Abstract
Background
Caveolin-1 (cav-1) is overexpressed by metastatic prostate cancer (PC) cells. Pre-operative serum cav-1 levels have been shown to be a prognostic marker for PC recurrence. This study evaluated the relationship between post-treatment serum cav-1 levels and single nucleotide polymorphisms (SNPs) in the cav-1 and -2 genes with risk of PC, aggressive PC, PC recurrence or death.
Methods
Two case-control studies of PC among men in Washington State were combined for this analysis. Cases (n=1,458) were diagnosed in 1993–96 or 2002–05 and identified via a SEER cancer registry. Age-matched controls (n=1,351) were identified via random digit dialing. Logistic regression assessed the relationship between exposures (19 haplotype-tagging SNPs from all subjects and post-treatment serum cav-1 levels from a sample of 202 cases and 226 controls) and PC risk and aggressive PC. Cox proportional hazards regression assessed the relationship between exposures and PC recurrence and death.
Results
Rs9920 in cav-1 was associated with an increased relative risk of overall PC (ORCT+CC=1.37, 95%CI=1.12, 1.68) and aggressive PC (ORCT+CC=1.57, 95%CI=1.20, 2.06), but not with PC recurrence or death. High post-treatment serum cav-1 levels were not associated with PC risk, aggressive PC, or PC-specific death, but approached a significant inverse association with PC recurrence (hazard ratio=0.69, 95%CI=0.47, 1.00).
Conclusions
We found modest evidence for an association with a variant in the cav-1 gene and risk of overall PC and aggressive PC, which merits further study. We found no evidence that higher post-treatment serum cav-1 is associated with risk of aggressive PC or adverse PC outcomes.
Keywords: prostate cancer, caveolin, case-control association study, single nucleotide polymorphism, biomarker, recurrence
Background
Prostate cancer (PC) remains the second leading cause of cancer-related deaths in US men [1]. The high mortality from PC is due in part to the inability of current biomarkers to predict which prostatic tumors will become life-threatening. Indeed, among patients undergoing radical prostatectomy (RP), an estimated 35% of patients will recur within 10 years [2–7]. New markers are needed to identify patients with more aggressive disease, who may most benefit from aggressive treatment. The protein caveolin-1 (cav-1) has been investigated as such a prognostic biomarker.
Cav-1 is a major structural component of caveolae, flask-shaped invaginations of the plasma membrane, which serve as a scaffold for signaling molecules related to cell adhesion, growth, and survival [8–10]. Evidence for an association between elevated cav-1 levels and PC has been found in both mouse and human studies [11–17]. Evidence has also been found for an association between elevated tissue cav-1 or pre-treatment serum cav-1 levels and metastatic disease [12], as well as with features of comparatively more aggressive prostate tumors, such as a higher Gleason score, elevated PSA levels, and early PSA-associated recurrence [18–21]. However, the relationship between post-treatment cav-1 levels and PC has not yet been evaluated.
The cav-1 and -2 genes are co-localized to 7q31.1, a highly conserved region that encompasses a known fragile site which is deleted or associated with loss of heterozygosity in a variety of human cancers, including cancer of the breast, colon, esophagus, head and neck, kidney, mouth, pancreas, prostate, ovary and stomach [10,22–23]. These types of data, as well as studies of cav-1 null mice, suggest cav-1 and -2 may function as tumor suppressor genes for some malignancies [24]. However, in PC, biochemical and genetic data support a tumor promoter function for cav-1 [15,17,22,25]. Yet no specific mutations associated with PC have been identified in the cav-1 or -2 genes.
We utilized data from two population-based PC case-control studies to examine the association between post-treatment cav-1 serum protein levels and single nucleotide polymorphisms (SNPs) in the cav-1 and -2 genes with the risk of PC, aggressive PC, and PC recurrence or death.
Methods
Study Population
Data from two population-based case-control studies of risk factors for PC among Caucasian and African-American men residing in King County, Washington were combined for the genetic analysis. The first study included 753 cases and 703 controls described previously [26]. Briefly, incident cases ascertained from the Seattle-Puget Sound Surveillance Epidemiology and End Results (SEER) cancer registry; they were diagnosed between January 1, 1993 and December 31, 1996, and were 40 to 64 years of age at diagnosis. The second study included 1,001 cases and 942 controls described previously [27]. Incident cases were also ascertained from the SEER cancer registry; they were diagnosed between January 1, 2002 and December 31, 2005, and were 35 to 74 years of age at diagnosis. Controls for both studies were men without a self-reported history of PC, recruited via random digit dialing (RDD) during the same ascertainment period and from the same underlying general population as the cases, and frequency matched to cases by five-year age groups. Among eligible subjects ascertained for the first study, 82% of cases and 75% of controls participated in the study interview, and of these participants, 84% of cases and 80% of controls provided a blood sample. Among eligible subjects ascertained for the second study, 75% of cases and 63% of controls participated in the study interview, and of these participants, 83% of cases and 84% of controls provided a blood sample. After combining these two studies, there were 1,457 PC cases and 1,351 controls with DNA available for the genetic analysis.
Background information including demographic and lifestyle factors, medical history, PC screening history, and family history of PC was collected from participants at interview. Clinical information such as Gleason score, tumor stage, serum prostate-specific antigen (PSA) level at diagnosis, and primary treatment was obtained from the SEER cancer registry. Vital status and underlying cause of death of cases has been ascertained on a regular basis through the SEER cancer registry, where the patient file is linked to the registry. For each deceased subject, a death certificate is requested from the state to confirm cause of death. In 2004, a follow-up survey was sent to 631 of the cases from the first study, 82% of whom responded, to assess secondary treatment(s) and evidence for PC recurrence or progression.
Because participants in the first study had adequate follow-up to assess recurrence (as defined in the statistical methods below), a sample of these men were selected for the serum protein analysis. Among participants who provided serum, the following were included: cases who had evidence of disease recurrence (n=83), cases who died of metastatic PC (n=37), cases who had pre- and post-treatment serum available (n=16), and a random sample of the remaining cases so that approximately half met the definition of aggressive disease (n=109) and half did not (n=93), for a total of 202 cases. “More aggressive” PC was defined by a Gleason score of 7(4+3) or 8–10, regional or distant tumor stage, or a diagnostic PSA value ≥20 ng/ml. On average, serum samples were obtained 10.5 months after diagnosis (range=2 to 44 months). Among the 570 eligible controls from the first study with serum available, 33 were subsequently diagnosed with PC according to a link to the SEER cancer registry in August 2005, and so were included as controls and to be analyzed separately. The remaining controls who reported no history of PC were randomly sampled and frequency matched to cases by five-year age groups (n=226). In addition, 5% (n=20) blind duplicates were included to evaluate the reliability of the serum assay.
Pre-treatment serum had been collected in a sub-study described previously [28]. The cav-1 levels in pre- and post-treatment serum were examined to evaluate whether cav-1 levels changed post-surgery, given most of the samples in this study were collected post-RP. Twelve (75%) of the 16 cases with pre- and post-treatment serum available received RP as their primary treatment. Among these cases, serum samples were obtained on average 17 days prior to radical prostatectomy (RP) (range 0 to 100 days) and 7 months after the RP (range 4 to 11 months). For the remaining cases (n=4), serum samples were obtained on average 3 months before and 14 months after diagnosis.
The Institutional Review Boards of the Fred Hutchinson Cancer Research Center and the National Human Genome Research Institute approved study procedures and materials, and written informed consent was obtained from all study participants.
Genetic Analysis
DNA samples were genotyped for 19 single nucleotide polymorphisms (SNPs) in the cav-1 and cav-2 genes for all cases and controls. The SNPs were selected using the Genome Variation Server (gvs.gs.washington.edu/gvs) to cover the genes as haplotype-tagging SNPs. The Applied Biosystems (ABI) SNPlex® Genotyping System was used for genotyping and proprietary GeneMapper® software was used for allele assignment (www.appliedbiosystems.com). Discrimination of the specific SNP allele was carried out with the ABI 3730×l DNA Analyzer and is based on the presence of a unique sequence assigned to the original allele-specific oligonucleotide. Quality control included genotyping of 144 blind duplicate samples distributed across all genotyping batches. There was >99% agreement between blinded samples. Each batch of DNA aliquots genotyped incorporated similar numbers of case and control samples, and laboratory personnel were blinded to the case-control status of samples. Genotype frequencies in cav-1 and cav-2 were evaluated among the Caucasian and African-American controls separately; all SNPs were consistent with the expected proportions under Hardy-Weinberg, except for rs17138765 among Caucasians (pexact=0.01), and so this SNP was removed from the analysis.
Protein analysis
Serum cav-1 levels were determined by the sandwich ELISA protocol described previously [19]. Costar microplate wells were coated with 0.5 µg cav-1 polyclonal antibody (Transduction Laboratories, San Diego, CA) and blocked with TBS buffer containing 1.5% bovine serum albumin and 0.05% v/v Tween 20. Serum samples, calibrators, and controls (50 µL) were added to the wells, and 50 µL TBS containing 0.5% v/v Tween 20 was added to each well. The plate was incubated at room temperature for 2 hours with shaking and after extensive washing, 100 µL horseradish peroxidase–conjugated cav-1 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) diluted 1:200 in blocking buffer was added to each well. The microplate was incubated for 90 minutes at room temperature with shaking, the wells were then washed extensively, and 100 µL 3,3’,5,5’-tetramethylbenzidine substrate solution (Sigma-Aldrich, St. Louis, MO) was added and the blue color was allowed to develop for 20 minutes in the dark. The reaction was stopped by adding 50 µL of 2 N H2SO4, and the absorbance was read at 450 nm using a microplate reader (Sunrise Microplate Reader, Tecan US, Inc., Charlotte, NC).
Statistical methods
Logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) to estimate the relative risk of PC among cases relative to controls for each SNP genotype. Polytomous logistic regression was used to calculate ORs and 95% CIs to estimate the relative risk of more aggressive and less aggressive PC (as defined above) relative to controls for each SNP genotype. Codominant and dominant genetic models were considered for each SNP. All models were adjusted for age, and tested for possible confounding by PC screening history or family history of PC. In addition, permuted p-values were calculated to adjust for multiple comparisons.
The Wilcoxan signed rank sum test was used to compare serum cav-1 levels in pre- and post-treatment samples. Logistic regression was used to calculate ORs and 95% CIs to estimate the relative risk of PC among cases relative to controls by cav-1 levels. Polytomous logistic regression was used to calculate ORs and 95% CIs to estimate the risk of more aggressive and less aggressive PC (as defined above) relative to controls for cav-1 levels. All models were adjusted for age, and tested for possible confounding by PC screening history or family history of PC.
