Abstract
Over-expression of heme binding proteins in E. coli often results in sub-optimal heme incorporation and the amount of heme-bound protein produced usually varies with the protein of interest. Complete heme incorporation is important for biochemical characterization, spectroscopy, structural studies, and for the production of homogeneous commercial proteins with high activity. We have determined that recombinant proteins expressed in E. coli often contain less than a full complement of heme because they rather are partially incorporated with free-base porphyrin. Porphyrin-incorporated proteins have similar spectral characteristics as the desired heme-loaded targets, and thus are difficult to detect, even in purified samples. We present a straightforward and inexpensive solution to this problem that involves the co-expression of native ferrochelatase with the protein of interest. The method is shown to be effective for proteins that contain either Cys- or His- ligated hemes.
Keywords: Heme proteins, Heme incorporation, Free base porphyrin, Ferrochelatase
Introduction
Heme proteins encompass a wide range of functions that include electron transfer, transport and storage of oxygen, production and sensing of nitric oxide, decomposition of reactive oxygen species, catalytic oxidation of substrates, signal transduction and control of gene expression [1–4]. Under incorporation of heme into recombinant proteins can adversely influence their characterization. Evaluation of enzymatic activity will be systematically underestimated if a significant proportion of the recombinant sample does not contain heme. Furthermore, the proteins devoid of heme may bind another non-native cofactor with a contaminating activity of its own. A pure protein sample is usually essential for structural characterization by techniques such as X-ray crystallography, as this method often requires a single, well-folded species for crystallization. Other methods of heme protein characterization are less sensitive to the level of heme incorporation, particularly if those methods directly detect the metallocofactor (e.g. UV/Vis, EPR, and Mossbauer spectroscopy) or rely on enzymatic activity. This is both good and bad in that under incorporation may not greatly affect the analysis, but heterogeneity in the sample may go undetected.
Incomplete heme incorporation into recombinant proteins has been a frequently encounter problem [5–10] and thus, techniques have been developed to improve heme loading [8–14]. During induction of recombinant protein expression from highly active vectors, such as those that employ the T7-polymerase, a population of protein will fold without the heme co-factor under conditions where folding outpaces heme delivery [15]. Supplementing the growth media with δ-amino levulinic acid (δ–ALA), a precursor in the C5 heme biosynthesis pathway, increases levels of heme biosynthesis and thereby heme incorporation into the target protein [8, 16, 17]. Increased heme biosynthesis rates through δ–ALA supplementation allows for complete heme incorporation into some proteins, but not others [11, 12, 18, 19].
Another technique for increasing heme incorporation into recombinant proteins involves supplying the bacteria with hemin in the growth media. However, most E. coli strains do not possess an efficient heme transport system, and thus uptake of hemin relies on diffusion through the cell membrane. As a result, hemin feeding is much more effective with strains that co-express heme transport genes from other gram-negative bacteria, along with the heme-protein of interest [10, 13, 14]. For example, co-expression of the heme transport system from P. shigelloides, which consists of the proteins Hug A/B/C/D, TonB, and Exb B/D, while also supplementing the growth media with hemin, results in higher amounts of the target holo-protein (in this case hemoglobin) [10]. A similar method [13] involves the co-expression of the heme receptor ChuA from E. coli. strain O157:H7 to enhance hemin. This latter method also shows a significant increase in the amount of heme-loaded protein generated, although in both cases, the ratio of holoprotein:apoprotein was not evaluated. Another approach utilizes the heme-permeability of E. coli strain RP523, which has the hem B, porphobilinogen synthase gene disrupted to prevent native heme synthesis. All heme and/or heme analogs are procured by the cells from the growth media and incorporation is nearly stoichiometric for proteins expressed in the cytoplasm (0.8–1.0 heme / heme analog per protein)[20].
Full incorporation of heme in recombinant proteins is also important for commercial applications. For example, the feasibility of employing recombinant human hemoglobin as an oxygen delivery pharmaceutical is limited by the yield of holoprotein that can be made in E. coli [10]. Some of the methods discussed above, while effective, require co-expression of several heme transport proteins, which could limit yields, and/or require addition of the heme cofactor. Herein, we put forth a straightforward and inexpensive method for high fidelity incorporation of heme into heme proteins that are over-expressed in E. coli. Co-expression of just one native protein, ferrochelatase (FC), in the presence of 60 µM δ-ALA (10 mg/L, ~$0.50 per liter of cell culture) is sufficient for 100% heme incorporation into three unrelated heme proteins derived from different organisms.
