Summary
Resistance to endocrine therapy is a major clinical problem in breast cancer. The role of ERα splice variants in endocrine resistance is largely unknown. We observed reduced protein expression of an N-terminally truncated ERα46 in endocrine- resistant LCC2, LCC9, and LY2 compared to MCF-7 breast cancer cells. Transfection of LCC9 and LY2 cells with hERα46 partially restored growth inhibition by TAM. Overexpression of hERα46 in MCF-7 cells reduced estradiol (E2)-stimulated endogenous pS2, Cyclin D1, nuclear respiratory factor-1 (NRF-1), and progesterone receptor transcription. Expression of oncomiR miR-21 was lower in TAM-resistant LCC9 and LY2 cells compared to MCF-7 cells. Transfection with ERα46 altered the pharmacology of E2 regulation of miR-21 expression from inhibition to stimulation, consistent with the hypothesis that hERα46 inhibits ERα activity. Established miR-21 targets PTEN and PDCD4 were reduced in ERα46-transfected, E2-treated MCF-7 cells. In conclusion, ERα46 appears to enhance endocrine responses by inhibiting selected ERα66 responses.
Keywords: estrogen receptor, tamoxifen, endocrine-resistance, splice variants, miR-21, gene regulation
1. Introduction
Breast cancer is the most common form of cancer diagnosed in women in the U.S. and the second leading cause of cancer-related death. Although survival has increased over the past decade, thanks to early detection and the use of tamoxifen (TAM) and aromatase inhibitors (AI) (Chia, et al. 2007), the molecular events leading to initial tumorigenesis and progression are complex and not completely understood. Lifetime estrogen exposure is widely accepted as a major risk factor for breast cancer development (Santen, et al. 2007). Estrogens promote cell replication by binding to estrogen receptors α and β (ERα and ERβ) and regulating the expression of genes and growth signaling pathways that increase cell proliferation (Mangelsdorf, et al. 1995). Cell-based studies indicate that ERβ inhibits ERα activity and may play a protective role in breast tumors (Behrens, et al. 2007; Williams, et al. 2007), but the role of ERβ agonists as therapeutics in breast cancer remains to be determined.
A great concern for women with breast cancer and their medical providers is that ~ 40% of initially ERα+ tumors become resistant to TAM and other endocrine therapies including AI (Ring and Dowsett 2004). A variety of interacting mechanisms are involved in endocrine-resistant breast cancer and full elucidation of these interacting mechanisms remains enigmatic (Clarke, et al. 2003; Lykkesfeldt 1996). Examples of mechanisms include: 1) overexpression of epidermal growth factor receptor (EGFR) and/or the oncogene HER-2/neu/ErbB2 (Dowsett 2001); 2) splice variants or point mutations in ERα (Herynk and Fuqua 2004); 3) alterations in the nuclear levels of ER coactivator or corepressor proteins, e.g., increased coactivator AIB1 (Louie, et al. 2004), or decreased corepressor NCoR (Girault, et al. 2003); 4) activation of MAPK (Fan, et al. 2009) and PI3K/AKT signaling pathways (Masri, et al. 2008); and increased expression of miR-221/222 which downregulate ERα post-transcriptionally (Miller, et al. 2008; Zhao, et al. 2008). Given that ERs and SERMs act through multiple cellular pathways, transformation from an endocrine -sensitive to a -resistant phenotype involves multiple genetic and epigenetic events in breast cancer cells (Achuthan, et al. 2001).
An N-terminal truncated splice variant of ERα called ERα46, lacking aa 1-173 which includes AF-1, a ligand-independent transactivation function that is regulated by phosphorylation (Lannigan 2003), was first identified and characterized as a dominant negative (DN) inhibitor of ERα activity in osteoblasts (Denger, et al. 2001). ERα46 heterodimerized with ERα and ERβ and bound EREs with higher affinity than the ERα homodimer in vitro (Denger et al. 2001). Overexpression of ERα46 inhibited MCF-7 breast cancer cell proliferation and inhibited E2-induced luciferase activity from a cyclin D1 promoter-reporter (Penot, et al. 2005). Overexpression of ERα46 and ERα66 in ERα-null MDA-MB-231 cells revealed that ERα46 inhibited basal transcription of the E2-regulated pS2 (TFF1) gene (Metivier, et al. 2004). Chromatin immunoprecipitation (ChIP) assays showed that unliganded (apo) ERα46 recruited components of the Sin3 corepressor (NCoR/SMRT) complex to the pS2 promoter and that addition of E2 displaced the corepressor complex, increased RNA pol II recruitment, and increased pS2 transcription (Metivier et al. 2004). Thus, apo-ERα46 appears to repress basal transcription of ER-responsive genes, but E2 may release ERα46 repression. The mechanisms regulating ERα46 splice variant expression are unknown, but nuclear levels of ERα46 protein increased with MCF-7 cell confluency (Penot et al. 2005). E2 was recently reported to increase ERα46 transcription in human macrophages, but not monocytes (Murphy, et al. 2009). In addition to its effects on genomic ERα activity, ERα46 has been identified as a plasma membrane-associated form of ERα that activates the c-Src-PI3K/Akt pathway in vascular endothelium (Kim and Bender 2005; Li, et al. 2003; Li, et al. 2007; Moriarty, et al. 2006).
Here we tested the hypothesis that ERα46 is reduced in TAM-resistant human breast cancer cells and that this reduction contributes to endocrine resistance. Because ERα46 is a dominant-negative effector of ERα66 activity (Denger et al. 2001; Metivier et al. 2004; Penot et al. 2005), we speculated that restoration of ERα46 expression would restore tamoxifen/antiestrogen-sensitivity to TAM-resistant breast cancer cells. Therefore, we examined the effect of transfection of ERα46 on basal and E2-regulated endogenous ERα target gene expression in E2-dependent and TAM-sensitive MCF-7 versus LCC9 and LY2 TAM-resistant breast cancer cell lines. Since E2 regulates microRNA expression (Klinge 2009) and specifically downregulates oncomiR miR-21 in MCF-7 cells through ERα (Wickramasinghe, et al. 2009), we examined how ERα46 affects the expression of miR-21 and its downstream mRNA targets, the tumor suppressors PDCD4 and PTEN. We report here that ERα46 expression was indeed reduced in the TAM-resistant breast cancer cell lines and that ERα46 activity opposes that of endogenous ER-regulated gene transcription in MCF-7, LCC9, and LY2 cells.
2. Materials and Methods
2.1. Chemicals
E2 and 4-hydroxytamoxifen (4-OHT) were purchased from Sigma (St. Louis, MO). ICI 182,780 was purchased from Tocris (Ellisville, MO).
2.2. Antibodies
Antibodies were purchased from the indicated suppliers: HC-20 for ERα from Santa Cruz Biotechnology (Santa Cruz, CA), ERβ from Upstate (cat #06-629, Lake Placid, NY), α-tubulin from LabVision (Fisher Scientific, Fremont, CA), β-actin from Sigma, Pdcd4 was Genetex (San Antonio, TX); Pten from Cell Signaling (Beverly, MA).
2.3. Cell Culture
MCF-7 (A) cells were purchased from American Type Tissue Collection (ATCC, Manassas, VA). MCF-7 (K) were obtained from Dr. Robert Pauley and Steven J. Santner of the Karmanos Cancer Institute. LCC9 and LY2 breast cancer cells are tamoxifen and raloxifene/ICI 182,780-resistant cell lines that were derived from MCF-7 cells and kindly provided by Dr. Robert Clarke of the Lombardi Cancer Ctr. of Georgetown University, School of Medicine (Bronzert, et al. 1985; Brunner, et al. 1997). Cells were maintained in IMEM supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen) and maintained in a humidified 37°C incubator containing 5% CO2.
2.4. Transient transfection
The ERα46 expression plasmid pCR3.1-hERα46 (+727/+2030) was kindly provided by Dr. Giles Flouriot (Penot et al. 2005). MCF-7, LCC9, and LY2 cells were transiently transfected with pCDNA3.1 parental vector or pCR3.1-hERα46 using FuGENE 6 from Roche (Indianapolis, IN) according to the instructions provided. 48 h post-transfection, cells were treated as indicated.