Cox proportional hazards regression was used to estimate hazards ratios and 95% CIs to assess the relationship between: (1) the SNPs found to be significantly associated with aggressive PC and disease recurrence, (2) cav-1 levels and PC recurrence or death from PC, and (3) PC diagnosis among men initially enrolled as controls who were subsequently diagnosed with PC. The analyses of recurrence were restricted to cases who were diagnosed with local or regional stage disease and either subsequently died of PC (prior to the follow-up survey) or completed a follow-up survey, which provided recurrence information and consent to obtain medical records. Medical record review confirmed self-reported recurrence in all but one case. Recurrence was defined as at least one of the following: positive bone scan, CT, MRI, or biopsy showing PC after primary treatment; biochemical failure after RP as primary therapy (serum PSA level ≥0.2 ng/mL); biochemical failure after radiation therapy (RT) as primary therapy (nadir PSA +2 ng/mL); rising PSA while on androgen deprivation therapy (ADT); or RT or ADT as secondary treatment. Time from diagnosis until recurrence was calculated as the difference between the date of diagnosis and the earliest date of evidence of recurrence: date of death from PC, date of recurrence or progression abstracted from medical records, date of recurrence from the follow-up survey, or, for those censored, the end of the year during which the follow-up survey was collected (December 31, 2005). The analyses of PC death included all cases. The censoring date for members last known to be alive was the date of the last vital status update from the cancer registry (December 1, 2008). The proportional hazards models were tested for possible confounding by age at diagnosis, PC screening history, or a family history of PC, and recalculated including only cases who received an RP as primary therapy.
Logistic regression was used to determine if the SNP in cav-1 found to be significantly associated with PC was also associated with high post-treatment serum cav-1 levels.
Because cav-1 levels tend not to be normally distributed, we used several measures of cav-1 to try to capture this variation. We considered cav-1 as a continuous variable, as a log-transformed continuous variable, as an ordered categorical variable using quintiles defined by the distribution of cav-1 among controls, and as a dichotomous variable where “high” cav-1 levels were those above the median level among controls.
Most analyses were performed in SAS® version 9.1.3 (SAS Institute, Cary, NC). Hardy-Weinberg equilibrium was calculated in STATA/SE® 10.0 for Windows (StataCorp, College Station, TX).
Results
Among the 1,458 cases and 1,351 controls included in the genetic study, a higher proportion of cases than controls were African-American (10.2% vs. 6.3%, respectively; Table 1), had a first-degree relative with PC (21.5% vs. 11.2%), and reported having a PSA or DRE screening test in the five years prior to diagnosis or reference date (89.2% and 86.4%, respectively).
Table 1.
Characteristics of prostate cancer cases and controls
| Characteristic | Genetic Study | Protein Sub-Study | ||||||
|---|---|---|---|---|---|---|---|---|
| Cases (n=1,458) |
Controls (n=1,351) |
Cases (n=202) |
Controls (n=226) |
|||||
| n | (%) | n | (%) | N | (%) | n | (%) | |
| Age at diagnosis/reference date | ||||||||
| 35–49 | 118 | (8.1) | 126 | (9.3) | 18 | (8.9) | 17 | (7.5) |
| 50–54 | 215 | (14.8) | 209 | (15.5) | 38 | (18.8) | 40 | (17.7) |
| 55–59 | 357 | (24.5) | 358 | (26.5) | 76 | (37.6) | 87 | (38.5) |
| 60–64 | 433 | (29.7) | 348 | (25.8) | 70 | (34.7) | 82 | (36.3) |
| 65–69 | 177 | (12.1) | 164 | (12.1) | -- | -- | -- | -- |
| 70–74 | 158 | (10.8) | 146 | (10.8) | -- | -- | -- | -- |
| Race | ||||||||
| Caucasian | 1,309 | (89.8) | 1,266 | (93.7) | 202 | (100.0) | 226 | (100.0) |
| African-American | 149 | (10.2) | 85 | (6.3) | -- | -- | -- | -- |
| First-degree relative with prostate cancer | ||||||||
| No | 1,145 | (78.5) | 1,200 | (88.8) | 173 | (85.6) | 215 | (95.1) |
| Yes | 313 | (21.5) | 151 | (11.2) | 29 | (14.4) | 11 | (4.9) |
| Screening historya | ||||||||
| None | 157 | (10.8) | 183 | (13.6) | 23 | (11.4) | 34 | (15.0) |
| DRE only | 258 | (17.7) | 518 | (38.3) | 42 | (20.8) | 114 | (50.4) |
| PSA | 1,043 | (71.6) | 650 | (48.1) | 137 | (67.8) | 78 | (34.5) |
| PSA valueb | ||||||||
| < 4.0 | 189 | (13.0) | 355 | (26.3) | 23 | (11.4) | 205 | (90.7) |
| 4.0–9.9 | 814 | (55.8) | 33 | (2.4) | 94 | (46.5) | 9 | (4.0) |
| 10.0–19.9 | 210 | (14.4) | 6 | (0.4) | 28 | (13.9) | 1 | (0.4) |
| ≥ 20.0 | 138 | (9.5) | 0 | (0.0) | 39 | (19.3) | 0 | (0.0) |
| Missing | 107 | (7.4) | 957 | (70.8) | 18 | (8.9) | 11 | (4.9) |
| Gleason score | ||||||||
| 2–4 | 72 | (4.9) | -- | -- | 12 | (5.9) | -- | -- |
| 5–6 | 741 | (50.8) | -- | -- | 84 | (41.6) | -- | -- |
| 7 (3+4) | 408 | (28.0) | -- | -- | 56 | (27.7) | -- | -- |
| 7 (4+3) | 91 | (6.2) | -- | -- | 13 | (6.4) | -- | -- |
| 8–10 | 140 | (9.6) | -- | -- | 36 | (17.8) | -- | -- |
| Missing | 6 | (0.4) | -- | -- | 1 | (0.5) | -- | -- |
| Stage at diagnosis | ||||||||
| Local | 1,141 | (78.3) | -- | -- | 120 | (59.4) | -- | -- |
| Regional | 280 | (19.2) | -- | -- | 67 | (33.2) | -- | -- |
| Distant | 37 | (2.5) | -- | -- | 15 | (7.4) | -- | -- |
| Primary treatment | ||||||||
| RP | 831 | (57.0) | -- | -- | 141 | (69.8) | -- | -- |
| RT | 412 | (28.3) | -- | -- | 34 | (16.8) | -- | -- |
| ADT | 72 | (4.9) | -- | -- | 19 | (9.4) | -- | -- |
| Other treatment | 5 | (0.3) | -- | -- | 1 | (0.5) | -- | -- |
| Active surveillance | 138 | (9.5) | -- | -- | 7 | (3.5) | -- | -- |
PSA=prostate-specific antigen; RP=radical prostatectomy; RT=radiation therapy; ADT=androgen deprivation therapy
Screening history within five years prior to diagnosis or reference date.
PSA at diagnosis for cases and measured at interview date for controls.
Genetic analysis
Among the 19 tagSNPs evaluated, one SNP in cav-1 (rs9920; ORCT+CC=1.37, 95%CI=1.12, 1.68) and no SNPs in cav-2 were associated with PC risk among Caucasians (Table 2). After adjusting for multiple comparisons using permutation p-values, rs9920 in cav-1 remained significant (p<0.05). Rs9920 was also associated with more aggressive PC among Caucasians (rs9920: ORCT+CC=1.57, 95%CI=1.20, 2.06; Table 3). The latter result remained the same when disease aggressiveness was defined with Gleason score alone. This SNP was not significantly associated with the risk of more aggressive versus less aggressive disease (p=0.12). The association with PC risk and with disease aggressiveness remained similar after adjustment for a first-degree relative with PC or having a PC screening test in the five years prior to reference date. Similar analyses in African-American men revealed no associations between any SNP genotypes and PC risk (Table 2) or disease aggressiveness (data not shown).
Table 2.