Materials and Methods
Co-expression of Ferrochelatase with gsNOS
FC and gsNOS were expressed from the same pACYCduet vector (Novagen). To clone FC, genomic DNA was extracted from E. coli BL21 (DE3) cells with the genomic DNA extraction kit from Epicenter. The FC gene was then PCR-amplified (Phusion polymerase, New England Biolabs) from the genomic DNA with primers that generated Nde1 and Xho1 sites at the 5’ and 3’ ends of the gene, respectively. A stop codon was introduced into the 3’ primer before the Xho1 site to prevent C-terminal attachment of the vector-supplied S-tag. The amplified FC gene was then cloned into the Nde1 and Xho1 sites in Multiple Cloning Site-2 of the pACYCduet vector. The gene for gsNOS was derived from a previous pET28a-gsNOS plasmid (21) by digesting the vector with Nco1 and Xho1 so as to include the His-tag and the thrombin cleavage site along with the coding sequence for gsNOS in the excised fragment. The Nco1-Xho1 fragment was then cloned into the pACYCduet-FC plasmid between the Nco1 and Sal1 sites. Sal1 and Xho1 produce compatible cohesive ends and thereby allow the His-tag, thrombin cleavage site and gsNOS fragment to be cloned between the Nco1 and Sal1 sites of Multiple cloning site-1 of the pACYCduet-FC plasmid. The resulting pACYCduet plasmid allows us over-expression of gsNOS with a cleavable His-tag and FC with no tag. GsNOS was expressed and purified as reported before [21]. Co-expression of gsNOS and FC was also performed similarly to expression of gsNOS alone, although, a lesser amount of δ-ALA was added at the time of induction (10mg/L versus 25 mg/L for gsNOS), and the growth media was supplemented with 100 µM FeCl3. The antibiotics, chloramphenicol (34 µg/L) and kanamycin (50 µg/L) were added to the growth media of pACYCduet-gsNOS-FC and pET28a-gsNOS plasmids respectively.
Co-expression of Ferrochelatase with BP450 and HBPAS
For co-expression of BP450 and HBPAS the same procedure was used. BP450 (NCBI: CBG70284) was cloned into pET151/D-TOPO (Invitrogen), a directional cloning vector with a N-terminal 6x His-Tag followed by a TEV cleavage site and an ampicillin selectable marker (provided by Sarah Barry, University of Warwick, UK). HBPAS (NCBI: NP_248866) was cloned into pET28a (Novagen) using NdeI and HindIII restriction sites, which included a N-terminal 6xHis-Tag followed by a Thrombin cleavage site and a kanamycin selectable maker. Competent E. coli BL21 (DE3) cells containing FC/pACYCduet were transformed with either BP450/pET151/D-TOPO or HBPAS/pET28. Cells were grown at 37°C in Luria broth containing 20 ug/mL Cm and 100 ug/L Amp (BP450) or 50 ug/L Kan (HBPAS) to an OD = 0.6–0.8. Prior to induction with IPTG, the temperature was reduced to 17°C and 25 mg/L δ-ALA was added to the growth media. Cells were harvested 18–20 hrs after induction. An identical procedure with cells lacking the FC plasmid was used to express BP450 and HBPAS without FC. Both proteins were purified using Ni-NTA (Qiagen) chromatography techniques following the manufacturer’s protocol. Furthermore, the proteins were purified to >95% purity using size exclusion chromatography after the removal of 6xHis.
Spectroscopy
Resonance Raman and UV-Visible spectra were recorded as described previously [22].
Materials
Sodium Chloride was obtained from Mallinkrodt, Ferric Chloride, IPTG and TRIS were from Fisher Scientific, Kanamycin and Chloramphenicol from USBiological. δ-ALA was obtained from Sigma-Aldrich.
Results and Discussion
GsNOS (Geobacillus stearothermophilus Nitric Oxide Synthase) is a thermophilic enzyme that forms a highly stable heme-oxygen complex [21] that has helped in identification of catalytic intermediates responsible for L-arginine oxidation to nitric oxide [23]. In the over-expression of heme proteins we routinely add the heme precursor δ-ALA to the growth media when protein production is induced. Such δ-ALA supplementation results in complete heme incorporation for two other bacterial NOS proteins: B. subtilis NOS [24] and D. radiodurans NOS [25]. However, in what follows, we show that gsNOS over-expressed and purified from E. coli consists of two species: native heme-containing gsNOS, and gsNOS with protoporphyrin IX (free-base porphyrin) bound instead of heme. We hypothesized that co-expression of ferrochelatase, the enzyme that metallates porphyrin would ameliorate this problem.