2.5. Protein Isolation
Whole cell extracts (WCE) were prepared in modified RIPA buffer (10mM sodium phosphate, pH 7.2; 1% NP-40; 1% Na-deoxycholate; 150mM NaCl; 2mM EDTA; 0.2mM Na3VO4; 50mM NaF; and 1μg/mL each of 3aprotinin, leupeptin, and pepstatin; Complete Protease Inhibitor (Roche); and 1mM PMSF). Protein concentrations were determined using the Bio-Rad DC Protein Assay (Bio-Rad Laboratories, Hercules, CA).
2.6. Western Blot Analysis
WCE (20, 30, or 40 μg protein, per Fig. legends) were separated on 10% polyacrylamide SDS gels and electroblotted onto PVDF membranes. Western blot was performed as described previously (Wickramasinghe et al. 2009). Immuno-reacting bands were visualized using HyGLO quick spray (Denville Scientific Inc, Metuchen, NJ) chemiluminescence reagent on Kodak BioMax ML film (Eastman Kodak, Rochester, NY). Membranes were stripped and reprobed for α-tubulin or β-actin for normalization. Resulting immunoblots were scanned into Adobe Photoshop 7.0 using a Microtek ScanMaker III scanner (Carson, CA). Un-Scan-It (Silk Scientific, Orem, UT) was used to quantitate the integrated optical densities (IOD) for each band. The IOD for each band was divided by concordant α-tubulin or β-actin IOD in the same blot. For comparison between experiments, the β-actin normalized pixel ratios for control-transfected, EtOH-treated MCF-7 cells was set to 1.
2.7. Cell Proliferation Assays
Cell proliferation was determined by bromodeoxyudridine (BrdU) incorporation using the BrdU ELISA assay from Roche as recommended by the manufacturer. Cells were plated in 96 well plates in phenol red-free IMEM supplemented with 3% dextran coated charcoal stripped FBS (DCC-FBS) for 24 h. Treatments (vehicle control, i.e., ethanol (EtOH), E2, 4-OHT, or ICI 182,780, alone or in combination) were added for 48 h. Within each experiment, each treatment was performed in quadruplicate and values were averaged. Values were compared to those in the wells treated with vehicle (EtOH) control which was set to 100. At least 4 separate experiments were performed for each cell line.
2.8. RNA Isolation and Quantitative Real Time RT-PCR (QRT-PCR)
RNA was extracted using Trizol reagent (Invitrogen), followed by purification on RNeasy columns (Qiagen, Valencia, CA). Total RNA was reverse transcribed using random hexamers and the High Capacity cDNA archive kit (PE Applied Biosystems, Foster City, CA). QIAquick PCR purification kit (Qiagen) was used to purify cDNA. Taqman primers and probes for ERα (ESR1), ERβ (ESR2), pS2 (TFF1), progesterone receptor (PR, PGR), NRF-1 (NRF1), PTEN, and PDCD4 and control genes 18S rRNA and GAPDH were purchased as Assays-on-Demand™ Gene Expression Products (PE Applied Biosystems). QRT-PCR was performed in the ABI PRISM 7900 SDS 2.1 (PE Applied Biosystems) using relative quantification with standard thermal cycler conditions. Analysis and fold differences were determined using the comparative CT method. Fold change was calculated from the ΔΔCT values with the formula 2−ΔΔCT and data are presented as relative to expression in EtOH-treated (control) MCF-7 or other cell lines unless otherwise indicated.
2.9. Quantitative real-time PCR (Q-PCR) analysis of miRNA expression
MCF-7, LCC9 and LY2 cells were transiently transfected with pcDNA3.1 or pcDNA3.1-ERα46 for 30 h and treated for 4 h with ethanol (EtOH, vehicle control), 10 nM E2, or 100 nM 4-OHT. MicroRNA was extracted using the Exiqon miRNA isolation kit and quantification of miR-21 was performed using Exiqon miRNA Assays according to manufacturer’s instructions (Exiqon, MA). SNORD38B, hsa-miR-765, and 5S rRNA were used for normalization of miRNA expression and 5S rRNA was selected for final normalization as described in the text. Analysis and fold differences were determined using the comparative threshold cycle (Ct) method (again using 5S). Fold change was calculated from the ΔΔCT values with the formula 2−ΔΔCT and data are presented as relative to expression in EtOH-treated (control) in each cell line.
3. Results
3.1 Expression of ERα66 and ERα46 in human breast cancer cell lines
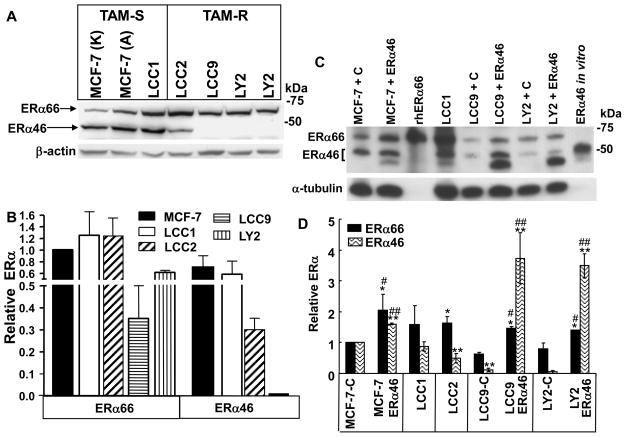
We used MCF-7 as a well-established E2-dependent, estrogen antagonist-sensitive breast cancer cell line and its derivatives E2-independent LCC1 and TAM-resistant LCC2, LCC9 and LY2 which are ERα positive (Bronzert et al. 1985; Brunner et al. 1997) to test the hypothesis that ERα46 expression is lower in TAM-resistant breast cancer cells compared to TAM-sensitive breast cancer cells. We noted no significant difference in ERα66 or ERα46 expression in MCF-7 from ATCC or the Karmanos Cancer Center (Fig. 1A, 1B) and subsequent experiments used MCF-7 cells purchased from ATCC. LCC1, LCC9 and LY2 cells differ in their derivation: LCC1 and LCC9 cells were derived from MCF-7 tumor xenografts grown in ovariectomized (ovex) nude mice or ovex mice treated with ICI 182,780, respectively, whereas LY2 cells were derived after step-wise treatment of MCF-7 cells with LY 117018 (Bronzert et al. 1985). Both are resistant to TAM and ICI 182,780. Western blots were performed using HC-20, an ERα antibody that was raised against a peptide in the C-terminus of ERα (Fig. 1A, 1B). TAM-resistant LCC2, LCC9, and LY2 breast cancer cell lines show lower ERα46 expression than the parental MCF-7 cell line (Fig. 1A, 1B). The levels of ERα66 and ERα46 in MCF-7 are similar to those previously reported (Penot et al. 2005).
Fig. 1. Expression of ERα66 and ERα46 in breast cancer cell lines.
Representative western blots for ERα protein expression using 30μg of WCE from the indicated TAM-sensitive and TAM-resistant breast cancer cell lines and the HC-20 ERα antibody (A). MCF-7 A is from ATCC and MCF-7 K is from the Karmanos Cancer Center. The bar graph summarizes ERα66 and ERα46 protein expression as the mean ± SEM of 4 separate experiments in which ERα66 and ERα46 were normalized by β-actin expression in that membrane (B). The levels of ERα66 and ERα46 were normalized to ERα66 expression in MCF-7 A cells between experiments for comparison with the ERα66 expression in MCF-7 A set to 1 for comparison. Western blot of ERα protein expression using 30μg of WCE from the indicated cell lines transfected with control vector (C) or ERα46 as indicated (C). rhERα66 was prepared from baculovirus-infected SF-21 cells as a standard (Kulakosky, et al. 2002). The bar graph (D) summarizes ERα66 and ERα46 protein expression in 7 different experiments. First, ERα66 and ERα46 were normalized by βactin or α-tubulin expression and then the levels of ERα66 and ERα46 were normalized to ERα66 and ERα46 in control-transfected MCF-7 cells (MCF-7-C, i.e., transfected with the pcDNA3.1 vector alone) which was set to 1 for comparison (D). Likewise, the –C after LCC9 and LY2 means that the cells were transfected with pCDNA3.1 parental plasmid. * Significantly different from ERα66 and ** ERα46 in MCF-7 control transfected cells, respectively. # and ## Significantly different from ERα66 or ERα46 in pcDNA3.1 control plasmid-transfected same cell line, respectively.