Prostate cancer risk associated with SNPs in cav-1 and cav-2, by race
| Genotype | Cases (n=1,457)a |
Controls (n=1,351)a |
ORb | 95% CI | Pc | |||
|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | |||||
|
Caucasians | ||||||||
|
Cav-1 | ||||||||
|
rs4236601, chr7:115949965 | ||||||||
| GG | 604 | (49.0) | 637 | (52.0) | 1.00 | Reference | ||
| GA | 537 | (21.8) | 500 | (40.8) | 1.13 | 0.96 | 1.34 | |
| AA | 92 | (3.7) | 89 | (7.3) | 1.10 | 0.80 | 1.50 | 0.32 |
| GA or AA | 629 | (51.0) | 589 | (48.0) | 1.13 | 0.96 | 1.32 | 0.13 |
|
rs926198, chr7:115954444 | ||||||||
| TT | 568 | (45.3) | 538 | (43.9) | 1.00 | Reference | ||
| CT | 537 | (42.9) | 539 | (43.9) | 0.94 | 0.80 | 1.12 | |
| CC | 148 | (11.8) | 150 | (12.2) | 0.94 | 0.73 | 1.22 | 0.77 |
| CT or CC | 685 | (54.7) | 689 | (56.2) | 0.94 | 0.80 | 1.11 | 0.47 |
|
rs10256914, chr7:115962947 | ||||||||
| TT | 676 | (53.2) | 684 | (55.0) | 1.00 | Reference | ||
| TC | 505 | (39.8) | 474 | (38.1) | 1.08 | 0.92 | 1.27 | |
| CC | 89 | (7.0) | 85 | (6.8) | 1.06 | 0.77 | 1.46 | 0.65 |
| TC or CC | 594 | (46.8) | 559 | (45.0) | 1.08 | 0.92 | 1.26 | 0.35 |
|
rs3807986, chr7:115965061 | ||||||||
| AA | 729 | (58.0) | 720 | (57.9) | 1.00 | Reference | ||
| GA | 455 | (36.2) | 457 | (36.8) | 0.98 | 0.83 | 1.16 | |
| GG | 72 | (5.7) | 66 | (5.3) | 1.09 | 0.77 | 1.55 | 0.85 |
| GA or GG | 527 | (42.0) | 523 | (42.1) | 1.00 | 0.85 | 1.17 | 0.95 |
|
rs959173, chr7:115969290 | ||||||||
| TT | 893 | (70.5) | 886 | (71.3) | 1.00 | Reference | ||
| CT | 335 | (26.4) | 328 | (26.4) | 1.02 | 0.85 | 1.22 | |
| CC | 39 | (3.1) | 29 | (2.3) | 1.35 | 0.82 | 2.20 | 0.49 |
| CT or CC | 374 | (29.5) | 357 | (28.7) | 1.04 | 0.88 | 1.24 | 0.63 |
|
rs3807989, chr7:115973477 | ||||||||
| GG | 436 | (34.5) | 421 | (33.9) | 1.00 | Reference | ||
| GA | 597 | (47.2) | 626 | (50.4) | 0.92 | 0.77 | 1.10 | |
| AA | 232 | (18.3) | 196 | (15.8) | 1.16 | 0.92 | 1.46 | 0.12 |
| GA or AA | 829 | (65.5) | 822 | (66.1) | 0.98 | 0.83 | 1.15 | 0.77 |
|
rs3801995, chr7:115977833 | ||||||||
| CC | 705 | (55.7) | 690 | (55.6) | 1.00 | Reference | ||
| CT | 474 | (37.4) | 479 | (38.6) | 0.97 | 0.82 | 1.14 | |
| TT | 87 | (6.9) | 72 | (5.8) | 1.20 | 0.86 | 1.67 | 0.46 |
| CT or TT | 561 | (44.3) | 551 | (44.4) | 1.00 | 0.85 | 1.17 | 0.98 |
|
rs1049314, chr7:115986931 | ||||||||
| CC | 847 | (67.2) | 863 | (69.3) | 1.00 | Reference | ||
| CA | 378 | (30.0) | 352 | (28.3) | 1.10 | 0.92 | 1.31 | |
| AA | 36 | (2.9) | 30 | (2.4) | 1.24 | 0.75 | 2.03 | 0.44 |
| CA or AA | 414 | (32.8) | 382 | (30.7) | 1.11 | 0.94 | 1.31 | 0.23 |
|
rs9920, chr7:115987328 | ||||||||
| TT | 993 | (78.3) | 1,031 | (83.0) | 1.00 | Reference | ||
| CT | 262 | (20.7) | 204 | (16.4) | 1.35 | 1.10 | 1.65 | |
| CC | 14 | (1.1) | 7 | (0.6) | 2.09 | 0.84 | 5.22 | 0.006 |
| CT or CC | 276 | (21.8) | 211 | (17.0) | 1.37 | 1.12 | 1.68 | 0.002 |
|
rs1049334, chr7:115987616 | ||||||||
| GG | 1,077 | (84.9) | 1,033 | (83.2) | 1.00 | Reference | ||
| GA | 186 | (14.7) | 203 | (16.3) | 0.88 | 0.71 | 1.09 | |
| AA | 6 | (0.5) | 6 | (0.5) | 0.96 | 0.31 | 3.00 | 0.50 |
| GA or AA | 192 | (15.1) | 209 | (16.8) | 0.88 | 0.71 | 1.09 | 0.24 |
|
rs1049337, chr7:115987823 | ||||||||
| CC | 675 | (53.7) | 625 | (50.4) | 1.00 | Reference | ||
| CT | 489 | (38.9) | 514 | (41.5) | 0.87 | 0.74 | 1.03 | |
| TT | 93 | (7.4) | 100 | (8.1) | 0.86 | 0.63 | 1.16 | 0.23 |
| CT or TT | 582 | (46.3) | 614 | (49.6) | 0.87 | 0.74 | 1.02 | 0.09 |
|
Cav-2 | ||||||||
|
rs17138767, chr7:115924913 | ||||||||
| AA | 1,076 | (85.3) | 1,041 | (84.0) | 1.00 | Reference | ||
| GA | 180 | (14.3) | 194 | (15.7) | 0.89 | 0.72 | 1.11 | |
| GG | 5 | (0.4) | 5 | (0.4) | 0.99 | 0.28 | 3.42 | 0.59 |
| GA or GG | 185 | (14.7) | 199 | (16.1) | 0.89 | 0.72 | 1.11 | 0.31 |
|
rs4730742, chr7:115925128 | ||||||||
| TT | 843 | (67.0) | 822 | (66.2) | 1.00 | Reference | ||
| GT | 381 | (30.3) | 379 | (30.5) | 0.98 | 0.82 | 1.16 | |
| GG | 35 | (2.8) | 40 | (3.2) | 0.86 | 0.54 | 1.37 | 0.80 |
| GT or GG | 416 | (33.0) | 419 | (33.8) | 0.97 | 0.82 | 1.14 | 0.68 |
|
rs8940, chr7:115933310 | ||||||||
| CC | 853 | (67.4) | 816 | (65.7) | 1.00 | Reference | ||
| CG | 377 | (29.8) | 379 | (30.5) | 0.95 | 0.80 | 1.13 | |
| GG | 35 | (2.8) | 47 | (3.8) | 0.72 | 0.46 | 1.13 | 0.32 |
| CG or GG | 412 | (32.6) | 426 | (34.3) | 0.92 | 0.78 | 1.09 | 0.35 |
|
rs4727833, chr7:115935144 | ||||||||
| CC | 352 | (27.8) | 313 | (25.2) | 1.00 | Reference | ||
| GC | 617 | (48.7) | 640 | (51.5) | 0.85 | 0.70 | 1.03 | |
| GG | 298 | (23.5) | 290 | (23.3) | 0.91 | 0.73 | 1.13 | 0.24 |
| GC or GG | 915 | (72.2) | 930 | (74.8) | 0.87 | 0.73 | 1.04 | 0.12 |
|
rs1052990, chr7:115935606 | ||||||||
| TT | 517 | (40.7) | 508 | (41.0) | 1.00 | Reference | ||
| GT | 591 | (46.6) | 590 | (47.6) | 0.98 | 0.83 | 1.16 | |
| GG | 161 | (12.7) | 142 | (11.5) | 1.12 | 0.87 | 1.45 | 0.59 |
| GT or GG | 752 | (59.3) | 732 | (59.0) | 1.01 | 0.86 | 1.18 | 0.92 |
|
rs6466578, chr7:115938105 | ||||||||
| TT | 897 | (71.0) | 879 | (70.8) | 1.00 | Reference | ||
| CT | 330 | (26.1) | 338 | (27.2) | 0.96 | 0.81 | 1.15 | |
| CC | 37 | (2.9) | 25 | (2.0) | 1.47 | 0.88 | 2.47 | 0.29 |
| CT or CC | 367 | (29.0) | 363 | (29.2) | 1.00 | 0.84 | 1.19 | 0.97 |
|
rs10262524, chr7:115938188 | ||||||||
| CC | 639 | (50.3) | 658 | (52.9) | 1.00 | Reference | ||
| CA | 538 | (52.9) | 495 | (42.4) | 1.12 | 0.95 | 1.32 | |
| AA | 93 | (7.3) | 90 | (7.2) | 1.07 | 0.79 | 1.46 | 0.38 |
| CA or AA | 631 | (49.7) | 585 | (47.1) | 1.11 | 0.95 | 1.30 | 0.18 |
|
African-Americans | ||||||||
|
Cav-1 | ||||||||
|
rs4236601, chr7:115949965 | ||||||||
| GG | 59 | (41.0) | 32 | (40.0) | 1.00 | Reference | ||
| GA | 64 | (44.4) | 35 | (43.8) | 1.02 | 0.54 | 1.93 | |
| AA | 21 | (14.6) | 13 | (16.3) | 0.91 | 0.39 | 2.16 | 0.97 |
| GA or AA | 85 | (59.0) | 48 | (60.0) | 0.99 | 0.55 | 1.79 | 0.98 |
|
rs926198, chr7:115954444 | ||||||||
| TT | 33 | (22.8) | 18 | (22.5) | 1.00 | Reference | ||
| CT | 81 | (55.9) | 41 | (51.3) | 1.27 | 0.61 | 2.66 | |
| CC | 31 | (21.4) | 21 | (26.3) | 0.78 | 0.34 | 1.81 | 0.41 |
| CT or CC | 112 | (77.2) | 62 | (77.5) | 1.08 | 0.54 | 2.16 | 0.83 |
|
rs10256914, chr7:115962947 | ||||||||
| TT | 93 | (64.1) | 51 | (63.8) | 1.00 | Reference | ||
| TC | 40 | (27.6) | 25 | (31.3) | 0.96 | 0.50 | 1.83 | |
| CC | 12 | (8.3) | 4 | (5.0) | 1.88 | 0.55 | 6.42 | 0.58 |
| TC or CC | 52 | (35.9) | 29 | (36.3) | 1.09 | 0.59 | 1.99 | 0.79 |
|
rs3807986, chr7:115965061 | ||||||||
| AA | 30 | (20.8) | 14 | (17.5) | 1.00 | Reference | ||
| GA | 62 | (43.1) | 40 | (50.0) | 0.79 | 0.36 | 1.76 | |