UV-Vis spectroscopy and SDS-PAGE analysis of gsNOS
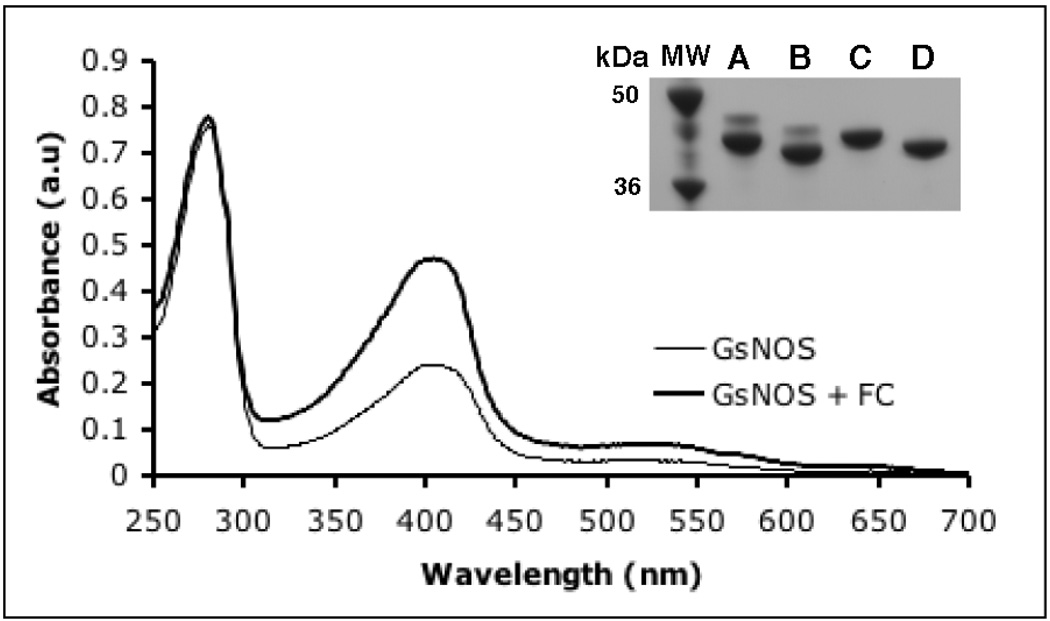
A UV-Vis spectroscopic analysis of gsNOS over-expressed in E. coli shows that the amount of heme incorporated with the protein changes from batch to batch, with the ratio of Soret peak height (403 nm) to protein peak height (280 nm) (Abs403/Abs280) varying between 0.25 – 0.40. Co-expression of FC with gsNOS increased Abs403/Abs280 to 0.6 i(Fig 1) in several (>3) different protein expression trials. Thus, gsNOS co-expressed with FC increases heme content of the protein in a consistent fashion.
Fig 1.
UV-Vis spectra of gsNOS expressed alone (thin line) and gsNOS expressed with FC (thick line). Co-expression of FC results in a substantial increase in heme content of gsNOS as measured by the Abs403/Abs280 ratio. This value saturates at 0.6, which indicates nearly complete heme incorporation. Inset: GsNOS expressed by itself results in two bands on SDS-PAGE (lane A), both of which shift on His-tag cleavage (lane B). GsNOS co-expressed with FC results in just one band (lane C), that shifts on His-tag cleavage (lane D) as expected.
Purified gsNOS, when over-expressed in E. coli with 25 mg/L δ-ALA added at the time of induction, results in two bands of ~42 kDa on SDS-PAGE (Fig 1, Inset, lane A). Both bands shift on His-tag cleavage (Fig 1, Inset, lane B), which indicates that both species represent recombinant gsNOS, with an intact N-terminus, but different gel mobilities. MALDI mass spectrometry of the sample shows only one sharp peak at a mass consistent with that of full-length gsNOS, which rules out proteolysis as the factor distinguishing the proteins represented in the two bands. Thus, the apparent difference in molecular weight on the gel of the two species (~3 kDa) may rather stem from a net charge difference. GsNOS, when co-expressed with FC from E. coli BL21 (DE3) cells, results in only a single species on SDS-PAGE (which corresponds to the lower band of the two observed previously; Fig 1 Inset, lane C). On His-tag cleavage, this band shifts as a single species (Fig 1, Inset, lane D). Protoporphyrin IX is the penultimate product in the heme biosynthesis pathway. FC catalyzes the last step in heme biosynthesis i.e insertion of an iron atom into protoporphyrin IX. Thus, the presence of two bands in the absence of FC co-expression suggests that protoporphyrin IX, rather than heme, has been incorporated into a substantial fraction of the sample.