3.2. Transfection of TAM-resistant LCC9 and LY2 cells with ERα46
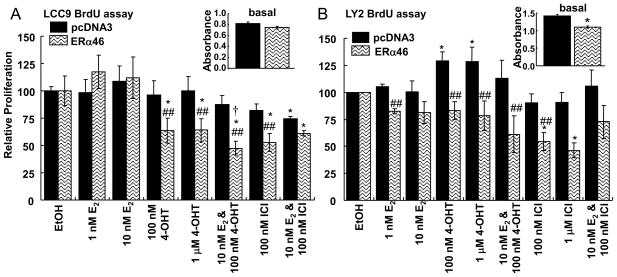
ERα46 is a dominant negative inhibitor of ERα66 function in transfected osteosarcoma SaOs cells (Denger et al. 2001) and overexpression of ERα46 in ER-negative rat PC12 pheochromocytoma cells did not stimulate thymidine incorporation in response to E2, indicating that ERα46 does not have ligand-dependent proliferative activity, at least in that cell line (Penot et al. 2005). If the observed decrease in ERα46 in the TAM-resistant LCC9 and LY2 cells shifts the balance of ERα46:ERα66 to a proliferative ERα66 state, then re-expression of ERα46 might be expected to inhibit ERα66-induced cell proliferation. LCC9 and LY2 cells were transfected with pCDNA3.1 parental vector or pCR3.1-ERα46 (Penot et al. 2005) expression vector. Transfected LCC9 and LY2 cells expressed ~3–4-fold higher ERα46 protein relative to vector-transfected cells (Fig. 1C and D). We observed only a 50% increase in ERα46 in transfected MCF-7 cells, but ERα46 transfection increased ERα66 ~ 2-fold. The reason for the ~15% increase in ERα46 protein in vector-transfected cells, including MCF-7, LCC9, and LY2 cells is unknown. It is possible that the CMV promoter in the pcDNA3.1 plasmid triggers an interferon-response in the cells (DeFilippis, et al. 2010) which has been reported to cause alternative splicing (Stoss, et al. 2000), in this case causing an increase in ERα46 expression, but studies examining this possibility are beyond the scope of the present study. ERα46 in the transfected LCC9 and LY2 cells was 4- and 3- fold higher than non-transfected MCF-7 and 2- and 1.3- fold higher than MCF-7 cells transfected with ERα46 (Fig. 1C and D). BrdU assays were performed to determine how ERα46 affects LCC9 and LY2 cell proliferation (Fig. 2A and B). Transfection with ERα46 reduced basal proliferation ~ 12% in LY2 cells (Inset in Fig. 2B), but had no effect on LCC9 basal proliferation. As anticipated, neither 4-OHT nor ICI inhibited LCC9 or LY2 cell proliferation. In fact, 4-OHT stimulated LCC9 cell proliferation. However, cells transfected with ERα46 were growth-inhibited by treatment with 4-OHT or ICI. Proliferation of LY2 cells transfected with ERα46 and treated with 1 nM E2 were also inhibited relative to the vector-transfected LY2 cells. Further, the relative proliferation of ERα46-transfected LY2 cells treated with 100 nM 4-OHT was significantly reduced by cotreatment with 10 nM E2. These data indicate that ERα46 has a ligand-dependent, anti-proliferative activity in these two TAM-resistant breast cancer cell lines. Thus, ERα46 appears necessary for antiestrogen-mediated inhibition of breast cancer cell proliferation since ERα46 transfection restored the ability of estrogen antagonists to repress the growth of these spontaneous endocrine-resistant breast cancer cells.
Fig. 2. Overexpression of ERα46 inhibits basal and ligand-activated cell proliferative responses in TAM-resistant LCC9 and LY2 cells.
Antiestrogen/TAM-resistant LCC9 (A) and LY2 (B) breast cells were transiently transfected with pCDNA3 parental vector or pCR3.1-hERα46 (Penot et al. 2005) for 24 h as described in Material and Methods. Cells were then treated as indicated and BrdU incorporation was measured after 48 h of treatment. Values are the average of 3 separate determinations ± SEM. The inset shows the actual absorbance value in a representative experiment for each cell line showing the higher proliferation of LY2 and the significant reduction in basal proliferation with ERα46 transfection in LY2 (B). * Significantly different from EtOH control, p < 0.05. ## Significantly different from pcDNA3-transfected control, p < 0.05. † Significantly different from 100 nM 4-OHT ERα46-transfected LY2, p < 0.05.
3.3. ERα46 inhibits E2-induced Cyclin D1 transcription in MCF-7 cells
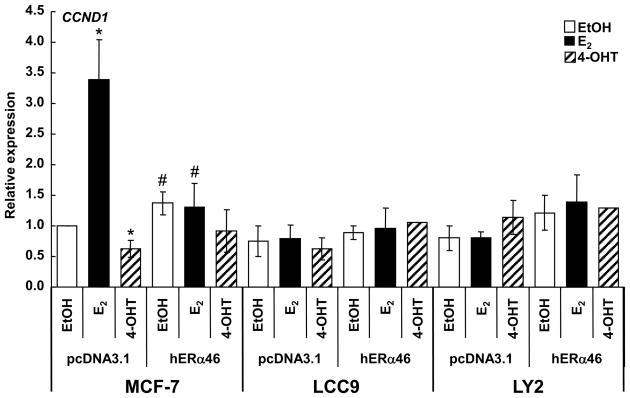
Since ERα46 overexpression in MCF-7 cells inhibited E2-induced luciferase reporter activity from the cyclin D1 promoter and inhibited cell cycle progression by blocking cells in G0/G1 (Penot et al. 2005), we examined how ERα46 transfection affected basal and E2-induced endogenous cyclin D1 in MCF-7, LCC9, and LY2 cells (Fig. 3). ERα46 slightly, but significantly, increased basal cyclin D1 expression, but blocked E2-induced cyclin D1 expression in MCF-7 cells. These data are in contrast to the suppression of basal luciferase activity from the cyclin D1-promoter luciferase reporter in transfected MCF-7 cells reported earlier (Penot et al. 2005). The difference between endogenous gene expression and cyclin D1 promoter activity is likely due to chromatin context and the role of E2-ERα interaction with an enhancer in the 3′UTR of the cyclin D1 gene that has a key role in E2-induced cyclin D1 transcription (Eeckhoute, et al. 2006). Cyclin D1 expression was significantly lower in LCC9 and LY2 cells compared to MCF-7 cells (Supplemental Fig. 1). When compared to EtOH treatment within each cell line, neither E2 nor 4-OHT regulated cyclin D1 transcription in LCC9 and LY2 cells (Fig. 3), findings commensurate with their endocrine-independent status. Further, and in contrast to results in MCF-7, transfection with ERα46 had no effect on cyclin D1 transcription in the TAM-resistant LCC9 or LY2 cells.
Fig 3. Overexpression of ERα46 inhibits E2-induced endogenous Cyclin D1 gene transcription in MCF-7 cells.
MCF-7 (ATCC), LCC9, and LY2 cells were transiently transfected with pCDNA3 parental vector or pCR3.1-hERα46 (Penot et al. 2005) for 30 h as described in Material and Methods. Cells were treated with EtOH, 10 nM E2, or 100 nM 4-OHT for 4 h. Q-RT-PCR analysis of cyclin D1 (CCND1) expression was normalized to 18S and the fold comparison was against EtOH for pcDNA 3.1-transfected cells within each cell line as described in Material and Methods. Values are the average ± SEM of three separate experiments. * Significantly different from EtOH control, p < 0.05. # Significantly different from pcDNA3-transfected control, p < 0.05.
