertion of a transgene downstream of the untranslated Ad5 TPL sequence enhances the expression of the transgene [37], either by increased translation during the late phase of infection [38-40] or due to more efficient transcription [41]. Ad TPL sequences have also been reported to improve mRNA export from the nucleus [42] and to abolish the required cap-binding protein complex during translation initiation [43, 44].
In our Ad5hr recombinants, the HIV env genes were placed in the deleted E3 region. Ad E3 genes encode some non-secreted proteins dedicated to control of various host immune responses (reviewed in [45]), and are not essential for the replication of the virus in cultured cells. Therefore, they are generally deleted in replication defective Ad vectors, in addition to the E1 genes, to create greater clone capacity. E3-encoded proteins include the adenovirus death protein (ADP) and at least six other proteins with varied effects including preventing lysis of Ad-infected cells by cytotoxic T lymphocytes (CTL) by down-modulation of MHC class I molecules and inhibition of the apoptosis process induced by tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL), TNF and Fas ligand [46, 47]. Robust expression of transgenes has been observed when they are inserted into the deleted E3 region, presumably driven via the major late promoter [48]. Taken together, removal of the E3 region may not only create space for a foreign gene, but also attenuate the resultant recombinant virus which will become more susceptible to host control. This effect is offset by the strong expression of the transgene. In terms of vaccine development, greater immune responses to the transgene as well as vector components may result, balanced by a shorter replicative capacity.
Upon intraperitoneal administration into Balb/C mice, both Ad5hr recombinants were immunogenic. Surprisingly, mice from group I immunized with Ad5hr-HIVTV1 developed better cellular immune responses not only to Env peptides of TV1 and consensus subtype C, but also to those of SF162 and consensus subtype B. Ad5hr-HIVSF162 also elicited cross-reactive immunity but overall gave the poorest levels of IFN-γ-secreting cells. Immunization with both recombinants did not improve overall immunity. Thus, the Ad5hr-HIVTV1 recombinant was more than adequate at eliciting immune responses specific for both subtype B and C envelopes.
Immunization using Ad vectored vaccines is aimed not only at eliciting cellular immunity, but also at priming antibody responses. Here, humoral immune responses were primed by the first immunization and induced by the second Ad administration, even without protein boosting. Binding antibodies with cross-reactivity for envelopes of subtypes A, B, C, E, and to a lesser extent for subtype D but not subtype O were elicited. In concert with the results of cellular immune assays, the highest antibody titers were induced by Ad5hr-HIVTV1 immunization. The data suggest that immunization with the Ad5hr-HIVTV1 recombinant might effectively prime antibody responses which could subsequently be boosted using envelope proteins of several different subtypes. A recently initiated study in rhesus macaques where the full potential of replication-competent Ad5 recombinants can be evaluated will first examine a homologous Env protein boost. Whether Ad5-HIVTV1 recombinant priming will result in strong neutralizing antibody after Env protein boosting will also be tested. This parameter was not evaluated in the mice immunized with Ad5hr-recombinants only, as we have previously observed that Env boosting is needed for elicitation of potent neutralizing antibodies [15].
Overall, while both recombinants were highly immunogenic in the murine model, this study showed that the Ad5hr-HIVTV1 recombinant vaccine was superior in eliciting more potent cellular and humoral immunity with similar cross reactivity. Replication-competent Ad vectors are limited in the amount of foreign DNA they can accommodate. If immunization with two different HIV envelope genes were required to elicit desired cross reactivity, two separate Ad-recombinants would be required. For future clinical applications, this would greatly increase both expense and quality control. The present findings suggest the single Ad5hr-HIVTV1 recombinant might be sufficient to induce cellular immunity and prime antibody responses against at least subtype B and C HIV envelopes. The recently initiated immunogenicity and protective efficacy studies in macaques are the next step in moving this approach forward. The findings will help determine if the HIVTV1 env insert is appropriate for use in a planned phase I clinical trial. While the Ad5hr vector is necessary for studies in macaques, the human vaccine will be based in a replication-competent Ad4 vector.