Resonance Raman and fluorescence analysis of gsNOS
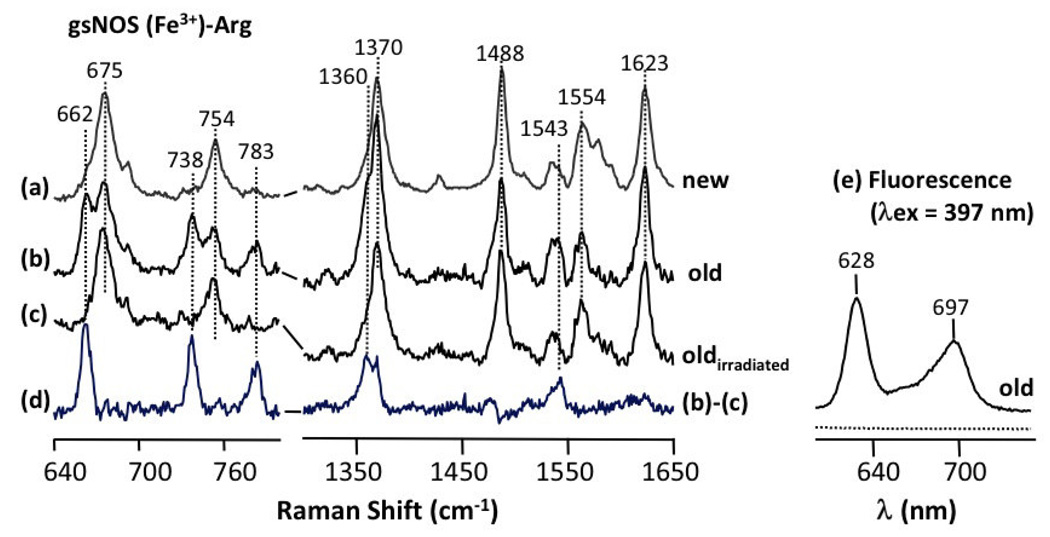
Confirmation of porphyrin incorporation into gsNOS came from resonance Raman studies of gsNOS in the presence of substrate L-arginine. A sample of gsNOS (expressed without FC) in the presence of substrate L-arginine, shows vibrational frequencies at 662 cm−1, 738 cm−1, 783 cm−1, 1360 cm−1 and 1543 cm−1, apart from the typical vibrational frequencies that have been previously observed [26, 27] for other NOSs (Fig 2b). These additional bands disappear after exposure to laser (Fig 2c) and the subsequent difference spectrum (before and after laser exposure) highlights the original, additional resonances (Fig 2d). The frequencies of these vibrations indicates the presence of free-base porphyrin in the protein sample [28, 29]. Furthermore, the enhanced photosensitivity of porphyrin compared to heme explains why the bands disappear after exposure [28, 29]. The presence of free base porphyrin is corroborated by the fluorescence spectrm of gsNOS (Fig 2e), which when measured with excitation at 397 nm shows definitive characteristics of un-metallated heme [30]. Iron-bound heme is not fluorescent when excited at this wavelength. Incorporation of free-base porphyrin suggests that the last step of heme biosynthesis i.e Fe metallation to heme, which is performed by FC, cannot keep pace with protein folding and porphyrin incorporation. Co-expression of FC from E. coli with gsNOS and addition of 10 mg/L of δ-ALA of (60 µM, ~ $0.50 per liter) generates a sample that is non-fluorescent and whose resonance Raman spectrum shows no evidence of porphyrin (Fig 2a). Thus, under these conditions, the protein is fully heme incorporated.
Fig 2.
Resonance Raman spectra of gsNOS from gsNOS co-expressed with FC (a), gsNOS expressed by itself (b) and (c). The spectrum in (b) is obtained with 3 mW of 413.1 nm laser excitation with an acquisition time of 5 min. The spectrum in (c) is for the same sample as (b), but obtained after prolonged irradiation with 42 mW of 413.1 nm laser for two hours. (d) shows the difference spectrum (b) – (c), which highlights spectral lines resulting from contamination by photosensitive protoporphyrin IX. The fluorescence spectrum in (e) obtained after a 397 nm excitation is representative of that of protoporphyrin IX bound to protein (see text) and is from same sample in (b).
We also tested if foregoing the addition of δ-ALA, while co-expressing FC would result in complete heme incorporation of gsNOS. The purified protein, which was checked for heme content by both SDS-PAGE and by UV-Vis spectroscopy had a higher degree of heme incorporation than under conditions of only adding δ-ALA (heme:protein ratio ~0.5); however, heme incorporation was not complete. Addition of a small amount of δ-ALA, (10 mg/L) is sufficient to make up for the slow rate of δ-ALA biosynthesis and produce fully incorporated protein in the presence of FC.