3.4. Overexpression of ERα46 decreases expression of estrogen-responsive genes in MCF-7 cells
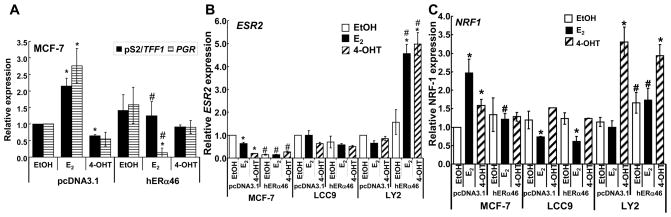
Overexpression of ERα46 in MCF-7 cells inhibited E2-induced endogenous pS2 (TFF1) and progesterone receptor (PR, PGR) transcription (Fig. 4A). The levels of E2-induced transcription of both pS2 and PR are consistent with other reports (Carreau, et al. 2008). LCC9 and LY2 cells did not express mRNA transcripts for either pS2 or PR and transfection with ERα46 had no effect on the transcription of either gene (data not shown). The lack of PR expression is consistent with the reported phenotypes of these cell lines (Bronzert et al. 1985; Brunner et al. 1997). Conversely, overexpression of ERα46 in MCF-7 cells reduced basal ERβ mRNA (Fig. 4B, Supplemental Fig. 2). The antiestrogen-resistant LCC9 and LY2 cells had lower ERβ mRNA expression relative to MCF-7 (Supplemental Fig. 2). ERβ mRNA expression did not change in ERα46-transfected LCC9 cells, but E2 and 4-OHT increased ERβ transcription ERα46-transfected LY2 cells, indicating cell line-specific differences in the ERβ transcriptional response to ERα46.
Fig. 4. Overexpression of ERα46 alters basal and E2-induced endogenous gene transcription in a gene- and cell- specific manner.
The cell lines indicated (MCF-7 (ATCC) in A; and MCF-7 (ATCC), LCC9, and LY2 in B and C) were transfected with pcDNA3.1 (vector) or pcDNA3.1-ERα46 as described in Materials and Methods. Thirty h after transfection, cells were treated with EtOH, 10 nM E2, or 100 nM 4-OHT for 4 h. Q-PCR was performed to measure pS2/TTF1 and PGR (A), ESR2 (ERβ) (B), and NRF1 (NRF-1) (C) mRNA expression. The fold comparison was against EtOH for pcDNA 3.1-transfected cells within each cell line as described in Material and Methods. Values are the average +/− SEM of three separate transfection experiments in which all determinations were performed in triplicate. * Significantly different from EtOH control, p < 0.05, # Significantly different from the same treatment in pcDNA3.1-transfected cells.
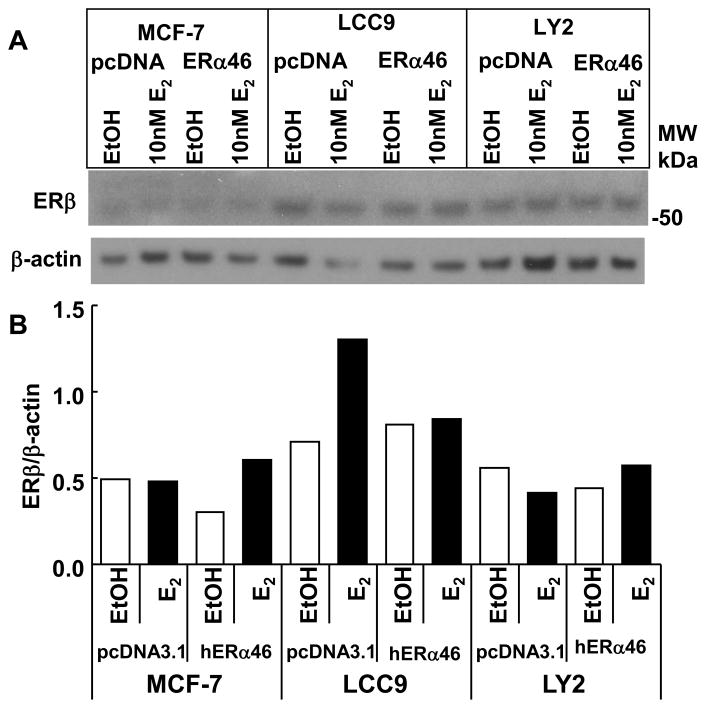
In contrast to the observed increase in ERβ mRNA in ERα46-transfected LY2 cells with E2 treatment (Fig 4B), no increase in ERβ protein was observed in LY2 cells (Fig. 5). To better visualize the ERβ bands in MCF-7 cells, a different exposure is shown in Supplemental Fig. 3. The differences in mRNA and protein levels are likely due to differences in treatment time: although cells were transfected with ERα46 for 24 h prior to ligand treatment for both gene and protein expression studies, mRNA and protein were analyzed 4 h and 24 h post-treatment, respectively. ERα46 reduced basal ERβ protein in MCF-7 cells and inhibited E2-induce increase in ERβ in LCC9 cells.
Fig. 5. Overexpression of ERα46 alters ERβ protein expression.
The indicated cell lines were transfected with pcDNA3.1 (control vector) or pcDNA3.1-hERα46 for 24 h and then treated with EtOH (vehicle control) or 10 nM E2 for 24 h. 20 μg WCE was separated by 10% SDS-PAGE and immunoblotted for ERβ (A) The membrane was stripped and reprobed for βactin. Quantitation of the data (B) are described in Materials and Methods. The blot is representative of 2 separate experiments showing similar results.
E2 and 4-OHT increased NRF-1 transcription in MCF-7 cells and overexpression of ERα46 blocked ligand-stimulated NRF-1 expression (Fig. 4C). E2 repressed NRF-1 transcription in LCC9 cells and overexpression of ERα46 had no effect on NRF-1 expression. In LY2 cells, 4-OHT increased NRF-1 transcription and overexpression of ERα46 increased basal NRF-1 expression while having no effect on the level of NRF-1 in 4-OHT-treated cells.
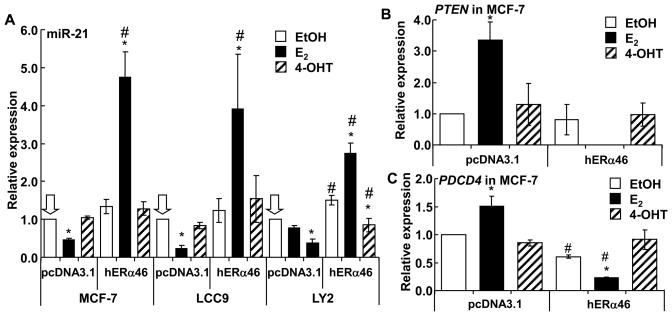
3.5. Overexpression of ERα46 allows E2-induced increase in miR-21 expression
We recently reported that E2 suppresses miR-21 expression in MCF-7 cells (Wickramasinghe et al. 2009). We examined how overexpression of ERα46 affected miR-21 transcription. For these experiments, we evaluated SNORD38B, hsa-miR-765, and 5S rRNA for normalization of miR-21 expression and 5S rRNA was selected for final normalization because SNORD38B and miR-765 were expressed at different levels in the 3 cell lines and their expression was altered by E2 and 4-OHT treatment (Supplemental Fig. 4A-C). miR-21 expression was lower in TAM-resistant LCC9 and LY2 cells compared to MCF-7 cells (Supplemental Fig. 4D). This observation agrees with reduced miR-21 in another TAM-resistance cell line derived from MCF-7 cells (Miller et al. 2008). E2 reduced miR-21 transcription in MCF-7 and LCC9 while the reduction in miR-21 in E2-treated LY2 cells was not statistically significant (Fig. 6A). 4-OHT reduced miR-21 in LY2 cells without affecting miR-21 expression in MCF-7 or LCC9 cells. ERα46 increased basal miR-21 in LY2, but not in MCF-7 or LCC9 cells. Overexpression of ERα46 in all three cell lines resulted in an increase in miR-21 in response to E2. In ERα46-transfected LY2 cells, 4-OHT increased miR-21 relative to untransfected LY2 cells, but the level of miR-21 was lower than that in the EtOH-treated, ERα46-transfected LY2 cells, so the relative expression was similar. Overall, these results indicate that ERα46 overexpression counteracts the E2-ERα66-mediated repression of miR-21 expression, suggesting that ERα AF-1 is important for E2-induced miR-21 repression. Further, there are cell line-specific differences in the regulation of miR-21 expression, implicating roles for other factors in regulating miR-21 expression..
Fig. 6. Overexpression of ERα46 stimulates E2-induced endogenous miR-21 gene transcription and reduces miR-21 targets PTEN and PDCD4 in MCF-7 cells.