| GG | 52 | (36.1) | 26 | (32.5) | 1.00 | 0.43 | 2.31 | 0.73 |
| GA or GG | 114 | (79.2) | 66 | (82.5) | 0.87 | 0.41 | 1.85 | 0.72 |
|
rs959173, chr7:115969290 | ||||||||
| TT | 52 | (35.9) | 31 | (38.8) | 1.00 | Reference | ||
| CT | 68 | (46.9) | 38 | (47.5) | 0.93 | 0.49 | 1.76 | |
| CC | 25 | (17.2) | 11 | (13.8) | 1.32 | 0.55 | 3.17 | 0.73 |
| CT or CC | 93 | (64.1) | 49 | (61.3) | 1.02 | 0.56 | 1.86 | 0.95 |
|
rs3807989, chr7:115973477 | ||||||||
| GG | 14 | (9.7) | 6 | (7.5) | 1.00 | Reference | ||
| GA | 64 | (44.1) | 39 | (48.8) | 0.83 | 0.28 | 2.51 | |
| AA | 67 | (46.2) | 35 | (43.8) | 1.01 | 0.33 | 3.08 | 0.81 |
| GA or AA | 131 | (90.3) | 74 | (92.5) | 0.91 | 0.32 | 2.65 | 0.87 |
|
rs3801995, chr7:115977833 | ||||||||
| CC | 59 | (40.7) | 28 | (35.0) | 1.00 | Reference | ||
| CT | 63 | (43.5) | 40 | (50.0) | 0.86 | 0.45 | 1.64 | |
| TT | 23 | (15.9) | 12 | (15.0) | 1.17 | 0.49 | 2.82 | 0.76 |
| CT or TT | 86 | (59.3) | 52 | (65.0) | 0.93 | 0.51 | 1.71 | 0.83 |
|
rs1049314, chr7:115986931 | ||||||||
| CC | 73 | (50.7) | 38 | (48.1) | 1.00 | Reference | ||
| CA | 56 | (38.9) | 36 | (46.6) | 0.74 | 0.40 | 1.38 | |
| AA | 15 | (10.4) | 5 | (6.3) | 1.63 | 0.53 | 5.03 | 0.35 |
| CA or AA | 71 | (49.3) | 41 | (51.9) | 0.85 | 0.48 | 1.53 | 0.59 |
|
rs9920, chr7:115987328 | ||||||||
| TT | 142 | (97.9) | 77 | (96.3) | 1.00 | Reference | ||
| CT | 3 | (2.1) | 3 | (3.8) | 0.46 | 0.08 | 2.60 | |
| CC | 0 | (0.0) | 0 | (0.0) | -- | -- | -- | 0.38 |
| CT or CC | 3 | (2.1) | 3 | (3.8) | 0.46 | 0.08 | 2.60 | 0.38 |
|
rs1049334, chr7:115987616 | ||||||||
| GG | 124 | (85.5) | 62 | (77.5) | 1.00 | Reference | ||
| GA | 21 | (14.5) | 18 | (22.5) | 0.74 | 0.35 | 1.55 | |
| AA | 0 | (0.0) | 0 | (0.0) | -- | -- | -- | 0.42 |
| GA or AA | 21 | (14.5) | 18 | (22.5) | 0.74 | 0.35 | 1.55 | 0.42 |
|
rs1049337, chr7:115987823 | ||||||||
| CC | 123 | (85.4) | 66 | (82.5) | 1.00 | Reference | ||
| CT | 21 | (14.6) | 14 | (17.5) | 0.77 | 0.35 | 1.68 | |
| TT | 0 | (0.0) | 0 | (0.0) | -- | -- | -- | 0.50 |
| CT or TT | 21 | (14.6) | 14 | (17.5) | 0.77 | 0.35 | 1.68 | 0.50 |
|
Cav-2 | ||||||||
|
rs17138767, chr7:115924913 | ||||||||
| AA | 136 | (94.4) | 72 | (91.1) | 1.00 | Reference | ||
| GA | 8 | (5.6) | 7 | (8.9) | 0.74 | 0.25 | 2.25 | |
| GG | 0 | (0.0) | 0 | (0.0) | -- | -- | -- | 0.60 |
| GA or GG | 8 | (5.6) | 7 | (8.9) | 0.74 | 0.25 | 2.25 | 0.60 |
|
rs4730742, chr7:115925128 | ||||||||
| TT | 106 | (73.6) | 60 | (75.0) | 1.00 | Reference | ||
| GT | 36 | (25.0) | 18 | (22.5) | 1.41 | 0.71 | 2.81 | |
| GG | 2 | (1.4) | 2 | (2.5) | 0.39 | 0.05 | 3.02 | 0.39 |
| GT or GG | 38 | (26.4) | 20 | (25.0) | 1.28 | 0.66 | 2.49 | 0.46 |
|
rs8940, chr7:115933310 | ||||||||
| CC | 97 | (66.9) | 52 | (65.0) | 1.00 | Reference | ||
| CG | 43 | (29.7) | 24 | (30.0) | 1.10 | 0.58 | 2.08 | |
| GG | 5 | (3.5) | 4 | (5.0) | 0.58 | 0.14 | 2.41 | 0.70 |
| CG or GG | 48 | (33.1) | 28 | (35.0) | 1.01 | 0.55 | 1.86 | 0.97 |
|
rs4727833, chr7:115935144 | ||||||||
| CC | 51 | (35.4) | 32 | (40.0) | 1.00 | Reference | ||
| GC | 74 | (51.4) | 38 | (47.5) | 1.29 | 0.69 | 2.42 | |
| GG | 19 | (13.2) | 10 | (12.5) | 1.13 | 0.44 | 2.90 | 0.73 |
| GC or GG | 93 | (64.6) | 48 | (60.0) | 1.26 | 0.69 | 2.29 | 0.45 |
|
rs1052990, chr7:115935606 | ||||||||
| TT | 54 | (37.2) | 29 | (36.3) | 1.00 | Reference | ||
| GT | 70 | (48.3) | 35 | (43.8) | 1.13 | 0.59 | 2.16 | |
| GG | 21 | (14.5) | 16 | (20.0) | 0.78 | 0.34 | 1.81 | 0.69 |
| GT or GG | 91 | (62.8) | 51 | (63.8) | 1.02 | 0.55 | 1.86 | 0.96 |
|
rs6466578, chr7:115938105 | ||||||||
| TT | 89 | (61.4) | 56 | (70.0) | 1.00 | Reference | ||
| CT | 50 | (34.5) | 21 | (26.3) | 1.52 | 0.80 | 2.89 | |
| CC | 6 | (4.1) | 3 | (3.8) | 0.83 | 0.18 | 3.90 | 0.42 |
| CT or CC | 56 | (38.6) | 24 | (30.0) | 1.42 | 0.77 | 2.63 | 0.26 |
|
rs10262524, chr7:115938188 | ||||||||
| CC | 59 | (40.7) | 33 | (41.3) | 1.00 | Reference | ||
| CA | 67 | (46.2) | 35 | (43.8) | 1.11 | 0.59 | 2.08 | |
| AA | 19 | (13.1) | 12 | (15.0) | 0.93 | 0.38 | 2.27 | 0.90 |
| CA or AA | 86 | (59.3) | 47 | (58.8) | 1.07 | 0.59 | 1.92 | 0.83 |
SNP=single nucleotide polymorphism; OR=odds ratio; CI=confidence interval
Total number of cases and controls vary by SNP due to missing genotype data.
Adjusted for age at reference date.
The first p-value is the test for trend using the codominant model; the second p-value is for the dominant model.
Table 3.
Disease aggressivenessaassociated with SNPs in the cav-1 and cav-2 genes (Caucasians only)
| Controls | Less Aggressive | More Aggressive | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (n=1,351)b | (n=966)b | (n=491)b | ||||||||||
| n | (%) | n | % | ORc | 95% CI | n | % | ORc | 95% CI | |||
|
Cav-1 | ||||||||||||
|
rs4236601, chr7:115949965 | ||||||||||||
| GG | 637 | (52.0) | 406 | (49.5) | 1.00 | Reference | 198 | (48.1) | 1.00 | Reference | ||
| GA | 500 | (40.8) | 352 | (42.9) | 1.11 | 0.92 | 1.33 | 185 | (44.9) | 1.19 | 0.94 | 1.50 |
| AA | 89 | (7.3) | 63 | (7.7) | 1.12 | 0.79 | 1.58 | 29 | (7.0) | 1.05 | 0.67 | 1.65 |
| GA or AA | 589 | (48.0) | 415 | (50.6) | 1.11 | 0.93 | 1.33 | 214 | (51.9) | 1.17 | 0.93 | 1.46 |
|
rs926198, chr7:115954444 | ||||||||||||
| TT | 538 | (43.9) | 362 | (43.3) | 1.00 | Reference | 206 | (49.4) | 1.00 | Reference | ||
| CT | 539 | (43.9) | 371 | (44.4) | 1.02 | 0.84 | 1.23 | 166 | (39.8) | 0.81 | 0.64 | 1.03 |
| CC | 150 | (12.2) | 103 | (12.3) | 1.03 | 0.77 | 1.37 | 45 | (10.8) | 0.79 | 0.54 | 1.14 |
| CT or CC | 689 | (56.2) | 474 | (56.7) | 1.02 | 0.85 | 1.22 | 211 | (50.6) | 0.81 | 0.65 | 1.01 |
|
rs10256914, chr7:115962947 | ||||||||||||
| TT | 684 | (55.0) | 572 | (53.5) | 1.00 | Reference | 103 | (52.6) | 1.00 | Reference | ||
| TC | 474 | (38.1) | 424 | (39.6) | 1.03 | 0.86 | 1.24 | 78 | (39.8) | 1.18 | 0.94 | 1.49 |
| CC | 85 | (6.8) | 74 | (6.9) | 1.03 | 0.72 | 1.47 | 15 | (7.7) | 1.13 | 0.72 | 1.76 |
| TC or CC | 559 | (45.0) | 387 | (45.7) | 1.03 | 0.87 | 1.23 | 207 | (48.9) | 1.17 | 0.94 | 1.47 |
|
rs3807986, chr7:115965061 | ||||||||||||
| AA | 720 | (57.9) | 474 | (56.6) | 1.00 | Reference | 255 | (60.9) | 1.00 | Reference | ||
| GA | 457 | (36.8) | 306 | (36.9) | 1.02 | 0.85 | 1.23 | 146 | (34.8) | 0.91 | 0.72 | 1.15 |
| GG | 66 | (5.3) | 54 | (6.5) | 1.25 | 0.86 | 1.83 | 18 | (4.3) | 0.79 | 0.46 | 1.35 |
| GA or GG | 523 | (42.1) | 363 | (43.4) | 1.05 | 0.88 | 1.25 | 164 | (39.1) | 0.89 | 0.71 | 1.12 |
|
rs959173, chr7:115969290 | ||||||||||||
| TT | 886 | (71.3) | 581 | (63.8) | 1.00 | Reference | 312 | (73.8) | 1.00 | Reference | ||
| CT | 328 | (26.4) | 236 | (28.0) | 1.10 | 0.90 | 1.34 | 99 | (23.4) | 0.87 | 0.67 | 1.12 |