FC-assisted heme incorporation for a bacterial P450 (BP450) and a heme-binding PAS protein (HBPAS)
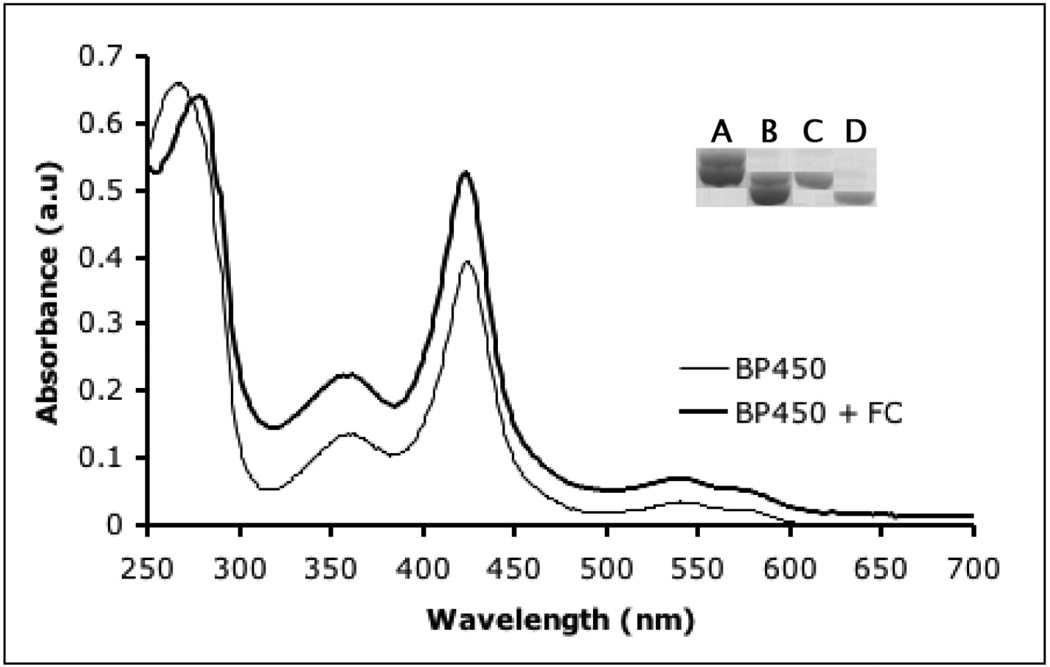
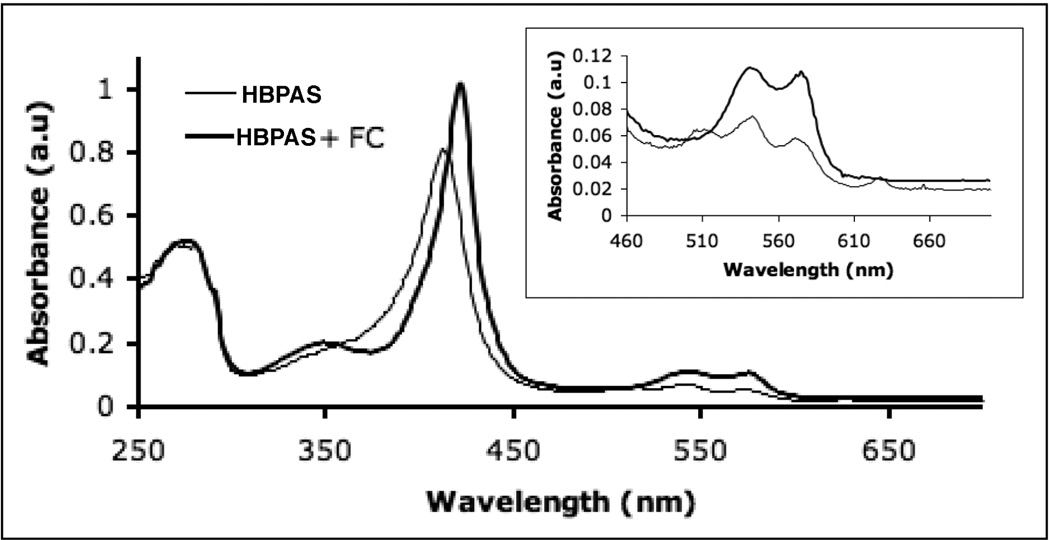
In addition to gsNOS, FC also increases heme content to saturating levels in two other unrelated proteins: BP450, a Cys-ligated heme protein and HBPAS: a Hisligated heme protein. Both of these proteins, when over-expressed in E. coli, are produced with partially heme incorporation. UV-Vis spectra of purified BP450 and BP450 co-expressed with FC are strikingly different, with the increased intensity of the Soret peak indicative of greater heme content in the material produced along with FC (Fig 3). When analyzed by SDS-PAGE, BP450 again also shows two bands (Fig 3, Inset, lane A), which both shift on His-tag cleavage (Fig 3, Inset, lane B). Like gsNOS, on co-expression with FC (Fig 3, Inset, lane C) BP450 produces only one band, which shifts on His-tag cleavage (Fig 3, Inset, lane D). In contrast, HBPAS always results in a single band on a SDS-PAGE gel (not shown), but shows absorption for four Q-bands in the UV-Vis spectrum when produced without FC (Fig 4). Pure heme proteins show only two such bands. The extra band(s) are representative of protein bound protoporphyrin IX [30]. Co-expression of FC results in an increase in the Abs(Soret)/Abs(280) and the extra Q-bands disappear (Fig 4). The fluorescence spectrum of HBPAS without co-expression of FC is similar to that of gsNOS expressed without FC (data not shown), but this fluorescence, attributable to protoporphyrin IX, also disappears on co-expression with FC.