Acknowledgements
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 consensus subtype B and subtype C Env (15-mer) peptides, complete sets. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Hall HI, Song R, Rhodes P, Prejean J, An Q, Lee LM, et al. Estimation of HIV incidence in the United States. JAMA. 2008;300(5):520–9. doi: 10.1001/jama.300.5.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Haase AT. The pathogenesis of sexual mucosal transmission and early stages of infection: obstacles and a narrow window of opportunity for prevention. AIDS. 2001;15(Suppl 1):S10–1. doi: 10.1097/00002030-200102001-00013. [DOI] [PubMed] [Google Scholar]
- [3].Holl V, Peressin M, Decoville T, Schmidt S, Zolla-Pazner S, Aubertin AM, et al. Nonneutralizing antibodies are able to inhibit human immunodeficiency virus type 1 replication in macrophages and immature dendritic cells. J Virol. 2006;80(12):6177–81. doi: 10.1128/JVI.02625-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Forthal DN, Landucci G, Cole KS, Marthas M, Becerra JC, Van Rompay K. Rhesus macaque polyclonal and monoclonal antibodies inhibit simian immunodeficiency virus in the presence of human or autologous rhesus effector cells. J Virol. 2006;80(18):9217–25. doi: 10.1128/JVI.02746-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Miller CJ, Genescà M, Abel K, Montefiori D, Forthal D, Bost K, et al. Antiviral antibodies are necessary for control of simian immunodeficiency virus replication. J Virol. 2007;81(10):5024–35. doi: 10.1128/JVI.02444-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Gómez-Román VR, Patterson LJ, Venzon D, Liewehr D, Aldrich K, Florese R, et al. Vaccine-elicited antibodies mediate antibody-dependent cellular cytotoxicity correlated with significantly reduced acute viremia in rhesus macaques challenged with SIVmac251. J Immunol. 2005;174(4):2185–9. doi: 10.4049/jimmunol.174.4.2185. [DOI] [PubMed] [Google Scholar]
- [7].Hidajat R, Xiao P, Zhou Q, Venzon D, Summers LE, Kalyanaraman VS, et al. Correlation of vaccine-elicited systemic and mucosal nonneutralizing antibody activities with reduced acute viremia following intrarectal simian immunodeficiency virus SIVmac251 challenge of rhesus macaques. J Virol. 2009;83(2):791–801. doi: 10.1128/JVI.01672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Florese RH, Demberg T, Xiao P, Kuller L, Larsen K, Summers LE, et al. Contribution of nonneutralizing vaccine-elicited antibody activities to improved protective efficacy in rhesus macaques immunized with tat/env compared with multigenic vaccines. J Immunol. 2009;182(6):3718–27. doi: 10.4049/jimmunol.0803115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Malkevitch NV, Robert-Guroff M. A call for replicating vector prime-protein boost strategies in HIV vaccine design. Expert Rev Vaccines. 2004;3(4 Suppl):S105–17. doi: 10.1586/14760584.3.4.s105. [DOI] [PubMed] [Google Scholar]
- [10].Patterson LJ, Robert-Guroff M. Replicating adenovirus vector prime/protein boost strategies for HIV vaccine development. Expert Opin Biol Ther. 2008;8(9):1347–63. doi: 10.1517/14712598.8.9.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wyand MS, Manson KH, Garcia-Moll M, Montefiori D, Desrosiers RC. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J Virol. 1996;70(6):3724–33. doi: 10.1128/jvi.70.6.3724-3733.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hofmann-Lehmann R, Vlasak J, Williams AL, Chenine AL, McClure HM, Anderson DC, et al. Live attenuated, nef-deleted SIV is pathogenic in most adult macaques after prolonged observation. AIDS. 2003;17(2):157–66. doi: 10.1097/00002030-200301240-00004. [DOI] [PubMed] [Google Scholar]
- [13].Klessig DF, Grodzicker T. Mutations that allow human Ad2 and Ad5 to express late genes in monkey cells map in the viral gene encoding the 72K DNA binding protein. Cell. 1979;17(4):957–66. doi: 10.1016/0092-8674(79)90335-0. [DOI] [PubMed] [Google Scholar]
- [14].Patterson LJ, Prince GA, Richardson E, Alvord WG, Kalyan N, Robert-Guroff M. Insertion of HIV-1 genes into Ad4ΔE3 vector abrogates increased pathogenesis in cotton rats due to E3 deletion. Virology. 2002;292(1):107–13. doi: 10.1006/viro.2001.1248. [DOI] [PubMed] [Google Scholar]