| CC | 29 | (2.3) | 27 | (3.2) | 1.43 | 0.84 | 2.44 | 12 | (2.8) | 1.19 | 0.60 | 2.37 |
| CT or CC | 357 | (28.7) | 263 | (31.2) | 1.12 | 0.93 | 1.36 | 111 | (26.2) | 0.89 | 0.70 | 1.15 |
|
rs3807989, chr7:115973477 | ||||||||||||
| GG | 421 | (33.9) | 279 | (33.1) | 1.00 | Reference | 157 | (37.3) | 1.00 | Reference | ||
| GA | 626 | (50.4) | 409 | (48.5) | 0.98 | 0.81 | 1.19 | 188 | (44.7) | 0.81 | 0.64 | 1.04 |
| AA | 196 | (15.8) | 156 | (18.5) | 1.21 | 0.93 | 1.57 | 76 | (18.1) | 1.06 | 0.77 | 1.47 |
| GA or AA | 822 | (66.1) | 565 | (66.9) | 1.03 | 0.86 | 1.25 | 264 | (62.7) | 0.87 | 0.69 | 1.10 |
|
rs3801995, chr7:115977833 | ||||||||||||
| CC | 690 | (55.6) | 466 | (55.3) | 1.00 | Reference | 239 | (56.5) | 1.00 | Reference | ||
| CT | 479 | (38.6) | 325 | (38.6) | 1.01 | 0.84 | 1.21 | 149 | (35.2) | 0.90 | 0.71 | 1.14 |
| TT | 72 | (5.8) | 52 | (6.2) | 1.08 | 0.74 | 1.57 | 35 | (8.3) | 1.45 | 0.94 | 2.23 |
| CT or TT | 551 | (44.4) | 377 | (44.7) | 1.01 | 0.85 | 1.21 | 184 | (43.5) | 0.97 | 0.77 | 1.21 |
|
rs1049314, chr7:115986931 | ||||||||||||
| CC | 863 | (69.3) | 569 | (67.7) | 1.00 | Reference | 278 | (66.0) | 1.00 | Reference | ||
| CA | 352 | (28.3) | 252 | (30.0) | 1.09 | 0.90 | 1.32 | 126 | (29.9) | 1.11 | 0.87 | 1.42 |
| AA | 30 | (2.4) | 19 | (2.3) | 0.96 | 0.53 | 1.72 | 17 | (4.0) | 1.83 | 0.99 | 3.38 |
| CA or AA | 382 | (30.7) | 271 | (32.3) | 1.08 | 0.90 | 1.30 | 143 | (34.0) | 1.17 | 0.92 | 1.48 |
|
rs9920, chr7:115987328 | ||||||||||||
| TT | 1,031 | (83.0) | 672 | (79.4) | 1.00 | Reference | 321 | (75.9) | 1.00 | Reference | ||
| CT | 204 | (16.4) | 165 | (19.5) | 1.25 | 1.00 | 1.57 | 97 | (22.9) | 1.54 | 1.18 | 2.03 |
| CC | 7 | (0.6) | 9 | (1.1) | 1.96 | 0.73 | 5.30 | 5 | (1.2) | 2.38 | 0.75 | 7.59 |
| CT or CC | 211 | (17.0) | 174 | (20.6) | 1.28 | 1.02 | 1.60 | 102 | (24.1) | 1.57 | 1.20 | 2.06 |
|
rs1049334, chr7:115987616 | ||||||||||||
| GG | 1,033 | (83.2) | 712 | (84.1) | 1.00 | Reference | 365 | (86.5) | 1.00 | Reference | ||
| GA | 203 | (16.3) | 131 | (15.5) | 0.93 | 0.73 | 1.19 | 55 | (13.0) | 0.77 | 0.56 | 1.06 |
| AA | 6 | (0.5) | 4 | (0.5) | 0.96 | 0.27 | 3.43 | 2 | (0.5) | 0.96 | 0.19 | 4.79 |
| GA or AA | 209 | (16.8) | 135 | (15.9) | 0.93 | 0.74 | 1.18 | 57 | (13.5) | 0.77 | 0.56 | 1.06 |
|
rs1049337, chr7:115987823 | ||||||||||||
| CC | 625 | (50.4) | 471 | (56.1) | 1.00 | Reference | 204 | (48.8) | 1.00 | Reference | ||
| CT | 514 | (41.5) | 313 | (37.3) | 0.80 | 0.67 | 0.97 | 176 | (42.1) | 1.04 | 0.82 | 1.31 |
| TT | 100 | (8.1) | 55 | (6.6) | 0.73 | 0.52 | 1.04 | 38 | (9.1) | 1.14 | 0.76 | 1.71 |
| CT or TT | 614 | (49.6) | 368 | (43.9) | 0.79 | 0.66 | 0.94 | 214 | (51.2) | 1.05 | 0.84 | 1.32 |
|
Cav-2 | ||||||||||||
|
rs17138767, chr7:115924913 | ||||||||||||
| AA | 1,041 | (84.0) | 715 | (84.9) | 1.00 | Reference | 361 | (86.2) | 1.00 | Reference | ||
| GA | 194 | (15.7) | 124 | (14.7) | 0.92 | 0.72 | 1.18 | 56 | (13.4) | 0.83 | 0.60 | 1.15 |
| GG | 5 | (0.4) | 3 | (0.4) | 0.88 | 0.21 | 3.71 | 2 | (0.5) | 1.20 | 0.23 | 6.23 |
| GA or GG | 199 | (16.1) | 127 | (15.1) | 0.92 | 0.72 | 1.17 | 58 | (13.8) | 0.84 | 0.61 | 1.15 |
|
rs4730742, chr7:115925128 | ||||||||||||
| TT | 822 | (66.2) | 565 | (67.3) | 1.00 | Reference | 278 | (66.2) | 1.00 | Reference | ||
| GT | 379 | (30.5) | 249 | (29.7) | 0.95 | 0.79 | 1.15 | 132 | (31.4) | 1.03 | 0.81 | 1.31 |
| GG | 40 | (3.2) | 25 | (3.0) | 0.91 | 0.55 | 1.53 | 10 | (2.4) | 0.74 | 0.37 | 1.51 |
| GT or GG | 419 | (33.8) | 274 | (32.7) | 0.95 | 0.79 | 1.14 | 142 | (33.8) | 1.00 | 0.79 | 1.27 |
|
rs8940, chr7:115933310 | ||||||||||||
| CC | 816 | (65.7) | 565 | (67.1) | 1.00 | Reference | 288 | (68.1) | 1.00 | Reference | ||
| CG | 379 | (30.5) | 253 | (30.1) | 0.96 | 0.79 | 1.17 | 124 | (29.3) | 0.93 | 0.73 | 1.18 |
| GG | 47 | (3.8) | 24 | (2.9) | 0.75 | 0.45 | 1.23 | 11 | (2.6) | 0.67 | 0.34 | 1.31 |
| CG or GG | 426 | (34.3) | 277 | (32.9) | 0.94 | 0.78 | 1.13 | 135 | (31.9) | 0.90 | 0.71 | 1.14 |
|
rs4727833, chr7:115935144 | ||||||||||||
| CC | 313 | (25.2) | 244 | (28.8) | 1.00 | Reference | 108 | (25.7) | 1.00 | Reference | ||
| GC | 640 | (51.5) | 413 | (48.8) | 0.82 | 0.67 | 1.01 | 204 | (48.5) | 0.92 | 0.70 | 1.20 |
| GG | 290 | (23.3) | 189 | (22.3) | 0.83 | 0.65 | 1.07 | 109 | (25.9) | 1.08 | 0.79 | 1.47 |
| GC or GG | 930 | (74.8) | 602 | (71.2) | 0.82 | 0.68 | 1.00 | 313 | (74.4) | 0.97 | 0.75 | 1.25 |
|
rs1052990, chr7:115935606 | ||||||||||||
| TT | 508 | (41.0) | 341 | (40.3) | 1.00 | Reference | 176 | (41.6) | 1.00 | Reference | ||
| GT | 590 | (47.6) | 399 | (47.2) | 1.01 | 0.83 | 1.21 | 192 | (45.4) | 0.94 | 0.74 | 1.19 |
| GG | 142 | (11.5) | 106 | (12.5) | 1.12 | 0.84 | 1.49 | 55 | (13.0) | 1.13 | 0.79 | 1.61 |
| GT or GG | 732 | (59.0) | 505 | (59.7) | 1.03 | 0.86 | 1.23 | 247 | (58.4) | 0.97 | 0.78 | 1.22 |
|
rs6466578, chr7:115938105 | ||||||||||||
| TT | 879 | (70.8) | 583 | (69.3) | 1.00 | Reference | 314 | (74.2) | 1.00 | Reference | ||
| CT | 338 | (27.2) | 231 | (27.5) | 1.03 | 0.85 | 1.26 | 99 | (23.4) | 0.83 | 0.64 | 1.07 |
| CC | 25 | (2.0) | 27 | (3.2) | 1.66 | 0.95 | 2.89 | 10 | (2.4) | 1.13 | 0.54 | 2.38 |
| CT or CC | 363 | (29.2) | 258 | (30.7) | 1.08 | 0.89 | 1.30 | 109 | (25.8) | 0.85 | 0.66 | 1.09 |
|
rs10262524, chr7:115938188 | ||||||||||||
| CC | 658 | (52.9) | 428 | (50.5) | 1.00 | Reference | 211 | (49.9) | 1.00 | Reference | ||
| CA | 495 | (39.8) | 356 | (42.0) | 1.11 | 0.93 | 1.33 | 182 | (43.0) | 1.15 | 0.91 | 1.44 |
| AA | 90 | (7.2) | 63 | (7.4) | 1.08 | 0.77 | 1.53 | 30 | (7.1) | 1.05 | 0.67 | 1.63 |
| CA or AA | 585 | (47.1) | 419 | (49.5) | 1.11 | 0.93 | 1.32 | 212 | (50.1) | 1.13 | 0.91 | 1.41 |
SNP=single nucleotide polymorphism; OR=odds ratio; CI=confidence interval
More aggressive PC is defined by a Gleason score of 7(4+3) or 8–10, regional or distant tumor stage, or a diagnostic PSA value ≥20 ng/ml.
Total number of cases and controls vary by SNP due to missing genotype data.
Adjusted for age at reference date.
Among the cases, the SNP rs9920 was not associated with early age at onset (rs9920: ORCT+CC=1.07, 95%CI=0.82, 1.40) nor with PC recurrence in Caucasians (rs9920: HRCT+CC=0.95, 95%CI=0.64, 1.42).
Protein analysis
“High” cav-1 levels were determined from the median level among controls (0.69 ng/ml). Using this cutoff value, the quality control of the blind duplicates resulted in 85% concordance. The distribution of cav-1 levels for both cases and controls was highly skewed, with the majority of values falling below 1.0 ng/ml and the rest stretching to almost 50 ng/ml.