Fig 3.
UV-Vis spectra of BP450 expressed by itself (thin line) and BP450 expressed with FC (thick line). Co-expression of FC results in a substantial increase in heme content of BP450 (AbsSoret/Abs280). Inset: BP450 expressed alone also results in two bands (lane A), both of which shift on His-tag cleavage (lane B). BP450 co-expressed with FC results in one band (lane C), which shifts on His-tag cleavage as expected (lane D).
Fig 4.
UV-Vis spectra of a Heme Binding PAS domain (HBPAS; His-ligated) when expressed by itself (thin line) shows four Q-band absorption peaks (inset). Co-expression with FC (thick line) increases the heme content and results in only two Q-bands (inset).
Conclusions
The production of δ-ALA production is rate-limiting step for heme biosynthesis [31–34] and δ-ALA synthesis is itself slowed by heme feedback inhibition. Thus, as has been well recognized [31–34], feeding with δ-ALA greatly aids recombinant heme protein production in E. coli. However, we have shown here that under conditions of augmentation with δ-ALA, ferrous iron insertion into protoporphyrin IX becomes rate-limiting. Co-expression with Ferrochelatase along with the addition of a small amount of δ-ALA, is sufficient to produce fully incorporated heme protein. This method is applicable for both Cys-ligated and His-ligated heme proteins. In the case of the two Cys-ligated proteins, porphyrin substitution could be observed on an SDS-PAGE gel as two closely spaced bands and also by fluorescence spectroscopy. In the one His-ligated heme protein example, UV-Visible and fluorescence spectra were effective indicators of insufficient porphyrin metallation, but only one band was observed by SDS-PAGE even with less than full porphyrin content. This is probably because the heme or porphyrin does not remain associated with the PAS protein during electrophoresis, unlike the other two cases. In conclusion, the simple and inexpensive method of co-expressing Ferrochelatase is effective at producing fully incorporated heme proteins in E. coli.
Acknowledgements
We thank S. Barry and G. Challis for providing the p450/pet151/D-TOPO vector and K. Watts for the HBPAS protein clone.
Abbreviations used
- δ-ALA
δ-aminolevulinic acid
- FC
Ferrochelatase
- gsNOS
Geobacillus stearothermophilus Nitric Oxide Synthase
- BP450
Bacterial P450 protein from S. turgidiscabies
- HBPAS
Heme binding PAS domain containing protein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions: B.R.C and J.S initiated studies; M.K, S.R.H and D.L.R designed and analyzed resonance raman experiments and discovered free base porphyrin as the contaminant; J.S and B.R.C designed and analyzed the heme-incorporation experiments using Ferrochelatase; B.P and M.V.A verified the method for BP450 and HBPAS; J.S and B.R.C wrote the manuscript.
Competing Interests Statement: The authors declare no competing financial interests.