MCF-7 (ATCC), LCC9, and LY2 cell lines were transfected with pcDNA3.1 (vector) or pcDNA3.1-ERα46 as described in Materials and Methods. Thirty h after transfection, cells were treated with EtOH, 10 nM E2, or 100 nM 4-OHT for 4 h. Q-PCR was performed to measure miR-21 normalized to 5S RNA (A). Q-PCR was performed to measure PTEN and PDCD4 relative to 18S in MCF-7 cells transfected and treated as indicated. Values are the average +/− SEM of three separate transfection experiments in which all determinations were performed in triplicate. * Significantly different from EtOH control, p < 0.05. # Significantly different from the same treatment in pcDNA3.1-transfected cells, p < 0.05.
3.6. Effect of ERα46 on endogenous miR-21 target genes in MCF-7 cells
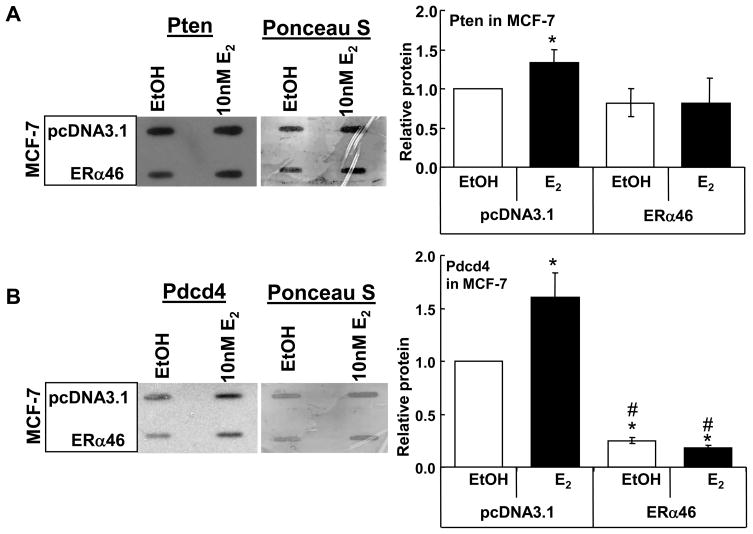
Since overexpression of ERα46 increased miR-21 expression with E2 in the three breast cancer cell lines cells, the effect of E2 on the mRNA and protein levels of endogenous miR-21-target genes PDCD4 and PTEN was examined by Q-PCR (Fig. 6B and C) and immunoblot (Fig. 7A and B). As expected based on the decrease in miR-21 with E2 in MCF-7 cells, and as previously reported (Wickramasinghe et al. 2009), E2 increased PDCD4 and PTEN mRNA (Fig. 6A and B) and Pten and Pdcd4 protein (Fig. 7A and B), results reflecting reduced miR-21 levels (Fig. 6A), thus increasing transcript stability. Conversely, overexpression of ERα46 reduced mRNA levels of PTEN and PDCD4 in E2-treated MCF-7 cells (Fig. 6B and C), results reflecting increased miR-21 (Fig. 6A), thus reducing transcript stability. The concordant reduction in Pten and Pdcd4 proteins is seen in Fig. 7A and B. Overall, these data indicate that E2 has opposite effects on miR-21 expression with ERα66 and ERα46, implicating a role for AF-1 in E2-induced miR-21 repression.
Fig. 7. ERα46 inhibits E2-induced increase Pten and Pdcd4 in MCF-7 cells.
MCF-7 (ATCC) cells were transfected with pcDNA3.1 or pcDNA3.1-ERα46. Twenty-four h after transfection, cells were treated with EtOH or 10 nM E2 for 24 h. 5 μg WCE were slot blotted onto PVDF membranes, stained with Ponceau S, washed and probed for Pten (A) or Pdcd4 (B). Values are the average +/− Std dev of duplicate samples. * Significantly different from EtOH control, p < 0.05, # Significantly different from the same treatment in pcDNA3.1-transfected cells.
4. Discussion
Because endocrine resistance is a major concern in breast cancer recurrence, metastases, and survival, numerous laboratories have invested major effort in elucidating the mechanisms by which cancer cells lose their sensitivity to antiestrogens. Multiple overlapping and interconnecting pathways involving ER function contribute in endocrine resistance (Clarke et al. 2003; Ring and Dowsett 2004). This study evaluated the hypothesis that a reduction in ERα46 expression in TAM-resistant breast cancer cells contributes to endocrine resistance. We focused our experiments on the effect of ERα46 overexpression on 4-OHT-responsiveness in LCC9 and LY2 cells, resistant to both TAM and Fulvestrant, because most breast cancer patients with ERα-positive tumors have historically been treated with tamoxifen as the first line of adjuvant therapy and only treated with fulvestrant at the time of relapse (Robertson, et al. 2004). ERα46 lacks AF-1 and has intact AF-1 (Flouriot, et al. 2000). Previous studies reported that ERα46 is a ligand-dependent transcriptional regulator that is a cell-specific manner depending on the relative strength of AF-1 and AF-2 in that cell line (Carreau et al. 2008; Flouriot et al. 2000; Penot et al. 2005). Here we report that ERα46 expression is reduced in TAM-resistant LCC2, LCC9, and LY2 breast cancer cells. A role for ERα46 in TAM-responsiveness was indicated by experiments in which restoration of ERα46 expression by transient transfection reduced basal LY2 cell proliferation and increased the ability of 4-OHT and ICI 182,780 to inhibit LCC9 and LY2 cell proliferation. These data are in agreement with a report that overexpression of ERα46 inhibits MCF-7 and PC12 cell proliferation (Penot et al. 2005) and HT-29 colon cancer cell proliferation (Jiang, et al. 2008) but are the first to examine ERα46 expression and activity in endocrine-resistant breast cancer cells. The observation that overexpression of ERα46 in LY2 cells caused the cells to be growth-inhibited by E2 and that E2 enhanced 4-OHT-induced inhibition of cell proliferation is reminiscent of earlier observations. This ‘reversal’ of E2 pharmacology in LY2 cells was seen previously with restoration of COUP-TFII expression in LY2 cells (Riggs, et al. 2006) and is similar to the model that Jordan has proposed for the third phase of antiestrogen resistance in breast cancer (Lewis-Wambi and Jordan 2009; Liu, et al. 2003). The inhibition of breast cancer cell proliferation by E2 involves E2-induced apoptosis, but the mechanism remains unknown (Lewis-Wambi and Jordan 2009).
This is, to our knowledge, the first study examining the impact of ERα46 on endogenous gene expression in MCF-7, LCC9, and LY2 breast cancer cells. As expected based on previous reports using E2-target gene promoter-luciferase reporter assays in transfected cells (Carreau et al. 2008; Penot et al. 2005), overexpression of ERα46 in MCF-7 cells inhibited E2-induced transcription of endogenous E2-regulated genes Cyclin D1/CCND1, NRF-1/NRF1, pS2/TFF1, and PR/PGR. However, in contrast to the finding that apoERα46 reduced basal pS2 expression in MDA-MB-231 cells (Metivier et al. 2004), we did not detect reduced basal pS2 or PR in MCF-7 cells. These findings implicate cell-line-specific mediators in differential responses to overexpression of ERα46. The inhibition of endogenous E2-induced CCND1 is also commensurate with the reduction in MCF-7 cell proliferation in ERα46-overexpressing cells (Penot et al. 2005). The LCC9 and LY2 tamoxifen-resistant cells did not express pS2/TFF1 or PR/PGR, and showed no E2-induced CCND1 or NRF1 transcription. NRF-1 is a transcription factor regulating mitochondrial biogenesis and function as well as a variety of cellular responses including protein synthesis, DNA replication and repair, and cell proliferation (Scarpulla 2008). Although forced overexpression of cyclin D1 rendered MCF-7 cells resistant to the antiestrogen arzoxifene (Zwart, et al. 2009) and overexpression of cyclin D1 is linked to TAM-resistance and STAT3 activation (Ishii, et al. 2008), endogenous cyclin D1 expression was lower in TAM-resistant LCC9 and LY2 cells and was not regulated by E2, 4-OHT, or by ERα46. Thus, inhibition of CCND1 or NRF-1 transcription does not appear to be involved in the ability of ERα46 to reduce LCC9 and LY2 cell proliferation in response to 4-OHT or ICI 182,780.