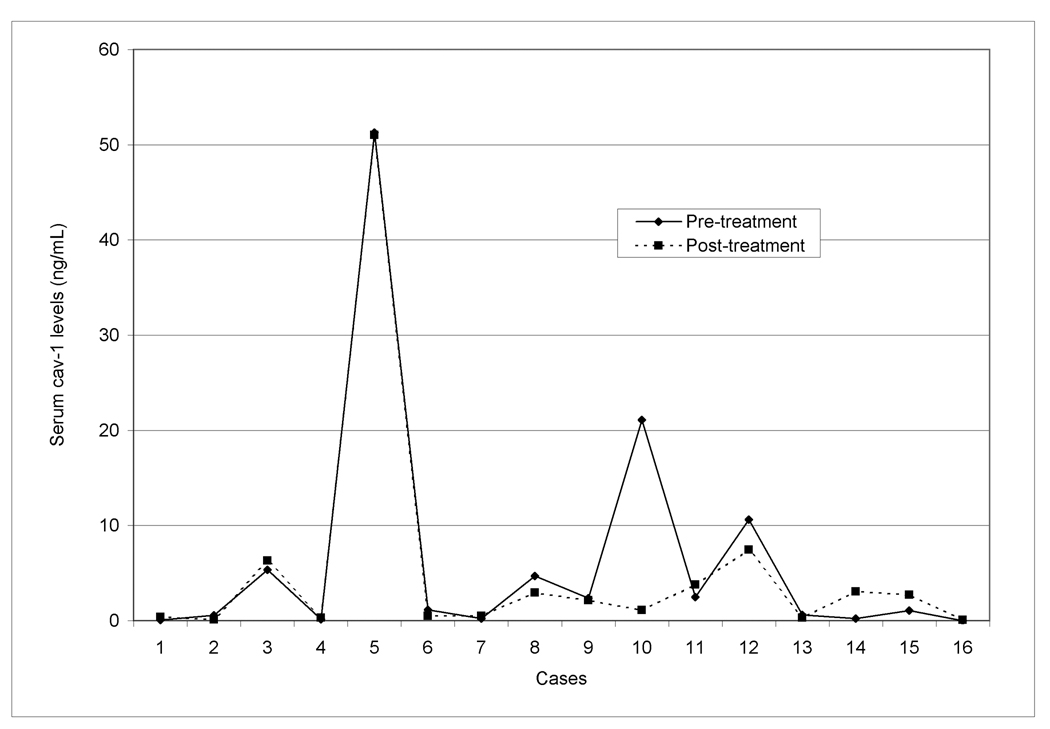
Serum cav-1 did not differ significantly between 16 patient samples taken prior to treatment (mean=6.4±13.2) and after treatment (mean=5.2±12.4; p=0.76). On average, cav-1 levels decreased 1.2 ng/ml from pre- to post-treatment levels; however, this change was heavily influenced by one subject with pre- and post-treatment cav-1 levels of 21.1 and 1.1, respectively, who did not undergo RP (subject 10 in Figure 1). Among the 12 cases with RP as primary therapy, cav-1 levels decreased on average 0.12 ng/ml. Overall, cav-1 levels did not differ by primary treatment: RP (n=143, mean=3.3 ± 6.3), radiation therapy (n=35, mean=3.1 ± 7.1), androgen deprivation therapy (n=17, mean=2.9 ± 7.3).
Figure 1.
The post-treatment cav-1 levels overall did not differ significantly between PC cases (mean=3.3 ± 6.5) and controls (mean=3.4 ± 7.3; OR=1.00, 95%CI=0.97, 1.03), nor between cases with more aggressive PC (mean=2.91 ± 6.7) or less aggressive PC (mean=3.7 ± 6.2) and controls (mean=3.4 ± 7.3; p=0.76; Table 4). Comparing the log of the cav-1 values, the proportion of cases and controls with high versus low cav-1 levels, or the quintiles of the cav-1 distribution did not distinguish between PC cases and controls or between more aggressive and less aggressive disease (Table 4). These estimates did not change substantially when adjusted for PC screening history or family history of PC.
Table 4.
| Cav-1 measure |
Controls (n=226) |
Less Aggressive (n=93) |
ORc | 95% CI | More Aggressive (n=109) |
ORc | 95% CI | ptrend | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | (SD) | Median | Mean | (SD) | Median | Mean | (SD) | Median | ||||||||
| Cav-1 | 3.36 | (7.27) | 0.69 | 3.71 | (6.22) | 0.81 | 1.01 | 0.97 | 1.04 | 2.91 | (6.70) | 0.85 | 0.99 | 0.96 | 1.03 | 0.76 |
| Ln of cav-1 | −0.27 | (1.86) | −0.38 | 0.08 | (1.75) | −0.21 | 1.11 | 0.97 | 1.28 | −0.26 | (1.77) | −0.17 | 1.00 | 0.88 | 1.14 | 0.27 |
| n | (%) | n | (%) | OR | 95% CI | n | (%) | OR | 95% CI | p | ||||||
| Low cav-1 | 34 | (15.0) | 10 | (10.8) | 1.00 | Ref | 96 | (88.1) | 1.00 | Ref | 0.37 | |||||
| High cav-1d | 192 | (85.0) | 83 | (89.3) | 1.29 | 0.79 | 2.10 | 13 | (11.9) | 1.34 | 0.84 | 2.12 | ||||
| n | (%) | n | (%) | OR | 95% CI | n | (%) | OR | 95% CI | p | ||||||
| Lowest | 45 | (19.9) | 12 | (12.9) | 1.00 | Ref | 16 | (14.7) | 1.00 | Ref | 0.36 | |||||
| 2nd | 45 | (19.9) | 21 | (22.6) | 1.76 | 0.77 | 4.02 | 22 | (20.2) | 1.33 | 0.62 | 2.87 | ||||
| 3rd | 45 | (19.9) | 22 | (23.7) | 1.84 | 0.81 | 4.18 | 24 | (22.0) | 1.46 | 0.68 | 3.12 | ||||
| 4th | 46 | (20.4) | 15 | (16.1) | 1.24 | 0.52 | 2.94 | 31 | (28.4) | 1.83 | 0.88 | 3.81 | ||||
| Highest | 45 | (19.9) | 23 | (24.7) | 1.91 | 0.85 | 4.31 | 16 | (14.7) | 0.99 | 0.44 | 2.22 | ||||
OR=odds ratio, adjusted for age at diagnosis/reference date; SD=standard deviation; CI=confidence interval
Caucasians only.
More aggressive PC is defined as regional or distant stage, or Gleason score 7 (4+3) or 8–10, or diagnostic PSA ≥20 ng/ml.
OR given a one unit (ng/ml) increase in cav-1 or the natural log of cav-1, respectively.
High cav-1 is defined by the median level among controls (0.69 ng/ml).
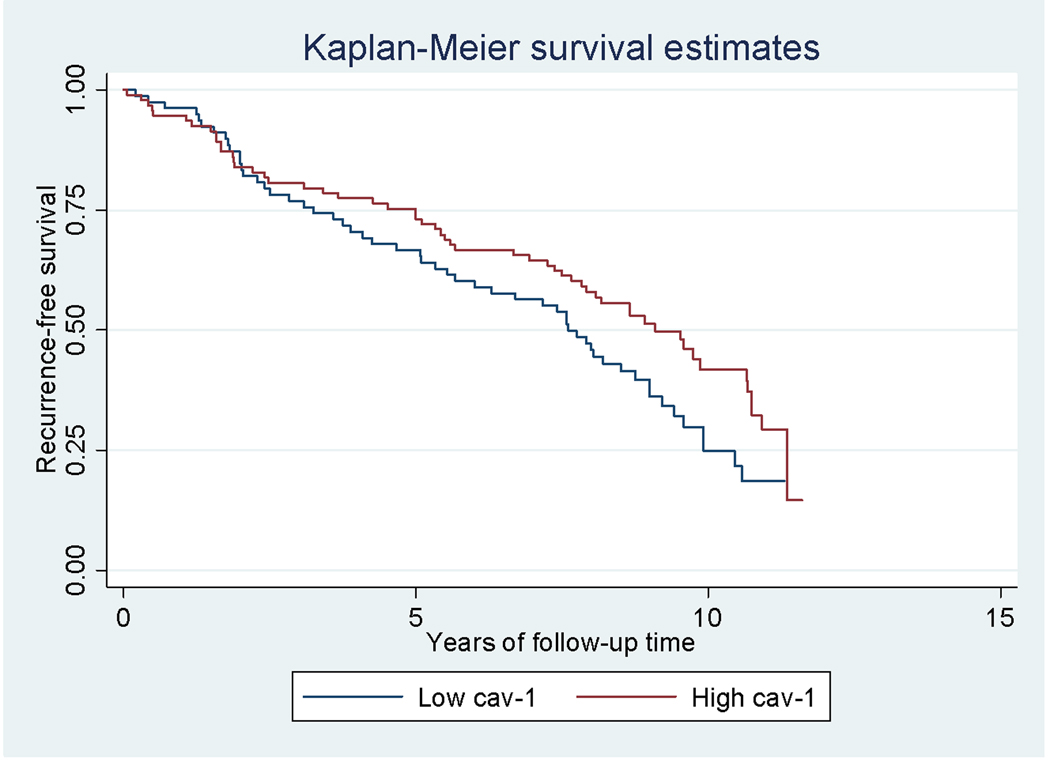
High post-treatment serum cav-1 levels approached a significant inverse association with PC recurrence (HR=0.69, 95%CI=0.47, 1.00; p=0.05; Figure 2). Serum cav-1 levels were not associated with PC-specific mortality (HR=0.94, 95%CI=0.50, 1.74). These estimates did not change substantially when the analysis was restricted to patients who received an RP as primary therapy. Among controls, cav-1 levels did not differ significantly between controls who developed PC during the follow-up period (n=33) and controls who remained disease-free (OR=0.97, 95%CI=0.89, 1.05). In addition, among controls, high cav-1 levels in serum were not associated with the purported risk allele in cav-1 (rs9920: ORCT vs. TT=1.09, 95%CI=0.56, 2.12).
Figure 2.
Discussion
Expression of the caveolin gene family, particularly cav-1, has been assessed in relationship to several human cancers. Cav-1 gene expression is down-regulated in human thyroid cancer and ovarian tumors, as well as mesenchymal sarcomas; cav-1 is over-expressed in bladder, esophageal, prostate, and thyroid tumors; and there are conflicting reports regarding the expression of cav-1 in breast, colon, kidney, lung, and pancreatic tumors [11,22,29–30]. In addition, high cav-1 expression is associated with progression for several malignancies, including cancer of the colon [31], kidney [32], bladder [33], lung [34], pancreas [35], ovary [36], and some types of breast cancer [37]. The level of cav-1 expression may depend on the tumor type and stage; for example, high cav-1 levels were reported in late or advanced squamous cell carcinoma [38] and in metastatic PC [12].