References
- 1.Gray HB, Winkler JR. Electron transfer in engineered heme enzymes. Faseb Journal. 1997;11:A781–A781. [Google Scholar]
- 2.Sono M, Roach MP, Coulter ED, Dawson JH. Heme-Containing Oxygenases. Chem Rev. 1996;96:2841–2888. doi: 10.1021/cr9500500. [DOI] [PubMed] [Google Scholar]
- 3.Alderton WK, Cooper CE, Knowles RG. Nitric oxide synthases: structure, function and inhibition. Biochem J. 2001;357:593–615. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perutz MF, Paoli M, Lesk AM. Fix L, a haemoglobin that acts as an oxygen sensor: signalling mechanism and structural basis of its homology with PAS domains. Chem. Biol. 1999;6:R291–R297. doi: 10.1016/s1074-5521(99)80121-5. [DOI] [PubMed] [Google Scholar]
- 5.Varadarajan R, Szabo A, Boxer SG. Cloning, Expression In Escherichia-Coli, And Reconstitution Of Human Myoglobin. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1985;82:5681–5684. doi: 10.1073/pnas.82.17.5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishikawa K, Sato M, Yoshida T. Expression Of Rat Heme Oxygenase In Escherichia-Coli As A Catalytically Active, Full-Length Form That Binds To Bacterial-Membranes. European Journal Of Biochemistry. 1991;202:161–165. doi: 10.1111/j.1432-1033.1991.tb16357.x. [DOI] [PubMed] [Google Scholar]
- 7.Smith AT, Santama N, Dacey S, Edwards M, Bray RC, Thorneley RNF, Burke JF. Expression Of A Synthetic Gene For Horseradish Peroxidase-C In Escherichia-Coli And Folding And Activation Of The Recombinant Enzyme With Ca-2+ And Heme. Journal Of Biological Chemistry. 1990;265:13335–13343. [PubMed] [Google Scholar]
- 8.Kery V, Elleder D, Kraus JP. Delta-Aminolevulinate Increases Heme Saturation And Yield Of Human Cystathionine Beta-Synthase Expressed In Escherichia-Coli. Archives Of Biochemistry And Biophysics. 1995;316:24–29. doi: 10.1006/abbi.1995.1005. [DOI] [PubMed] [Google Scholar]
- 9.Varnado CL, Hertwig KM, Thomas R, Roberts JK, Goodwin DC. Properties of a novel periplasmic catalase-peroxidase from Escherichia coli O157: H7. Archives Of Biochemistry And Biophysics. 2004;421:166–174. doi: 10.1016/j.abb.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 10.Graves PE, Henderson DP, Horstman MJ, Solomon BJ, Olson JS. Enhancing stability and expression of recombinant human hemoglobin in E-coli: Progress in the development of a recombinant HBOC source. Biochimica Et Biophysica Acta-Proteins And Proteomics. 2008;1784:1471–1479. doi: 10.1016/j.bbapap.2008.04.012. [DOI] [PubMed] [Google Scholar]
- 11.Weickert MJ, Doherty DH, Best EA, Olins PO. Optimization of heterologous protein production in Escherichia coli. Current Opinion In Biotechnology. 1996;7:494–499. doi: 10.1016/s0958-1669(96)80051-6. [DOI] [PubMed] [Google Scholar]
- 12.Shen TJ, Ho NT, Simplaceanu V, Zou M, Green BN, Tam MF, Ho C. Production Of Unmodified Human Adult Hemoglobin In Escherichia-Coli. Proceedings Of The National Academy Of Sciences Of The United States Of America. 1993;90:8108–8112. doi: 10.1073/pnas.90.17.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varnado CL, Goodwin DC. System for the expression of recombinant hemoproteins in Escherichia coli. Protein Expression And Purification. 2004;35:76–83. doi: 10.1016/j.pep.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Varnado C, Olson J, Goodwin D. Expression of recombinant hemoproteins in E. coli using a heme protein expression system. Biophysical Journal. 2007:384A–384A. [Google Scholar]
- 15.Weickert MJ, Pagratis M, Curry SR, Blackmore R. Stabilization of apoglobin by low temperature increases yield of soluble recombinant hemoglobin in Escherichia coli. Applied And Environmental Microbiology. 1997;63:4313–4320. doi: 10.1128/aem.63.11.4313-4320.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pesce A, Nardini M, Dewilde S, Geuens E, Yamauchi K, Ascenzi P, Riggs AF, Moens L, Bolognesi M. The 109 residue nerve tissue minihemoglobin from Cerebratulus lacteus highlights striking structural plasticity of the alpha-helical globin fold. Structure. 2002;10:725–735. doi: 10.1016/s0969-2126(02)00763-3. [DOI] [PubMed] [Google Scholar]
- 17.Summerford CM, Pardanani A, Betts AH, Poteete AR, Colotti G, Royer WE. Bacterial Expression Of Scapharca Dimeric Hemoglobin - A Simple-Model System For Investigating Protein Cooperativity. Protein Engineering. 1995;8:593–599. doi: 10.1093/protein/8.6.593. [DOI] [PubMed] [Google Scholar]