In contrast, to our results in ERα-expressing breast cancer cells, overexpression of ERα46 in ERα/PGR/ERBB3-negative MDA-MB-231 breast cancer cells increased E2-stimulated pS2 expression (Metivier et al. 2004). This difference is likely due to the activity levels of AF-1 in MCF-7 versus MDA-MB-231 cells. ERα46 lacks the N-terminal A/B domain and thus has no AF-1 (Flouriot et al. 2000). AF-1 has higher activity than the ligand-activated C-terminal AF-2 in more differentiated cell lines including MCF-7, and LCC9 and LY2 cells which retain ERα expression, although they show reduced E2-regulated transcriptional responses as seen here and reported previously (Riggs et al. 2006). In contrast, AF-2 predominates in dedifferentiated and undifferentiated cell lines including MDA-MB-231 and HeLa cells (Flouriot et al. 2000; Metivier, et al. 2002; Metivier, et al. 2002; Penot et al. 2005).
To our knowledge, this is the first report that E2-ERα46 increases transcription of the oncomiR miR-21 and thus reduces the transcript and protein expression of miR-21 target genes PDCD4 and PTEN in MCF-7 human breast cancer cells. The data shown here demonstrate opposite effects of endogenous ERα66 and ERα46 in mediating E2-regulation of miR-21 transcription. Thus, AF-1 function in ERα66 results in E2-induced miR-21 inhibition whereas the absence of AF-1 in ERα46 allows E2-induced miR-21 transcription. Further detailed mechanistic studies are in progress to parse the roles of AF-1 and AF-2 in ERα-regulation of miR-21 transcription are in progress. Given the established role of miR-21 as a key regulator in oncogenesis (Selcuklu, et al. 2009), the increase in miR-21 in response to E2 in ERα46-expressing breast cancer cells appears to contradict results from the cell proliferation studies showing that ERα46 caused LY2 cells to be growth inhibited by E2. Differences in time course of the experiments is one possible explanation. However, miR-21 also downregulates CDC25A, negatively regulates G1-S transition, and participates in DNA damage-induced G2-M checkpoint control in colon cancer cells (Wang, et al. 2009). Thus, despite the observation that miR-21 was the most significantly up-regulated miRNA in breast tumor biopsies (Sempere, et al. 2007), the precise regulation and role of miR-21 in TAM-resistant breast cancer requires further investigation.
Since its identification in 1996, the role of ERβ in breast cancer has remained enigmatic with some studies indicating that ERβ acts as a tumor suppressor by antagonizing ERα activity (Behrens et al. 2007; Paruthiyil, et al. 2004; Pinton, et al. 2009; Sotoca Covaleda, et al. 2008; Williams, et al. 2008), while other studies oppose this model (Speirs, et al. 1999; Speirs, et al. 1999). Thus, while the increase in ERβ mRNA expression seen with E2 and 4-OHT treatment of ERα46 transfected TAM-resistant LY2 cells correlates with reduced basal, E2, and 4-OHT–regulated proliferation, we did not detect an increase in ERβ protein 24 h post treatment. Future studies will be required to assay the time course of ERβ mRNA and protein stability to parse the mechanism connecting these findings. For example, mRNA and protein stability may be altered by ERα46 and E2 treatment in LY2. Although no one has examined the half-life of ERβ in MCF-7, LCC9, and LY2 cells, ERβ mRNA half-life was ~ 18 h in rat granulosa cells (Guo, et al. 2001). Altered ERβ mRNA stability in the ERα46-transfected LY2 cells may result from differences in E2-ERα46-regulated miRNA expression, i.e., upregulation of miRNAs that reduce ESR2 message levels, thus reducing ERβ protein. Given our findings of altered miR-21 expression and the observation that no one has yet identified miRNAs regulating ERβ expression, this idea appears worthy of follow-up experiments. Although 2 common SNPs were identified in the 3′UTR of the human ESR2 gene, they did not influence ERβ mRNA stability in a heterologous HEK-293 cell system (Putnik, et al. 2009). Likewise, since unliganded ERβ represses ERα-mediated MCF-7 cell proliferation, closer examination of the time course of ERβ mRNA and protein expression in ERa46-transfected cells will provide insight into ERβ’s apparent antagonist activity (Levy, et al. 2010).
Interest in the role of ERα splice variants in breast cancer has intensified recently with the identification of ERα36 (Fowler, et al. 2009). ERα36 is produced by alternative splicing at the 3′ end, yielding a 36 kDa protein lacking both AF-1 and AF-2; further, the last 138 aa in the C-terminal F-domain of ERα66 are replaced with a unique 22 aa sequence (Shi, et al. 2009). Coexpression of ERα66 and ERα36 in human breast tumors was associated with shorter disease-free survival (Shi et al. 2009). When transfected in ER-null HEK293 cells, ERα36, like ERα46, was a dominant negative inhibitor of ERα66 transcriptional activity (Wang, et al. 2006) ERα36 localized to the cytoplasm and plasma membrane of HEK293 cells and treatment with E2, 4-OHT, or ICI 182,780 stimulated MAPK phosphorylation (Wang et al. 2006). ERα66-positive/ERα36-positive breast cancers have been speculated to be resistant to tamoxifen (Fowler et al. 2009), but this remains to be determined.
An [3H]Tamoxifen-aziridine-bound protein of 43kDa was increased in crude tumor homogenates prepared from hormone refractory breast tumors and separated sucrose density gradients (Piccart, et al. 1991). How this relates to ERα46 as described here can not be determined. Further, there was no confirmation of the identity of the [3H]Tamoxifen-aziridine-bound fragment and no mechanistic follow-up experiments were performed on this observation until the present study. Interestingly, the expression of the [3H]Tamoxifen-aziridine-bound 43 kDa variant seems to be connected with a 36kDa ERa variant in aggressive breast tumors (Trivedi et al Breast cancer Res. Treat 40:231–241 1996). Again, how these
In summary, we report for the first time that ERα46 expression is reduced in tamoxifen-resistant breast cancer cell lines and that overexpression of ERα46 results in a restoration of TAM-inhibition of cell proliferation. ERα46 overexpression results in transcriptional responses opposite that of endogenous ERα66 in MCF-7 and the two antiestrogen-resistant breast cancer cell lines (LCC9 and LY2). For example, miR-21 is down-regulated in response to E2 in an ERα-dependent manner, but upregulated by E2-ERα46. Furthermore, this stimulation of miR-21 correlates with up-regulation of miR-21 targets: tumor suppressors PDCD4 and PTEN. The identification of miR-21 as a miRNA target of ERα46 regulation may offer a role for ERα46 in inhibiting disease progression.
Supplementary Material
Acknowledgments
We thank Dr. Gilles Flouriot for providing the pCR3.1-hERα46 plasmid and Dr. Robert Clarke for providing the LCC1, LCC2, LCC9, and LY2 cells.