- [15].Peng B, Wang LR, Gómez-Román VR, Davis-Warren A, Montefiori DC, Kalyanaraman VS, et al. Replicating rather than nonreplicating adenovirus-human immunodeficiency virus recombinant vaccines are better at eliciting potent cellular immunity and priming high-titer antibodies. J Virol. 2005;79(16):10200–9. doi: 10.1128/JVI.79.16.10200-10209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Malkevitch N, Patterson LJ, Aldrich K, Richardson E, Alvord WG, Robert-Guroff M. A replication competent adenovirus 5 host range mutant-simian immunodeficiency virus (SIV) recombinant priming/subunit protein boosting vaccine regimen induces broad, persistent SIV-specific cellular immunity to dominant and subdominant epitopes in Mamu-A*01 rhesus macaques. J Immunol. 2003;170(8):4281–9. doi: 10.4049/jimmunol.170.8.4281. [DOI] [PubMed] [Google Scholar]
- [17].Patterson LJ, Malkevitch N, Pinczewski J, Venzon D, Lou Y, Peng B, et al. Potent, persistent induction and modulation of cellular immune responses in rhesus macaques primed with Ad5hr-simian immunodeficiency virus (SIV) env/rev, gag, and/or nef vaccines and boosted with SIV gp120. J Virol. 2003;77(16):8607–20. doi: 10.1128/JVI.77.16.8607-8620.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Patterson LJ, Malkevitch N, Venzon D, Pinczewski J, Gómez-Román VR, Wang L, Kalyanaraman VS, Markham PD, Robey FA, Robert-Guroff M. Protection against mucosal simian immunodeficiency virus SIV(mac251) challenge by using replicating adenovirus-SIV multigene vaccine priming and subunit boosting. J Virol. 2004;78(5):2212–21. doi: 10.1128/JVI.78.5.2212-2221.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Malkevitch NV, Patterson LJ, Aldrich MK, Wu Y, Venzon D, Florese RH, Kalyanaraman VS, Pal R, Lee EM, Zhao J, Cristillo A, Robert-Guroff M. Durable protection of rhesus macaques immunized with a replicating adenovirus-SIV multigene prime/protein boost vaccine regimen against a second SIVmac251 rectal challenge: role of SIV-specific CD8+ T cell responses. Virology. 2006;353(1):83–98. doi: 10.1016/j.virol.2006.05.012. [DOI] [PubMed] [Google Scholar]
- [20].Demberg T, Florese RH, Heath MJ, Larsen K, Kalisz I, Kalyanaraman VS, et al. A replication-competent adenovirus-human immunodeficiency virus (Ad-HIV) tat and Ad-HIV env priming/Tat and envelope protein boosting regimen elicits enhanced protective efficacy against simian/human immunodeficiency virus SHIV89.6P challenge in rhesus macaques. J Virol. 2007;81(7):3414–27. doi: 10.1128/JVI.02453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Patterson LJ, Beal J, Demberg T, Florese RH, Malkevich N, Venzon D, et al. Replicating adenovirus HIV/SIV recombinant priming alone or in combination with a gp140 protein boost results in significant control of viremia following a SHIV89.6P challenge in Mamu-A*01 negative rhesus macaques. Virology. 2008;374(2):322–37. doi: 10.1016/j.virol.2007.12.037. [DOI] [PubMed] [Google Scholar]
- [22].Bogers WM, Davis D, Baak I, Kan E, Hofman S, Sun Y, et al. Systemic neutralizing antibodies induced by long interval mucosally primed systemically boosted immunization correlate with protection from mucosal SHIV challenge. Virology. 2008;382(2):217–25. doi: 10.1016/j.virol.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J Exp Med. 1997;185(4):621–8. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shankarappa R, Margolick JB, Gange SJ, Rodrigo AG, Upchurch D, Farzadegan H, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73(12):10489–502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105(21):7552–7. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Natuk RJ, Chanda PK, Lubeck MD, Davis AR, Wilhelm J, Hjorth R, et al. Adenovirus-human immunodeficiency virus (HIV) envelope recombinant vaccines elicit high-titered HIV-neutralizing antibodies in the dog model. Proc Natl Acad Sci USA. 1992;89(16):7777–81. doi: 10.1073/pnas.89.16.7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lubeck MD, Natuk RJ, Myagkikh M, Kalyan N, Aldrich K, Sinangil F, et al. Long-term protection of chimpanzees against high-dose HIV-1 challenge induced by immunization. Nature Med. 1997;3(6):651–8. doi: 10.1038/nm0697-651. [DOI] [PubMed] [Google Scholar]
- [28].Zhao J, Lou Y, Pinczewski J, Malkevitch N, Aldrich K, Kalyanaraman VS, et al. Boosting of SIV-specific immune responses in rhesus macaques by repeated administration of Ad5hr-SIVenv/rev and Ad5hr-SIVgag recombinants. Vaccine. 2003;21(25-26):4022–35. doi: 10.1016/s0264-410x(03)00266-4. [DOI] [PubMed] [Google Scholar]
- [29].Buge SL, Richardson E, Alipanah S, Markham P, Cheng S, Kalyan N, et al. An adenovirus-simian immunodeficiency virus env vaccine elicits humoral, cellular, and mucosal immune responses in rhesus macaques and decreases viral burden following vaginal challenge. J Virol. 1997;71(11):8531–41. doi: 10.1128/jvi.71.11.8531-8541.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Srivastava IK, Kan E, Sun Y, Sharma VA, Cisto J, Burke B, et al. Comparative evaluation of trimeric envelope glycoproteins derived from subtype C and B HIV-1 R5 isolates. Virology. 2008;372(2):273–90. doi: 10.1016/j.virol.2007.10.022. [DOI] [PubMed] [Google Scholar]