- 18.Weickert MJ, Apostol I. High-fidelity translation of recombinant human hemoglobin in Escherichia coli. Appl Environ Microbiol. 1998;64:1589–1593. doi: 10.1128/aem.64.5.1589-1593.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weickert MJ, Pagratis M, Glascock CB, Blackmore R. A mutation that improves soluble recombinant hemoglobin accumulation in Escherichia coli in heme excess. Applied And Environmental Microbiology. 1999;65:640–647. doi: 10.1128/aem.65.2.640-647.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodward JJ, Martin NI, Marletta MA. An Escherichia coli expression-based method for heme substitution. Nat Methods. 2007;4:43–45. doi: 10.1038/nmeth984. [DOI] [PubMed] [Google Scholar]
- 21.Sudhamsu J, Crane BR. Structure and reactivity of a thermostable prokaryotic nitric-oxide synthase that forms a long-lived oxy-heme complex. J Biol Chem. 2006;281:9623–9632. doi: 10.1074/jbc.M510062200. [DOI] [PubMed] [Google Scholar]
- 22.Kabir M, Sudhamsu J, Crane BR, Yeh SR, Rousseau DL. Substrate-Ligand Interactions in Geobacillus stearothermophilus Nitric Oxide Synthase. Biochemistry. 2008 doi: 10.1021/bi801491e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davydov RH, Sudhamsu J, Lees NS, Crane BR, Hoffman BM. EPR and ENDOR characterization of the reactive intermediates in the generation of NO by cryoreduced oxy-nitric oxide synthase from G. stearothermophilus. J. Am. Chem. Soc. 2009;131:14493–14507. doi: 10.1021/ja906133h. [DOI] [PubMed] [Google Scholar]
- 24.Pant K, Bilwes AM, Adak S, Stuehr DJ, Crane BR. Structure of a nitric oxide synthase heme protein from Bacillus subtilis. Biochemistry. 2002;41:11071–11079. doi: 10.1021/bi0263715. [DOI] [PubMed] [Google Scholar]
- 25.Buddha MR, Tao T, Parry RJ, Crane BR. Regioselective nitration of tryptophan by a complex between bacterial nitric-oxide synthase and tryptophanyl-tRNA synthetase. J Biol Chem. 2004;279:49567–49570. doi: 10.1074/jbc.C400418200. [DOI] [PubMed] [Google Scholar]
- 26.Santolini M, Roman M, Stuehr DJ, Mattioli TA. Resonance Raman study of Bacillus subtilis NO synthase-like protein: Similarities and differences with mammalian NO synthases. Biochemistry. 2006;45:1480–1489. doi: 10.1021/bi051710q. [DOI] [PubMed] [Google Scholar]
- 27.Rousseau DL, Li D, Couture M, Yeh SR. Ligand-protein interactions in nitric oxide synthase. Journal Of Inorganic Biochemistry. 2005;99:306–323. doi: 10.1016/j.jinorgbio.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 28.Blackwood ME, Rush TS, Romesberg F, Schultz PG, Spiro TG. Alternative modes of substrate distortion in enzyme and antibody catalyzed ferrochelation reactions. Biochemistry. 1998;37:779–782. doi: 10.1021/bi972616f. [DOI] [PubMed] [Google Scholar]
- 29.Lu Y, Sousa A, Franco R, Mangravita A, Ferreira GC, Moura I, Shelnutt JA. Binding of protoporphyrin IX and metal derivatives to the active site of wild-type mouse ferrochelatase at low porphyrin-to-protein ratios. Biochemistry. 2002;41:8253–8262. doi: 10.1021/bi025569m. [DOI] [PubMed] [Google Scholar]
- 30.Lozovaya GI, Masinovsky Z, Sivash AA. Protoporphyrin-Ix As A Possible Ancient Photosensitizer - Spectral And Photochemical Studies. Origins Of Life And Evolution Of The Biosphere. 1990;20:321–330. [Google Scholar]
- 31.Ades IZ. Heme Production In Animal-Tissues - The Regulation Of Biogenesis Of Delta-Aminolevulinate Synthase. International Journal Of Biochemistry. 1990;22:565–578. doi: 10.1016/0020-711x(90)90032-x. [DOI] [PubMed] [Google Scholar]
- 32.Woodard SI, Dailey HA. Regulation Of Heme-Biosynthesis In Escherichia-Coli. Archives Of Biochemistry And Biophysics. 1995;316:110–115. doi: 10.1006/abbi.1995.1016. [DOI] [PubMed] [Google Scholar]
- 33.Gibson SL, Havens JJ, Metz L, Hilf R. Is delta-aminolevulinic acid dehydratase rate limiting in heme biosynthesis following exposure of cells to delta-aminolevulinic acid? Photochemistry And Photobiology. 2001;73:312–317. doi: 10.1562/0031-8655(2001)073<0312:iaadrl>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 34.Heinemann IU, Jahn M, Jahn D. The biochemistry of heme biosynthesis. Archives Of Biochemistry And Biophysics. 2008;474:238–251. doi: 10.1016/j.abb.2008.02.015. [DOI] [PubMed] [Google Scholar]