A number of studies have investigated the relationship between cav-1 expression and PC, particularly as a possible biomarker for more aggressive disease. Using mouse and human PC cell lines, cav-1 expression was found to be elevated in tissues with primary adenocarcinomas compared to normal epithelial tissue [11]. Using androgen-insensitive mouse PC cells, cav-1 expression was shown to induce androgen sensitivity [12]. Cav-1 immunoreactivity has been associated with a higher Gleason score, positive surgical margins, metastases to lymph nodes, and a shorter time to disease recurrence in three studies [18,20–21]. In a retrospective study of 142 clinically confined PC specimens obtained from RP, higher cav-1 immunoreactivity was noted among African-American men than in white-American men [13]. Using mouse and human PC cell lines, Li and colleagues found a reduction in lymph node and lung metastases when cav-1 expression was suppressed [39], and androgen-insensitive PC cells were found to secrete cav-1, which may stimulate viability and growth in PC cells that do not express cav-1 [19]. Cav-1 expression has been found to maintain the oncogenic activities of Akt in PC cells [15], and proangiogenic effects of cav-1 have been demonstrated in mouse and human PC cells [17]. Williams et al. interbred cav-1 (−/−) null mice with TRAMP mice (that spontaneously develop advanced prostate cancer) and found loss of cav-1 reduced progression to and extent of metastatic disease [40]. And recent data show that specific tumor suppressor microRNAs suppress the oncogenic activities of cav-1 and other genes in human PC cells [25].
In 2003, Tahir and colleagues developed the immunoassay for determination of serum cav-1 levels that was used in the current study [16]. Using this assay in pre-treatment serum, Tahir et al. found the median cav-1 level in 102 PC cases with clinically localized disease (0.46 ng/ml) was significantly higher than that in 81 healthy control men (normal digital rectal examinations and serum PSA levels ≤ 1.5 ng/ml over a period of 2 years) (0.32 ng/ml; p=0.05) [16]. In the current study, the median cav-1 levels in the post-treatment serum of cases (0.82 ng/ml) was higher than in controls (0.69 ng/ml), but did not attain statistical significance (p=0.90). In 2006, using serum collected before RP from 419 PC cases, Tahir et al. found that high pre-treatment levels of cav-1 in serum were associated with a shorter time to biochemical failure (defined as a serum PSA level of ≥ 0.2 ng/mL on two consecutive measurements) (HR=2.78; 95% CI 1.00, 7.70; p=0.05) [14]. High pre-treatment serum cav-1 levels were those ≥ 0.13 ng/ml, a cutoff the authors determined using the minimum p-value method [41]. In the current study, the association with PC recurrence was not significant when 0.13 ng/ml was used to define high cav-1 values (HR=0.71, 95% CI=0.41, 1.23; p=0.23); however, when the median level for the controls in our dataset (0.69 ng/ml) was used to define high cav-1 levels, the association approached statistical significance (HR=0.68; 95%CI 0.46, 0.99; p=0.04), but in the opposite direction as previously reported.
The comparison of results from this study to prior studies of serum cav-1 levels may be limited since this study used post-treatment serum. The biological and clinical significance of pre- and post-treatment serum cav-1 levels may differ, given that a patient’s condition post-treatment may represent a different “clinical state” [42]. Removal of the prostate, RT, or ADT could change the levels of serum cav-1, although to our knowledge no prior published data have compared pre- and post-treatment serum cav-1 levels to confirm these possibilities. Furthermore, it is not known at what time before treatment maximum cav-1 levels could be captured – several weeks before surgery, as in this study, or immediately before surgery, as in previous studies. We attempted to address this concern at least partially by comparing pre- and post-treatment cav-1 levels in 16 patients who had serum samples taken, on average, 17 days (range 0 to 100 days) prior to RP and 7 months (range 4 to 11 months) after RP. We found no significant change between pre- and post-treatment cav-1 levels in these patients (Figure 1). In addition, the distribution of cav-1 in the post-treatment samples (mean=3.28; median=0.82; range=0.0–45.9 ng/ml) is similar to the distribution of cav-1 in pre-treatment samples used in a previous study (mean=4.52; median=1.01; range=0.0–156.7 ng/ml)[14]. The sample size may not have been large enough to detect a significant difference between pre- and post-treatment samples. Likewise, the study population may not have been large enough to detect true associations.
Despite the numerous studies showing an association between PC and cav-1 expression, no specific mutations associated with PC have been identified in the cav-1 gene [10,43], although a point mutation in cav-1 has been associated with breast cancer [44,45]. We found one tagSNP, rs9920 in cav-1, to be associated not only with PC risk but also with aggressive disease. This SNP is not in a repetitive region [46]. The cav-1 gene is on the long arm of chromosome 7, in a region associated with tumor suppression and with loss of heterozygosity in several types of cancer [10]. For each of the 19 SNPs, we calculated 12 significance tests, so one would expect about 11 results might be due solely to chance. However, rs9920 remained significant based on a permutated p-value. Because the same SNP was significant in both the PC risk analysis and the analysis of aggressive disease, this lends strength to the result. We found no SNPs in cav-2 to be associated with PC risk nor with risk of aggressive disease, including rs8940, the only SNP in cav-2 to be found associated with PC in previous studies [47].
There are several strengths to this study. Unlike previous studies, the data used for this analysis were from two population-based case-control studies, which means men with all grades and stages of disease, and who received a range of initial treatments, were included. In addition, we have over 10 years of patient follow-up to evaluate recurrence and progression. Clinical and patient information was available for potential confounders and effect modifiers. In addition, we utilized nonparametric methods and several measures of cav-1 to try to capture the non-normal distribution of serum cav-1 levels.
Conclusion
Evidence from previous studies indicates that pre-treatment serum cav-1 may be a biomarker for defining men with more aggressive PC. However, this population-based study found no evidence that higher post-treatment serum cav-1 is associated with risk of more aggressive PC or risk for adverse PC outcomes. Larger studies of the differences between pre- and post-treatment cav-1 levels are needed to further define the biological and clinical significance of pre- versus post-treatment serum cav-1 levels in PC. We did find evidence for an association with a gene variant in cav-1 in relation to risk of overall PC and more aggressive disease, which also merits further study.
Acknowledgements
We are grateful to the men who participated in these studies; without their help, this work would not be possible. We also thank Ilir Agalliu, Anna Frolov-Schroeder, Sarah Holt, Ulrike Peters, Brandon Pierce, and Claudia Salinas for helpful suggestions.
This work was supported by grants RO1 CA56678, RO1 CA082664, RO1 CA092579, R01 CA50588, and P50 CA097186 from the National Cancer Institute, with additional support from the Fred Hutchinson Cancer Research Center and the Intramural Program of the National Human Genome Research Institute.
References
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008 Mar-Apr;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.D’Amico AV, Chen MH, Roehl KA, Catalona WJ. Identifying patients at risk for significant versus clinically insignificant postoperative prostate-specific antigen failure. J Clin Oncol. 2005;23:4975–4979. doi: 10.1200/JCO.2005.08.904. [DOI] [PubMed] [Google Scholar]
- 3.Han M, Partin AW, Zahurak M, Piantadosa S, Epstein JI, Walsh PC. Biochemical (Prostate Specific Antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169:517–523. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- 4.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281:1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 5.Catalona WJ, Smith DS. Cancer recurrence and survival rates after anatomic radical retropubic prostatectomy for prostate cancer: intermediate-term results. J Urol. 1998;160:2428. doi: 10.1097/00005392-199812020-00012. [DOI] [PubMed] [Google Scholar]
- 6.Ohori M, Goad JR, Wheeler TM, Eastham JA, Thompson TC, Scardino PT. Can radical prostatectomy alter the progression of poorly differentiated prostate cancer? J Urol. 1994 Nov;152(5 Pt 2):1843–1849. doi: 10.1016/s0022-5347(17)32398-4. [DOI] [PubMed] [Google Scholar]
- 7.Zietman AL, Edelstein RA, Coen JJ, Babayan RK, Krane RJ. Radical prostatectomy for adenocarcinoma of the prostate: the influence of preoperative and pathologic findings on biochemical disease-free outcome. Urology. 1994 Jun;43(6):828–833. doi: 10.1016/0090-4295(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 8.Anderson RG. The caveolae membrane system. Annu Rev Biochem. 1998;67:199–225. doi: 10.1146/annurev.biochem.67.1.199. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto T, Schlegel A, Scherer PE, Lisanti MP. Caveolins, a family of scaffolding proteins for organizing “preassembled signaling complexes” at the plasma membrane. J Biol Chem. 1998 Mar 6;273(10):5419–5422. doi: 10.1074/jbc.273.10.5419. [DOI] [PubMed] [Google Scholar]
- 10.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004 Oct;84(4):1341–1379. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 11.Yang CP, Galbiati F, Volonte D, Horwitz SB, Lisanti MP. Upregulation of caveolin-1 and caveolae organelles in Taxol-resistant A549 cells. FEBS Lett. 1998 Nov 20;439(3):368–372. doi: 10.1016/s0014-5793(98)01354-4. [DOI] [PubMed] [Google Scholar]