Footnotes
Disclosure statement:
None of the authors have anything to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Achuthan R, Bell SM, Roberts P, Leek JP, Horgan K, Markham AF, MacLennan KA, Speirs V. Genetic events during the transformation of a tamoxifen-sensitive human breast cancer cell line into a drug-resistant clone. Cancer Genet Cytogenet. 2001;130:166–72. doi: 10.1016/s0165-4608(01)00475-7. [DOI] [PubMed] [Google Scholar]
- Behrens D, Gill JH, Fichtner I. Loss of tumourigenicity of stably ER[beta]-transfected MCF-7 breast cancer cells. Molecular and Cellular Endocrinology. 2007;274:19–29. doi: 10.1016/j.mce.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Bronzert DA, Greene GL, Lippman ME. Selection and characterization of a breast cancer cell line resistant to the antiestrogen LY 117018. Endocrinology. 1985;117:1409–17. doi: 10.1210/endo-117-4-1409. [DOI] [PubMed] [Google Scholar]
- Brunner N, Boysen B, Jirus S, Skaar TC, Holst-Hansen C, Lippman J, Frandsen T, Spang-Thomsen M, Fuqua SA, Clarke R. MCF7/LCC9: an antiestrogen-resistant MCF-7 variant in which acquired resistance to the steroidal antiestrogen ICI 182,780 confers an early cross-resistance to the nonsteroidal antiestrogen tamoxifen. Cancer Res. 1997;57:3486–3493. [PubMed] [Google Scholar]
- Carreau C, Flouriot G, Bennetau-Pelissero C, Potier M. Enterodiol and enterolactone, two major diet-derived polyphenol metabolites have different impact on ER[alpha] transcriptional activation in human breast cancer cells. The Journal of Steroid Biochemistry and Molecular Biology. 2008;110:176–185. doi: 10.1016/j.jsbmb.2008.03.032. [DOI] [PubMed] [Google Scholar]
- Chia SK, Speers CH, D’Yachkova Y, Kang A, Malfair-Taylor S, Barnett J, Coldman A, Gelmon KA, O’Reilly SE, Olivotto IA. The impact of new chemotherapeutic and hormone agents on survival in a population-based cohort of women with metastatic breast cancer. Cancer. 2007;110:973–9. doi: 10.1002/cncr.22867. [DOI] [PubMed] [Google Scholar]
- Clarke R, Liu MC, Bouker KB, Gu Z, Lee RY, Zhu Y, Skaar TC, Gomez B, O’Brien K, Wang Y, Hilakivi-Clarke LA. Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene. 2003;22:7316–39. doi: 10.1038/sj.onc.1206937. [DOI] [PubMed] [Google Scholar]
- DeFilippis VR, Alvarado D, Sali T, Rothenburg S, Fruh K. Human Cytomegalovirus Induces the Interferon Response via the DNA Sensor ZBP1. J Virol. 2010;84:585–598. doi: 10.1128/JVI.01748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denger S, Reid G, Kos M, Flouriot G, Parsch D, Brand H, Korach KS, Sonntag-Buck V, Gannon F. ERalpha Gene Expression in Human Primary Osteoblasts: Evidence for the Expression of Two Receptor Proteins. Mol Endocrinol. 2001;15:2064–77. doi: 10.1210/mend.15.12.0741. [DOI] [PubMed] [Google Scholar]
- Dowsett M. Overexpression of HER-2 as a resistance mechanism to hormonal therapy for breast cancer. Endocr Relat Cancer. 2001;8:191–5. doi: 10.1677/erc.0.0080191. [DOI] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P, Yue W, Wang J-P, Aiyar S, Li Y, Kim T-H, Santen RJ. Mechanisms of Resistance to Structurally Diverse Antiestrogens Differ under Premenopausal and Postmenopausal Conditions: Evidence from in Vitro Breast Cancer Cell Models. Endocrinology. 2009;150:2036–2045. doi: 10.1210/en.2008-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G, Brand H, Denger S, Metivier R, Kos M, Reid G, Sonntag-Buck V, Gannon F. Identification of a new isoform of the human estrogen receptor-alpha (hER-alpha) that is encoded by distinct transcripts and that is able to repress hER-alpha activation function 1. Embo J. 2000;19:4688–700. doi: 10.1093/emboj/19.17.4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler AM, Santen RJ, Allred DC. “Dwarf” Estrogen Receptor in Breast Cancer and Resistance to Tamoxifen. J Clin Oncol. 2009;27:3413–3415. doi: 10.1200/JCO.2009.21.8776. [DOI] [PubMed] [Google Scholar]
- Girault I, Lerebours F, Amarir S, Tozlu S, Tubiana-Hulin M, Lidereau R, Bieche I. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin Cancer Res. 2003;9:1259–66. [PubMed] [Google Scholar]
- Guo C, Savage L, Sarge KD, Park-Sarge O-K. Gonadotropins Decrease Estrogen Receptor-{beta} Messenger Ribonucleic Acid Stability in Rat Granulosa Cells. Endocrinology. 2001;142:2230–2237. doi: 10.1210/endo.142.6.8102. [DOI] [PubMed] [Google Scholar]
- Herynk MH, Fuqua SAW. Estrogen Receptor Mutations in Human Disease. Endocr Rev. 2004;25:869–898. doi: 10.1210/er.2003-0010. [DOI] [PubMed] [Google Scholar]
- Ishii Y, Waxman S, Germain D. Tamoxifen Stimulates the Growth of Cyclin D1-Overexpressing Breast Cancer Cells by Promoting the Activation of Signal Transducer and Activator of Transcription 3. Cancer Res. 2008;68:852–860. doi: 10.1158/0008-5472.CAN-07-2879. [DOI] [PubMed] [Google Scholar]
- Jiang HP, Teng RY, Wang Q, Zhang X, Wang HH, Cao J, Teng LS. Estrogen receptor alpha variant ERalpha46 mediates growth inhibition and apoptosis of human HT-29 colon adenocarcinoma cells in the presence of 17beta-oestradiol. Chin Med J (Engl) 2008;121:1025–31. [PubMed] [Google Scholar]
- Kim KH, Bender JR. Rapid, estrogen receptor-mediated signaling: why is the endothelium so special? Sci STKE. 2005:pe28. doi: 10.1126/stke.2882005pe28. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen Regulation of MicroRNA Expression. Current Genomics. 2009;10:169–183. doi: 10.2174/138920209788185289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakosky PC, Jernigan SC, McCarty MA, Klinge CM. Response element sequence regulates estrogen receptor alpha and beta affinity and activity. J Mol Endocrinol. 2002;29:137–52. doi: 10.1677/jme.0.0290137. [DOI] [PubMed] [Google Scholar]
- Lannigan DA. Estrogen receptor phosphorylation. Steroids. 2003;68:1–9. doi: 10.1016/s0039-128x(02)00110-1. [DOI] [PubMed] [Google Scholar]
- Levy N, Paruthiyil S, Zhao X, Vivar OI, Saunier EF, Griffin C, Tagliaferri M, Cohen I, Speed TP, Leitman DC. Unliganded estrogen receptor-[beta] regulation of genes is inhibited by tamoxifen. Molecular and Cellular Endocrinology. 2010;315:201–207. doi: 10.1016/j.mce.2009.08.030. [DOI] [PubMed] [Google Scholar]
- Lewis-Wambi JS, Jordan VC. Estrogen regulation of apoptosis: how can one hormone stimulate and inhibit? Breast Cancer Res. 2009;11:206. doi: 10.1186/bcr2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Haynes MP, Bender JR. Plasma membrane localization and function of the estrogen receptor alpha variant (ER46) in human endothelial cells. Proc Natl Acad Sci U S A. 2003;100:4807–12. doi: 10.1073/pnas.0831079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Hisamoto K, Kim KH, Haynes MP, Bauer PM, Sanjay A, Collinge M, Baron R, Sessa WC, Bender JR. Variant estrogen receptor c-Src molecular interdependence and c-Src structural requirements for endothelial NO synthase activation. Proceedings of the National Academy of Sciences. 2007;104:16468–16473. doi: 10.1073/pnas.0704315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Lee E-S, Gajdos C, Pearce ST, Chen B, Osipo C, Loweth J, McKian K, De Los Reyes A, Wing L, Jordan VC. Apoptotic Action of 17{beta}-Estradiol in Raloxifene-Resistant MCF-7 Cells In Vitro and In Vivo. J Natl Cancer Inst. 2003;95:1586–1597. doi: 10.1093/jnci/djg080. [DOI] [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen H-W. ACTR/AIB1 Functions as an E2F1 Coactivator To Promote Breast Cancer Cell Proliferation and Antiestrogen Resistance. Mol Cell Biol. 2004;24:5157–5171. doi: 10.1128/MCB.24.12.5157-5171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykkesfeldt AE. Mechanisms of tamoxifen resistance in the treatment of advanced breast cancer. Acta Oncol. 1996;35(Suppl 5):9–14. doi: 10.3109/02841869609083961. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Phung S, Wang X, Wu X, Yuan Y-C, Wagman L, Chen S. Genome-Wide Analysis of Aromatase Inhibitor-Resistant, Tamoxifen-Resistant, and Long-Term Estrogen-Deprived Cells Reveals a Role for Estrogen Receptor. Cancer Res. 2008;68:4910–4918. doi: 10.1158/0008-5472.CAN-08-0303. [DOI] [PubMed] [Google Scholar]