- [31].Kothe DL, Decker JM, Li Y, Weng Z, Bibollet-Ruche F, Zammit KP, et al. Antigenicity and immunogenicity of HIV-1 consensus subtype B envelope glycoproteins. Virology. 2007;360(1):218–34. doi: 10.1016/j.virol.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kothe DL, Li Y, Decker JM, Bibollet-Ruche F, Zammit KP, Salazar MG, et al. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology. 2006;352(2):438–49. doi: 10.1016/j.virol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- [33].Cheng-Mayer C, Weiss C, Seto D, Levy JA. Isolates of human immunodeficiency virus type 1 from the brain may constitute a special group of the AIDS virus. Proc Natl Acad Sci U S A. 1989;86(21):8575–9. doi: 10.1073/pnas.86.21.8575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cheng-Mayer C, Quiroga M, Tung JW, Dina D, Levy JA. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathogenicity, and CD4 antigen modulation. J Virol. 1990;64(9):4390–8. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lian Y, Srivastava I, Gómez-Román VR, Zur Megede J, Sun Y, Kan E, et al. Evaluation of envelope vaccines derived from the South African subtype C human immunodeficiency virus type 1 TV1 strain. J Virol. 2005;79(21):13338–49. doi: 10.1128/JVI.79.21.13338-13349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Haas J, Park EC, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6(3):315–24. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- [37].Sheay W, Nelson S, Martinez I, Chu TH, Bhatia S, Dornburg R. Downstream insertion of the adenovirus tripartite leader sequence enhances expression in universal eukaryotic vectors. Biotechniques. 1993;15(5):856–62. [PubMed] [Google Scholar]
- [38].Logan J, Shenk T. Adenovirus tripartite leader sequence enhances translation of mRNAs late after infection. Proc Natl Acad Sci U S A. 1984;81(12):3655–9. doi: 10.1073/pnas.81.12.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Berkner KL, Sharp PA. Effect of the tripartite leader on synthesis of a non-viral protein in an adenovirus 5 recombinant. Nucleic Acids Res. 1985;13(3):841–57. doi: 10.1093/nar/13.3.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kaufman RJ. Identification of the components necessary for adenovirus translational control and their utilization in cDNA expression vectors. Proc Natl Acad Sci U S A. 1985;82(3):689–93. doi: 10.1073/pnas.82.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Alonso-Caplen FV, Katze MG, Krug RM. Efficient transcription, not translation, is dependent on adenovirus tripartite leader sequences at late times of infection. J Virol. 1988;62(5):1606–16. doi: 10.1128/jvi.62.5.1606-1616.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Huang W, Flint SJ. The tripartite leader sequence of subgroup C adenovirus major late mRNAs can increase the efficiency of mRNA export. J Virol. 1998;72(1):225–35. doi: 10.1128/jvi.72.1.225-235.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dolph PJ, Racaniello V, Villamarin A, Palladino F, Schneider RJ. The adenovirus tripartite leader may eliminate the requirement for cap-binding protein complex during translation initiation. J Virol. 1988;62(6):2059–66. doi: 10.1128/jvi.62.6.2059-2066.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Dolph PJ, Huang JT, Schneider RJ. Translation by the adenovirus tripartite leader: elements which determine independence from cap-binding protein complex. J Virol. 1990;64(6):2669–77. doi: 10.1128/jvi.64.6.2669-2677.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Horwitz MS. Adenovirus immunoregulatory genes and their cellular targets. Virology. 2001;279(1):1–8. doi: 10.1006/viro.2000.0738. [DOI] [PubMed] [Google Scholar]
- [46].Horwitz MS. Function of adenovirus E3 proteins and their interactions with immunoregulatory cell proteins. J Gene Med. 2004;6(Suppl 1):S172–83. doi: 10.1002/jgm.495. [DOI] [PubMed] [Google Scholar]
- [47].Lichtenstein DL, Toth K, Doronin K, Tollefson AE, Wold WS. Functions and mechanisms of action of the adenovirus E3 proteins. Int Rev Immunol. 2004;23(1-2):75–111. doi: 10.1080/08830180490265556. [DOI] [PubMed] [Google Scholar]
- [48].Le LP, Le HN, Nelson AR, Matthews DA, Yamamoto M, Curiel DT. Core labeling of adenovirus with EGFP. Virology. 2006;351(2):291–302. doi: 10.1016/j.virol.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]