- 12.Nasu Y, Timme TL, Yang G, Bangma CH, Li L, Ren C, Park SH, DeLeon M, Wang J, Thompson TC. Suppression of caveolin expression induces androgen sensitivity in metastatic androgen-insensitive mouse prostate cancer cells. Nat Med. 1998 Sep;4(9):1062–1064. doi: 10.1038/2048. [DOI] [PubMed] [Google Scholar]
- 13.Yang G, Addai J, Ittmann M, Wheeler TM, Thompson TC. Elevated caveolin-1 levels in African-American versus white-American prostate cancer. Clin Cancer Res. 2000 Sep;6(9):3430–3433. [PubMed] [Google Scholar]
- 14.Tahir SA, Frolov A, Hayes TG, Mims MP, Miles BJ, Lerner SP, Wheeler TM, Ayala G, Thompson TC, Kadmon D. Preoperative serum caveolin-1 as a prognostic marker for recurrence in a radical prostatectomy cohort. Clin Cancer Res. 2006 Aug 15;12(16):4872–4875. doi: 10.1158/1078-0432.CCR-06-0417. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Ren CH, Tahir SA, Ren C, Thompson TC. Caveolin-1 maintains activated Akt in prostate cancer cells through scaffolding domain binding site interactions with and inhibition of serine/threonine protein phosphatases PP1 and PP2A. Mol Cell Biol. 2003 Dec;23(24):9389–9404. doi: 10.1128/MCB.23.24.9389-9404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahir SA, Ren C, Timme TL, Gdor Y, Hoogeveen R, Morrisett JD, Frolov A, Ayala G, Wheeler TM, Thompson TC. Development of an immunoassay for serum caveolin-1: a novel biomarker for prostate cancer. Clin Cancer Res. 2003;9:3653–3659. [PubMed] [Google Scholar]
- 17.Tahir SA, Yang G, Goltsov AA, Watanabe M, Tabata K, Addai J, Fattahel MA, Kadmon D, Thompson TC. Tumor cell-secreted caveolin-1 has proangiogenic activities in prostate cancer. Cancer Res. 2008 Feb 1;68(3):731–739. doi: 10.1158/0008-5472.CAN-07-2668. [DOI] [PubMed] [Google Scholar]
- 18.Yang G, Truong LD, Wheeler TM, Thompson TC. Caveolin-1 expression in clinically confined human prostate cancer: a novel prognostic marker. Cancer Res. 1999 Nov 15;59(22):5719–5723. [PubMed] [Google Scholar]
- 19.Tahir SA, Yang G, Ebara S, Timme TL, Satoh T, Li L, Goltsov A, Ittmann M, Morrisett JD, Thompson TC. Secreted caveolin-1 stimulates cell survival/clonal growth and contributes to metastasis in androgen-insensitive prostate cancer. Cancer Res. 2001 May 15;61(10):3882–3885. [PubMed] [Google Scholar]
- 20.Karam JA, Lotan Y, Roehrborn CG, Ashfaq R, Karakiewicz PI, Shariat SF. Caveolin-1 overexpression is associated with aggressive prostate cancer recurrence. Prostate. 2007 May 1;67(6):614–622. doi: 10.1002/pros.20557. [DOI] [PubMed] [Google Scholar]
- 21.Goto T, Nguyen BP, Nakano M, Ehara H, Yamamoto N, Deguchi T. Utility of Bcl-2, P53, Ki-67, and caveolin-1 immunostaining in the prediction of biochemical failure after radical prostatectomy in a Japanese population. Urology. 2008 Jul;72(1):167–171. doi: 10.1016/j.urology.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Williams TM, Lisanti MP. Caveolin-1 in oncogenic transformation, cancer, and metastasis. Am J Physiol Cell Physiol. 2005 Mar;288(3):C494–C506. doi: 10.1152/ajpcell.00458.2004. [DOI] [PubMed] [Google Scholar]
- 23.Engelman JA, Zhang XL, Lisanti MP. Genes encoding human caveolin-1 and −2 are co-localized to the D7S522 locus (7q31.1), a known fragile site (FRA7G) that is frequently deleted in human cancers. FEBS Lett. 1998 Oct 9;436(3):403–410. doi: 10.1016/s0014-5793(98)01134-x. [DOI] [PubMed] [Google Scholar]
- 24.Razani B, Woodman SE, Lisanti MP. Caveolae: from cell biology to animal physiology. Pharmacol Rev. 2002 Sep;54(3):431–467. doi: 10.1124/pr.54.3.431. [DOI] [PubMed] [Google Scholar]
- 25.Gandellini P, Folini M, Longoni N, Pennati M, Binda M, Colecchia M, Salvioni R, Supino R, Moretti R, Limonta P, Valdagni R, Daidone MG, Zaffaroni N. miR-205 Exerts Tumor-Suppressive Functions in Human Prostate through Down-regulation of Protein Kinase Cepsilon. Cancer Res. 2009 Feb;:24. doi: 10.1158/0008-5472.CAN-08-2894. [DOI] [PubMed] [Google Scholar]
- 26.Stanford JL, Wicklund KG, McKnight B, Daling JR, Brawer MK. Vasectomy and risk of prostate cancer. Cancer Epidemiol Biomarkers Prev. 1999 Oct;8(10):881–886. [PubMed] [Google Scholar]
- 27.Agalliu I, Salinas CA, Hansten PD, Ostrander EA, Stanford JL. Statin use and risk of prostate cancer: results from a population-based epidemiologic study. Am J Epidemiol. 2008 Aug 1;168(3):250–260. doi: 10.1093/aje/kwn141. Epub 2008 Jun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newcomer LM, King IB, Wicklund KG, Stanford JL. The association of fatty acids with prostate cancer risk. Prostate. 2001 Jun 1;47(4):262–268. doi: 10.1002/pros.1070. [DOI] [PubMed] [Google Scholar]
- 29.Lavie Y, Fiucci G, Liscovitch M. Upregulation of caveolin in multidrug resistant cancer cells: functional implications. Adv Drug Deliv Rev. 2001 Jul 28;49(3):317–323. doi: 10.1016/s0169-409x(01)00144-2. [DOI] [PubMed] [Google Scholar]
- 30.Joshi B, Strugnell SS, Goetz JG, Kojic LD, Cox ME, Griffith OL, Chan SK, Jones SJ, Leung SP, Masoudi H, Leung S, Wiseman SM, Nabi IR. Phosphorylated caveolin-1 regulates Rho/ROCK-dependent focal adhesion dynamics and tumor cell migration and invasion. Cancer Res. 2008 Oct 15;68(20):8210–8220. doi: 10.1158/0008-5472.CAN-08-0343. [DOI] [PubMed] [Google Scholar]
- 31.Patlolla JM, Swamy MV, Raju J, Rao CV. Overexpression of caveolin-1 in experimental colon adenocarcinomas and human colon cancer cell lines. Oncol Rep. 2004 May;11(5):957–963. [PubMed] [Google Scholar]
- 32.Joo HJ, Oh DK, Kim YS, Lee KB, Kim SJ. Increased expression of caveolin-1 and microvessel density correlates with metastasis and poor prognosis in clear cell renal cell carcinoma. BJU Int2004. Feb;93(3):291–296. doi: 10.1111/j.1464-410x.2004.04604.x. [DOI] [PubMed] [Google Scholar]
- 33.Rajjayabun PH, Garg S, Durkan GC, Charlton R, Robinson MC, Mellon JK. Caveolin-1 expression is associated with high-grade bladder cancer. Urology. 2001 Nov;58(5):811–814. doi: 10.1016/s0090-4295(01)01337-1. [DOI] [PubMed] [Google Scholar]
- 34.Ho CC, Huang PH, Huang HY, Chen YH, Yang PC, Hsu SM. Up-regulated caveolin-1 accentuates the metastasis capability of lung adenocarcinoma by inducing filopodia formation. Am J Pathol. 2002 Nov;161(5):1647–1656. doi: 10.1016/S0002-9440(10)64442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuoki M, Miyamoto M, Kato K, Hiraoka K, Oshikiri T, Nakakubo Y, Fukunaga A, Shichinohe T, Shinohara T, Itoh T, Kondo S, Katoh H. Impact of caveolin-1 expression on prognosis of pancreatic ductal adenocarcinoma. Br J Cancer. 2002 Nov 4;87(10):1140–1144. doi: 10.1038/sj.bjc.6600619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davidson B, Goldberg I, Givant-Horwitz V, Nesland JM, Berner A, Bryne M, Risberg B, Kopolovic J, Kristensen GB, Tropé CG, van de Putte G, Reich R. Caveolin-1 expression in ovarian carcinoma is MDR1 independent. Am J Clin Pathol. 2002 Feb;117(2):225–234. doi: 10.1309/u40r-1bn4-6kj3-bdg3. [DOI] [PubMed] [Google Scholar]
- 37.Van den Eynden GG, Van Laere SJ, Van der Auwera I, Merajver SD, Van Marck EA, van Dam P, Vermeulen PB, Dirix LY, van Golen KL. Overexpression of caveolin-1 and −2 in cell lines and in human samples of inflammatory breast cancer. Breast Cancer Res Treat. 2006 Feb;95(3):219–228. doi: 10.1007/s10549-005-9002-1. [DOI] [PubMed] [Google Scholar]
- 38.Yoo SH, Park YS, Kim HR, Sung SW, Kim JH, Shim YS, Lee SD, Choi YL, Kim MK, Chung DH. Expression of caveolin-1 is associated with poor prognosis of patients with squamous cell carcinoma of the lung. Lung Cancer. 2003 Nov;42(2):195–202. doi: 10.1016/s0169-5002(03)00287-3. [DOI] [PubMed] [Google Scholar]
- 39.Li L, Yang G, Ebara S, Satoh T, Nasu Y, Timme TL, Ren C, Wang J, Tahir SA, Thompson TC. Caveolin-1 mediates testosterone-stimulated survival/clonal growth and promotes metastatic activities in prostate cancer cells. Cancer Res. 2001 Jun 1;61(11):4386–4392. [PubMed] [Google Scholar]
- 40.Williams TM, Hassan GS, Li J, Cohen AW, Medina F, Frank PG, Pestell RG, Di Vizio D, Loda M, Lisanti MP. Caveolin-1 promotes tumor progression in an autochthonous mouse model of prostate cancer: genetic ablation of Cav-1 delays advanced prostate tumor development in tramp mice. J Biol Chem. 2005 Jul 1;280(26):25134–25145. doi: 10.1074/jbc.M501186200. [DOI] [PubMed] [Google Scholar]
- 41.Kadmon D, Tahir S, Li L, Thompson The role of secreted caveolin-1 in prostate cancer progression [abstract]; 13th SPORE Investigators’ Workshop program; Jul 9–12; Washington, D.C.. 2005. pp. 207–208. Abstract nr 218. [Google Scholar]
- 42.Scher HI, Heller G. Clinical states in prostate cancer: toward a dynamic model of disease progression. Urology. 2000 Mar;55(3):323–327. doi: 10.1016/s0090-4295(99)00471-9. [DOI] [PubMed] [Google Scholar]
- 43.Hurlstone AF, Reid G, Reeves JR, Fraser J, Strathdee G, Rahilly M, Parkinson EK, Black DM. Analysis of the CAVEOLIN-1 gene at human chromosome 7q31.1 in primary tumours and tumour-derived cell lines. Oncogene. 1999 Mar 11;18(10):1881–1890. doi: 10.1038/sj.onc.1202491. [DOI] [PubMed] [Google Scholar]
- 44.Hayashi K, Matsuda S, Machida K, Yamamoto T, Fukuda Y, Nimura Y, Hayakawa T, Hamaguchi M. Invasion activating caveolin-1 mutation in human scirrhous breast cancers. Cancer Res. 2001 Mar 15;61(6):2361–2364. [PubMed] [Google Scholar]
- 45.Li T, Sotgia F, Vuolo MA, Li M, Yang WC, Pestell RG, Sparano JA, Lisanti MP. Caveolin-1 mutations in human breast cancer: functional association with estrogen receptor alpha-positive status. Am J Pathol. 2006 Jun;168(6):1998–2013. doi: 10.2353/ajpath.2006.051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.SeattleSNPs [homepage on the internet. Seattle, WA: NHLBI Program for Genomic Applications; [cited 2008]. Available from: http://pga.gs.washington.edu. [Google Scholar]
- 47.Burmester JK, Suarez BK, Lin JH, Jin CH, Miller RD, Zhang KQ, Salzman SA, Reding DJ, Catalona WJ. Analysis of candidate genes for prostate cancer. Hum Hered. 2004;57(4):172–178. doi: 10.1159/000081443. [DOI] [PubMed] [Google Scholar]