- Metivier R, Gay FA, Hubner MR, Flouriot G, Salbert G, Gannon F, Kah O, Pakdel F. Formation of an hER alpha-COUP-TFI complex enhances hER alpha AF-1 through Ser118 phosphorylation by MAPK. Embo J. 2002;21:3443–53. doi: 10.1093/emboj/cdf344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Carmouche RP, Hubner MR, Reid G, Denger S, Manu D, Brand H, Kos M, Benes V, Gannon F. Transcriptional complexes engaged by apo-estrogen receptor-{alpha} isoforms have divergent outcomes. EMBO J. 2004;23:3653–3666. doi: 10.1038/sj.emboj.7600377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Stark A, Flouriot G, Hubner MR, Brand H, Penot G, Manu D, Denger S, Reid G, Kos M, Russell RB, Kah O, Pakdel F, Gannon F. A dynamic structural model for estrogen receptor-alpha activation by ligands, emphasizing the role of interactions between distant A and E domains. Mol Cell. 2002;10:1019–32. doi: 10.1016/s1097-2765(02)00746-3. [DOI] [PubMed] [Google Scholar]
- Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, Jacob S, Majumder S. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27(Kip1) J Biol Chem. 2008;283:29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty K, Kim KH, Bender JR. Estrogen Receptor-Mediated Rapid Signaling. Endocrinology. 2006;147:5557–5563. doi: 10.1210/en.2006-0729. [DOI] [PubMed] [Google Scholar]
- Murphy AJ, Guyre PM, Wira CR, Pioli PA. Estradiol regulates expression of estrogen receptor ERalpha46 in human macrophages. PLoS ONE. 2009;4:e5539. doi: 10.1371/journal.pone.0005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC. Estrogen Receptor beta Inhibits Human Breast Cancer Cell Proliferation and Tumor Formation by Causing a G(2) Cell Cycle Arrest. Cancer Res. 2004;64:423–428. doi: 10.1158/0008-5472.can-03-2446. [DOI] [PubMed] [Google Scholar]
- Penot G, Le Peron C, Merot Y, Grimaud-Fanouillere E, Ferriere F, Boujrad N, Kah O, Saligaut C, Ducouret B, Metivier R, Flouriot G. The Human Estrogen Receptor-{alpha} Isoform hER{alpha}46 Antagonizes the Proliferative Influence of hER{alpha}66 in MCF7 Breast Cancer Cells. Endocrinology. 2005;146:5474–5484. doi: 10.1210/en.2005-0866. [DOI] [PubMed] [Google Scholar]
- Piccart MJ, Muquardt C, Bosman C, Pirotte P, Veenstra S, Grillo F, Leclercq G. Comparison of tritiated estradiol and tamoxifen aziridine for measurement of estrogen receptors in human breast cancer cytosols. J Natl Cancer Inst. 1991;83:1553–9. doi: 10.1093/jnci/83.21.1553. [DOI] [PubMed] [Google Scholar]
- Pinton G, Brunelli E, Murer B, Puntoni R, Puntoni M, Fennell DA, Gaudino G, Mutti L, Moro L. Estrogen Receptor-{beta} Affects the Prognosis of Human Malignant Mesothelioma. Cancer Res. 2009;69:4598–4604. doi: 10.1158/0008-5472.CAN-08-4523. [DOI] [PubMed] [Google Scholar]
- Putnik M, Zhao C, Gustafsson JA, Dahlman-Wright K. Effects of two common polymorphisms in the 3′ untranslated regions of estrogen receptor beta on mRNA stability and translatability. BMC Genet. 2009;10:55. doi: 10.1186/1471-2156-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs KA, Wickramasinghe NS, Cochrum RK, Watts MB, Klinge CM. Decreased Chicken Ovalbumin Upstream Promoter Transcription Factor II Expression in Tamoxifen-Resistant Breast Cancer Cells. Cancer Res. 2006;66:10188–10198. doi: 10.1158/0008-5472.CAN-05-3937. [DOI] [PubMed] [Google Scholar]
- Ring A, Dowsett M. Mechanisms of tamoxifen resistance. Endocr Relat Cancer. 2004;11:643–658. doi: 10.1677/erc.1.00776. [DOI] [PubMed] [Google Scholar]
- Robertson JF, Erikstein B, Osborne KC, Pippen J, Come SE, Parker LM, Gertler S, Harrison MP, Clarke DA. Pharmacokinetic profile of intramuscular fulvestrant in advanced breast cancer. Clin Pharmacokinet. 2004;43:529–38. doi: 10.2165/00003088-200443080-00003. [DOI] [PubMed] [Google Scholar]
- Santen RJ, Boyd NF, Chlebowski RT, Cummings S, Cuzick J, Dowsett M, Easton D, Forbes JF, Key T, Hankinson SE, Howell A, Ingle J. Critical assessment of new risk factors for breast cancer: considerations for development of an improved risk prediction model. Endocr Relat Cancer. 2007;14:169–187. doi: 10.1677/ERC-06-0045. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Nuclear Control of Respiratory Chain Expression by Nuclear Respiratory Factors and PGC-1-Related Coactivator. Annals of the New York Academy of Sciences. 2008;1147:321–334. doi: 10.1196/annals.1427.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcuklu SD, Donoghue MTA, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochemical Society Transactions. 2009;037:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, Wells W, Kauppinen S, Cole CN. Altered MicroRNA Expression Confined to Specific Epithelial Cell Subpopulations in Breast Cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- Shi L, Dong B, Li Z, Lu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Wang Z, Xie Y. Expression of ER-{alpha}36, a Novel Variant of Estrogen Receptor {alpha}, and Resistance to Tamoxifen Treatment in Breast Cancer. J Clin Oncol. 2009;27:3423–3429. doi: 10.1200/JCO.2008.17.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotoca Covaleda AM, van den Berg H, Vervoort J, van der Saag P, Strom A, Gustafsson J-A, Rietjens I, Murk AJ. Influence of Cellular ER{alpha}/ER{beta} Ratio on the ER{alpha}-Agonist Induced Proliferation of Human T47D Breast Cancer Cells. Toxicol Sci. 2008;105:303–311. doi: 10.1093/toxsci/kfn141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs V, Malone C, Walton DS, Kerin MJ, Atkin SL. Increased expression of estrogen receptor beta mRNA in tamoxifen- resistant breast cancer patients. Cancer Res. 1999;59:5421–5424. [PubMed] [Google Scholar]
- Speirs V, Parkes AT, Kerin MJDSW, Carleton PJ, Fox JN, Atkin SL. Coexpression of estrogen receptor alpha and beta: Poor prognostic factors in human breast cancers? Cancer Res. 1999;59:525–528. [PubMed] [Google Scholar]
- Stoss O, Stoilov P, Daoud R, Hartmann AM, Olbrich M, Stamm S. Misregulation of pre-mRNA splicing that causes human diseases. Concepts and therapeutic strategies. Gene Therapy and Molecular Biology. 2000;5:9–30. [Google Scholar]
- Wang P, Zou F, Zhang X, Li H, Dulak A, Tomko RJ, Jr, Lazo JS, Wang Z, Zhang L, Yu J. microRNA-21 Negatively Regulates Cdc25A and Cell Cycle Progression in Colon Cancer Cells. Cancer Res. 2009;69:8157–8165. doi: 10.1158/0008-5472.CAN-09-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. A variant of estrogen receptor-{alpha}, hER-{alpha}36: Transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. PNAS. 2006:0603339103. doi: 10.1073/pnas.0603339103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe N, Manavalan T, Dougherty S, Riggs K, Li Y, Klinge C. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009;37:2584–2595. doi: 10.1093/nar/gkp117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2007 doi: 10.1038/sj.onc.1210712. [DOI] [PubMed] [Google Scholar]
- Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA. A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene. 2008;27:1019–32. doi: 10.1038/sj.onc.1210712. [DOI] [PubMed] [Google Scholar]
- Zhao J-J, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ. MicroRNA-221/222 negatively regulates ERalpha and associates with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zwart W, Rondaij M, Jalink K, Sharp ZD, Mancini MA, Neefjes J, Michalides R. Resistance to Antiestrogen Arzoxifene Is Mediated by Overexpression of Cyclin D1. Mol Endocrinol. 2009;23:1335–1345. doi: 10.1210/me.2008-0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.